Abstract
Visual arrestin-1 plays a key role in the rapid and reproducible shutoff of rhodopsin signaling. Its highly selective binding to light-activated phosphorylated rhodopsin is an integral part of the functional perfection of rod photoreceptors. Structure-function studies revealed key elements of the sophisticated molecular mechanism ensuring arrestin-1 selectivity and paved the way to the targeted manipulation of the arrestin-1 molecule to design mutants that can compensate for congenital defects in rhodopsin phosphorylation. Arrestin-1 self-association and light-dependent translocation in photoreceptor cells work together to keep a constant supply of active rhodopsin-binding arrestin-1 monomer in the outer segment. Recent discoveries of arrestin-1 interaction with other signaling proteins suggest that it is a much more versatile signaling regulator than previously thought, affecting the function of the synaptic terminals and rod survival. Elucidation of the fine molecular mechanisms of arrestin-1 interactions with rhodopsin and other binding partners is necessary for the comprehensive understanding of rod function and for devising novel molecular tools and therapeutic approaches to the treatment of visual disorders.
Keywords: arrestin, rhodopsin, signal shutoff, photoreceptors, protein-protein interactions
1. Introduction
The paradigm of the receptor-initiated signaling cascade consisting of receptor (rhodopsin, Rh), G protein (transducin, Td), and effector (cGMP phosphodiesterase, PDE), was developed through studies of light-induced signaling in rod photoreceptors (Fung et al., 1981). This was long before it was appreciated that animals have a large family of G protein-coupled receptors (GPCRs or 7TMRs), of which the founding member is rhodopsin. Receptor phosphorylation as a means of its regulation was also discovered in the visual system (Liebman and Pugh, 1980) before it became clear that kinases specifically phosphorylating active receptors (G protein-coupled receptor kinases, or GRKs) “prepare” the receptor for high-affinity arrestin binding (Gurevich and Gurevich, 2004). The first member of the arrestin family, arrestin-11, was discovered twice: first as S-antigen causing uveitis (Wacker et al., 1977), then as a 48 kDa protein that binds light-activated rhodopsin (Kuhn et al., 1984). Shortly thereafter, Pfister et al (Pfister et al., 1985; Pfister et al., 1984) established that both are one and the same protein. The name “arrestin” was proposed after seminal studies by Dr. Kuhn and co-workers demonstrated that the binding of this protein to light-activated phosphorylated rhodopsin (P-Rh*) inhibits PDE activation (Wilden et al., 1986). The first non-visual GRK (Benovic et al., 1989) and arrestin (Lohse et al., 1990), first termed β-adrenergic receptor kinase and β-arrestin, respectively, for their ability to turn off signaling by the β2-adrenergic receptor, were cloned soon thereafter. The demonstration that both can actually quench rhodopsin signaling (Lohse et al., 1992) suggested that the two-step signal shutoff, phosphorylation of active receptor followed by arrestin binding, is a fairly universal mechanism of GPCR regulation (Benovic et al., 1987). Years of homology cloning and subsequent genome projects revealed that this type of regulation is specific for animals, and that the first GRKs and “true” phosphoreceptor-binding arrestins appeared very early in pre-Metazoan evolution (Gurevich and Gurevich, 2006a; Gurevich et al., 2011).
Most mammals have seven GRK subtypes, of which GRK1 (rhodopsin kinase) and GRK7 are specific for photoreceptors, with the former expressed in both rods and cones, and the latter found exclusively in cones (Lorenz et al., 1991; Shichi and Somers, 1978; Weiss et al., 1998; Weller et al., 1975). GRK1 was shown to be crucial for timely signal shutoff in both types of photoreceptors (Chen et al., 1999; Cideciyan et al., 1998). However, the fact that mice and some other nocturnal rodents with rod-dominated vision lack GRK7 in photoreceptors prevents the use of genetically modified mice to definitively determine its role in visual signaling. Mammals have four arrestin subtypes. Arrestin-1 is expressed at very high levels in both rods (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006) and cones (Nikonov et al., 2008a), whereas the cone-specific arrestin-4 (Craft et al., 1994; Murakami et al., 1993) constitutes only ~2% of the total arrestin complement in cone photoreceptors (Nikonov et al., 2008b). Nonetheless, both arrestins significantly contribute to rapid recovery of cones (Nikonov et al., 2008b).
2. The mechanics of the arrestin-rhodopsin interaction
Arrestin-1 (called 48 kDa protein at the time) was identified as one of the proteins that, similar to transducin, selectively binds light-activated rhodopsin (Kuhn, 1978). However, it soon became clear that, unlike transducin, arrestin preferentially binds phosphorylated rhodopsin (Kuhn et al., 1984). In the rod, rhodopsin exists in multiple functional forms: inactive unphosphorylated (Rh), active unphosphorylated (Rh*), inactive phosphorylated (P-Rh), and active phosphorylated (P-Rh*). In addition, light-activated forms decay to opsin (Ops) and phospho-opsin (P-Ops). Thus, any mechanistic model of the arrestin-rhodopsin interaction first and foremost must explain arrestin specificity for a single functional form of rhodopsin, P-Rh*, which ensures timely shutoff of the photoresponse (Mendez et al., 2000; Xu et al., 1997).
2.1. What makes arrestin-1 specific for P-Rh*?
Obviously, to be selective for one particular functional form of rhodopsin, arrestin-1 must be able to go through a selection process in which it “tests” the functional status of rhodopsin. This implies that arrestin-1 must be able to bind with low affinity to all forms of rhodopsin and then, if it turns out not to be the appropriate one, to be able to dissociate quickly. To make experimental testing of this idea feasible, an extremely sensitive binding assay was needed. Luckily, arrestin-1 (and other members of the family) can be expressed in functional form in a cell-free translation system using rabbit reticulocyte lysate (Gurevich and Benovic, 1992; Gurevich et al., 1995). The beauty of this system is that amino acids are supplied exogenously along with added mRNA, so that labeled proteins are produced from radiolabeled amino acids. Because the synthesized protein is the only labeled macromolecule in the mix, it can be used for binding assays without further purification. Moreover, the sensitivity of the assay can be increased at will by using amino acids with higher specific activity or by using several radiolabeled amino acids instead of one. This makes accurate measurement of femtomolar binding feasible (Gurevich and Benovic, 1993), allowing fairly precise quantification of both high- and low-affinity interactions with P-Rh* and non-preferred functional forms of rhodopsin, respectively. In contrast, earlier methods, based on detecting bound arrestin-1 as the fraction pelleted with rhodopsin-containing membranes (Kuhn et al., 1984; Palczewski et al., 1991a) or on spectral detection of Meta II rhodopsin formed in the presence of arrestin-1 (Schleicher et al., 1989), are essentially “all-or-nothing” assays that readily detect arrestin-1 binding to P-Rh* but not to any other functional form of rhodopsin. Besides, by its nature extra-Meta II assay cannot be used for dark forms of rhodopsin or opsin.
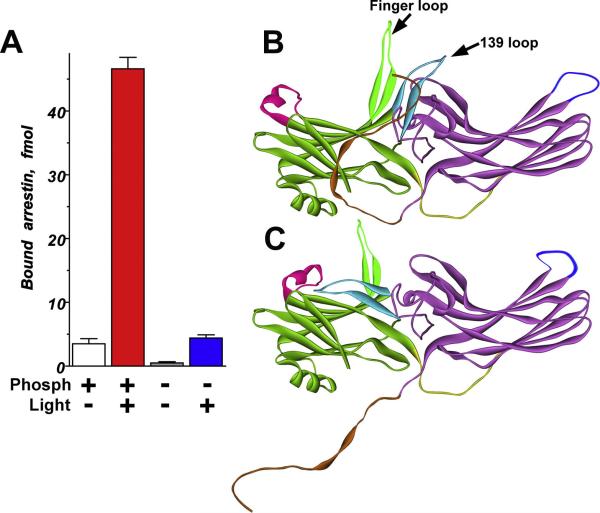
The direct binding assay with radiolabeled arrestin-1 was validated by the demonstration that it yields high binding to the preferred arrestin-1 target P-Rh*, (Kuhn et al., 1984), and virtually no binding to dark Rh (Gurevich and Benovic, 1992). Once these controls were in place, the assay was used to show for the first time that arrestin-1 specifically binds dark P-Rh and Rh*, albeit at much lower levels (Fig.1A) (Gurevich and Benovic, 1992). This was the first indication that arrestin-1 can recognize rhodopsin activation and phosphorylation independently of each other and implied that arrestin-1 must have at least two separate receptor-binding elements, one specifically interacting with the active rhodopsin conformation, and the other with the rhodopsin-attached phosphates. Although two-site interactions are usually cooperative, this is hardly sufficient to account for the 10-20-fold higher binding to P-Rh* than to dark P-Rh or Rh* (Fig. 1A).
Fig. 1. Arrestin-1 selectivity for P-Rh* is achieved by a multi-step binding mechanism.
A. Arrestin-1 demonstrates many times higher binding to P-Rh* than to inactive P-Rh or Rh*. Low-affinity interactions with dark P-Rh or Rh* are mediated by arrestin-1 elements that specifically recognize receptor-attached phosphates or the active state of rhodopsin, respectively. These elements act as phosphate and active conformation sensors, respectively. Only P-Rh* can engage both sites. Simultaneous activation of both sensors allows arrestin-1 transition from the basal (B) to the active high-affinity receptor-binding state (C). This transition involves the release of the arrestin C-tail (light brown), relatively small shifts in the relative positions of the N-domain (green) and C-domain (purple), significant rearrangement of flexible loops on the receptor-binding surface: “finger loop” (light green), “139 loop” (light blue), “344 loop” (dark blue), and “163 loop” (pink), and is limited by the length of the inter-domain hinge (yellow).
The first mechanistic explanation of arrestin-1 selectivity, the sequential multi-site binding model, was proposed in 1993 (Gurevich and Benovic, 1993). On the basis of the observed selectivity profile, the model (reviewed in (Gurevich and Gurevich, 2004)) posits that arrestin-1 first binds rhodopsin either via its elements that specifically interact with the light-activated rhodopsin conformation, or via residues that directly bind rhodopsin-attached phosphates. These interactions mediate the observed low-affinity binding to Rh* and dark P-Rh, respectively. The latter also mediates arrestin-1 binding to phosphoopsin (Gurevich and Benovic, 1993). If the phosphates or the active state turn out to be the only “attraction”, then arrestin-1 binds with low affinity and then dissociates quickly. However, when arrestin-1 encounters P-Rh*, and only in this case, the other site also finds its target on the same rhodopsin molecule. Simultaneous engagement of both primary sites relieves conformational constraints and allows arrestin transition into the high-affinity rhodopsin-binding state, bringing additional arrestin elements into contact with rhodopsin. This transition involves significant conformational rearrangements in the arrestin-1 molecule (Fig. 1B,C). The new contact surface provides extra energy of the interaction, which accounts for the much greater affinity for P-Rh*. As an added bonus, the model also explains why, in the end, arrestin-1 dissociates: when P-Rh* decays to phosphoopsin, the elements that specifically engaged light-activated rhodopsin no longer have their partners, so that arrestin-1 returns to its basal conformation. The model predicts that arrestin-1 can hold onto phosphoopsin only via its phosphate-binding residues, so that reduced affinity for this form would allow faster dissociation. Indeed, arrestin-1 release was previously shown to be required for rhodopsin dephosphorylation (Palczewski et al., 1989).
The model yields several experimentally testable predictions. First, it implies a significant conformational change in the arrestin-1 molecule during rhodopsin binding. This predicts that the arrestin-1 structure must be compatible with global conformational rearrangements. Second, the model predicts that the basal conformation of arrestin-1 is constrained by several intra-molecular interactions, the elimination of which can enhance its binding to the non-preferred forms of rhodopsin, Rh* and dark P-Rh. Third, in the model the primary binding sites on arrestin-1 function as “sensors”, translating interaction with corresponding parts of P-Rh* into the relief of conformational constraints. This predicts that mutagenesis of certain residues can relieve these constraints without the receptor. For example, turning the phosphate sensor “on” by mutations of certain phosphate-binding residues should create mutants that do not require rhodopsin-attached phosphates for activation, but would bind with high affinity to any active form of rhodopsin, Rh* and P-Rh*. The first prediction was consistent with the high Arrhenius activation energy reported earlier (Schleicher et al., 1989). The second was supported by a previous report that the deletion of the arrestin-1 C-terminus increases its binding to dark P-Rh (Palczewski et al., 1991a) and Rh* (Gurevich and Benovic, 1992). The third broke completely new ground, predicting the feasibility of designing activation- or phosphorylation-independent mutants that would bind with high affinity to any phosphorylated or activated form of rhodopsin, respectively, and specifying particular structural features that WT arrestin must have to make this possible.
2.2. Structural predictions before the crystal
The function of the arrestin C-tail as an inhibitor of its binding to non-preferred forms of rhodopsin, dark P-Rh (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993; Palczewski et al., 1991a) and Rh* (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993) was suggested based on the binding profiles of arrestins where this element was removed by proteolysis (Palczewski et al., 1991a) or mutagenesis (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993). Subsequent studies showed that the deletion of the first 16 amino acids in the N-terminus similarly reduces arrestin selectivity, and that the effects of N- and C-terminal deletions are non-additive (Gurevich et al., 1994). The net positive charge of the N-terminus, the high negative charge of the C-tail, and the fact that heparin, a sulphated carbohydrate with multiple negative charges, appears to substitute for the arrestin C-tail by increasing the selectivity of truncated arrestin for P-Rh*, led to the idea that the arrestin termini interact (Gurevich et al., 1994). The interaction between arrestin N- and C-termini was proposed to ensure arrestin selectivity for P-Rh* by preventing its binding to non-preferred forms of rhodopsin (Gurevich et al., 1994). Extensive mutagenesis of the C-terminus of arrestin-1 revealed that multiple non-redundant mechanisms prevent arrestin binding to dark P-Rh and Rh*. These are mediated by several negative charges, one positive charge, and three hydrophobic residues in the arrestin C-tail (Gurevich, 1998). These data along with an earlier finding that both heparin and P-Rh* make the C-tail more exposed to proteases (Palczewski et al., 1991c), suggested that receptor-induced C-tail release is part of arrestin transition into the active high-affinity binding state.
The first success in the search for putative sensors in arrestin was also achieved before the crystal structure became available. Previous findings that arrestin requires multiple rhodopsin-attached phosphates for high-affinity binding (Gurevich and Benovic, 1993; Wilden et al., 1986) suggested that an arrestin element with multiple positive charges must be involved in phosphate binding. Assuming that these are clustered in the linear sequence, bovine arrestin-1 has only one obvious candidate, residues 163-176, carrying six positive charges, five of which were conserved in all arrestins cloned at the time (Gurevich and Benovic, 1995). The model predicts that the elimination of a residue that simply binds phosphates should reduce the binding to any phosphorylated form, dark P-Rh and P-Rh*, without affecting Rh* binding. In contrast, mutation of the phosphate sensor was expected to “activate” arrestin-1, enhancing its binding to Rh*, whereas its effects on P-Rh and P-Rh* binding were impossible to predict. The substitution of five of the charges in the 163-176 element with neutral hydrophilic residues, K163S, K166S, K167S, R171Q, and K176S, duly decreased P-Rh* binding without affecting the Rh* interaction (Gurevich and Benovic, 1995). The sixth mutation, R175N, yielded a most peculiar phenotype: the binding to Rh* was dramatically increased, virtually to the level of WT arrestin-1 binding to P-Rh*, matching the predicted result of the activation of the proposed phosphate sensor perfectly (Gurevich and Benovic, 1995). However, R175N also caused a concomitant significant increase in P-Rh* binding, which was hard to reconcile with the idea that Arg175 interacts with phosphates and therefore contributes to the overall binding energy. A simpler tool was used to sort this out: arrestin-(1-191) binds dark P-Rh and Rh* with low affinity and demonstrates somewhat higher binding to P-Rh*, consistent with a simple cooperative two-site interaction, yet its binding does not involve any conformational changes that complicate analysis of the data (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993). In the context of this unsophisticated mini-arrestin, the R175N mutation reduced the binding to P-Rh* and did not affect the Rh* interaction, just like the neutralization of other positive charges in this region (Gurevich and Benovic, 1995). These data proved that Arg175 indeed binds phosphates, identifying this residue as a key part of the phosphate sensor.
The simplest possible model explaining the R175N phenotype was proposed: in the basal state of arrestin, Arg175 is likely engaged in an intra-molecular interaction with a negatively charged partner. Receptor-attached phosphates neutralize its charge, disrupting this interaction, which then serves as a signal for the rest of the arrestin molecule that the phosphates are in place. The R175N mutation achieves the same result without phosphates, “tricking” arrestin into treating any rhodopsin as phosphorylated, making activation sufficient to trigger high-affinity binding (Gurevich and Benovic, 1995). Mechanistically, this model suggests that putative interaction of Arg175 with a negative charge must stabilize the basal state, whereas its disruption allows arrestin transition into a distinct “active” conformation. This idea received further support from experiments replacing Arg175 with every possible residue (Gurevich and Benovic, 1997). This study showed that the most conservative substitution preserving the charge, R175K, does not activate arrestin-1. Negatively charged residues in this position in full-length arrestin-1 yield maximum increase in Rh* binding, and virtually all other residues have qualitatively similar, although smaller “activating” effects. Importantly, in the context of truncated arrestin-1-(1-191) the effect of various residues correlated well with their expected ability to bind the phosphates: positive charges Arg, Lys, and His yielded the highest P-Rh* binding, followed by residues with H-bonding capability, whereas bulky hydrophobic and negatively charged residues yielded similar low binding levels. These data confirmed that Arg175 directly binds a negatively charged moiety, i.e., phosphate, upon arrestin-1 binding to the phosphoreceptor. The disruption of Arg175 interaction with an internal negative charge in arrestin-1 underlies its activation, enabling its binding to unphosphorylated Rh*, as well as a truncated form of rhodopsin, 329G-Rh*, where the C-terminus with all phosphorylation sites have been proteolytically removed (Gurevich and Benovic, 1997). Direct demonstration using purified proteins in vitro that the R175E mutant effectively quenches Rh* signaling, whereas WT arrestin-1 does not (Gray-Keller et al., 1997), confirmed the functional significance of this element of the phosphate sensor in arrestin-1. Thus, two intra-molecular interactions: between the arrestin-1 termini and between Arg175 and an internal negatively charged partner, were proposed to stabilize the basal state of arrestin before the crystal structure was solved.
2.3. Arrestin structure as the key to its function
The first crystal structure showed that arrestin-1 is a virtually all-β-strand two-domain molecule (Granzin et al., 1998). Importantly, Arg175 was found at the inter-domain interface within a group of five charged residues, three of which were aspartic acids (Asp30, Asp 296, Asp303) suitably positioned to play the role of the intra-molecular negatively charged Arg175 partners proposed earlier (Gurevich and Benovic, 1997; Gurevich et al., 1995). However, no part of the C-tail appeared to be resolved at 3.5 Å, so the ideas regarding its role and proximity to the N-terminus remained untested.
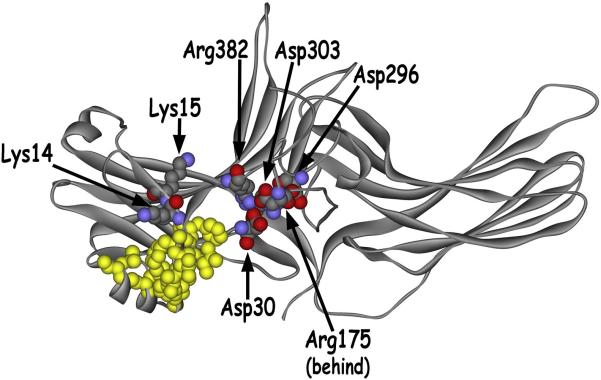
The second higher resolution (2.8 Å) structure (Hirsch et al., 1999) proved more informative. It confirmed that arrestin-1 is an elongated two-domain molecule, with relatively few contacts between the domains, making it perfectly designed for the global conformational rearrangements predicted by the sequential multi-site binding model (Gurevich and Benovic, 1993). It confirmed that Arg175 is part of the arrangement of five charged residues in the middle of the molecule (Fig. 2). This element was termed the polar core, because these residues are largely solvent-excluded, in contrast to the majority of charged residues in most soluble proteins that are localized on the surface. It also revealed part of the C-tail in close contact with the N-domain via a three-element interaction with the N-terminal β-strand I and α-helix I (Fig. 2), confirming the hypothesis that the two arrestin-1 termini interact (Gurevich et al., 1994). Importantly, it provided a plausible mechanism whereby rhodopsin-attached phosphates can facilitate the observed release of the arrestin-1 C-tail (Palczewski et al., 1991c). It turned out that one of the positive charges in the polar core is supplied by the C-tail residue Arg382 that was earlier shown to contribute to arrestin-1 selectivity for P-Rh* (Gurevich, 1998). This side chain was misidentified as Lys2 in the first structure, which was shown not to affect arrestin-1 specificity by follow-up mutagenesis (Vishnivetskiy et al., 1999). Since the interaction of receptor-attached phosphates with Arg175 would destabilize the polar core, correct identification of Arg382 suggested that one effect of the phosphates was to promote the release of the C-tail. Both structures identified the same three aspartates in the proximity of Arg175, and subsequent exhaustive mutagenesis showed that Asp296 is the main intra-molecular partner of Arg175. The reversal of either charge by R175E or D296R mutation yields essentially the same phenotype, greatly enhanced binding to Rh* (Vishnivetskiy et al., 1999). Importantly, simultaneous reversal of both charges, which reconstructs the salt bridge in an alternative configuration, was shown to suppress Rh* binding, restoring high selectivity for P-Rh*. Collectively, these data proved that the salt bridge between Arg175 and Asp296 is the main phosphate sensor in arrestin-1.
Fig. 2. Critical inter-domain interactions disrupted during arrestin-1 activation by rhodopsin-attached phosphates.
The arrestin-1 crystal structure (Hirsch et al., 1999) shows that the polar core, localized at the inter-domain interface, stabilizes the relative orientation of the two arrestin-1 domains. The polar core consists of five charged solvent-excluded residues (Asp30, Arg175, Asp296, Asp303, Arg382 shown as CPK models) that form a network of interactions and includes the salt bridge between Arg175 and Asp296 that serves as the main phosphate sensor. The C-tail folds back on the N-domain, where it participates in the three-element interaction with β-strand I and α-helix I. Bulky hydrophobic residues mediating this interaction are shown in yellow as CPK models. Two adjacent positive charges in β-strand I (Lys14 and Lys15, shown as CPK models) bind receptor-attached phosphates. Their engagement by phosphates and consequent change in orientation contributes to the destabilization of the three-element interaction, with subsequent release of the arrestin-1 C-tail (see Fig. 1). Destabilization of both interactions constitutes part of the arrestin-1 activation mechanism, facilitating its transition into a high-affinity rhodopsin-binding state.
The crystal also revealed a “three-element interaction” involving three hydrophobic residues in the C-tail (Phe375, Val376, and Phe377), Val11-Ile12-Phe13 in the N-terminal β-strand I, and leucines 103, 107, and 111 aligned on the same side of α-helix I (Fig. 2) (Hirsch et al., 1999). The latter likely contributes more to the anchoring of the C-tail to the body of the N-domain than the interaction of Arg382 with polar core residues. However, the structure per se did not identify a clear mechanism whereby receptor-attached phosphates disrupt this three-element interaction to promote release of the arrestin C-tail upon P-Rh* binding (Palczewski et al., 1991c). A subsequent study showed that, as expected, direct disruption of the hydrophobic three-element interaction by alanine substitutions of the hydrophobic residues on the C-tail (3A), on β-strand I, or on α-helix I promotes arrestin-1 binding to Rh* (Vishnivetskiy et al., 2000). Unexpectedly, this study found that alanine substitutions of Lys14 and especially Lys15 dramatically reduce arrestin-1 binding to P-Rh*, and simultaneous substitution of both residues virtually prevents binding completely (Vishnivetskiy et al., 2000). The effect of other substitutions suggested that the two lysines likely bind receptor-attached phosphates: K14R and K15R with a conserved positive charge worked essentially like WT arrestin-1, whereas charge reversal mutations K14E and K15E reduced the binding to P-Rh* to an even greater extent than the corresponding alanine substitutions. In fact, K15E still remains the most disabling point mutation ever described in arrestin-1. Interestingly, the same K14A and K15A substitutions in the context of the “pre-activated” phosphorylation-independent arrestin-1 mutants either with a destabilized polar core (R175E and D296R), or a detached (3A) or deleted (1-378) C-tail had virtually no effect on P-Rh* and Rh* binding (Vishnivetskiy et al., 2000). These data suggested that these two highly exposed lysines bind the phosphates first, subsequently “delivering” them to the half-buried Arg175 (Fig. 2). Thus, their function is vital in the context of WT arrestin-1, but becomes dispensable when the polar core is already disrupted or when arrestin-1 is pre-activated by the forced detachment or deletion of the C-tail. Considering that Lys14 and Lys15 are localized on the same β-strand as residues Val11-Ile-12-Phe13 which mediate the three-element interaction, and that their side chains point in opposite directions (Fig. 2), it stands to reason that the approach of phosphates from any direction would force at least one of them to shift or flip over. This would destabilize β-strand I, likely moving adjacent bulky hydrophobic residues out of a position favorable for interaction with the C-tail and α-helix (Vishnivetskiy et al., 2000). This model provides the missing mechanistic connection between phosphate binding and C-tail release and explains why phosphates are no longer required when the arrestin C-tail is deleted by mutagenesis (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993), alternative splicing (Smith et al., 1994), or detached by the 3A mutation (Gurevich, 1998). Thus, the correct crystal structure supplied a plausible reason why the substitution of polar core residue Arg382 with an uncharged Asn increases Rh* binding much less than the triple alanine substitution of Phe375-Val376-Phe377 (3A mutation) (Gurevich, 1998) and why the deletion of N-terminal residues 2-16 enhances arrestin binding to Rh* (Gurevich and Benovic, 1993; Gurevich et al., 1994).
The mechanics of the putative activation sensor in arrestin that detects the active state of the receptor are not well established. Indirect evidence indicates that the rest of the inter-domain surface is involved (Hanson and Gurevich, 2006), since its destabilization by mutations enhances binding to dark P-Rh, but the effects are far less impressive that those of manipulation of the phosphate sensor. Interestingly, non-visual arrestin-2 and -3 demonstrate less than a two-fold differential in binding to inactive P-Rh and P-Rh*, as well as to active and inactive phosphorylated forms of their cognate non-visual receptors (Gurevich et al., 1995; Gurevich et al., 1993), as if the activation sensor in these subtypes is already halfway “on”. This issue still needs to be addressed experimentally.
2.4. Implications of arrestin activation by receptor-attached phosphates
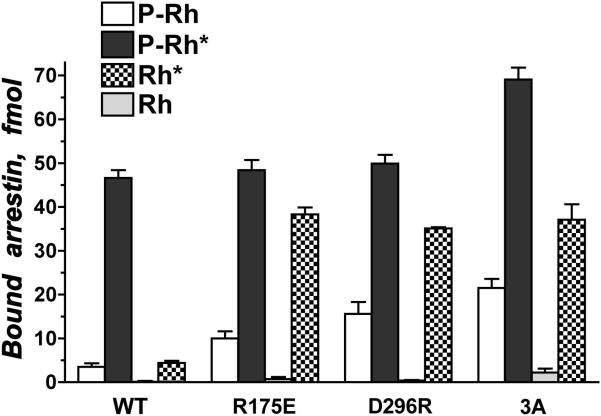
The evidence suggests that the two best-described conformational rearrangements in arrestin, the “melting” of the polar core and the release of the C-tail, are induced by receptor-attached phosphates. The effects of the mutations suggest that the mechanism of phosphate action on the two lysines in β-strand I and the polar core is purely electrostatic, i.e., any spatially concentrated negative charge should be able to “activate” arrestin. Indeed, highly negatively charged molecules, such as the polysulfated carbohydrate heparin or inositol-hexaphosphate, have been shown to engage arrestin residues that bind the phosphates (Zhuang et al., 2010), compete with P-Rh* for arrestin binding (Gurevich et al., 1994; Palczewski et al., 1991b), and induce the release of the arrestin C-tail (Palczewski et al., 1991c). The measurements of the mobility of individual C-tail residues by NMR (Zhuang et al., 2010) and of distances between the C-tail and other parts of the molecule by EPR (Hanson et al., 2006b; Vishnivetskiy et al., 2010) indicate that, in contrast to the basal state, in rhodopsin-bound arrestin-1 the C-tail does not have a fixed position and is simply “flopping” around. The level of Rh* binding to pre-activated mutants (Fig. 3) (Gurevich, 1998; Gurevich and Benovic, 1995; Gurevich and Benovic, 1997; Vishnivetskiy et al., 1999; Vishnivetskiy et al., 2000) also indicates that direct interaction with the phosphates contributes relatively little to the overall energy of arrestin binding. The ability of an exogenous rhodopsin C-terminal peptide carrying seven phosphates (Puig et al., 1995) or a phosphate mimic like heparin (Gurevich et al., 1994) to stimulate arrestin-1 binding to Rh* strongly supports this idea.
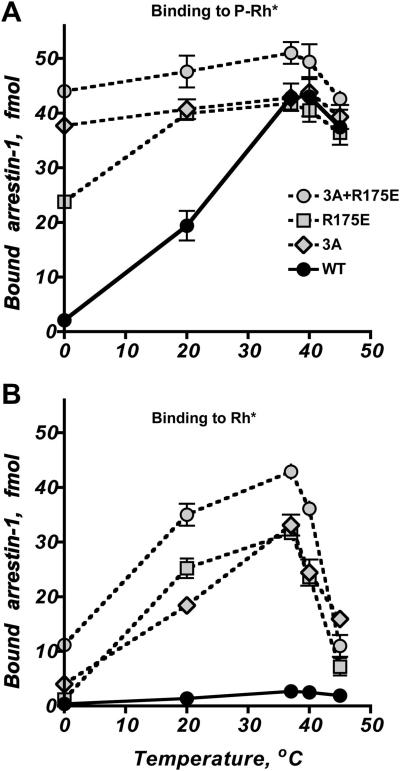
Fig. 3. Specific mutations in arrestin-1 pre-activate the phosphate sensor, yielding mutants with high affinity for active unphosphorylated Rh*.
Targeted disruption of the salt bridge between Arg175 and Asp296 by charge reversal mutations R175E or D296R, as well as forced detachment of the arrestin-1 C-tail by triple alanine substitution of bulky hydrophobic residues that anchor it to the body of the N-domain (3A) yield partially activated arrestin-1 mutants with greatly increased binding to Rh*, and to a lesser extent, dark P-Rh. These data reveal the molecular mechanism of arrestin-1 activation and demonstrate the potential of targeted manipulation of arrestin-1 selectivity for different functional forms of rhodopsin.
The mechanism of arrestin activation by receptor-attached phosphates suggests that arrestin cannot be particularly sensitive to the sequence context of phosphorylated residues. Indeed, both in vitro (Vishnivetskiy et al., 2007) and in vivo (Mendez et al., 2000) arrestin requires three phosphates on rhodopsin for high-affinity binding, but does not care which particular residues out of six (mouse) or seven (bovine) serines and threonines are phosphorylated. The extra sites can serve as backups and additional targets for rhodopsin kinase (GRK1) that simply increase the probability that GRK1 binding to active rhodopsin will result in phosphate transfer, thereby facilitating rhodopsin shutoff (Caruso et al.). This property also explains why arrestin-1 effectively shuts off signaling by cone opsin that is transgenically expressed in rods (Shi et al., 2007) and endogenously expressed in cones, where ~98% of the total arrestin complement is represented by arrestin-1 (Nikonov et al., 2008a). The mechanism of phosphate sensing is conserved in arrestin-21, -3, and -4 (Carter et al., 2005; Celver et al., 2002; Gurevich et al., 1997; Kovoor et al., 1999; Sutton et al., 2005). In fact, the ability of arrestin to be activated by phosphates regardless of the surrounding sequence allows only two non-visual subtypes in vertebrates to terminate signaling by hundreds of G protein-coupled receptors (GPCRs). This mechanism must have appeared early in evolution: the polar core residues and hydrophobics mediating the three-element interaction are remarkably conserved in all known arrestins (Gurevich and Gurevich, 2006a). The resulting broad receptor specificity also appears to be conserved: a single arrestin works with the hundreds of GPCRs present in the round worm C. elegans (Palmitessa and Benovic, 2010; Palmitessa et al., 2005), in the proto-chordate Ciona intestinalis the same arrestin acts as visual in the eyes of mobile larva and as non-visual in the blind sessile adult (Nakagawa et al., 2002), and a single non-visual subtype in Drosophila (kurtz) serves all non-sensory GPCRs (Ge et al., 2006; Roman et al., 2000).
2.5. The rhodopsin-binding surface and receptor specificity
Several groups using a wide range of methods have identified arrestin-1 elements involved in rhodopsin binding. Deletion mutagenesis showed that multiple parts of the N- and C-terminal halves of the molecule are involved and tentatively excluded ~40 C-terminal residues (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993). Lysine acetylation, hydrogen/deuterium exchange, and arrestin-1/2 chimeras were used to establish that many parts of arrestin-1 become shielded in the arrestin-rhodopsin complex (Gurevich et al., 1995; Gurevich et al., 1993; Ohguro et al., 1994). Site-directed mutagenesis identified multiple positively charged arrestin-1 residues interacting with receptor-attached phosphates (Gurevich and Benovic, 1995; Gurevich and Benovic, 1997; Sutton et al., 2005). The elucidation of the arrestin-1 crystal structure revealed that it is an elongated molecule consisting of two cup-like domains (Granzin et al., 1998; Hirsch et al., 1999). All previously identified rhodopsin-binding elements mapped to the concave sides of the two domains (Fig. 4), identifying the side of the molecule that faces the receptor in the complex (Gurevich and Gurevich, 2004). The two arrestin peptides that prevent its binding to rhodopsin by competing with full-length arrestin-1 (Pulvermuller et al., 2000) were also localized on this surface. Recent studies of the effects of the systematic mutagenesis of arrestin-1 surface charges on its binding to different functional forms of rhodopsin (Hanson and Gurevich, 2006) and analysis of changes in the mobility of spin labels on the arrestin-1 surface by dark P-Rh and P-Rh* (Hanson et al., 2006b; Vishnivetskiy et al., 2010) confirmed that the concave sides of the arrestin-1 domains constitute the rhodopsin-binding surface. The latter study also identified one of the loops in the central crest of the molecule (“finger loop”, residues 68-78; Fig. 1B) as the key player in the interaction. Several positions in this loop showed the most dramatic immobilization upon dark P-Rh binding, and an even greater loss of mobility upon binding to P-Rh* (Hanson et al., 2006b).
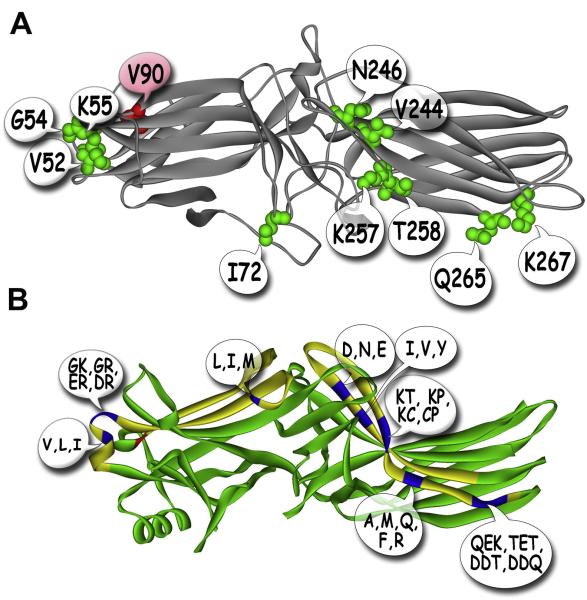
Fig. 4. Receptor specificity of arrestins is encoded in the structure of the receptor-binding surface.
A. View of the receptor-binding surface. Four exposed residues on the concave side of the N-domain (Val52, Gly54, Lys55, Ile72) and six in the C-domain (Val244, Asn246, Lys257, Thr258, Gln265, Lys267) determine arrestin-1 preference for P-Rh*. Val90 (shown in red) makes the β-strand sandwich of the N-domain more rigid, ensuring strict receptor specificity of arrestin-1. Ser86 or Ala87 replace Val90 in non-visual arrestin-2 and -3, respectively, ensuring broad receptor specificity of these subtypes. The replacement of homologous arrestin-2 residues with those derived from arrestin-1 also fully reverses arrestin-2 receptor preference, yielding a mutant with very high binding to P-Rh* and low binding to other GPCRs (Vishnivetskiy et al., 2011). B. Side view of the arrestin-2 crystal structure (Han et al., 2001), where the elements that determine its receptor preference are shown in yellow, key receptor discriminator residues within these elements are shown in blue, and buried Ser86 is shown in red. In over 600 million years of arrestin evolution, very few residues (out of 20 possible) occupy the positions responsible for receptor preference (shown in quote bubbles) (Gurevich and Gurevich, 2010a).
The fact that all animals have hundreds of different GPCRs and only a few arrestin subtypes (Gurevich and Gurevich, 2006a) raises the issue of receptor specificity. This becomes particularly intriguing considering that arrestin-1 and -4 are expressed only in photoreceptors and shut off opsins, whereas the two non-visual arrestins in vertebrates and one in Drosophila regulate an incredible variety of other GPCRs. In vitro, arrestin-1 shows remarkable preference for P-Rh* over other active phosphorylated receptors, whereas non-visual arrestin-2 and -3 demonstrate much lower binding to P-Rh* (Gurevich et al., 1995; Gurevich et al., 1993). This suggested that the construction of arrestin-1/2 chimeras is the most straightforward approach to the identification of arrestin elements responsible for receptor specificity. The exchange of elements between arrestin-1 and -2 should increase the binding to the receptor preferred by the donor and reduce the binding to the preferred target of the acceptor arrestin (Vishnivetskiy et al., 2004). This study identified two elements, one comprising β-strands V and VI (with adjacent loops) in the N-domain (residues 49-90), the other β-strands XV and XVI in the C-domain (residues 237-268) (Fig. 4). The exchange of both pieces between arrestin-1 and -2 completely reversed their receptor preference even though each chimera retained ~95% of the original sequence because of high homology (Vishnivetskiy et al., 2004). Subsequent reversal mutagenesis of non-conserved residues identified ten that play a key role in receptor discrimination: four in the N-domain (V52, G54, K55, I72) and six in the C-domain (V244, N246, K257, T258, Q265, K267) (Fig. 4A) (Vishnivetskiy et al., 2011). The introduction of these residues into corresponding positions in arrestin-2 yielded a mutant that binds P-Rh* at least as well as arrestin-1. Interestingly, the replacement of these residues with alanines in arrestin-1, -2, and -3 essentially killed their ability to bind their cognate receptors, indicating that receptor discriminator residues are also key drivers of the interaction (Vishnivetskiy et al., 2011). Interestingly, in at least 600 million years of arrestin evolution (Gurevich and Gurevich, 2006a) only a few different residues can be found in all key positions that determine receptor preference (Fig. 4B).
It is a wonder why arrestin-1 needs to be receptor specific at all considering that it is found virtually exclusively in rod and cone photoreceptors where it is expressed at very high levels (Hanson et al., 2007b; Nikonov et al., 2008a; Song et al., 2011; Strissel et al., 2006), along with huge amounts of rhodopsin and cone opsins, respectively (Pugh and Lamb, 2000). For example, arrestin-4 binds opsins and other GPCRs essentially as well as non-visual arrestins in vitro (Sutton et al., 2005), suggesting that its specificity is determined only by its selective expression in cones (Chan et al., 2007; Nikonov et al., 2008a). The difference is in absolute concentrations: there is >2 mM arrestin-1 in rods (Kim et al., 2011a; Song et al., 2011) and >0.6 mM in cones, where it represents ~98% of total arrestin complement (Nikonov et al., 2008a) (whereas arrestin-4 is expressed at a much lower level (Nikonov et al., 2008a)). Ignoring the outer segment, the rest of the photoreceptor is a normal neuron, with a variety of GPCRs regulating synaptic transmission at the terminals. In contrast to arrestin-2 and –3, arrestin-1 does not have a clathrin-binding site localized in the C-terminus (Goodman et al., 1996), and therefore does not effectively support GPCR internalization. Since at millimolar concentrations even low affinity is sufficient for binding, it is likely that evolution made arrestin-1 highly selective for rhodopsin to prevent it from unduly interfering with the trafficking of non-visual GPCRs in synaptic terminals.
2.6. The stoichiometry of the arrestin-rhodopsin complex
Many GPCRs appear to oligomerize at least during some stages of their life cycle, suggesting that receptor monomers, dimers, and/or larger oligomers could play distinct functional roles (Gurevich and Gurevich, 2008b; Milligan, 2009, 2010). Rhodopsin in disc membranes was observed to form multi-molecule arrays by freeze-fracture electron microscopy (Roof and Heuser, 1982). Dramatic images of these arrays generated by atomic force microscopy in mica-adsorbed disc membranes twenty years later (Fotiadis et al., 2003) sparked a debate whether the rhodopsin monomer or dimer is the functional unit (Chabre and le Maire, 2005; Fotiadis et al., 2006). In view of these data, rhodopsin diffusion in the OS discs was measured again, with surprisingly disparate results. In Xenopus OS rhodopsin diffusing slow enough to account for these huge arrays was not detected (Wang et al., 2008), whereas in salamander (Ambystoma), frog (Rana), toad (Bufo), and Gecko rods a fraction of rhodopsin, that varied for unknown reasons from virtually zero to 100% in different cells, was found to be essentially immobile (Govardovskii et al., 2009). The authors of the latter study presented a pretty compelling argument why large rhodopsin arrays are unlikely to contribute to signaling, and hypothesized that if the assembly of rhodopsin into these arrays is regulated, it likely serves to remove it from the active pool to reduce light sensitivity of the rod (Govardovskii et al., 2009). In contrast, rhodopsin dimer would be easily accessible for transducin and other binding partners, and its expected diffusion is only ~40% slower than that of the monomer, putting the difference well within experimental error and leaving the dimer as a possible actor. Members of three protein families preferentially bind active GPCRs: G proteins, GRKs, and arrestins. Modeling the crystal structures of rhodopsin in its inactive (Li et al., 2004; Palczewski et al., 2000) and active form (Choe et al., 2011; Park et al., 2008; Scheerer et al., 2008; Standfuss et al., 2011) with those of heterotrimeric transducin (Lambright et al., 1996 ), GRK1 (Singh et al., 2008), and arrestin-1 (Granzin et al., 1998; Hirsch et al., 1999) suggests that all of these interaction partners can potentially dock with a rhodopsin dimer if turned in a particular way (Fotiadis et al., 2006). While modeling without follow-up experimentation does not provide answers, it can raise valid questions.
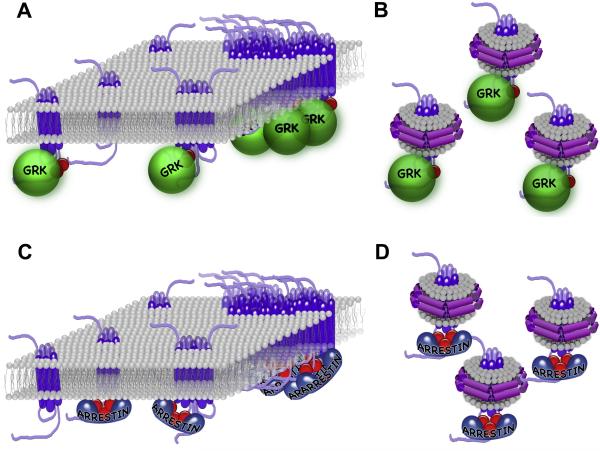
Since there is no reason to believe that any GPCR, including rhodopsin, performs all functions in the same oligomerization state (Gurevich and Gurevich, 2008a), the possible role of rhodopsin dimers in interactions with transducin, GRK1 (rhodopsin kinase), and arrestin-1 are separate issues that have recently progressed from heated discussions to experimental testing. Monomeric solubilized rhodopsin has been shown to activate transducin at the maximum rate allowed by diffusion (Ernst et al., 2007). Rhodopsin reconstituted as a monomer into lipid bilayer nanoparticles has also been shown to effectively activate transducin by three different groups (Banerjee et al., 2008; Bayburt et al., 2007; Whorton et al., 2008). Moreover, nanoparticles containing two rhodopsin molecules were found to be half as efficient as the monomer (Bayburt et al., 2007). Other class A GPCRs, such as the β2-adrenergic receptor (Whorton et al., 2007) and the neurotensin NTS1 receptor (White et al., 2007) were also shown to effectively couple to their cognate G proteins as monomers, and less so as a dimer (White et al., 2007). Thus, G proteins do not need more than a single receptor for activation. If oligomers play any role in G protein interaction, it is most likely for the inhibition of productive coupling. Similarly, GRK1 was shown to phosphorylate monomeric rhodopsin in nanoparticles at least as efficiently as rhodopsin in native disc membranes (Fig. 5B) (Bayburt et al., 2011).
Fig. 5. Monomeric rhodopsin is the physiologically relevant target of GRK1 and arrestin-1.
A,C. Rhodopsin in native disc membranes, as well as other GPCRs, can form dimers and other higher order oligomers. Although oligomeric forms of class A GPCRs tend to be very transient, with a sub-second half-life ((Fonseca and Lambert, 2009; Hern et al., 2010; Kasai et al., 2011; Lan et al., 2011); reviewed in (Gurevich and Gurevich, 2008b; Lambert, 2010)), their existence raises the question of whether monomers or dimers are biologically relevant partners of G proteins, GRKs, and arrestins. To determine the form of rhodopsin that is phosphorylated by GRK1 and binds arrestin, rhodopsin was reconstituted into nanodiscs as a monomer and its phosphorylation by purified GRK1 and the subsequent ability of monomeric P-Rh* to bind arrestin-1 were tested (Bayburt et al., 2011). The results showed that monomeric Rh* in nanodiscs is phosphorylated by GRK1 at least as efficiently as rhodopsin in disc membranes (B), and monomeric P-Rh* in nanodiscs bound arrestin-1 with physiologically relevant affinity (KD ~4 nM) and with the same 1:1 stoichiometry observed in vivo and in vitro in disc membranes (D). Along with evidence that monomeric Rh* efficiently activates transducin (Banerjee et al., 2008; Bayburt et al., 2007; Ernst et al., 2007; Whorton et al., 2008), these data demonstrate that the rhodopsin monomer is the functional unit at all steps of signaling and inactivation.
Transducin and GRK1 are present in rods at much lower levels than rhodopsin (Pugh and Lamb, 2000), whereas arrestin-1 is expressed at ~0.8:1 ratio to rhodopsin (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006). Since massive arrestin-1 translocation to the outer segment (OS) in the light (see section 4) is driven by its binding to rhodopsin (Nair et al., 2005), this phenomenon can be exploited to determine the stoichiometry of the interaction in living animals. The study of the limits of arrestin-1 translocation in genetically modified mice expressing arrestin-1 at different ratios to rhodopsin (from ~0.4 to 2.7), showed that the amount of arrestin-1 that can move to the OS is determined by the molar rhodopsin content of this compartment, reaching up to 0.83 of the latter (Hanson et al., 2007b). Obviously, this parameter would never exceed 0.5 if two rhodopsins were required to bind a single arrestin-1. However, one cannot just assume that rhodopsin is the only protein in the OS that binds arrestin-1. Therefore, the same arrestin-1/rhodopsin ratios were used in experiments with two pure proteins in vitro, which yielded the same result: arrestin-1 saturates rhodopsin at a 1:1 ratio (Hanson et al., 2007b). Still, one could argue that since arrestin-1 oligomerizes (see section 3), the binding of an arrestin dimer to a rhodopsin dimer could account for the data. This issue was addressed by experimental assessment of the functional capabilities of arrestin-1 oligomers, which showed that only monomeric arrestin-1 can bind rhodopsin (Hanson et al., 2007c). Collectively these data clearly showed that each rhodopsin molecule binds its own arrestin, but did not rule out a possible role for the rhodopsin dimer in the process. In all of the in vivo and in vitro experiments described above, rhodopsin was in native disc membranes where it had the opportunity to oligomerize (Fig. 5A,C). Thus, one could easily picture rhodopsin dimer binding two, rather than one, molecules of arrestin-1. Recently two different groups used monomeric rhodopsin in nanoparticles and showed that a single rhodopsin molecule is sufficient for arrestin binding (Bayburt et al., 2011; Tsukamoto et al., 2010). Moreover, the affinity of arrestin-1 for monomeric P-Rh* was found to be at least as high as in native membranes (apparent KD ~3-4 nM), with the same 1:1 binding stoichiometry (Bayburt et al., 2011) (Fig.5D). Interestingly, the same 1:1 stoichiometry of arrestin translocation to photoactivated rhodopsin was recently reported in Drosophila (Satoh et al., 2010), indicating that this mechanism is conserved in evolution. Thus, arrestin-1 requires a single P-Rh* molecule for physiologically relevant high-affinity binding. It makes perfect sense that the same monomeric form of rhodopsin that activates transducin to initiate signal transduction also serves as a substrate for GRK1, and ultimately is targeted by arrestin-1 to terminate the signaling. This molecular mechanism is consistent with the established single photon sensitivity of the rod (Baylor et al., 1979): since one photon can only isomerize 11-cis-retinal in a single rhodopsin molecule, it stands to reason that an individual active rhodopsin is the signaling unit, as well as the substrate of GRK1 and the target of arrestin-1 (Fig. 5).
Rods only work as photoreceptors in relatively dim light (Burns and Arshavsky, 2005). In bright daylight when cones take over, rods essentially exist in a survival mode until dusk. At least two mechanisms apparently prevent “useless” signaling in bright light: the bulk of transducin leaves the OS (Sokolov et al., 2002) while arrestin-1 moves in and binds rhodopsin (Nair et al., 2005). However, mice do not express enough arrestin-1 to block every rhodopsin molecule (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006). A recent study suggests that at high bleaching levels each molecule of arrestin-1 may be binding more than a single rhodopsin (Sommer et al., 2011). It appears that arrestin binding of only one P-Rh* with nanomolar affinity is necessary to quench signaling, and that this binding manifests itself by stabilization of the Meta II state (Schleicher et al., 1989). In contrast, arrestin-1 that binds two rhodopsins demonstrates much lower affinity (Sommer et al., 2011), suggesting that this complex is structurally distinct. The study by Sommer et al is important as the first experimental demonstration that anything other than 1:1 arrestin:rhodopsin binding is even possible. If arrestin can prevent more than one rhodopsin from signaling in bright light, this would help the rod get maximum benefit from its limited supply of arrestin-1. Indeed, rods in Arr1+/− mice that express about half the normal arrestin complement (Doan et al., 2009; Gross and Burns, 2010; Song et al., 2009b; Song et al., 2011; Xu et al., 1997) or mice expressing even lower levels of arrestin-1 (Song et al., 2011) are not as healthy as in wild type animals. No photoreceptor loss was detected at 16 weeks, but OS shortening, particularly in the peripheral retina, appears to become progressively worse with decreasing arrestin-1 expression (Song et al., 2011).
2.7. The shape of “active” receptor-bound arrestin
An unusually high Arrhenius activation energy for arrestin-1 binding to P-Rh* was the first indication that a global conformational change likely accompanies this process (Schleicher et al., 1989). Since the C-terminus of P-Rh*-bound arrestin-1 was shown to become more susceptible to proteolysis (Palczewski et al., 1991c), its release was the first conformational rearrangement to be examined. Site-directed spin labeling followed by continuous wave (CW) electron paramagnetic resonance (EPR) or double electron-electron resonance (DEER) allows direct measurements of inter-spin distances up to 20 Å and 60 Å, respectively (Altenbach et al., 2008; Hanson et al., 2007c; Hubbell et al., 2003). Thus, this method is perfectly suited for the detection of conformational rearrangements by measuring the distances between the same pairs of residues in free and rhodopsin-bound arrestin. To this end, a cysteine-less functional arrestin base mutant was generated (Hanson et al., 2006b). Pairs of cysteines were then introduced in desired positions and chemically modified with a spin label. Distance measurements between spin labels in the C-tail and other participants of the three-element interaction, β-strand I and α-helix I, yielded direct proof that the arrestin-1 C-tail actually moves significantly from its basal position (Hanson et al., 2006b; Vishnivetskiy et al., 2010), as earlier indirect evidence implied (Gurevich and Benovic, 1992; Palczewski et al., 1991c). However, additional conformational changes in arrestin-1 must occur since p44 (a splice variant of arrestin-1 lacking the C-tail) binding to P-Rh* still retains about half the activation energy (Pulvermuller et al., 1997).
The discovery of the polar core between the two arrestin domains (Hirsch et al., 1999) along with evidence that it must be destabilized by phosphates to allow high-affinity binding (Vishnivetskiy et al., 1999) indicated that rearrangement of the polar core is one of these additional conformational transitions. As evidenced by greatly enhanced binding to the non-preferred forms of rhodopsin (particularly Rh*), the polar core is destabilized by the charge reversal of Arg175 (R175E) (Gurevich and Benovic, 1997), and the C-tail is effectively detached from the body of the molecule by the triple alanine substitution of Phe375, Val376, and Phe377 (3A mutation) (Gurevich, 1998). In contrast to WT arrestin-1, these mutants show greatly increased binding to P-Rh* at low temperatures (Fig. 6A). Interestingly, the binding of both 3A and the 3A+R175E combination mutant to P-Rh* at 0°C is almost as high as at 37°C, with the binding of R175E in between that of WT and 3A. These data suggest that C-tail detachment reduces the energy barrier more than the destabilization of the polar core, which does not appear to add much to the 3A mutation (Fig. 6A). However, a significant difference between these three mutants is revealed by their interactions with Rh*: at 0°C the R175E, 3A, and 3A+R175E mutants show 4%, 12%, and 26% of the maximum binding achieved at 37°C, respectively (Fig. 6B). Thus, all three mutants still have an energy barrier for transition into the high-affinity binding state, and even simultaneous disruption of both stabilizing interactions does not completely eliminate it.
Fig. 6. The molecular basis of the high Arrhenius energy of arrestin activation.
A. The binding of WT arrestin-1 to P-Rh* has a high activation energy (Schleicher et al., 1989), and therefore can be inhibited by reduced temperature. Binding at low temperature is dramatically increased by the destabilization of the polar core (R175E mutation), forced detachment of the C-tail (3A mutation), or a combination of both (3A+R175E), indicating that these mutations significantly reduce the arrestin-1 activation energy. B. The binding of pre-activated mutants to Rh* is significantly lower at 20°C and especially at 0°C than at 37°C. Even the combination of 3A and R175E mutations, relieving both known conformational constraints in arrestin-1, yields arrestin-1 with four-fold lower binding to Rh* at 0°C than at 37°C. Thus, additional conformational constraints contribute to the activation energy of arrestin-1.
Since the three-element interaction that anchors the C-tail and the polar core both support the basal orientation of the two arrestin-1 domains (Fig. 2), it was suggested that their destabilization by rhodopsin-attached phosphates allows domain movement improving the fit between arrestin and P-Rh*. The finding that increasing deletions in the inter-domain hinge (Fig. 1B,C) progressively reduce the ability of arrestin-1 (Vishnivetskiy et al., 2002) and other arrestins (Hanson et al., 2007a) to bind P-Rh* is consistent with this idea. However, direct comparison of intra-molecular distances by DEER between two spin labels in free and P-Rh*-bound arrestin-1 revealed only a small shift of the two domains (Kim et al., 2011b), suggesting that other rearrangements in the molecule must take place.
Several loops in all arrestin subtypes are quite flexible and assume distinct conformations in different crystal forms or in monomers within crystal oligomers (Han et al., 2001; Hirsch et al., 1999; Sutton et al., 2005; Zhan et al., 2011). One such loop is the “finger loop” (residues 68-78; Fig. 1B,C) in the center of the receptor-binding surface: it exists in either a “bent” or “extended” conformation in the same crystal (Hirsch et al., 1999) and is clearly involved in receptor binding (Dinculescu et al., 2002; Feuerstein et al., 2009; Hanson et al., 2006b; Hanson and Gurevich, 2006; Pulvermuller et al., 2000; Sommer et al., 2007; Vishnivetskiy et al., 2004). Spin label in positions 72, 74, or 75 in this loop is dramatically immobilized upon dark P-Rh binding, with further immobilization upon rhodopsin activation to P-Rh*(Hanson et al., 2006b), placing these residues in the binding interface. Moreover, based on changes in the quenching of a monobromobimane fluorophore in the finger loop by tryptophan in positions 148 and 298, it was proposed that this loop shifts from a bent to an extended conformation upon P-Rh* binding (Sommer et al., 2007). A peptide mimicking this loop (residues 67-77) binds Rh* and P-Rh* with similar affinity, stabilizes the Metarhodopsin II photo-intermediate, and was shown by solution NMR to assume an α-helical conformation upon binding (Feuerstein et al., 2009). However, measurements of distances between this loop and multiple places in free arrestin-1 by DEER suggest that its basal conformation in solution is intermediate between bent and extended, and although upon binding P-Rh* its position shifts, it never reaches the fully extended state observed in some monomers in the crystal (Kim et al., 2011b).
Spin label in position 139 on an adjacent loop (Fig. 1B,C) showed peculiar behavior: it was immobilized upon binding to dark P-Rh, but reverted to high mobility similar to that in free arrestin-1 when in complex with P-Rh* (Hanson et al., 2006b). This suggests that the “139 loop” participates in low-affinity arrestin-1 binding to P-Rh, but not in the final high-affinity binding to P-Rh*, and moves during the transition from one state to the other. Indeed, DEER measurements show that this loop moves significantly upon P-Rh* binding, dramatically changing its position in the complex (Fig. 1B,C) (Kim et al., 2011b). Similarly, spin label in position 244 was immobilized upon P-Rh binding, but increased mobility in complex with P-Rh* even greater than in free arrestin-1. This also suggests that this element participates in the “pre-docking” of arrestin to dark P-Rh, but not in high-affinity P-Rh* binding (Hanson et al., 2006b). Considering that this residue is localized in a fairly rigid part of the C-domain, these data suggest that the whole arrestin molecule might be oriented somewhat differently when bound to P-Rh, as compared to P-Rh*.
Systematic measurements of 30 intra-molecular distances in free and P-Rh*-bound arrestin-1 revealed multiple changes in the bound form and the overall shape of “active” arrestin (Kim et al., 2011b). The data excluded a large “clam shell” movement of the two domains, indicating that concerted movement of multiple flexible loops helps arrestin to mold itself onto rhodopsin (Fig. 1B,C). Even though the conformation of light-activated rhodopsin has also been established (Choe et al., 2011; Park et al., 2008; Scheerer et al., 2008; Standfuss et al., 2011), the elucidation of the structure of the arrestin-rhodopsin complex requires a lot of additional information that can be obtained by arrestin-rhodopsin distance measurements, co-crystallization, or both: while crystallography yields detailed structural information, EPR and DEER reveal the dynamics of the interaction.
2.8. Why do cones need a special arrestin subtype?
While invertebrates apparently use the same two arrestins in all photoreceptors (Hyde et al., 1990 ; Smith et al., 1990), the specialized cone-specific form (arrestin-4) appeared fairly early in vertebrate lineage and evolved independently of arrestin-1 in fish and beyond (Gurevich and Gurevich, 2006a). Even taking into account that only 2-3% of photoreceptors in mice are cones, the expression level of arrestin-4 in the mouse retina is still much lower than expected (Chan et al., 2007). Cell-by-cell quantification based on immunostaining suggested that only ~2% of the arrestin complement in cones is represented by arrestin-4, whereas the remainder is arrestin-1 (formerly known as rod arrestin) (Nikonov et al., 2008a). Functional studies of mouse rods transgenically expressing cone opsin instead of rhodopsin showed that arrestin-1 quenches cone opsin signaling perfectly well (Shi et al., 2007). Collectively, these data appear to suggest that vertebrates do not need arrestin-4 and could have simply used arrestin-1 in both photoreceptor subtypes, yet its persistence through hundreds of millions of years of evolution clearly suggests that there must be a reason for its existence. Structurally, arrestin-4 (Sutton et al., 2005) is very similar to arrestin-1 (Hirsch et al., 1999) and both non-visual subtypes, arrestin-2 (Han et al., 2001) and arrestin-3 (Zhan et al., 2011). However, two functional features make arrestin-4 unique and likely explain its existence in vertebrate cones. First, in contrast to arrestin-1 (Bayburt et al., 2011; Pulvermuller et al., 1997; Schleicher et al., 1989; Zhang et al., 1997) and non-visual arrestins (Gurevich et al., 1995; Gurevich et al., 1997; Gurevich et al., 1993), which form relatively stable complexes with their cognate receptors by binding them with high affinity, the complex of arrestin-4 with cone opsin is very transient (Sutton et al., 2005). Because cones function in bright light, where there is a high rate of photobleaching, the rapid return of cone opsins back into the functional pool must be important. Cone opsins decay much faster than rhodopsin, which speeds up their cycle of bleaching, loss of all-trans-retinal, and regeneration with 11-cis-retinal, making arrestin-mediated shutoff less crucial than it is in rods (Nikonov et al., 2008a). Nonetheless, short lifetime of the arrestin-4-opsin complex likely also contributes to high speed of this process. Second, unlike arrestin-1 that robustly self-associates (see section 3) and non-visual arrestins that form oligomers in the presence of physiological concentrations of IP6 (Hanson et al., 2008b; Milano et al., 2006; Storez et al., 2005), arrestin-4 is a constitutive monomer (Hanson et al., 2008b). Since only monomeric arrestin can bind receptors (Hanson et al., 2007c), this makes the whole pool of arrestin-4 in cones immediately available. Based on the self-association constants of arrestin-1 in different mammalian species (Hanson et al., 2007c; Kim et al., 2011a), at physiological concentrations only 4.4%, 2.3%, and 1.5% of arrestin-1 exists in the monomeric form in mouse, bovine, and human rods, respectively. Thus the addition of an extra 2% of "ready-to-go" constitutively monomeric arrestin-4 may be quite significant.
These data suggest the following scenario for the interplay between two visual arrestins in cones (Gurevich and Gurevich, 2010b). Invariably monomeric arrestin-4 constitutes about half of the pool of active monomeric arrestin in cones and is predominantly used at lower light levels. Since it rapidly dissociates from cone opsin, arrestin-4 quickly reenters the active pool and is constantly reused. As the rate of pigment bleaching increases, the proportion of active opsin quenched by arrestin-1 also progressively increases. Thus, in brighter light a larger fraction of opsin is quenched by arrestin-1, and therefore taken out of action for longer periods of time. This process would progressively reduce photon catch, likely contributing to the virtually limitless light adaptability of cones (Burns and Arshavsky, 2005; Burns and Pugh, 2010). This increasingly ties up the less “precious” arrestin-1 (the supply of which is ~50-fold larger (Nikonov et al., 2008a)), whereas due to rapid recycling of arrestin-4, cones never run short of active arrestin. Although this model appears plausible, it needs to be tested experimentally. In particular, light adaptation of mouse cones lacking arrestin-4 needs to be carefully compared to that of WT cones, especially at very high light intensities.
2.9. Non-visual arrestins: conserved mechanism without perfection
Arrestin-1 binding to rhodopsin served as a model to elucidate the basic mechanisms of the arrestin-receptor interaction (Gurevich and Gurevich, 2004; Gurevich et al., 2007). Key residues mediating the intra-molecular interactions that stabilize the basal conformation of arrestin are conserved remarkably well in evolution (Gurevich and Gurevich, 2006a). The overall fold and key structural features of non-visual arrestin-2 (Han et al., 2001; Milano et al., 2002) and arrestin-3 (Zhan et al., 2011) are virtually identical to those of arrestin-1 (Hirsch et al., 1999). These arrestins also appear to employ the same mechanism of activation via binding to receptor-attached phosphates, so that mutations destabilizing the polar core or three-element interaction also yield phosphorylation-independent mutants (Celver et al., 2002; Gurevich et al., 1997; Kovoor et al., 1999; Pan et al., 2003). Yet, the hallmark of arrestin-1, its remarkable selectivity toward the active phosphorylated form of the receptor, is much less significant for non-visual arrestins: the difference in binding levels to the active and inactive phosphoreceptor is usually less than two-fold, and between the active phosphorylated and unphosphorylated receptor is less than five-fold (Gurevich et al., 1995; Gurevich et al., 1993). Thus, non-visual arrestins behave as partially “pre-activated” arrestin-1 mutants, suggesting that they are more conformationally loose and that the energy barrier for their transition into the active state is likely much lower. At least one structural feature shared by non-visual forms is consistent with this notion: Val90 (Fig. 4), which stabilizes the β-sandwich of the N-domain via interactions with multiple hydrophobic partners in arrestin-1, is replaced with Ser86 or Ala87 in arrestin-2 or -3, respectively. The V90S mutation in arrestin-1 makes it more promiscuous, enabling much higher binding to the non-preferred M2 muscarinic receptor than that of WT arrestin-1 (Han et al., 2001). Arrestin-3 is the less selective of the two non-visual subtypes, which appears to be partially accounted for by one “pre-melted” β-strand in the receptor-binding β-sheet in the C-domain (Zhan et al., 2011).
2.10. Invertebrate arrestins: rule breakers?
Flies express two arrestins in photoreceptors: Arr2 is the main isoform, whereas Arr1 represents ~20% of the total (Smith et al., 1990). Although Gprk1, Drosophila kinase that closely resembles mammalian GRK2/3 (Gurevich et al., 2011), was tentatively identified as rhodopsin kinase, and its elimination was reported to slow down rhodopsin inactivation (Lee et al., 2004), in two fly species, Calliphora (Plangger et al., 1994) and Drosophila (Kiselev et al., 2000; Liu et al., 2008; Vinós et al., 1997), Arr2 was shown to bind rhodopsin regardless of its phosphorylation. Since Arr2 readily interacts with a truncated form of rhodopsin lacking all phosphorylation sites and flies expressing this mutant display normal kinetics of photoresponse inactivation (Kiselev et al., 2000; Liu et al., 2008; Vinós et al., 1997), its phosphorylation-independent binding appears to be proven beyond a reasonable doubt. While we do not have any structures of invertebrate arrestins, every residue implicated in phosphate-dependent activation of vertebrate arrestins is conserved in fly Arr2 (Gurevich and Gurevich, 2006a). Thus, Arr2 must be activated by Rh* itself, without the help of receptor-attached phosphates. This is not entirely unprecedented in the arrestin family (reviewed in (Gurevich and Gurevich, 2006b)). In some cases, negatively charged residues in the receptor sequence appear to act as phosphate substitutes, as has been described for the luteinizing hormone/choriogonadotropin receptor (Mukherjee et al., 2002), human chemokine decoy receptor D6 (Galliera et al., 2004), and leukotriene B4 receptor (Jala et al., 2005). In addition, negatively charged lipid head-groups were recently shown to facilitate arrestin-1 activation by P-Rh* (Bayburt et al., 2011). Thus, it appears likely that the negative charges on fly rhodopsin and/or on associated lipids act in lieu of receptor-attached phosphates, promoting Arr2 transition into its active receptor-binding state.
In contrast to both mammalian visual arrestins, fly Arr2 is phosphorylated in a light-dependent manner even faster than rhodopsin (Yamada et al., 1990). Arr2 phosphorylation is not necessary for rhodopsin binding but appears to be required for its dissociation from inactivated rhodopsin (Alloway and Dolph, 1999; Liu et al., 2008). Similar to mammalian arrestin-1 (Palczewski et al., 1989), fly Arr2 shields the phosphates on rhodopsin, preventing its dephosphorylation (Byk et al., 1993; Vinós et al., 1997).
3. The biological role of arrestin-1 self-association
In rod and cone photoreceptors, arrestin-1 is the second most abundant signaling protein after the corresponding opsins (Hanson et al., 2007b; Nikonov et al., 2008a; Song et al., 2011; Strissel et al., 2006). In the dark, when the bulk of arrestin-1 resides in the inner segments (IS), cell body, and synaptic terminals (see section 4), its concentration in these compartments is expected to be >2 mM (Gurevich et al., 2007; Kim et al., 2011a; Song et al., 2011). The concentration of virtually all signaling proteins in rods is several orders of magnitude higher than in any other cell in the body (Pugh and Lamb, 2000). However, only arrestin-1 robustly self-associates at physiological concentrations, forming dimers and tetramers (Hanson et al., 2007c; Imamoto et al., 2003; Schubert et al., 1999). Although the oligomers were assumed to be storage forms, for many years no coherent ideas for the biological role of arrestin-1 oligomerization were even proposed.
Curiously, self-association of S-antigen (as arrestin-1 was called at the time) was discovered in 1977 (Wacker et al., 1977), before its specific binding to P-Rh* (Kuhn et al., 1984) and its role in quenching rhodopsin signaling (Wilden et al., 1986) were established. This phenomenon was largely ignored until twenty years later, when both crystal structures revealed virtually identical tetramers (Granzin et al., 1998; Hirsch et al., 1999). Subsequent studies confirmed arrestin-1 self-association in solution and discussed the possible dimers assuming that the interaction interfaces found in the crystal tetramer were physiologically relevant (Imamoto et al., 2003; Schubert et al., 1999).
3.1. Structure and function of arrestin-1 oligomers
Surprisingly, spin label immobilization by sister subunits and inter-molecular distance measurements of the solution tetramer using DEER demonstrated that its shape is nothing like the one observed in the crystal (Hanson et al., 2007c). Importantly, by showing that all inter-subunit distances disappeared upon the addition of enough P-Rh* to bind all arrestin-1 (Hanson et al., 2007c), this study provided the first direct evidence that only monomeric arrestin-1 binds the receptor while oligomers do not. This effect is specific for rhodopsin, as the addition of microtubules, which also bind arrestin-1 (Hanson et al., 2006a; Nair et al., 2004), does not affect inter-subunit distances, suggesting that microtubules bind monomer, dimer, and tetramer with comparable affinity (Hanson et al., 2007c).
The first investigation of arrestin-1 self-association equilibrium constants using small angle X-ray scattering led to the conclusion that the process is best described by a monomer-dimer-tetramer equilibrium (Imamoto et al., 2003). Tetramerization was found to be cooperative, with KD,dimer > KD,tetramer, so that as soon as the dimer forms, it has high probability of assembling into a tetramer (Imamoto et al., 2003). However, the values of the equilibrium constants could not be determined with high precision because the X-ray wavelength is comparable to the dimensions of the tetramer, so data analysis required certain assumptions regarding its shape. The use of visible light with a much longer wavelength made these assumptions unnecessary. Analysis by multi-angle light scattering confirmed the cooperativity of tetramer formation by bovine arrestin-1 and yielded KD,dimer = 37.2±0.2 μM and KD,tetramer = 7.4±0.1 μM (Hanson et al., 2007c). In the dark, no more than ~15% of arrestin-1 is in the OS, whereas ~85% resides in the IS and cell body (Song et al., 2011; Strissel et al., 2006) (see section 4), with predicted concentrations of ~300 μM and ~2 mM, respectively. However, due to self-association the concentration of free monomer in both compartments is less than 100 μM (Kim et al., 2011a).
The direct inter-subunit distance measurements of arrestin in solution (Hanson et al., 2007c) showed that the solution oligomer was structurally different from the crystal. To determine the actual shape of the solution tetramer, a follow-up study used protein docking and the experimental distance restraints to generate possible structures. Putative models were tested by measuring additional distances between protomers and disulfide bond formation between cysteines engineered in positions predicted to come within ~5A in the tetramer. The model that yielded the correct predictions was ultimately validated by targeted disruption of inter-subunit interactions, resulting in the construction of constitutively monomeric mutant arrestin-1-(F85A,F197A) (Hanson et al., 2008a).
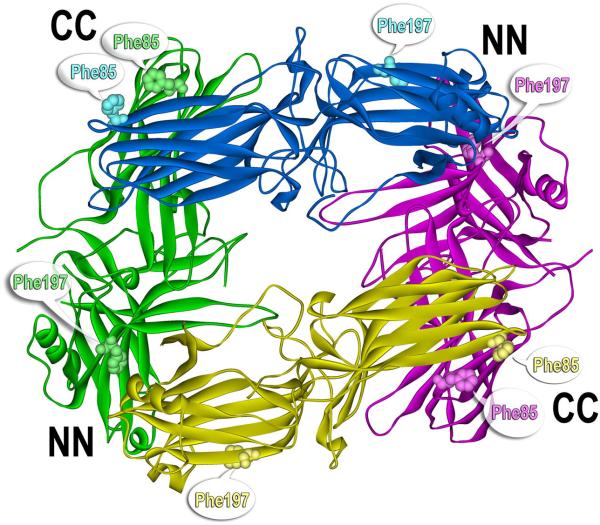
The solution tetramer turned out to be a virtually symmetrical closed diamond with two N-N and two C-C interfaces (Fig. 7). This structure readily explains why WT arrestin-1 can recruit self-association-deficient mutants into the tetramer: the disruption of one side of a single interface cannot prevent the interaction of the other domain with another protomer. The structure also makes it clear why oligomeric forms cannot bind P-Rh*: well-defined receptor-binding surfaces on the concave sides of both domains (see section 2.5) of all molecules in the tetramer and both possible dimers are shielded by sister subunits (Fig. 7).
Fig. 7. The structure of the solution tetramer of bovine arrestin-1.
Inter-subunit distance measurements, mutagenesis, and the formation of inter-subunit disulfide bonds showed that the solution tetramer is dramatically different from the one observed in the crystal (Hanson et al., 2008a; Hanson et al., 2007c). The solution tetramer is virtually symmetrical, has a closed diamond shape with two N-domain-N-domain (NN) and two C-domain-C-domain (CC) inter-subunit interfaces. The receptor-binding concave sides of both domains are shielded by sister subunits in the tetramer and both possible dimers, explaining why only monomeric arrestin-1 can bind rhodopsin (Hanson et al., 2007c). Phe86 and Phe197 (shown as CPK models in all four subunits) play a key role in the stabilization of the NN and CC interfaces, respectively. Double alanine substitution in bovine (F86A, F197A) and mouse (F87A, F198A) arrestin-1 disrupts both interfaces, yielding constitutively monomeric mutants (Kim et al., 2011a). Thus, the shape of the solution tetramer is likely conserved between species.
3.2. Arrestin-1 self-association in different species
The dynamics of arrestin-1 self-association was characterized using bovine protein, whereas the biological functions of arrestin-1 in rods (Cleghorn et al., 2011; Mendez et al., 2000; Song et al., 2011; Xu et al., 1997) and cones (Nikonov et al., 2008a) are largely studied in mice. If self-association has a specific functional role, one would expect this feature of arrestin-1 to be conserved in different species. A recent study (Kim et al., 2011a) that compared bovine, mouse, and human arrestin-1 yielded interesting and somewhat unexpected results. While all three arrestins were shown to form dimers and tetramers, the absolute values of the self-association constants turned out to be very different. No cooperativity was detected for mouse arrestin-1, where KD,dimer and KD,tetramer are virtually the same (57.5±0.6 μM and 63.1±2.6 μM, respectively). Despite the dramatic difference in equilibrium constants, the overall structure of the mouse tetramer is likely very similar to that of the bovine protein since alanine substitution of phenylalanines 86 and 198 (homologues of bovine Phe85 and Phe197; Fig. 7) also completely abolished its self-association (Kim et al., 2011a). Importantly, in both bovine and mouse arrestin-1 these mutations did not affect binding to P-Rh* or microtubules, indicating that key self-association residues are conserved to preserve this function, rather than being a by-product of the conservation of rhodopsin- or microtubule-binding elements (Kim et al., 2011a). Human arrestin-1 is quite different from both bovine and mouse homologues: it has much higher propensity to dimerize (KD,dimer = 2.95±0.02 μM), whereas its tetramerization is significantly weaker (KD,tetramer = 224±5 μM).
Arrestin-1 localization in dark-adapted rods has only been quantitatively determined in mice, where no more than ~15% resides in the OS, and the rest is distributed between IS, cell body, and synaptic terminals (Song et al., 2011; Strissel et al., 2006). Based on the 3 mM rhodopsin concentration in the OS (Pugh and Lamb, 2000) and the measured arrestin-1:rhodopisn ratio in this compartment, the absolute arrestin-1 concentration in mouse OS in the dark is ~300 μM (Song et al., 2011). Virtually the same concentration was found in dark-adapted frog rod OS (Hamm and Bownds, 1986), which has a very different size and morphology, suggesting that arrestin concentration is likely similar in various vertebrate species. Based on this assumption (that clearly needs experimental validation), the 0.8:1 arrestin-1:rhodopsin ratio (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006), and the measured self-association constants, the concentration of the active monomer in mouse, bovine, and human OS is calculated as ~53 μM, 28 μM, and 15 μM, respectively (Kim et al., 2011a). Although we certainly need experimental measurements of arrestin-1 in the OS of different species, these numbers are rather suggestive: nocturnal mice that rely more on rod vision, have the highest monomer concentration, whereas diurnal humans that predominantly use cones have the lowest. Considering that quick arrestin-1 binding to P-Rh*, which is rate-limited by the concentration of active monomer (see section 5), is a prerequisite for rapid and reproducible photoresponse shutoff (Doan et al., 2006; Mendez et al., 2000), one is tempted to speculate that rods with higher concentration of monomeric arrestin-1 are more precise “photon counters”, a function that is likely to be more biologically important for nocturnal animals. This idea requires experimental testing, such as measurements of the kinetics of single photon responses in different species and/or in mice expressing widely different levels of arrestin-1.
3.3. Why does arrestin-1 self-associate?
All signaling proteins are expressed in photoreceptors at concentrations that are orders of magnitude higher than in “normal” neurons: 3 mM rhodopsin, 0.3 mM transducin and 0.3 mM arrestin-1 in the dark-adapted OS, as well as ~2 mM arrestin-1 in the cell body (Pugh and Lamb, 2000; Song et al., 2011). Yet arrestin-1 is the only protein that has a special inactive oligomeric storage form. This mechanism seems counter-intuitive, but since it is conserved in at least three mammalian species (Kim et al., 2011a), it must have biological meaning. Although this issue needs to be addressed experimentally via expression of constitutively monomeric mutants (Kim et al., 2011a) in vivo, a recent study using a different arrestin-1 mutant provided interesting clues. The enhanced arrestin-1 mutant, 3A, that binds unphosphorylated Rh* with much higher affinity than WT protein, was transgenically expressed in mice in an attempt to compensate for deficits in rhodopsin phosphorylation (see section 7) (Song et al., 2009b). Mutants of this type have decreased ability to self-associate (Hanson et al., 2008a; Hanson et al., 2007c; Schubert et al., 1999). While moderate expression of arrestin-1-3A proved beneficial, at high (more than double WT) levels it induced rapid photoreceptor degeneration (Song et al., 2009b). Importantly, despite massive loss of rods within weeks, the outer segments of the remaining photoreceptors looked longer and healthier than in arrestin knockout animals (Song et al., 2009b), suggesting that the mutant inflicts damage elsewhere, likely in the cell body. One obvious difference between moderate and high expressors of arrestin-1-3A is the predicted concentration of the monomer, which in the latter case is expected to significantly exceed WT levels. These data provide indirect evidence that excess of monomeric arrestin-1 is harmful, although the molecular mechanisms of this phenomenon remain unclear. This would easily explain why arrestin-1 needs to self-associate: oligomerization allows the rod to have the huge supply of arrestin-1 that is necessary to prevent useless signaling by rhodopsin in bright light, while keeping the concentration of the monomer at a fairly safe low level in the dark. This also provides another good reason for cones to have only a small fraction of their arrestin complement in the form of constitutively monomeric arrestin-4 (Hanson et al., 2008b), while keeping the rest safely stored as oligomeric arrestin-1 (Nikonov et al., 2008a). However plausible, this idea needs to be tested experimentally, since arrestin-1-3A mutant differs from WT arrestin-1 in other important aspects. It was shown to bind P-Rh*, Rh*, dark P-Rh, and microtubules with higher affinity (Gurevich, 1998; Nair et al., 2005), and its interactions with many other proteins (see section 6) were never directly compared to the WT.
4. Light-dependent arrestin-1 translocation in rods
Photoreceptors are neurons with a unique design: they have a specialized signaling compartment, the outer segment (OS), connected to the rest of the cell via a narrow cilium. This provides an opportunity to modulate the gain of signaling by changing the effective concentrations of the proteins involved simply by moving them in and out of the OS, rather than by changing their expression like most other cells do. Rods must have enormous concentrations of transducin (~0.3 mM (Pugh and Lamb, 2000)), arrestin (~2 mM (Song et al., 2011; Strissel et al., 2006)) and receptor kinase (GRK1/rhodopsin kinase, (Pugh and Lamb, 2000)) to achieve single photon sensitivity. Under these circumstances, protein translocation, rather than degradation and energy-intensive de novo synthesis on this massive scale, appears to be the only mechanism of signaling modulation by varying protein concentration that is economically feasible in rods.
4.1. Arrestin-1 and transducin move in a light-dependent fashion
Arrestin-1 translocation to the OS in bright light and to the inner segment (IS) and cell body in the dark was discovered in 1985 (Broekhuyse et al., 1985). The light-dependent movement of transducin in the opposite direction reported soon after (Brann and Cohen, 1987; Philp et al., 1987) suggested that this mechanism might be involved in signaling regulation: in the dark, when rods need maximum sensitivity, virtually all transducin is present in the rhodopsin-containing OS, whereas the concentration of arrestin-1, which quenches the signaling, is relatively low; in brighter light, when sensitivity needs to be reduced, these proteins change places, presumably reducing gain and facilitating shutoff. These findings were independently confirmed by several groups (Mendez et al., 2003; Nair et al., 2005; Organisciak et al., 1991; Whelan and McGinnis, 1988), but challenged by others, who proposed that epitope masking, rather than physical protein translocation, accounts for the “movement” detected by immunohistochemical methods (Roof and Heth, 1988). The development of a method to tangentially section the flattened retina, which provides the ability to obtain virtually uncontaminated samples of different photoreceptor compartments and compare the distribution of proteins that move and those that certainly do not (e.g., rhodopsin in the OS and mitochondrial proteins in the IS), unambiguously settled this issue: arrestin-1 (Strissel et al., 2006), transducin (Sokolov et al., 2002) moving with the help of phosducin (Sokolov et al., 2004), and recoverin (Strissel et al., 2005) actually redistribute within photoreceptors, although translocation of some proteins (e.g., growth factor receptor-bound protein 14 (Rajala et al., 2009)), does not appear to play a role in rhodopsin signaling. Importantly, the depletion of transducin from the OS in the light was quantitatively correlated with the reduced gain of phototransduction (Sokolov et al., 2002). However, these results raised an interesting question: if transducin redistribution fully accounts for changes in the efficiency of rhodopsin signaling, why do the other proteins need to move?
4.2. The rates and mechanisms of protein movement
Orderly movement of arrestin-1 and transducin in opposite directions in response to light was initially assumed to involve active transport. Careful measurement of the translocation rates, which were found to be quite different for each of these two proteins, was interpreted as a confirmation of this idea (Elias et al., 2004). The reported involvement of microtubules (that often serve as “rails” for active protein transport) in vertebrate rods (McGinnis et al., 2002; Peterson et al., 2005), and of kinesin-dependent transport of arrestins associated with phosphoinositide-containing vesicles in Drosophila (Lee and Montell, 2004; Lee et al., 2003) also appeared to support this notion. Obviously, active transport consumes energy, so the real test of this hypothesis required proof that translocation does not occur in energy-depleted photoreceptors. Unexpectedly, it was shown that arrestin translocates in both directions with essentially normal speed in KCN-treated explanted mouse retinas that have a ~100-fold reduced ATP content (Nair et al., 2005). Moreover, transducin movement from the cell body to the OS in the dark also did not seem to be affected in this system (Nair et al., 2005). However, the reverse movement of transducin requires GTP binding and subunit dissociation, so even if no active transport is involved, this movement could not be observed in energy-starved photoreceptors. With considerable ingenuity, this problem was soon solved: a non-hydrolyzable GTP analogue that activates transducin but cannot serve as a source of energy, and a peptide that dissociates transducin α- and βγ-subunits without activation were both used to show that transducin movement from the OS in the light is completely energy-independent (Rosenzweig et al., 2007). Collectively, these results suggested that the translocation of both arrestin-1 and transducin in rods can be explained by interaction-restricted diffusion (Slepak and Hurley, 2008). Diffusion-controlled arrestin movement was also proposed in Drosophila by a group that did not observe the requirement for kinesin for this process in fly photoreceptors (Satoh and Ready, 2005). However, several recent studies in Xenopus tadpoles have implicated energy-dependent active transport (Orisme et al., 2010; Reidel et al., 2008). As this issue appears controversial, the expected energy cost of translocating transducin, expressed at a ~1:10 ratio to rhodopsin (Hamm and Bownds, 1986; Pugh and Lamb, 2000) and arrestin-1, expressed at an 8:10 ratio to rhodopsin (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006), into (or out of) a mouse rod OS with a length of ~25 μm (Shen et al., 2010) needs to be calculated. In a mouse rod containing ~108 rhodopsins (Pugh and Lamb, 2000), ~10 million transducin α-subunits, ~10 million βγ-complexes, and ~80 million arrestin-1 molecules need to be moved. Even assuming the most generous scenario in which arrestin-1 is only transported as a tetramer, the number for arrestin-1 cargo units is still ~20 million. Molecular motors involved in active transport consume at least one molecule of ATP per 4-8 nm of movement (Gennerich and Vale, 2009). Disregarding the fact that both of the most commonly used molecular motors, kinesin and dynein, virtually never move exclusively in one direction for long distances, the movement of 40 million units of cargo for >25 μm would require at least 125-250 billion molecules of ATP, or 0.21-0.42 pmoles, with the total weight of 126-252 pg For comparison, the OS volume is ~ 40 μm3, i.e., its weight is ~ 40 pg. Thus, simple energy considerations suggest that active transport of arrestin-1 and transducin is virtually impossible even in a relatively small mouse rod, let alone a much bigger frog photoreceptor.
However, active gating, where only small amounts of ATP are consumed to open/close a gate while the proteins still move by diffusion, does not place an enormous demand on energy. One study of arrestin translocation in mice (Strissel et al., 2006) and another in Drosophila (Satoh et al., 2010) suggest that arrestin movement towards rhodopsin is triggered by a light stimulus, consistent with the idea of active gating. The finding that arrestin-1 translocation in Xenopus rods can be induced by the activators of phospholipase C and protein kinase C (Orisme et al., 2010) is also consistent with this model. Further experimentation is necessary for comprehensive elucidation of the mechanisms controlling arrestin-1 movement.
4.3. How does arrestin-1 stay put in the light and dark?
Arrestin-1 is visibly concentrated in different rod compartments in the light and dark (McGinnis et al., 2002; Nair et al., 2005; Strissel et al., 2006). Careful comparison of the distribution of GFP (which is obviously equilibrated within the cell), with that of arrestin-GFP in Xenopus photoreceptors clearly showed that arrestin-1 is at disequilibrium in the cell (Peet et al., 2004). Thus, if transport is involved, most of the time it would have to work against diffusion and a steep concentration gradient. One possible solution is to anchor arrestin-1 to immobile proteins. In the OS, the only possible candidate is rhodopsin: it is a proven arrestin-1 binding partner, and there is ~20% molar access of rhodopsin over arrestin-1 in the WT mouse rod (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006). Indeed, several lines of evidence suggest that arrestin-1 stays in the OS in bright light because it is bound to rhodopsin. First, in GRK1 knockout mice, where rhodopsin cannot be phopshorylated and converted to the high-affinity arrestin-1 target, arrestin-1 still translocates to the OS in bright light but its movement out of the OS in the dark is significantly accelerated (Mendez et al., 2003; Nair et al., 2005), consistent with its much lower affinity for Rh* (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993). The same phenomenon is observed in energy-starved retinas, where rhodopsin is not phosphorylated because GRK1 does not have enough ATP (Nair et al., 2005). Moreover, the overall amount of arrestin-1 that can translocate to the OS in the light is determined by the amount of rhodopsin present in this compartment, so that in all genetically modified mouse lines expressing more arrestin-1 than rhodopsin, its translocation to the OS is incomplete, with the fraction remaining in the cell body determined by the molar excess of arrestin-1 (Hanson et al., 2007b).
The discovery of low-affinity arrestin-1 binding to microtubules (Hanson et al., 2007a; Hanson et al., 2006a; Nair et al., 2004) suggested that the cytoskeleton might serve as the “parking space” for arrestin-1 in the cell body in the dark. Arrestin-1 association with microtubule bundles extending along the whole length of the OS was detected by electron microscopy (Nir and Ransom, 1992, 1993). The pattern of arrestin-1 distribution in the dark-adapted rod is consistent with this idea: it is highly enriched in the IS, perinuclear space, and synaptic terminals (Broekhuyse et al., 1985; Hanson et al., 2007b; Nair et al., 2005), i.e., in the compartments where microtubules are particularly abundant (Eckmiller, 2000). Indeed, two structurally distinct arrestin-1 mutants with higher affinity for microtubules were shown to translocate slower than WT protein from the cell body to the OS in the light (Nair et al., 2005). Every αβ-tubulin dimer can bind arrestin-1 (Hanson et al., 2007a), likely providing sufficient space to “park” all arrestin-1 (Hanson et al., 2007b). Interestingly, while only arrestin-1 monomer can bind rhodopsin (Hanson et al., 2007c), monomers, dimers, and tetramers each appear to bind microtubules fairly well (Hanson et al., 2007c). In fact, the calculations of the distribution of the monomers and oligomers of arrestin-1 in the mouse rod suggest that the order of potency of microtubule binding is likely tetramer > dimer > monomer (Kim et al., 2011a). This makes perfect sense biologically considering that in the dark the concentration of arrestin-1 in the cell body approaches 2 mM, so that the great majority of it exists as a tetramer (Kim et al., 2011a).
Together, these data suggest the simplest and the least energy-demanding model for arrestin-1 translocation in vertebrate rods. In the dark, in the absence of high-affinity interaction partners, arrestin-1 is largely bound to microtubules, which concentrates it in the compartments where cytoskeleton is most abundant. The concentrations of free monomers, dimers, and tetramers of arrestin-1 are equalized throughout the cytoplasm by diffusion, which actually ensures its presence in the microtubule-poor OS. Light generates Rh*, P-Rh*, and the latter decays into phospho-opsin, all of which have much higher affinity for arrestin-1 than microtubules (Nair et al., 2005). Arrestin-1 binding to rhodopsin reduces its concentration in the OS. The concentration of free arrestin-1 is equalized by diffusion in response, translating into its net movement to the OS. The more arrestin-1 is “consumed” by rhodopsin binding, the greater the overall translocation, so that in bright light, when most of the rhodopsin is bleached, virtually all arrestin-1 concentrates in the OS and stays there in the complex with rhodopsin. In the dark, rhodopsin is no longer bleached, and its regeneration and dephosphorylation generates dark Rh, the only form of rhodopsin with lower affinity for arrestin-1 than microtubules (Nair et al., 2005). Arrestin release from rhodopsin in the OS and its binding to the cytoskeleton in the cell body creates a gradient, so that now diffusion moves it away from the OS. This mechanism translocates arrestin-1 to the appropriate compartments without spending any energy, which is a great advantage considering the enormous energy demands of other processes involved in rod signaling (Linton et al., 2010).
Although this model appears logical, it cannot be considered proven. Its main caveat is that the amount of polymerized tubulin in the inner segment and the body of the rod is unknown. Thus, it would have to be seriously reconsidered if it turns out that the molar amount of αβ-tubulin dimers in microtubules is less than expected molar amount of arrestin-1 tetramers in this compartment in the dark.
4.4. Why does arrestin-1 need to move?
While the translocation of transducin from the OS in the light was directly linked to reduced gain of phototransduction (Sokolov et al., 2002), there is no experimental evidence for the physiological role of arrestin-1 translocation. This is hardly surprising: both interaction-restricted diffusion (Slepak and Hurley, 2008) and reversible self-association (Kim et al., 2011a) are essentially buffering mechanisms maintaining a fairly constant concentration of free active arrestin-1 monomer in the OS at all light levels. This raises the question of why the bulk of arrestin-1 resides in the dark-adapted rod away from its primary binding target. Simply keeping it out of the way is one possible reason: the dark OS is packed with signaling proteins required for proper response to low light levels: transducin, phosphodiesterase, GRK1, recoverin, RGS9, phosducin, guanylyl cyclase, GCAPs, etc. Monomeric arrestin-1 at ~ 50 μM (achieved at ~300 μM total arrestin) is adequate for rapid shutoff. Thus, there is no need to increase its concentration 8-fold by keeping the whole complement of arrestin-1 in the OS, which would increase the viscosity of the cytoplasm and slow down the diffusion of signaling proteins and second messengers. In fact, the illumination that triggers arrestin-1 translocation to the OS is beyond the normal dynamic range of rods, suggesting that this process contributes to rod maintenance, rather than to the regulation of signaling.
4.5. Light adaptation or rod survival?
The same logic applies to the translocation of transducin and other proteins that move in a light-dependent fashion in rods. The level of light that induces their movement is in the cone range, and diurnal animals actually use cones as photoreceptors at this illumination level. However, rods have an important task during bright daylight: they have to survive until dusk, when they will be needed again as the main source of visual information. The strongest argument against the role of protein translocation in light/dark adaptation is timing. The fast and slow phases of light adaptation have time constants of 1 s and 9 s, respectively (Calvert and Makino, 2002), whereas significant redistribution of transducin, arrestin, and other proteins in the rod takes many minutes (Elias et al., 2004).
In contrast, the movement of arrestin-1 to the OS upon prolonged illumination makes perfect sense as a survival mechanism. In bright light arrestin-1 largely resides in the OS bound to rhodopsin, apparently preventing unnecessary signaling. The removal of transducin from the OS likely further decreases the signaling generated by any active rhodopsin molecule that is not blocked by arrestin-1 for a short period of time. Thus, the time course of protein movement in bright light is more consistent with a role in rod survival than in light adaptation. Similarly, the rate of arrestin-1 return to the cell body and of transducin movement back to the OS matches the rate of dimming of natural light at dusk, making rods ready to function at night.
Again, however logical it sounds, this model has not been proven. Although light or dark adaptation with the kinetics matching protein translocation rates was not reported so far, this does not mean that such slow processes do not exist. These issues need to be addressed experimentally.
5. The functional role of arrestin-1 concentration in the dark-adapted outer segment
Most physiological studies of rod signaling, both electroretinography (ERG) and single cell recording, use dark-adapted photoreceptors. In mammalian rods, rhodopsin shutoff appears to be faster than transducin inactivation, therefore the latter rate-limits the recovery (Krispel et al., 2006). The upper limit of active rhodopsin lifetime was determined in animals where transducin shutoff was accelerated by the over-expression of RGS9, and was estimated to be < 60 ms (Krispel et al., 2006) or even as low as ~30 ms (Burns and Pugh, 2009; Gross and Burns, 2010). Considering that rhodopsin is inactivated by a two-step mechanism, where GRK1 has to attach three phosphates before high-affinity arrestin binding becomes possible (Mendez et al., 2000; Vishnivetskiy et al., 2007), a sufficiently high arrestin-1 concentration in the OS that prevents its binding from becoming rate-limiting is vital. However, its precise measurement has proven to be tricky, and existing estimates vary widely.
5.1. Quantification of arrestin-1 distribution in the dark-adapted rod
The first comprehensive quantification of signaling proteins in the dark-adapted OS, reported in frogs, suggested that there is one arrestin-1 per ten rhodopsins (Hamm and Bownds, 1986). For a long time this value was assumed to represent the total expression ratio in the rod. Several recent studies in mice each reported an ~0.8:1 arrestin-1:rhodopsin ratio in mouse rods (Hanson et al., 2007b; Song et al., 2011; Strissel et al., 2006), that will be used here in subsequent calculations. The fraction of total arrestin in the dark-adapted OS was estimated to be 2-4% by immunohistochemistry (Hanson et al., 2007b; Nair et al., 2005). However, these estimates involve several assumptions that cannot be tested experimentally. First, they make the unlikely assumption that the antibodies have identical accessibility to arrestin-1 in different compartments, where it can be monomeric, dimeric, tetrameric, free or bound to rhodopsin, microtubules, and other proteins (see section 6). Second, despite the extremely wide range of concentrations in the rod, the intensity of immunostaining is assumed to correlate linearly with arrestin-1 concentration. Theoretically, tangential sectioning of flattened retinas followed by the measurement of protein content in different fractions by Western blot is the best possible method of quantifying the distribution. Protein estimates from Western blots are only truly quantitative and reliable when a calibration curve generated with known amounts of purified protein (preferably supplemented with a standard amount of retinal protein from the corresponding knockout mice), is created and measurements are made using only the linear range (Hanson et al., 2007b; Nikonov et al., 2008a). While tangential sectioning worked beautifully on rat retinas (Sokolov et al., 2002), the much smaller mouse retinas proved problematic: the amount of material was insufficient to detect arrestin-1 in dark-adapted OS (Strissel et al., 2006), even though it is known to be present there. This necessitated complicated calculations that yielded a much higher estimate, ~9% of the total (Strissel et al., 2006). The only existing alternative was purification of dark-adapted OS and measurement of the arrestin-1:rhodopsin ratio (to take into account inevitable yield variability) in this preparation compared to whole retina homogenate (Song et al., 2011). The key problem with this approach is possible contamination by the adjacent inner segment, where arrestin-1 is highly concentrated in the dark. As rod mitochondria are virtually exclusively localized in the IS, the extent of IS contamination can be measured using a mitochondrial marker (preferably a membrane protein, like COX IV, rather than cytochrome C which can easily leak out). However, because mitochondria are also abundant in the neurons of the inner nuclear layer of vascular retinas (such as mouse) (Bentmann et al., 2005; Stone et al., 2008), their distribution in the retina must be taken into account. This method, along with the realistic (but not absolutely proven) assumption that the rod IS contains about half of the retinal mitochondria, yielded an even higher estimate of the arrestin-1 fraction in the OS, ~13-15% in WT mice (Song et al., 2011). Since a similar or even somewhat greater fraction of total arrestin-1 was found in several mouse lines expressing different levels of arrestin (Song et al., 2011), we will use this estimate. Interestingly, it predicts ~300 μM total arrestin-1 in mouse OS (Song et al., 2011), which is remarkably similar to the value reported for frog using OS purification (Hamm and Bownds, 1986). It is also worth noting that, if one takes into account inevitable uncertainties involved, the estimates of 9% (Strissel et al., 2006) and 13% (Song et al., 2011) are remarkably close and do not dramatically change quantitative predictions.
5.2. How much arrestin-1 is sufficient for rod function?
Every protein in every cell is expressed at a particular level that is strictly controlled by the rates of synthesis and degradation. Although the biological role of protein expression level is rarely tested experimentally, several studies have been done in rods and the results have been illuminating. Reduced rhodopsin expression facilitates photoresponse and recovery (Calvert et al., 2001), whereas its over-expression slows down the response (Wen et al., 2009). RGS9 over-expression facilitates recovery, proving that transducin inactivation rate-limits the recovery kinetics in rods (Gross and Burns, 2010; Krispel et al., 2006). Varying by ~2-fold using WT and hemizygous knockout animals of GRK1 and arrestin-1 expression on the background of the RGS9 over-expressing line, permitted evaluation of the timing of even faster events involved in signaling shutoff, rhodopsin phosphorylation by GRK1 and arrestin-1 binding (Gross and Burns, 2010). Similar experiments with ARR1+/− and GRK1+/− mice performed without increased RGS9 proved harder to interpret (Doan et al., 2009).
The first comparison of mice expressing normal and very low arrestin-1 levels (Burns et al., 2006) proved very informative. The authors compared transgenic mice expressing on Arr1−/− background full-length arrestin-1 (a.k.a. p48) at ~ 140% and a truncated form p44 at ~6% of WT level. The latter was constructed by analogy with the short splice variant of bovine arestin-1 (Smith et al., 1994), which was previously shown in vitro to bind Rh* with higher affinity than full-length protein (Pulvermuller et al., 1997). The authors did not detect expected differences in the ability of p44 and p48 to quench signaling by unphosphorylated Rh*: p48 was shown to be more efficient in this regard, likely due to much higher expression level (Burns et al., 2006). This in vivo result is consistent with specific arrestin-1 binding to Rh* previously measured in vitro (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993). The most unexpected finding was that just 6% of WT arrestin-1 level supports normal kinetics of single photon and dim flash responses, indirectly demonstrating that rhodopsin shutoff does not rate-limit the recovery in this case (Burns et al., 2006). However, at higher flash intensities low expressors significantly deviated from WT animals, demonstrating extremely long recovery approximating that of Arr1−/− animals after the brightest flashes (Burns et al., 2006). This suggested that the overall amount of arrestin-1 matters when a significant proportion of it is “consumed” by binding to numerous molecules of light-activated rhodopsin. It appears that a higher proportion of p44 than p48 was found in dark-adapted OS in this study, as p44 expressed at ~6% of WT arrestin-1, but its content in the OS was ~12% of WT (Burns et al., 2006).
The effects of an extremely wide range of arrestin-1 expression levels, from ~4% to 220% of WT, on functional performance and the health of photoreceptors were recently tested (Song et al., 2011). Measurements of arrestin-1 in purified dark-adapted OS in these lines showed that arrestin-1 content in the OS changes linearly with total expression in mice with arrestin-1 expressed at supra-physiological levels, but is much less than proportional in under-expressing animals. OS content of arrestin-1 in mice expressing 4% and 12% of WT levels is only 2% and 4% of that found in WT animals (Song et al., 2011). This study yielded a few additional unexpected findings. First, even mice with extremely low arrestin-1 levels were strikingly different from arrestin-1 knockout animals: their rods had longer OS and performed virtually normally even beyond the physiologically relevant range of light intensities, up to 2,500 photoisomerizations per rod. However, when the sum of rhodopsin molecules activated by consecutive flashes came close to the total arrestin-1 content in the rod OS, functional performance deteriorated, approximating that of rods lacking arrestin-1 (Song et al., 2011), in perfect agreement with earlier findings in mice expressing short isoform of arrestin-1 at 6% of WT level (Burns et al., 2006). As expected, this “breakdown point” was twice as high in mice with 4% of the normal arrestin-1 content in the OS, as compared to animals with 2%. Importantly, these functional data provided an independent measure of the arrestin-1 content in dark-adapted OS, which was in remarkable agreement with the Western blot measurements in purified rod OS (Song et al., 2011).
Another notable finding of this study was that rods with 4% of the normal arrestin-1 content in the OS demonstrated essentially a WT rate of photoresponse recovery, whereas rods with 2% recovered much slower (Song et al., 2011), suggesting that there is a threshold of arrestin-1 concentration in OS below which the rate of recovery decreases. Recovery is measured by double-flash ERG, where the first flash desensitizes the rod, and the second flash, delivered at different time intervals after the first, measures the extent of recovery (Birch et al., 1995; Hetling and Pepperberg, 1999; Lyubarsky and Pugh, 1996; Pepperberg et al., 1997; Pepperberg et al., 1992). To test whether this threshold depends on the intensity of the desensitizing flash, in a follow-up study the latter was varied from 160 to 2,500 photoisomerizations per rod (Cleghorn et al., 2011). Unexpectedly, it turned out that it does not: rods with 100%, 50%, and 4% of normal arrestin-1 in the OS demonstrated virtually the same recovery rates, which were ~2.5-fold slower after 2,500 than after 160 photoisomerizations in all cases. Rods with 2% of the normal arrestin-1 level in the OS were different at all intensities. In this line, the rate of recovery showed a much more significant change with increasing intensity of the desensitizing flash: at 160 photoisomerizations/rod it was only two times slower than normal, whereas at 2,500 it became 28-fold slower (Cleghorn et al., 2011). These results cannot be explained simply by arrestin-1 depletion in the low expressors: 2% of the normal arrestin-1 content in the OS translates into a 7.6 μM concentration, or 200,000 molecules per OS (Cleghorn et al., 2011), i.e., orders of magnitude greater than the 160 or even 2,500 active rhodopsins produced by the desensitizing flash. Thus, the simplest explanation of this phenomenon is that there are two functionally distinct pools of arrestin-1 in the OS, one of which, roughly equal to 2% of WT arrestin-1 content in the OS, is not immediately available for quenching rhodopsin (Cleghorn et al., 2011). Theoretically, since only monomeric arrestin-1 can bind rhodopsin, oligomer dissociation could account for the delayed generation of active monomer. However, the dimerization and tetramerization constants of mouse arrestin-1 are Kd dimer = 57.5 μM and Kd tetramer = 63.1 μM (Kim et al., 2011a), which translates into dissociation rates of ~12 ms per step. Thus, active monomer can be generated even from tetramer within ~24 ms, i.e., much faster than the observed recovery in the lowest expressors. However, the OS contains several bundles of microtubules localized along the plasma membrane (Eckmiller, 2000; McGinnis et al., 2002), which were previously shown to contain associated arrestin-1 by electron microscopy (Nir and Ransom, 1992, 1993). Considering the OS diameter is ~ 1.4 μm (Shen et al., 2010) and arrestin-1 bound to these microtubules has to diffuse up to 0.7 μm to reach photoactivated rhodopsin, this would explain the delay of several seconds. These considerations indicate that microtubule-bound arrestin-1 is the most likely candidate for the pool with limited availability, suggesting that in WT OS at least 2% of arrestin-1 content is in this pool (Cleghorn et al., 2011). The physiological significance of this heterogeneity of arrestin-1 in the OS remains to be elucidated.
Interestingly, moderate arrestin-1 over-expression improved functional performance: it accelerated rod recovery after bright flashes and slightly improved light sensitivity. It also turned out that all deviations from WT expression level adversely affected photoreceptor well-being, as reflected in the shortening of the OS, particularly in the peripheral retina of mice expressing much more or much less arrestin-1 than WT. Thus, the level of arrestin-1 in wild type rods likely represents a compromise, ensuring good functional performance along with the preservation of photoreceptor morphology and long-term survival (Song et al., 2011).
5.3. The affinity of arrestin-1 for rhodopsin
Obviously, arrestin-1 affinity for P-Rh* is a critical parameter, but reported measurements are surprisingly controversial. Arrestin-1, like transducin, preferentially binds active Metarhodopsin II, and therefore stabilizes this state. The first measurements took advantage of this phenomenon (Pulvermuller et al., 1997; Schleicher et al., 1989). The KD determined by the extra-Meta II assay reported by Dr. Hofmann’s lab ranged from 50 nM (Schleicher et al., 1989) to 25 nM (Pulvermuller et al., 1997). The limitations of this assay are that it only works at fairly low non-physiological temperatures <14°C, and at a non-physiological pH 8.0 that favors Metarhodopsin I, making the amount of extra-Meta II greater and therefore easier to measure (Sommer et al., 2011). Considering that the KD of arrestin-1 for the non-cognate phosphorylated active β2-adrenergic receptor and M2 muscarinic cholinergic receptor were determined to be 2 nM and 10 nM, respectively (Gurevich et al., 1995), the reported affinity of arrestin for P-Rh* did not seem right. Indeed, measurement under more physiological conditions yielded a KD of ~1 nM (Zhang et al., 1997). This issue was recently revisited by measuring the KD of arrestin-1 for monomeric P-Rh* in nanodiscs, yielding a value of ~4 nM at 25°C (Bayburt et al., 2011). Taking the temperature difference into account, this is consistent with the 25-50 nM affinity observed at 4°C (Pulvermuller et al., 1997; Schleicher et al., 1989).
Physiology provides useful limits for arrestin-1 affinity for P-Rh*. It is well established that to ensure high fidelity of rhodopsin shutoff, arrestin-1 stays bound longer than it takes rhodopsin to decay into opsin. Thermal decay of Rh* in rats and mice in vivo was shown to take ~ 50-60 s (Cone and Cobbs, 1969; Shi et al., 2007; Xu et al., 1997). Assuming that the minimum half-life of the arrestin-1-P-Rh* complex must be greater than this (i.e., several minutes), and taking into account that t1/2 = 6.93*10−7M*s/KD, one can estimate that the highest KD (i.e., the lowest affinity) compatible with physiology is ~3-5 nM. This number is close to the 4 nM measured for monomeric P-Rh* (Bayburt et al., 2011). Thus, one can be fairly confident that the true KD of this interaction in the photoreceptor is equal to or lower than 4 nM.
Arrestin-1 binds P-Rh and Rh* substantially better than inactive Rh in vitro (Gurevich and Benovic, 1992; Gurevich and Benovic, 1993). At the very low physiological levels of rhodopsin activation when rods operate as photoreceptors, the activation of a single rhodopsin molecule actually results in phosphorylation of many more rhodopsins, a phenomenon termed high-gain phopshorylation (Binder et al., 1990; Binder et al., 1996). By taking advantage of differences in spectral sensitivity, it was recently conclusively demonstrated in vivo that the activation of a cone opsin transgenically expressed in rods leads to the phosphorylation of rhodopsin, and vice versa (Shi et al., 2005). These data proved that phosphorylation of “bystander” inactive photopigment is real, and it can be confidently assumed that rods contain a certain level of inactive phosphorhodopsin, P-Rh. In another study it was shown that arrestin-1 could compete with GRK1 for Rh* (Doan et al., 2009). Moreover, arrestin-1 does have a very low affinity even for inactive Rh (Gurevich and Benovic, 1993), which needs to be taken into account considering that the Rh concentration in the OS is ~ 3 mM (Pugh and Lamb, 2000). Thus, in order to elucidate a realistic picture of arrestin-1 dynamics in the OS, precise measurements of arrestin-1 affinities for all functional forms of rhodopsin must be performed.
5.4. The functional role of arrestin-1 concentration
With the fastest response, sub-second recovery, and surprisingly low noise (toad and monkey rhodopsin spontaneously activates only once in 500-1,000 years (Baylor et al., 1980; Field et al., 2005)), rod photoreceptors harbor the most perfect GPCR-driven signaling system. Extremely rapid shutoff of light-activated rhodopsin is a necessary part of this perfection. Rhodopsin, like most GPCRs, is turned off by a two-step mechanism: phosphorylation by GRK1 followed by arrestin-1 binding. The active rhodopsin lifetime in mice in vivo is estimated to be very short, < 60 ms (Krispel et al., 2006), or ~ 30 ms (Gross and Burns, 2010). Part of this time window elapses while GRK1 attaches three phosphates, the minimum number necessary for tight arrestin-1 binding (Mendez et al., 2000; Vishnivetskiy et al., 2007). Thus, the concentration of arrestin-1 must be high enough to encounter P-Rh* very soon after it emerges. Since dimer dissociation would create a delay of ~ 12 ms in mice (see 5.2) that rods cannot functionally afford, the photoreceptors must have a high concentration of active arrestin-1 monomer ready to bind rhodopsin without delay (Hanson et al., 2007c). Arrestin-1 concentration in the dark-adapted OS is ~ 300 μM (Song et al., 2011). The self-association constants of mouse arrestin-1 (Kim et al., 2011a) predict that ~ 300 μM total arrestin-1 yields ~50 μM monomer. With an on-rate constant of ~106 (Bayburt et al., 2011), this translates into an on-rate of ~50 s−1. This means that arrestin-1 encounters each rhodopsin 50 times per second, or every 20 ms. While this number does not directly contradict the 30 ms active rhodopsin lifetime, it leaves only ~ 10 ms for GRK1 to attach multiple phosphates, suggesting that GRK1 is likely processive (i.e., it attaches more than one phosphate per encounter with Rh*). This important issue needs to be addressed experimentally. Alternatively, the concentration of monomeric arrestin-1 in the OS may be higher than self-association alone predicts: even very low affinity binding to dark Rh can significantly shift the equilibrium. If the KD of arrestin-1 for Rh is higher than Kd dimer (i.e., the affinity for Rh is lower), the delay necessary for dissociation from dark Rh could be much shorter than 12 ms needed for dimer dissociation. For example, at a hypothetical arrestin KD = 0.3 mM for Rh (which would still make Rh concentration in the OS ten times higher than the KD, i.e., essentially saturating), the delay would only be ~2.3 ms. Arrestin-1 affinity for dark Rh needs to be measured to estimate the possible contribution of this mechanism to the increase of the on-rate. When its affinity for dark Rh is measured, one can calculate the concentration of all forms of arrestin-1 in the OS using a simple system of equations describing all equilibria and gauge the limitations on active rhodopsin lifetime imposed by arrestin-1 concentration.
5.5. Modeling of rhodopsin shutoff and photoresponse kinetics
The parameters of the biochemical reactions underlying the visual amplification cascade in rods were established using purified proteins in a test tube. Thus, it was only natural that in the early attempts to model single photon response, the rod outer segment was treated as a homogeneous “well-stirred” volume. However, this modeling produced a conundrum: the randomness of the encounters between signaling proteins predicted fairly high variability of the single photon response (SPR), whereas the measured coefficient of variation (CV) of mouse SPR turned out to be much lower, ~37% (Doan et al., 2006). The main contributor to this variability is the variation of active rhodopsin lifetime, determined by the stochastic nature of its inactivation via multiple encounters with GRK1 and subsequently arrestin-1. Thus, the first attempts to explain the lower than expected SPR variability focused on the number of these steps (Rieke and Baylor, 1998). Simple statistical considerations led to the conclusion that if the number of these steps is sufficiently high, their randomness evens out, significantly reducing the variability of the active rhodopsin lifetime. The first estimate of the number of inactivation steps necessary to produce the observed variability was 10-20 (Rieke and Baylor, 1998). This number did not seem realistic: mouse rhodopsin has only six phosphorylation sites, such that even if all of them are required, together with subsequent arrestin-1 binding, the number of steps cannot exceed seven. Moreover, only three rhodopsin-attached phosphates were found to be necessary for high-affinity arrestin-1 binding in vivo (Mendez et al., 2000) and in vitro (Vishnivetskiy et al., 2007), bringing the most probable number of inactivation steps to four. Even if the somewhat contrived notion that GRK1 binding and phosphate transfer are two separate steps (Doan et al., 2009) is accepted, this only returns the number to seven (i.e. still much too low to account for CV = 37% (Doan et al., 2006)). In addition, this logic is only valid if all steps in rhodopsin inactivation contribute equally to the generation of the active effector, cGMP phosphodiesterase (PDE) (Caruso et al., 2010). The discovery of the Ca2+-dependent inhibition of GRK1 via recoverin (Klenchin et al., 1995) suggested that the first step, before the transfer of the first phosphate, is much longer than the others, while biochemical experiments showed that the activity of Rh* towards transducin progressively decreases with increasing number of attached phosphates (Gibson et al., 2000). Collectively, these data mean that unphosphorylated Rh* activates the majority of PDE* produced during the SPR in the first step, completely invalidating the idea that a high number of inactivation steps can reduce the variability. Indeed, careful analysis of SPR variability and the role of Ca2+-mediated feedback in its suppression in toad rods suggested that known feedback mechanisms can fully account for observed variation without invoking unrealistic number of inactivation steps (Whitlock and Lamb, 1999). This study was the first to emphasize the role of rhodopsin interactions with different proteins and highly localized changes in Ca2+ and GTP in the area of the rod adjacent to the disc with activated Rh* (Whitlock and Lamb, 1999). The authors found that even in the extreme case of stochastic one-step Rh* inactivation biochemically realistic model predicts SPR variability closely matching experimental values (Whitlock and Lamb, 1999). Subsequent analysis of SPR variability in mammalian rods (Field and Rieke, 2002) generated very interesting data but was concluded with essentially circular argument: if all steps of rhodopsin inactivation equally contribute to the photoresponse, the multiplicity of inactivation steps accounts for observed reproducibility much better than any alternative model (Field and Rieke, 2002). Indeed, this beautiful model perfectly describes an imaginary rod where all rhodopsin inactivation steps contribute equally to the SPR, but not a real rod where a substantial body of evidence suggests the opposite.
Next breakthrough came when the rod OS was mathematically described in more realistic terms using a spatially and temporally resolved model (Bisegna et al., 2008). The OS cytoplasm has an extremely intricate shape, with ~800 narrow inter-discal spaces where PDE activated by Rh* hydrolyzes cGMP, and a thin layer around the stack of discs near the plasma membrane where cGMP-gated channels are localized (Fig. 8). Moreover, cGMP is replenished by guanylyl cyclase (GC), which is tightly regulated via GCAPs by cytoplasmic Ca2+ that enters through cGMP-gated channels and needs to reach the cyclase before it can have any effect. A spatio-temporal model of rod signaling revealed that the numbers of active PDE* produced are much more variable than the current suppression on the outer shell of the OS, demonstrating that the diffusion of the second messengers in the OS cytoplasm acts as an effective variability suppressor (Bisegna et al., 2008). The model, populated with experimentally determined parameters of the mouse rod (Shen et al., 2010), was recently tested for its ability to reproduce the photoresponse in rods expressing rhodopsin with a variable number of phosphorylation sites (Caruso et al., 2010), which was earlier described experimentally (Mendez et al., 2000). In this case, multi-step rhodopsin inactivation was explicitly modeled by a series of Bernulli trials based on biochemically realistic assumptions (Caruso et al., 2010). The results were very clear: the number of rhodopsin inactivation steps affects SPR variability only in an unrealistic “well-stirred” OS, and is essentially irrelevant when cGMP and Ca2+ diffuse in the cytoplasm of the shape found in rods (Caruso et al., 2010). This is hardly unexpected considering that the complex shape of the photoreceptor cytoplasm with its stacks of photopigment-containing discs is very similar in vertebrate and invertebrate photoreceptors separated by hundreds of millions of years of independent evolution.
Fig. 8. The intricate shape of the rod cytoplasm and highly localized signaling suppress the variability of the single photon response.
The biochemical signal amplification cascade Rh*-Td-PDE, and Rh* inactivation by GRK1 phosphorylation followed by arrestin-1 binding both involve stochastic processes, yet the single photon response in rods, measured as the light-induced change of current at the plasma membrane, shows much better reproducibility than expected. Theoretically, a large number of inactivation steps could serve as variability suppressor (Rieke and Baylor, 1998), but only if the contribution of all steps to Td and PDE activation is roughly equal. Since this is not the case, and as rhodopsin is likely inactivated in only four steps (three phosphorylations and arrestin-1 binding), other mechanisms must ensure high reproducibility of the response. A spatiotemporal mathematical model shows that the variability is suppressed by highly localized signaling initiated by a single Rh*.The biochemical changes occur within a narrow slice of the rod around the activated disc due to the complex shape of the rod cytoplasm that prevents signal spreading (Bisegna et al., 2008; Caruso et al., 2011). Photoresponse is “standardized” by the fact that PDE rapidly depletes cGMP in the vicinity of the disc containing active Rh*, so that PDE generated by the unusually long-lived Rh* has little substrate to hydrolyze, and therefore increases the variation less than it would in a well-stirred model. High cooperativity of the effect of cGMP concentration on channel opening probability further reduces the variability, ensuring that current suppression translates contiguous cGMP changes into a highly localized and essentially “all-or-nothing” channel response. The recovery phase is also highly localized. Moreover, channel closure results in an immediate local drop in Ca2+, which is replaced with the constantly present Mg2+ on GCAPs, converting them into activators of guanylate cyclase (GC) (Dizhoor et al., 2010). The ensuing rapid synthesis of cGMP replenishes its local concentration, opening the channels and returning the rod to the initial state. The activation of one rhodopsin by a single photon in dark-adapted rod suppresses 3-5% of dark current, which is equivalent to channel closure in the membrane surrounding 24-40 discs (out of ~800 in mouse rod). To make illustration of the local nature of the single photon response possible, each disc shown here represents multiple discs in the real OS. Key molecules involved in signaling and recovery are shown below the schematics.
Interestingly, further mathematical dissection of rod signaling suggests that, in addition to diffusion, several other factors play a role in stabilizing the SPR: the highly non-linear relationships (nH >> 1) between cGMP concentration and the probability of channel opening, along with non-linear relationship between intracellular Ca2+ concentration and guanylyl cyclase activity, and the highly localized cGMP hydrolysis by PDE activated by a single Rh* (Fig. 8) (Caruso et al., 2011). Simply put, localized PDE activity in the vicinity of discs with active Rh* rapidly depletes local cGMP, so that additional PDE molecules activated by longer-lived Rh* have little extra cGMP to hydrolyze (Fig. 8). Next, due to the high cooperativity of Na+/Ca2+ channel regulation by cGMP, the channels operate locally in an almost “all-or-nothing” mode, so that cGMP changes only in a relatively small part of the concentration range affect channel opening, thus making additional cGMP changes less effective. Finally, a local drop in Ca2+ results in localized GCAP-dependent activation of guanylyl cyclase, which also operates in an almost “all-or-nothing’ regime thereby reducing the functional consequences of possible biochemical excesses due to inevitable variations of the lifetime of individual Rh* molecules. Collectively, these features significantly reduce the variations of current suppression due to variability of the active Rh* lifetime (Caruso et al., 2011).
6. Arrestin-1 interactions with other signaling proteins
Both non-visual arrestins were reported to interact with hundreds of non-receptor signaling proteins (Xiao et al., 2007), ranging from components of the internalization machinery, such as clathrin (Goodman et al., 1996), AP2 (Laporte et al., 1999), and N-ethylmaleimide-sensitive factor (NSF) (McDonald et al., 1999) to MAP kinases activating JNK3 (McDonald et al., 2000), ERK1/2 (Luttrell et al., 2001), and p38 (Bruchas et al., 2006) and several E3 ubiquitin ligases (Ahmed et al., 2011; Bhandari et al., 2007; Shenoy et al., 2001). Compared to these riches, arrestin-1, which was believed to bind rhodopsin exclusively, looked like a poor relative. However, this “single-mindedness” of arrestin-1 did not appear plausible considering the >50% homology (Gurevich and Gurevich, 2006a) and remarkable structural similarity of all four vertebrate arrestins (Han et al., 2001; Hirsch et al., 1999; Sutton et al., 2005; Zhan et al., 2011).
6.1. Are visual arrestins one-trick ponies?
In fact, the first demonstrated binding partner of arrestin-1 (called S-antigen at the time) was arrestin-1 itself: its self-association (section 3) was discovered in 1977 (Wacker et al., 1977), well before its binding to rhodopsin. The next non-receptor binding partner, polymerized tubulin, was discovered only recently (Nair et al., 2004). Subsequent studies showed that arrestin-1 binding to microtubules is intimately involved in its light-dependent translocation (Nair et al., 2005) (section 4), that arrestin-1 binds both polymerized tubulin and αβ-dimers directly (Hanson et al., 2007a), and that the arrestin-1 molecule undergoes a distinct conformational change upon microtubule binding (Hanson et al., 2006a). These discoveries opened the floodgates: arrestin-1 interactions with MAP kinases JNK3 (Song et al., 2006), ERK2 (Coffa et al., 2011; Hanson et al., 2007a), Ca2+-liganded calmodulin (Wu et al., 2006), E3 ubiquitin ligases Mdm2 (Hanson et al., 2007a; Song et al., 2006) and parkin (Ahmed et al., 2011), as well as NSF (Huang et al., 2010) and enolase (Smith et al., 2011) were discovered in rapid succession. In contrast to non-visual arrestins, where many of the putative interactions were proposed on the basis of co-immunoprecipitation (which cannot prove that the interaction is direct rather than mediated by other proteins), arrestin-1 binding to microtubules (Hanson et al., 2006a), calmodulin (Wu et al., 2006), NSF (Huang et al., 2010), ERK2 (Coffa et al., 2011), enolase (Smith et al., 2011), and parkin (Ahmed et al., 2011) were conclusively shown to be direct in experiments with purified proteins. The affinities of some interactions were even actually measured (Ahmed et al., 2011; Hanson et al., 2007a; Wu et al., 2006). Based on these findings, one can be sure that in terms of versatility arrestin-1 is a signaling regulator that can hold its own against its non-visual cousins. The discovery of additional proteins interacting with arrestin-1 is expected.
Interestingly, the binding sites of microtubules and calmodulin on arrestin-1 overlap with the rhodopsin “footprint”, making arrestin-1 interactions with each of these partners mutually exclusive (Nair et al., 2005; Wu et al., 2006). In contrast, Mdm2 and ERK2 appear to interact with the non-receptor-binding side of arrestin-1, which allows it to mobilize these proteins to the cytoskeleton, reducing overall ERK2 activity in the cell and directing Mdm2 towards microtubule-associated proteins (Hanson et al., 2007a). It is worth noting that the cone-specific arrestin-4 also binds JNK3, Mdm2 (Song et al., 2007), MKK4, ASK1 (Fig. 9), parkin (Ahmed et al., 2011), and microtubules (Hanson et al., 2007a), but does not mobilize ERK2 or Mdm2 to the cytoskeleton (Hanson et al., 2007a).
Fig. 9. Arrestin-1 interacts with all three kinases in the ASK1-MKK4-JNK3 signaling module.
A, C. A nuclear exclusion assay demonstrates arrestin-1 interactions with JNK3 and ASK1. JNK3 (A) and ASK1 with an engineered nuclear localization sequence (NLS) (C) predominantly localize to the nucleus. Arrestin-1 has internal nuclear export signals that ensure its cytoplasmic localization. Co-expressed arrestin-1 relocalizes JNK3 and ASK1-NLS from the nucleus to the cytoplasm, similar to non-visual arrestin-2 and arrestin-3, as well as cone arrestin-4. Lower panels in A and C show the fraction of cells where more JNK3 (A) or ASK1-NLS (C) was localized in the cytoplasm than in the nucleus (C>N). Thus, arrestin-1 binds JNK3 and ASK1 (and that in complex the nuclear export signal of arrestin-1 is stronger than the NLS of JNK3 or ASK1). B. Co-immunoprecipitation shows arrestin-1 interaction with MKK4 in cells. Flag-tagged arrestins are immunoprecipitated by anti-Flag antibodies. Co-expressed HA-tagged MKK4 co-immunoprecipitates with arrestins, but not in control samples where arrestins were not expressed (last lane).
6.2. The functional consequences of arrestin-1 binding to non-receptor partners
Physiological expression levels are the obvious difference between arrestin subtypes: while arrestin-2 and –3 are present in the cytoplasm of neurons at sub-micromolar concentrations (Gurevich et al., 2002, 2004), and arrestin-4 is present at low micromolar levels in cones (Nikonov et al., 2008a), intracellular concentrations of arrestin-1 range from high micromolar (Nikonov et al., 2008a) to millimolar (Kim et al., 2011a; Song et al., 2011) in cones and rods, respectively. These concentrations exceed those of the most abundant non-visual subtype, arrestin-2, by ~4 orders of magnitude. This huge difference puts clear limitations on what arrestin-1 binding to non-receptor partners can and cannot do.
First, for MAP kinases arrestins apparently act as scaffolds to bring together the three kinases of a particular signaling module, such as ASK1-MKK4-JNK3 (McDonald et al., 2000). Similar to non-visual arrestins (Song et al., 2009a), arrestin-1 binds all three kinases in this cascade (Fig. 9). However, binding per se does not necessarily lead to productive scaffolding even in case of non-visual arrestins. For example, it was recently shown that several arrestin-3 mutants that bind the kinases in ASK1-MKK4-JNK3 module as well as WT arrestin-3 or even better, do not organize productive complexes that facilitate JNK3 phosphorylation (Seo et al., 2011). Moreover, even when an arrestin can organize a productive complex, the functional outcome of the ability to bind kinases in the same signaling module depends on the relative concentrations of scaffolding protein and the kinases it is supposed to scaffold (Levchenko et al., 2000; Locasale and Chakraborty, 2008). Simply put, when the number of scaffolding molecules is lower than the number of proteins they scaffold, they promote co-localization of the latter, thereby facilitating signaling. However, when the number of scaffolds significantly exceeds the number of enzyme molecules it can bring together, the majority of scaffold molecules would have only one of the several kinases bound, making the probability of their interaction and productive signal transduction lower than in the absence of the scaffold. Thus, non-visual arrestins, expressed at levels comparable to those of MAP kinases, can promote signaling by recruiting several kinases into a signaling complex. In contrast, arrestin-1 at concentrations several orders of magnitude higher than that of MAP kinases, can only act in a dominant-negative manner, pulling these kinases apart and blocking signal transduction. In fact, it was shown that over-expressed arrestin-1, as well as arrestin-2 and –3, recruit ERK2 and reduce its activity in the cell (Hanson et al., 2007a).
However, this logic does not apply when the abundance of at least one of the interaction partners is comparable to that of arrestin-1. For example, arrestin-1 over-expressed in HEK293 cells at a level sufficient to suppress ERK2 activity recruits Mdm2 to microtubules (which are very abundant in the cell) and increases ubiquitination of microtubule-associated proteins (Hanson et al., 2007a). Thus, the ability of arrestin-1 to bind ubiquitin ligases Mdm2 (Hanson et al., 2007a; Song et al., 2006) and parkin (Ahmed et al., 2011) likely translates into its role in the regulation of ubiquitination of cytoskeletal proteins highly abundant in the cell body, and possibly other binding partners. This aspect of arrestin-1 function needs to be tested experimentally in photoreceptor cells. Arrestin-1 has also been shown to bind enolase and affect its activity (Smith et al., 2011). However, the role of this interaction in photoreceptors also remains to be elucidated.
So far, arrestin-1 binding to only one non-rhodopsin partner, NSF, has been shown to affect rod function (Huang et al., 2010). In the photoreceptor synapse, NSF participates in vesicle exocytosis and endocytosis that retrieves vesicle membrane and synaptic proteins from the plasma membrane. Arrestin-1 binds to the junction between the N-terminal and the first ATPase domain of NSF in an ATP-dependent manner, and enhances both ATPase and disassembly activities of NSF (Huang et al., 2010). It was shown that in arrestin-1 knockout retinas the expression levels of NSF and several other synaptic proteins were markedly reduced, which resulted in a substantial decrease in exocytosis. Thus, arrestin-1 binding to NSF modulates normal synaptic function in mouse photoreceptors (Huang et al., 2010).
Arrestin-1 specifically binds Ca2+-liganded, but not free calmodulin (Wu et al., 2006). The affinity of this interaction appears low (~ 7 μM), but the concentration of arrestin-1 in rod and cone photoreceptors is very high (Hanson et al., 2007b; Nikonov et al., 2008a; Song et al., 2011; Strissel et al., 2006). Considering that rods contain much less calmodulin than arrestin-1 (Pugh and Lamb, 2000), and that even “high” Ca2+ concentrations in dark-adapted photoreceptors are still sub-micromolar, it seems likely that most of the Ca2+-liganded calmodulin exists in complex with arrestin-1. It is unclear whether this interaction serves to localize Ca2+-calmodulin to particular cellular compartments or to prevent Ca2+-calmodulin interactions with other proteins in the cell.
7. Enhanced arrestin-1 and a compensational approach to gene therapy
Mutations in >100 different genes have been shown to cause a variety of visual disorders, including several forms of retinal degeneration, that affect ~1 in 3,000 people (Smith et al., 2009). Mechanistically, there are two distinct kinds of genetic disorders, associated with either loss or gain of function of the affected protein. Although it is still technically challenging, conceptually the path to dealing with loss-of-function mutations is fairly straightforward: cDNA encoding functional protein under the appropriate promoter has to be introduced into affected cells. Recently, the potential of gene replacement therapy using recombinant non-pathogenic adeno-associated virus as a vector was demonstrated in human patients with Leber congenital amaurosis (reviewed in (Roy et al., 2010; Smith et al., 2009)). The same approach is likely to be effective in Oguchi disease patients with loss-of-function mutations in the arrestin-1 or GRK1 gene (Fuchs et al., 1995; Yamamoto et al., 1997). However, the situation is much more complicated for gain-of-function mutations: the loss of phosphorylation sites abolishes the ability of normal GRK1 and arrestin-1 to quench rhodopsin signaling, and this genetic error leads to retinal degeneration in humans (Apfelstedt-Sylla et al., 1993; Kim et al., 1993; Restagno et al., 1993). These mutations are dominant: rapid shutoff of rhodopsin generated by the other perfectly normal allele does not prevent the excessive signaling by the defective protein. Gain-of-function mutations resulting in excessive signaling by other GPCRs underlie about a dozen disorders, ranging from dwarfism, hypo- and hyper-thyroidism, to several forms of cancer (reviewed in (Schöneberg et al., 2004)). Theoretically, one could design a ribozyme specifically destroying mutant mRNA while preserving the normal one transcribed from the intact allele. However, this has to be achieved with exceptional precision since the two messages may differ by as little as one base (as in the case of frame-shift mutations (Apfelstedt-Sylla et al., 1993)), and virtually 100% efficacy because even low expression of unquenchable rhodopsin kills photoreceptors (Chen et al., 1995). In this situation compensational gene therapy appears to be a viable alternative: counter-balancing a mutant receptor that signals too much with an arrestin mutant that quenches the signaling more readily than wild type. Indeed, it was recently demonstrated that in rods with defective rhodopsin phosphorylation, the expression of an enhanced arrestin-1 that binds unphosphorylated Rh* with high affinity (3A mutant) improves functional performance, accelerates the recovery, and promotes photoreceptor survival (Song et al., 2009b).
This study also revealed the limitations of existing “off-the-shelf” enhanced arrestin-1 mutants: in GRK1−/− rods, the arrestin-1-3A mutant reduced the recovery half-time from ~18 s in animals expressing wild type arrestin-1 to ~6 s, whereas at the same strength of desensitizing flash normal wild type rods recovered with half-time of ~400 ms (Song et al., 2009b). Thus, compensated phosphorylation-deficient rods recovered much slower than wild type, suggesting that additional redesign of the rhodopsin-binding surface to improve the affinity for Rh* is necessary. Nonetheless, this proof-of-principle study demonstrated for the first time the potential of compensational gene therapy, which has to be further perfected to address various disorders associated with gain-of-function mutations in rhodopsin, non-visual GPCRs, and possibly other signaling proteins (reviewed in (Gurevich and Gurevich, 2010a)). The work of Song et al. also clearly demonstrated that the mechanisms of protein-protein interactions that need to be targeted for therapeutic purposes must be elucidated in minute molecular detail to ensure efficient compensation.
8. Future directions
Although structurally and functionally arrestin-1 is one of the most studied vertebrate proteins, many important aspects of its biology still remain to be elucidated. The range of its non-rhodopsin interaction partners is only beginning to emerge, and in most cases the physiological role of these interactions remains unclear. The biological function of the robust self-association conserved among mammalian species needs to be conclusively established. Both the exact mechanism of its light-dependent translocation and its functional significance need to be clarified. Even the mechanism of its interaction with rhodopsin is not yet understood in the detail necessary to design enhanced mutants for therapeutic purposes. The combination of further structural, biophysical, and biochemical studies with rigorous experimentation in living animals is necessary to answer many remaining questions. This work is clearly worth the effort: thorough understanding of every aspect of arrestin-1 function and the underlying fine molecular mechanisms will enable the development of arrestin-based molecular tools with enormous therapeutic potential.
Acknowledgements
The authors are grateful to our collaborators who made the studies described here possible. In particular, the authors would like to thank the labs of Drs. Paul B. Sigler, Wayne L. Hubbell, Jeannie Chen, Marie E. Burns, Emmanuele DiBenedetto, Heidi Hamm, and Candice Klug. Funded by NIH grants EY011500, GM077561, and GM081756 (VVG), NS045117 and NS065868 (EVG), and P30 core grant in vision research EY008126 (to Vanderbilt University).
Footnotes
We use systematic names of arrestin proteins: arrestin-1 (a.k.a. visual or rod arrestin, 48 kDa protein, or S-antigen), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin).
References
- Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry. 2011:3749–3763. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloway PG, Dolph PJ. A role for the light-dependent phosphorylation of visual arrestin. Proc Natl Acad Sci U S A. 1999;96:6072–6077. doi: 10.1073/pnas.96.11.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci U S A. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfelstedt-Sylla E, Kunisch M, Horn M, Ruther K, Gerding H, Gal A, Zrenner E. Ocular findings in a family with autosomal dominant retinitis pigmentosa and a frameshift mutation altering the carboxyl terminal sequence of rhodopsin. Br J Ophthalmol. 1993;77:495–501. doi: 10.1136/bjo.77.8.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Huber T, Sakmar TP. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Vishnivetskiy SA, McLean M, Morizumi T, Huang C.-c., Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Rhodopsin monomer is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. J Physiol. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- Baylor DA, Matthews G, Yau KW. Two components of electrical dark noise in toad retinal rod outer segments. J Physiol. 1980;309:591–621. doi: 10.1113/jphysiol.1980.sp013529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovic JL, DeBlasi A, Stone WC, Caron MG, Lefkowitz RJ. Beta-adrenergic receptor kinase: primary structure delineates a multigene family. Science. 1989;246:235–240. doi: 10.1126/science.2552582. [DOI] [PubMed] [Google Scholar]
- Benovic JL, Kuhn H, Weyand I, Codina J, Caron MG, Lefkowitz RJ. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein) Proc Nat Acad Sci USA. 1987;84:8879–8882. doi: 10.1073/pnas.84.24.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann A, Schmidt M, Reuss S, Wolfrum U, Hankeln T, Burmester T. Divergent distribution in vascular and avascular mammalian retinae links neuroglobin to cellular respiration. J Biol Chem. 2005;280:20660–20665. doi: 10.1074/jbc.M501338200. [DOI] [PubMed] [Google Scholar]
- Bhandari D, Trejo J, Benovic JL, Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- Binder BM, Biernbaum MS, Bownds MD. Light activation of one rhodopsin molecule causes the phosphorylation of hundreds of others. A reaction observed in electropermeabilized frog rod outer segments exposed to dim illumination. J Biol Chem. 1990;265:15333–15340. [PubMed] [Google Scholar]
- Binder BM, O’Connor TM, Bownds MD, Arshavsky VY. Phosphorylation of non-bleached rhodopsin in intact retinas and living frogs. J Biol Chem. 1996;271:19826–19830. doi: 10.1074/jbc.271.33.19826. [DOI] [PubMed] [Google Scholar]
- Birch DG, Hood DC, Nusinowitz S, Pepperberg DR. Abnormal activation and inactivation mechanisms of rod transduction in patients with autosomal dominant retinitis pigmentosa and the pro-23-his mutation. Invest Ophthalmol Vis Sci. 1995;36:1603–1614. [PubMed] [Google Scholar]
- Bisegna P, Caruso G, Andreucci D, Shen L, Gurevich VV, Hamm HE, DiBenedetto E. Diffusion of the second messengers in the cytoplasm acts as a variability suppressor of the single photon response in vertebrate phototransduction. Biophys J. 2008;94:3363–3383. doi: 10.1529/biophysj.107.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann MR, Cohen LV. Diurnal expression of transducin mRNA and translocation of transducin in rods of rat retina. Science. 1987;235:585–587. doi: 10.1126/science.3101175. [DOI] [PubMed] [Google Scholar]
- Broekhuyse RM, Tolhuizen EF, Janssen AP, Winkens HJ. Light induced shift and binding of S-antigen in retinal rods. Curr Eye Res. 1985;4:613–618. doi: 10.3109/02713688508999993. [DOI] [PubMed] [Google Scholar]
- Bruchas MR, Macey TA, Lowe JD, Chavkin C. Kappa opioid receptor activation of p38 MAPK is GRK3- and arrestin-dependent in neurons and astrocytes. J Biol Chem. 2006;281:18081–18089. doi: 10.1074/jbc.M513640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns ME, Arshavsky VY. Beyond counting photons: trials and trends in vertebrate visual transduction. Neuron. 2005;48:387–401. doi: 10.1016/j.neuron.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Burns ME, Mendez A, Chen CK, Almuete A, Quillinan N, Simon MI, Baylor DA, Chen J. Deactivation of phosphorylated and nonphosphorylated rhodopsin by arrestin splice variants. J Neurosci. 2006;26:1036–1044. doi: 10.1523/JNEUROSCI.3301-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns ME, Pugh EN., Jr. Lessons from photoreceptors: turning off g-protein signaling in living cells. Physiology (Bethesda) 2010;25:72–84. doi: 10.1152/physiol.00001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns ME, Pugh ENJ. RGS9 concentration matters in rod phototransduction. Biophys J. 2009;97:1538–1547. doi: 10.1016/j.bpj.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byk T, Bar-Yaacov M, Doza YN, Minke B, Selinger Z. Regulatory arrestin cycle secures the fidelity and maintenance of the fly photoreceptor cell. Proc Natl Acad Sci U S A. 1993;90:1907–1911. doi: 10.1073/pnas.90.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Govardovskii VI, Krasnoperova N, Anderson RE, Lem J, Makino CL. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 2001;411:90–94. doi: 10.1038/35075083. [DOI] [PubMed] [Google Scholar]
- Calvert PD, Makino CL. The time course of light adaptation in vertebrate retinal rods. Adv Exp Med Biol. 2002;514:37–60. doi: 10.1007/978-1-4615-0121-3_3. [DOI] [PubMed] [Google Scholar]
- Carter JM, Gurevich VV, Prossnitz ER, Engen JR. Conformational differences between arrestin2 and pre-activated mutants as revealed by hydrogen exchange mass spectrometry. J Mol Biol. 2005;351:865–878. doi: 10.1016/j.jmb.2005.06.048. [DOI] [PubMed] [Google Scholar]
- Caruso G, Bisegna P, Andreucci D, Lenoci L, Gurevich VV, Hamm HE, DiBenedetto E. Identification of key factors that reduce the variability of the single photon response. Proc Nat Acad Sci USA. 2011 doi: 10.1073/pnas.1018960108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso G, Bisena P, Lenoci L, Andreucci D, Gurevich VV, Hamm HE, DiBenedetto E. Kinetics of rhodopsin inactivation and its role in regulating recovery and reproducibility of rod photoresponse. PLoS Computational Biology. 2010;6:e1001031. doi: 10.1371/journal.pcbi.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J. Biol. Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- Chabre M, le Maire M. Monomeric G-protein-coupled receptor as a functional unit. Biochemistry. 2005;44:9395–9403. doi: 10.1021/bi050720o. [DOI] [PubMed] [Google Scholar]
- Chan S, Rubin WW, Mendez A, Liu X, Song X, Hanson SM, Craft CM, Gurevich VV, Burns ME, Chen J. Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest. Ophthalmol. Vis. Sci. 2007;48:1968–1075. doi: 10.1167/iovs.06-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Nat Acad Sci USA. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Mechanisms of rhodopsin inactivation in vivo as revealed by a COOH-terminal truncation mutant. Science. 1995;267:374–377. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauß N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Zhao X, Nielsen L, Khani SC, Jacobson SG, Palczewski K. Null mutation in the rhodopsin kinase gene slows recovery kinetics of rod and cone phototransduction in man. Proc Natl Acad Sci U S A. 1998;95:328–333. doi: 10.1073/pnas.95.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleghorn WM, Tsakem EL, Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Progressive reduction of its expression in rods reveals two pools of arrestin-1 in the outer segment with different roles in photoresponse recovery. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022797. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa S, Breitman M, Spiller BW, Gurevich VV. A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry. 2011;50 doi: 10.1021/bi200745k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone RA, Cobbs W.H.r. Rhodopsin cycle in the living eye of the rat. Nature. 1969;221:820–822. doi: 10.1038/221820a0. [DOI] [PubMed] [Google Scholar]
- Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. J Biol Chem. 1994;269:4613–4619. [PubMed] [Google Scholar]
- Dinculescu A, McDowell JH, Amici SA, Dugger DR, Richards N, Hargrave PA, Smith WC. Insertional mutagenesis and immunochemical analysis of visual arrestin interaction with rhodopsin. J Biol Chem. 2002;277:11703–11708. doi: 10.1074/jbc.M111833200. [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Olshevskaya EV, Peshenko IV. Mg2+/Ca2+ cation binding cycle of guanylyl cyclase activating proteins (GCAPs): role in regulation of photoreceptor guanylyl cyclase. Mol Cell Biochem. 2010;334:117–124. doi: 10.1007/s11010-009-0328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Azevedo AW, Hurley JB, Rieke F. Arrestin competition influences the kinetics and variability of the single-photon responses of mammalian rod photoreceptors. J Neurosci. 2009;29:11867–11879. doi: 10.1523/JNEUROSCI.0819-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Mendez A, Detwiler PB, Chen J, Rieke F. Multiple phosphorylation sites confer reproducibility of the rod’s single-photon responses. Science. 2006;313:530–533. doi: 10.1126/science.1126612. [DOI] [PubMed] [Google Scholar]
- Eckmiller MS. Microtubules in a rod-specific cytoskeleton associated with outer segment incisures. Vis Neurosci. 2000;17:711–722. doi: 10.1017/s0952523800175054. [DOI] [PubMed] [Google Scholar]
- Elias RV, Sezate SS, Cao W, McGinnis JF. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha-transducin in mouse photoreceptor cells. Mol Vis. 2004;10:672–681. [PubMed] [Google Scholar]
- Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc. Natl. Acad. Sci. USA. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein SE, Pulvermüller A, Hartmann R, Granzin J, Stoldt M, Henklein P, Ernst OP, Heck M, Willbold D, Koenig BW. Helix formation in arrestin accompanies recognition of photoactivated rhodopsin. Biochemistry. 2009;48:10733–10742. doi: 10.1021/bi900544p. [DOI] [PubMed] [Google Scholar]
- Field GD, Rieke F. Mechanisms regulating variability of the single photon responses of mammalian rod photoreceptors. Neuron. 2002;35:733–747. doi: 10.1016/s0896-6273(02)00822-x. [DOI] [PubMed] [Google Scholar]
- Field GD, Sampath AP, Rieke F. Retinal processing near absolute threshold: from behavior to mechanism. Annu Rev Physiol. 2005;67:491–514. doi: 10.1146/annurev.physiol.67.031103.151256. [DOI] [PubMed] [Google Scholar]
- Fonseca JM, Lambert NA. Instability of a class a G protein-coupled receptor oligomer interface. Mol Pharmacol. 2009;75:1296–1299. doi: 10.1124/mol.108.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr. Opin. Struct. Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10:360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- Fung BK, Hurley JB, Stryer L. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc Natl Acad Sci U S A. 1981;78:152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliera E, Jala VR, Trent JO, Bonecchi R, Signorelli P, Lefkowitz RJ, Mantovani A, Locati M, Haribabu B. beta-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–25597. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- Ge H, Krishnan P, Liu L, Krishnan B, Davis RL, Hardin PE, Roman G. A Drosophila nonvisual arrestin is required for the maintenance of olfactory sensitivity. Chem Senses. 2006;31:49–62. doi: 10.1093/chemse/bjj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SK, Parkes JH, Liebman PA. Phosphorylation modulates the affinity of light-activated rhodopsin for G protein and arrestin. Biochemistry. 2000;39:5738–5749. doi: 10.1021/bi991857f. [DOI] [PubMed] [Google Scholar]
- Goodman OB, Jr., Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Govardovskii VI, Korenyak DA, Shukolyukov SA, Zueva LV. Lateral diffusion of rhodopsin in photoreceptor membrane: a reappraisal. Mol Vis. 2009;15:1717–1729. [PMC free article] [PubMed] [Google Scholar]
- Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- Gray-Keller MP, Detwiler PB, Benovic JL, Gurevich VV. Arrestin with a single amino acid sustitution quenches light-activated rhodopsin in a phosphorylation0independent fasion. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- Gross OP, Burns ME. Control of rhodopsin’s active lifetime by arrestin-1 expression in mammalian rods. J Neurosci. 2010;30:3450–3457. doi: 10.1523/JNEUROSCI.5391-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Benovic JL, Gurevich VV. Arrestin2 and arrestin3 are differentially expressed in the rat brain during postnatal development. Neuroscience. 2002;109:421–436. doi: 10.1016/s0306-4522(01)00511-5. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Benovic JL, Gurevich VV. Arrestin2 expression selectively increases during neural differentiation. J Neurochem. 2004;91:1404–1416. doi: 10.1111/j.1471-4159.2004.02830.x. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Gurevich VV. Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006a;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Tesmer JJG, Mushegian A, Gurevich EV. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharm Ther. 2011 doi: 10.1016/j.pharmthera.2011.08.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Cell-free expression of visual arrestin. Truncation mutagenesis identifies multiple domains involved in rhodopsin interaction. J Biol Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin: Sequential multisite binding ensures strict selectivity towards light-activated phosphorylated rhodopsin. J. Biol. Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin: diverse functional roles of positively charged residues within the phosphorylation-recignition region of arrestin. J. Biol. Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Mechanism of phosphorylation-recognition by visual arrestin and the transition of arrestin into a high affinity binding state. Mol Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Chen C-Y, Kim CM, Benovic JL. Visual arrestin binding to rhodopsin: Intramolecular interaction between the basic N-terminus and acidic C-terminus of arrestin may regulate binding selectivity. J Biol Chem. 1994;269:8721–8727. [PubMed] [Google Scholar]
- Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharm Ther. 2006b;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends. Neurosci. 2008a;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. How and why do GPCRs dimerize? Trends Pharmacol Sci. 2008b;29:234–240. doi: 10.1016/j.tips.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. Custom-designed proteins as novel therapeutic tools? The case of arrestins. Expert Rev Mol Med. 2010a;12:e13. doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. Phototransduction: inactivation in cones. Encyclopedia of the Eye. 2010b;3:370–374. [Google Scholar]
- Gurevich VV, Hanson SM, Gurevich EV, Vishnivetskiy SA. How Rod Arrestin Achieved Perfection: Regulation of its Availability and Binding Selectivity. In: Fliesler SJ, Kisselev O, editors. Signal transduction in the retina. CRC Press; Boca Raton, FL: 2007. pp. 55–88. [Google Scholar]
- Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J Biol Chem. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Richardson RM, Kim CM, Hosey MM, Benovic JL. Binding of wild type and chimeric arrestins to the m2 muscarinic cholinergic receptor. J Biol Chem. 1993;268:16879–16882. [PubMed] [Google Scholar]
- Hamm HE, Bownds MD. Protein complement of rod outer segments of frog retina. Biochemistry. 1986;25:4512–4523. doi: 10.1021/bi00364a010. [DOI] [PubMed] [Google Scholar]
- Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song X, Nair KS, Slepak VZ, Klug CS, Gurevich VV. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007a doi: 10.1016/j.jmb.2007.02.053. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Dawson ES, Francis DJ, Van Eps N, Klug CS, Hubbell WL, Meiler J, Gurevich VV. A model for the solution structure of the rod arrestin tetramer. Structure. 2008a;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 2006a;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci U S A. 2006b;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc Nat Acad Sci USA. 2007b;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Van Eps N, Francis DJ, Altenbach C, Vishnivetskiy SA, Arshavsky VY, Klug CS, Hubbell WL, Gurevich VV. Structure and function of the visual arrestin oligomer. EMBO J. 2007c;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Vishnivetskiy SA, Hubbell WL, Gurevich VV. Opposing effects of inositol hexakisphosphate on rod arrestin and arrestin2 self-association. Biochemistry. 2008b;47:1070–1075. doi: 10.1021/bi7021359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JE, Lazareno S, Molloy JE, Birdsall NJ. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc Natl Acad Sci U S A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetling JR, Pepperberg DR. Sensitivity and kinetics of mouse rod flash responses determined in vivo from paired-flash electroretinograms. J Physiol. 1999;516:593–609. doi: 10.1111/j.1469-7793.1999.0593v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- Huang SP, Brown BM, Craft CM. Visual Arrestin 1 acts as a modulator for N-ethylmaleimide-sensitive factor in the photoreceptor synapse. J Neurosci. 2010;30:9381–9391. doi: 10.1523/JNEUROSCI.1207-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- Hyde DR, Mecklenburg KL, Pollock JA, Vihtelic TS, Benzer S. Twenty Drosophila visual system cDNA clones: one is a homolog of human arrestin. Proc Natl Acad Sci U S A. 1990;87:1008–1012. doi: 10.1073/pnas.87.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Concentration-dependent tetramerization of bovine visual arrestin. Biophys J. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jala VR, Shao WH, Haribabu B. Phosphorylation-independent beta-arrestin translocation and internalization of leukotriene B4 receptors. J Biol Chem. 2005;280:4880–4887. doi: 10.1074/jbc.M409821200. [DOI] [PubMed] [Google Scholar]
- Kasai RS, Suzuki KG, Prossnitz ER, Koyama-Honda I, Nakada C, Fujiwara TK, Kusumi A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Hanson SM, Vishnivetskiy SA, Song X, Cleghorn WM, Hubbell WL, Gurevich VV. Robust self-association is a common feature of mammalian visual arrestin-1. Biochemistry. 2011a;50:2235–2242. doi: 10.1021/bi1018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vishnivetskiy SA, Van Eps N, Zhan X, Cleghorn WM, Alexander N, Hanson SM, Meiler J, Gurevich VV, Hubbell WL. Conformation of receptor-bound arrestin-1: a site-directed spin labeling study. ASBMB meeting.2011b. pp. 750–759. [Google Scholar]
- Kim RY, al-Maghtheh M, Fitzke FW, Arden GB, Jay M, Bhattacharya SS, Bird AC. Dominant retinitis pigmentosa associated with two rhodopsin gene mutations. Leu-40-Arg and an insertion disrupting the 5′-splice junction of exon 5. Arch Ophthalmol. 1993;111:1518–1524. doi: 10.1001/archopht.1993.01090110084030. [DOI] [PubMed] [Google Scholar]
- Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, Ranganathan R. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Klenchin VA, Calvert PD, Bownds MD. Inhibition of rhodopsin kinase by recoverin. Further evidence for a negative feedback system in phototransduction. J Biol Chem. 1995;270:16147–16152. doi: 10.1074/jbc.270.27.16147. [DOI] [PubMed] [Google Scholar]
- Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent b-arrestin mutants with constitutive activity in cells. J. Biol. Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Kuhn H. Light-regulated binding of rhodopsin kinase and other proteins to cattle photoreceptor membranes. Biochemistry. 1978;17:4389–4395. doi: 10.1021/bi00614a006. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- Lambert NA. GPCR dimers fall apart. Sci Signal. 2010;3:pe12. doi: 10.1126/scisignal.3115pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Lan TH, Kuravi S, Lambert NA. Internalization dissociates β2-adrenergic receptors. PLoS One. 2011;6:e17361. doi: 10.1371/journal.pone.0017361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson s.S.G., Caron MG, Barak LS. The 2-adrenergic receptor/arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Nat Acad Sci USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Montell C. Light-dependent translocation of visual arrestin regulated by the NINAC myosin III. Neuron. 2004;43:95–103. doi: 10.1016/j.neuron.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Xu H, Kang LW, Amzel LM, Montell C. Light adaptation through phosphoinositide-regulated translocation of Drosophila visual arrestin. Neuron. 2003;39:121–132. doi: 10.1016/s0896-6273(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Xu H, Montell C. Rhodopsin kinase activity modulates the amplitude of the visual response in Drosophila. Proc Natl Acad Sci U S A. 2004;101 doi: 10.1073/pnas.0402205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci U S A. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Liebman PA, Pugh EN., Jr. ATP mediates rapid reversal of cyclic GMP phosphodiesterase activation in visual receptor membranes. Nature. 1980;287:734–736. doi: 10.1038/287734a0. [DOI] [PubMed] [Google Scholar]
- Linton JD, Holzhausen LC, Babai N, Song H, Miyagishima KJ, Stearns GW, Lindsay K, Wei J, Chertov AO, Peters TA, Caffe R, Pluk H, Seeliger MW, Tanimoto N, Fong K, Bolton L, Kuok DL, Sweet IR, Bartoletti TM, Radu RA, Travis GH, Zagotta WN, Townes-Anderson E, Parker E, Van der Zee CE, Sampath AP, Sokolov M, Thoreson WB, Hurley JB. Flow of energy in the outer retina in darkness and in light. Proc Natl Acad Sci U S A. 2010;107:8599–8604. doi: 10.1073/pnas.1002471107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Satoh AK, Postma M, Huang J, Ready DF, Hardie RC. Ca2+-dependent metarhodopsin inactivation mediated by calmodulin and NINAC myosin III. Neuron. 2008;59:778–789. doi: 10.1016/j.neuron.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Chakraborty AK. Regulation of signal duration and the statistical dynamics of kinase activation by scaffold proteins. PLoS Comput Biol. 2008;4:e1000099. doi: 10.1371/journal.pcbi.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J Biol Chem. 1992;267:8558–8564. [PubMed] [Google Scholar]
- Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- Lorenz W, Inglese J, Palczewski K, Onorato JJ, Caron MG, Lefkowitz RJ. The receptor kinase family: primary structure of rhodopsin kinase reveals similarities to the beta-adrenergic receptor kinase. Proc Natl Acad Sci U S A. 1991;88:8715–8719. doi: 10.1073/pnas.88.19.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci U S A. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubarsky AL, Pugh EN., Jr. Recovery phase of the murine rod photoresponse reconstructed from electroretinographic recordings. J Neurosci. 1996;16:563–571. doi: 10.1523/JNEUROSCI.16-02-00563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- McDonald PH, Cote NL, Lin FT, Premont RT, Pitcher JA, Lefkowitz RJ. Identification of NSF as a beta-arrestin1-binding protein. Implications for beta2-adrenergic receptor regulation. J Biol Chem. 1999;274:10677–10680. doi: 10.1074/jbc.274.16.10677. [DOI] [PubMed] [Google Scholar]
- McGinnis JF, Matsumoto B, Whelan JP, Cao W. Cytoskeleton participation in subcellular trafficking of signal transduction proteins in rod photoreceptor cells. J Neurosci Res. 2002;67:290–297. doi: 10.1002/jnr.10120. [DOI] [PubMed] [Google Scholar]
- Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, Chen J. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Mendez A, Lem J, Simon M, Chen J. Light-dependent translocation of arrestin in the absence of rhodopsin phosphorylation and transducin signaling. J Neurosci. 2003;23:3124–3129. doi: 10.1523/JNEUROSCI.23-08-03124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J. Biol. Chem. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- Milligan G. G protein-coupled receptor hetero-dimerization: contribution to pharmacology and function. Br J Pharmacol. 2009;158:5–14. doi: 10.1111/j.1476-5381.2009.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G. The role of dimerisation in the cellular trafficking of G-protein-coupled receptors. Curr Opin Pharmacol. 2010;10:23–29. doi: 10.1016/j.coph.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Gurevich VV, Preninger A, Hamm HE, Bader M-F, Fazleabas AT, Birnbaumer L, Hunzicker-Dunn M. Aspartic acid 564 in the third cytoplasmic loop of luteinizing hormone/choriogonadotropin receptor is crucial for phosphorylation-independent interaction with arrestin2. J Biol Chem. 2002;277:17916–17927. doi: 10.1074/jbc.M110479200. [DOI] [PubMed] [Google Scholar]
- Murakami A, Yajima T, Sakuma H, McLaren MJ, Inana G. X-arrestin: a new retinal arrestin mapping to the X chromosome. FEBS Lett. 1993;334:203–209. doi: 10.1016/0014-5793(93)81712-9. [DOI] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, Slepak VZ. Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Orii H, Yoshida N, Jojima E, Horie T, Yoshida R, Haga T, Tsuda M. Ascidian arrestin (Ci-arr), the origin of the visual and nonvisual arrestins of vertebrate. Eur J Biochem. 2002;269:5112–5118. doi: 10.1046/j.1432-1033.2002.03240.x. [DOI] [PubMed] [Google Scholar]
- Nikonov SS, Brown BM, Davis JA, Zuniga FI, Bragin A, Pugh EN, Jr, Craft CM. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008a;59:462–474. doi: 10.1016/j.neuron.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonov SS, Brown BM, Davis JA, Zuniga FI, Bragin A, Pugh ENJ, Craft CM. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008b;59:462–474. doi: 10.1016/j.neuron.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir I, Ransom N. S-antigen in rods and cones of the primate retina: different labeling patterns are revealed with antibodies directed against specific domains in the molecule. J Histochem Cytochem. 1992;40:343–352. doi: 10.1177/40.3.1372630. [DOI] [PubMed] [Google Scholar]
- Nir I, Ransom N. Ultrastructural analysis of arrestin distribution in mouse photoreceptors during dark/light cycle. Exp Eye Res. 1993;57:307–318. doi: 10.1006/exer.1993.1129. [DOI] [PubMed] [Google Scholar]
- Ohguro H, Palczewski K, Walsh KA, Johnson RS. Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 1994;3:2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organisciak DT, Xie A, Wang H-M, Jiang Y-L, Darrow RM, Donoso LA. Adaptive changes in visual cell transduction protein levels: effect of light. Exp Eye Res. 1991;53:773–779. doi: 10.1016/0014-4835(91)90113-s. [DOI] [PubMed] [Google Scholar]
- Orisme W, Li J, Goldmann T, Bolch S, Wolfrum U, Smith WC. Light-dependent translocation of arrestin in rod photoreceptors is signaled through a phospholipase C cascade and requires ATP. Cell Signal. 2010;22:447–456. doi: 10.1016/j.cellsig.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Buczyłko J, Imami NR, McDowell JH, Hargrave PA. Role of the carboxyl-terminal region of arrestin in binding to phosphorylated rhodopsin. J Biol Chem. 1991a;266:15334–15339. [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, LeTrong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Palczewski K, McDowell H, Jakes S, Ingebritsen TS, Hargrave PA. Regulation of rhodopsin dephosphorylation by arrestin. J. Biol. Chem. 1989;264:15770–15773. [PubMed] [Google Scholar]
- Palczewski K, Pulvermüller A, Buczylko J, Gutmann C, Hofmann KP. Binding of inositol phosphates to arrestin. FEBS Lett. 1991b;295:195–199. doi: 10.1016/0014-5793(91)81416-6. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991c;266:18649–18654. [PubMed] [Google Scholar]
- Palmitessa A, Benovic JL. Arrestin and the multi-PDZ domain-containing protein MPZ-1 interact with phosphatase and tensin homolog (PTEN) and regulate Caenorhabditis elegans longevity. J. Biol. Chem. 2010;285:15187–15200. doi: 10.1074/jbc.M110.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmitessa A, Hess HA, Bany IA, Kim YM, Koelle MR, Benovic JL. Caenorhabditus elegans arrestin regulates neural G protein signaling and olfactory adaptation and recovery. J. Biol. Chem. 2005;280:24649–24662. doi: 10.1074/jbc.M502637200. [DOI] [PubMed] [Google Scholar]
- Pan L, Gurevich EV, Gurevich VV. The nature of the arrestin x receptor complex determines the ultimate fate of the internalized receptor. J Biol Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- Peet JA, Bragin A, Calvert PD, Nikonov SS, Mani S, Zhao X, Besharse JC, Pierce EA, Knox BE, Pugh EN., Jr. Quantification of the cytoplasmic spaces of living cells with EGFP reveals arrestin-EGFP to be in disequilibrium in dark adapted rod photoreceptors. J Cell Sci. 2004;117:3049–3059. doi: 10.1242/jcs.01167. [DOI] [PubMed] [Google Scholar]
- Pepperberg DR, Birch DG, Hood DC. Photoresponses of human rods in vivo derived from paired-flash electroretinograms. Vis Neurosci. 1997;14:73–82. doi: 10.1017/s0952523800008774. [DOI] [PubMed] [Google Scholar]
- Pepperberg DR, Cornwall MC, Kahlert M, Hofmann KP, Jin J, Jones GJ, Ripps H. Light-dependent delay in the falling phase of the retinal rod photoresponse. Vis Neurosci. 1992;8:9–18. doi: 10.1017/s0952523800006441. [DOI] [PubMed] [Google Scholar]
- Peterson JJ, Orisme W, Fellows J, McDowell JH, Shelamer CL, Dugger DR, Smith WC. A role for cytoskeletal elements in the light-driven translocation of proteins in rod photoreceptors. Invest Ophthalmol Vis Sci. 2005;46:3988–3998. doi: 10.1167/iovs.05-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister C, Chabre M, Plouet J, Tuyen VV, De Kozak Y, Faure JP, Kühn H. Retinal S antigen identified as the 48K protein regulating light-dependent phosphodiesterase in rods. Science. 1985;228:891–893. doi: 10.1126/science.2988124. [DOI] [PubMed] [Google Scholar]
- Pfister C, Dorey C, Vadot E, Mirshahi M, Deterre P, Chabre M, Faure JP. Identification of the so-called 48 K protein that interacts with illuminated rhodopsin in retinal rods, and the retinal S antigen, inductor of experimental autoimmune uveoretinitis. C R Acad Sci III. 1984;299:261–265. [PubMed] [Google Scholar]
- Philp NJ, Chang W, Long K. Light-stimulated protein movement in rod photoreceptor cells of the rat retina. FEBS Lett. 1987;225:127–132. doi: 10.1016/0014-5793(87)81144-4. [DOI] [PubMed] [Google Scholar]
- Plangger A, Malicki D, Whitney M, Paulsen R. Mechanism of arrestin 2 function in rhabdomeric photoreceptors. J Biol Chem. 1994;269:26969–26975. [PubMed] [Google Scholar]
- Pugh EN, Jr., Lamb TD. Phototransduction in vertebrate rods and cones: Molecular mechanisms of amplification, recovery and light adaptation. In: Stavenga DG, DeGrip WJ, Pugh EN Jr., editors. Handbook of Biological Physics. Molecular Mechanisms in Visual Transduction. Elsevier; Amsterdam: 2000. pp. 183–255. [Google Scholar]
- Puig J, Arendt A, Tomson FL, Abdulaeva G, Miller R, Hargrave PA, McDowell JH. Synthetic phosphopeptide from rhodopsin sequence induces retinal arrestin binding to photoactivated unphosphorylated rhodopsin. FEBS Lett. 1995;362:185–188. doi: 10.1016/0014-5793(95)00225-x. [DOI] [PubMed] [Google Scholar]
- Pulvermuller A, Maretzki D, Rudnicka-Nawrot M, Smith WC, Palczewski K, Hofmann KP. Functional differences in the interaction of arrestin and its splice variant, p44, with rhodopsin. Biochemistry. 1997;36:9253–9260. doi: 10.1021/bi970772g. [DOI] [PubMed] [Google Scholar]
- Pulvermuller A, Schroder K, Fischer T, Hofmann KP. Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J. Biol. Chem. 2000;275:37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- Rajala A, Daly RJ, Tanito M, Allen DT, Holt LJ, Lobanova ES, Arshavsky VY, Rajala RV. Growth factor receptor-bound protein 14 undergoes light-dependent intracellular translocation in rod photoreceptors: functional role in retinal insulin receptor activation. Biochemistry. 2009;48:5563–5572. doi: 10.1021/bi9000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidel B, Goldmann T, Giessl A, Wolfrum U. The translocation of signaling molecules in dark adapting mammalian rod photoreceptor cells is dependent on the cytoskeleton. Cell Motil Cytoskeleton. 2008;65:785–800. doi: 10.1002/cm.20300. [DOI] [PubMed] [Google Scholar]
- Restagno G, Maghtheh M, Bhattacharya S, Ferrone M, Garnerone S, Samuelly R, Carbonara A. A large deletion at the 3′ end of the rhodopsin gene in an Italian family with a diffuse form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1993;2:207–208. doi: 10.1093/hmg/2.2.207. [DOI] [PubMed] [Google Scholar]
- Rieke F, Baylor DA. Origin of reproducibility in the responses of retinal rods to single photons. Biophys J. 1998;75:1836–1857. doi: 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, He J, Davis RL. kurtz, a novel nonvisual arrestin, is an essential neural gene in Drosophila. Genetics. 2000;155:1281–1295. doi: 10.1093/genetics/155.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DJ, Heth CA. Expression of transducin in retinal rod photoreceptor outer segments. Science. 1988;241:845–847. doi: 10.1126/science.3136548. [DOI] [PubMed] [Google Scholar]
- Roof DJ, Heuser JE. Surfaces of rod photoreceptor disk membranes: integral membrane components. J Cell Biol. 1982;95:487–500. doi: 10.1083/jcb.95.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig DH, Nair KS, Wei J, Wang Q, Garwin G, Saari JC, Chen CK, Smrcka AV, Swaroop A, Lem J, Hurley JB, Slepak VZ. Subunit dissociation and diffusion determine the subcellular localization of rod and cone transducins. J Neurosci. 2007;27:5484–5494. doi: 10.1523/JNEUROSCI.1421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Stein L, Kaushal S. Ocular gene therapy: an evaluation of recombinant adeno-associated virus-mediated gene therapy interventions for the treatment of ocular disease. Hum Gene Ther. 2010;21:915–927. doi: 10.1089/hum.2010.041. [DOI] [PubMed] [Google Scholar]
- Satoh AK, Ready DF. Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Curr Biol. 2005;15:1722–1733. doi: 10.1016/j.cub.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Satoh AK, Xia H, Yan L, Liu CH, Hardie RC, Ready DF. Arrestin translocation is stoichiometric to rhodopsin isomerization and accelerated by phototransduction in Drosophila photoreceptors. Neuron. 2010;67:997–1008. doi: 10.1016/j.neuron.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- Schöneberg T, Schulz A, Biebermann H, Hermsdorf T, Römpler H, Sangkuhl K. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol Ther. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. Visual arrestin activity may be regulated by self-association. J Biol Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- Seo J, Tsakem EL, Breitman M, Gurevich VV. Identification of arrestin-3-specific residues necessary for JNK3 kinase activation. J Biol Chem. 2011;286 doi: 10.1074/jbc.M111.260448. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Caruso G, Bisegna P, Andreucci D, Gurevich VV, Hamm HE, Dibenedetto E. Dynamics of mouse rod phototransduction and its sensitivity to variation of key parameters. IET Syst Biol. 2010;4:12–32. doi: 10.1049/iet-syb.2008.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- Shi G, Yau KW, Chen J, Kefalov VJ. Signaling properties of a short-wave cone visual pigment and its role in phototransduction. J Neurosci. 2007;27:10084–10093. doi: 10.1523/JNEUROSCI.2211-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi GW, Chen J, Concepcion F, Motamedchaboki K, Marjoram P, Langen R, Chen J. Light causes phosphorylation of nonactivated visual pigments in intact mouse rod photoreceptor cells. J Biol Chem. 2005;280:41184–41191. doi: 10.1074/jbc.M506935200. [DOI] [PubMed] [Google Scholar]
- Shichi H, Somers RL. Light-dependent phosphorylation of rhodopsin. Purification and properties of rhodopsin kinase. J Biol Chem. 1978;253:7040–7046. [PubMed] [Google Scholar]
- Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepak VZ, Hurley JB. Mechanism of light-induced translocation of arrestin and transducin in photoreceptors: interaction-restricted diffusion. IUBMB Life. 2008;60:2–9. doi: 10.1002/iub.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Bainbridge JW, Ali RR. Prospects for retinal gene replacement therapy. Trends Genet. 2009;25:156–165. doi: 10.1016/j.tig.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Smith DP, Shieh BH, Zuker CS. Isolation and structure of an arrestin gene from Drosophila. Proc Natl Acad Sci U S A. 1990;87:1003–1007. doi: 10.1073/pnas.87.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WC, Bolch S, Dugger DR, Li J, Esquenazi I, Arendt A, Benzenhafer D, McDowell JH. Interaction of arrestin with enolase1 in photoreceptors. Invest Ophthalmol Vis Sci. 2011;52:1832–1840. doi: 10.1167/iovs.10-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WC, Milam AH, Dugger D, Arendt A, Hargrave PA, Palczewski K. A splice variant of arrestin. Molecular cloning and localization in bovine retina. J Biol Chem. 1994;269:15407–15410. [PubMed] [Google Scholar]
- Sokolov M, Lyubarsky AL, Strissel KJ, Savchenko AB, Govardovskii VI, Pugh EN, Jr., Arshavsky VY. Massive light-driven translocation of transducin between the two major compartments of rod cells: a novel mechanism of light adaptation. Neuron. 2002;34:95–106. doi: 10.1016/s0896-6273(02)00636-0. [DOI] [PubMed] [Google Scholar]
- Sokolov M, Strissel KJ, Leskov IB, Michaud NA, Govardovskii VI, Arshavsky VY. Phosducin facilitates light-driven transducin translocation in rod photoreceptors. Evidence from the phosducin knockout mouse. J Biol Chem. 2004;279:19149–19156. doi: 10.1074/jbc.M311058200. [DOI] [PubMed] [Google Scholar]
- Sommer ME, Farrens DL, McDowell JH, Weber LA, Smith WC. Dynamics of arrestin-rhodopsin interactions: loop movement is involved in arrestin activation and receptor binding. J Biol Chem. 2007;282:25560–25568. doi: 10.1074/jbc.M702155200. [DOI] [PubMed] [Google Scholar]
- Sommer ME, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286:7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009a;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Gurevich EV, Gurevich VV. Cone arrestin binding to JNK3 and Mdm2: conformational preference and localization of interaction sites. J Neurochem. 2007;103:1053–1062. doi: 10.1111/j.1471-4159.2007.04842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their "inactive" conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Enhanced Arrestin Facilitates Recovery and Protects Rod Photoreceptors Deficient in Rhodopsin Phosphorylation. Curr Biol. 2009b;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Arrestin-1 expression in rods: balancing functional performance and photoreceptor health. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, van Driel D, Valter K, Rees S, Provis J. The locations of mitochondria in mammalian photoreceptors: relation to retinal vasculature. Brain Res. 2008;1189:58–69. doi: 10.1016/j.brainres.2007.10.083. [DOI] [PubMed] [Google Scholar]
- Storez H, Scott MG, Issafras H, Burtey A, Benmerah A, Muntaner O, Piolot T, Tramier M, Coppey-Moisan M, Bouvier M, Labbé-Jullié C, Marullo S. Homo- and hetero-oligomerization of beta-arrestins in living cells. J Biol Chem. 2005;280:40210–40215. doi: 10.1074/jbc.M508001200. [DOI] [PubMed] [Google Scholar]
- Strissel KJ, Lishko PV, Trieu LH, Kennedy MJ, Hurley JB, Arshavsky VY. Recoverin undergoes light-dependent intracellular translocation in rod photoreceptors. J Biol Chem. 2005;280:29250–29255. doi: 10.1074/jbc.M501789200. [DOI] [PubMed] [Google Scholar]
- Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal Structure of Cone Arrestin at 2.3Å: Evolution of Receptor Specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Sinha A, Dewitt M, Farrens DL. Monomeric Rhodopsin Is the Minimal Functional Unit Required for Arrestin Binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinós J, Jalink K, Hardy RW, Britt SG, Zuker CS. A G protein-coupled receptor phosphatase required for rhodopsin function. Science. 1997;277:687–690. doi: 10.1126/science.277.5326.687. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Francis DJ, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, Gurevich VV. The role of arrestin alpha-helix I in receptor binding. J. Mol. Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. Transition of arrestin in the active receptor-binding state requires an extended interdomain hinge. J. Biol. Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J. Biol. Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J. Biol. Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. An additional phosphate-binding element in arrestin molecule: implications for the mechanism of arrestin activation. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- Wacker WB, Donoso LA, Kalsow CM, Yankeelov JA, Jr., Organisciak DT. Experimental allergic uveitis. Isolation, characterization, and localization of a soluble uveitopathogenic antigen from bovine retina. J Immunol. 1977;119:1949–1958. [PubMed] [Google Scholar]
- Wang Q, Zhang X, Zhang L, He F, Zhang G, Jamrich M, Wensel TG. Activation-dependent hindrance of photoreceptor G protein diffusion by lipid microdomains. J Biol Chem. 2008;283:30015–30024. doi: 10.1074/jbc.M803953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ER, Raman D, Shirakawa S, Ducceschi MH, Bertram PT, Wong F, Kraft TW, Osawa S. The cloning of GRK7, a candidate cone opsin kinase, from cone- and rod-dominant mammalian retinas. Mol Vis. 1998;4:27. [PubMed] [Google Scholar]
- Weller M, Virmaux N, Mandel P. Light-stimulated phosphorylation of rhodopsin in the retina: the presence of a protein kinase that is specific for photobleached rhodopsin. Proc Natl Acad Sci U S A. 1975;72:381–385. doi: 10.1073/pnas.72.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen XH, Shen L, Brush RS, Michaud N, Al-Ubaidi MR, Gurevich VV, Hamm HE, Lem J, Dibenedetto E, Anderson RE, Makino CL. Overexpression of rhodopsin alters the structure and photoresponse of rod photoreceptors. Biophys J. 2009;96:939–950. doi: 10.1016/j.bpj.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan JP, McGinnis JF. Light-dependent subcellular movement of photoreceptor proteins. J Neurosci Res. 1988;20:263–270. doi: 10.1002/jnr.490200216. [DOI] [PubMed] [Google Scholar]
- White JF, Grodnitzky J, Louis JM, Trinh LB, Shiloach J, Gutierrez J, Northup JK, Grisshammer R. Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12199–12204. doi: 10.1073/pnas.0705312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock GG, Lamb TD. Variability in the time course of single photon responses from toad rods: termination of rhodopsin’s activity. Neuron. 1999;23:337–351. doi: 10.1016/s0896-6273(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, Jastrzebska B, Park PSH, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J. Biol. Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden U, Hall SW, Kühn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proceedings of the National Academy of Sciences. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, Gurevich VV. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J Mol Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, Lefkowitz RJ. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci U S A. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–509. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- Yamada T, Takeuchi Y, Komori N, Kobayashi H, Sakai Y, Hotta Y, Matsumoto H. A 49-kilodalton phosphoprotein in the Drosophila photoreceptor is an arrestin homolog. Science. 1990;248:483–486. doi: 10.1126/science.2158671. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 1997;15:175–178. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual arrestins. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sports CD, Osawa S, Weiss ER. Rhodopsin phosphorylation sites and their role in arrestin binding. J. Biol. Chem. 1997;272:14762–14768. doi: 10.1074/jbc.272.23.14762. [DOI] [PubMed] [Google Scholar]
- Zhuang T, Vishnivetskiy SA, Gurevich VV, Sanders CR. Elucidation of IP6 and heparin interaction sites and conformational changes in arrestin-1 by solution NMR. Biochemistry. 2010:10473–10485. doi: 10.1021/bi101596g. [DOI] [PMC free article] [PubMed] [Google Scholar]