Summary
The Na+/Ca2+ exchanger CALX promotes Ca2+ efflux in Drosophila sensory neuronal cells to facilitate light-mediated Ca2+ homeostasis. CALX activity is negatively regulated by specific Ca2+ interaction within its two intracellular Ca2+ regulatory domains CBD1 and CBD2, yet how the Ca2+ binding is converted to molecular motion to operate the exchanger is unknown. Here we report crystal structures of the entire Ca2+ regulatory domain CBD12 from two alternative splicing isoforms, CALX, 1.1 and 1.2, exhibiting distinct regulatory Ca2+-dependency. The structures show an open V-shaped conformation with four Ca2+ ions bound on the CBD domain interface, confirmed by LRET analysis. The structures together with Ca2+ binding analysis support that the Ca2+ inhibition of CALX is achieved by interdomain conformational changes induced by Ca2+ binding at CBD1. The conformational difference between the two isoforms also indicates that alternative splicing adjusts the interdomain orientation angle to modify the Ca2+ regulatory property of the exchangers.
Introduction
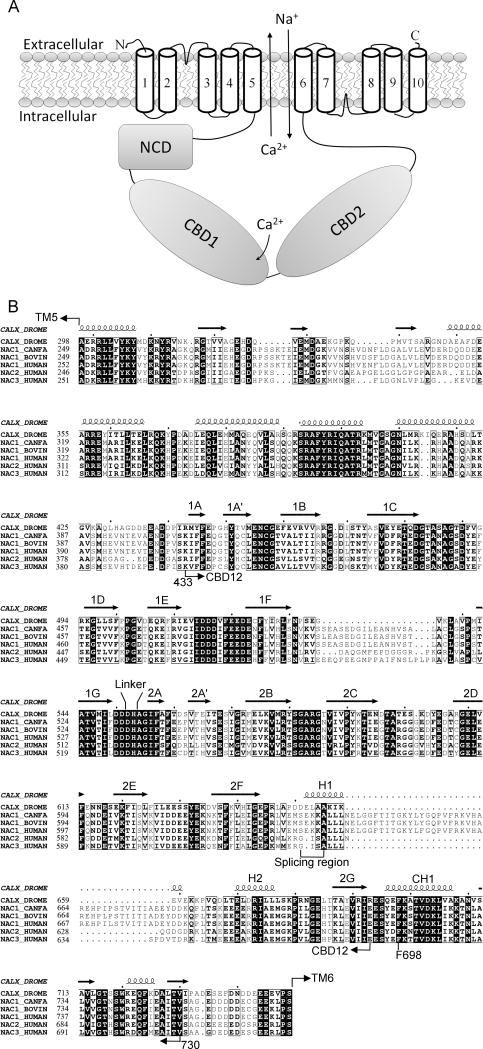
Sodium-calcium exchangers (NCXs) catalyze Ca2+ flux across the plasma membrane by utilizing a counter-Na+ electrochemical gradient. NCXs play a major role in Ca2+ homeostasis in many tissues. The cardiac exchanger NCX1 facilitates cardiac contractibility on the myocytic membrane (Bers, 2001); whereas NCX3 is involved in excitation-relaxation coupling in neurons (Blaustein et al., 1996). CALX, a NCX homolog found in Drosophila sensory neurons, is responsible for Ca2+ efflux after light-induced Ca2+ stimulation and the photoreceptor cell cascade ([Hryshko et al., 1996], [Schwarz et al., 1997] and [Wang et al., 2005]). NCXs share a conserved structural model: ten transmembrane helices (TMs) are predicted to form a Na+/Ca2+ exchange domain, along with a large intracellular regulatory region of about 500 amino acid residues between TM 5 and 6 (Figure 1A & 1B) ([Schwarz et al., 1997] and [Nicoll et al., 2002]).
Figure 1. Conserved conformation of Na+/Ca2+ exchanger CALX.
A. Topological model of a Na+/Ca2+ exchanger. NCD represents the not-characterized fragment between TM5 and CBD1. B. Sequence alignment of the intracellular domain of Na+/Ca2+ exchangers from Drosphilia CALX (UniPort entry: CALX_DROME), canine NCX1 (NAC1_CANFA), bovine NCX1 (NAC1_BOVIN), and human NCX1 (NAC1_HUMAN), NCX2 (NAC2_HUMAN) and NAC1 (NAC3_HUMAN). Conserved residues are in rectangles. Identical residues are in black. Secondary structure motifs and the splicing region are for CALX and conserved throughout the family. Seven β-strands from CBD1 or CBD2 are labeled as 1A-1G or 2A-2G.
Regulation by intracellular Ca2+ (Ca2+i) is one of the most important features of the NCX proteins (Hilgemann, 1990). Two specific Ca2+ binding domains, CBD1 and CBD2, have been identified within the intracellular region (Hilge et al., 2006). Ca2+ interaction with the two CBDs modulates Na+/Ca2+ exchange activity of NCX in response to [Ca2+i] change ([Matsuoka et al., 1995] and [Besserer et al., 2007]). In spite of high sequence homology within the intracellular regions (Figure 1B), CALX exhibits a unique negative Ca2+ regulatory property in contrast to the positive effect of other characterized NCXs (Hryshko et al., 1996). Our recent X-ray crystallographic analyses of individual CBD1 and CBD2 domains from the CALX 1.1 isoform showed that only CBD1 is a functional Ca2+ binding domain ([Wu et al., 2010] and [Wu et al., 2009]). It has a nearly identical 4-Ca2+ binding conformation to that of canine NCX1 (Nicoll et al., 2006), further supporting the importance of CBD1 in the Ca2+ regulatory mechanism of NCXs.
A plausible model for the Ca2+ regulatory mechanism of CALX is that Ca2+ binding on CBD1 induces a protein conformational change in the TM domain where exchange activity resides. However, Ca2+ interaction with CBD1 from CALX or NCX1 results in a protein conformational change limited to the local Ca2+ binding site, mainly the 1E-1F loop of CBD1 (the loop between the β strands 1E and 1F), arguing overall protein conformational change is not required for the Ca2+ regulation ([Wu et al., 2010] and [Johnson et al., 2006]). NCXs have a very short linker predicted between CBD1 and CBD2 (Figure 1B). Several functional studies have suggested that CBD1-CBD2 interaction is crucial for Ca2+ regulation of the NCX proteins ([Matsuoda et al., 1995], [Dyck et al., 1998] and [Giladi et al., 2010]). Despite the fact that CBD2 of CALX is not a Ca2+ binding domain (Wu et al., 2009), a single mutation G555P at CBD2 near the linker region of CALX completely abolished Ca2+ regulation (Dyck et al., 1998). The hypothesis has been raised that Ca2+ regulation of CALX is achieved through an inter-domain conformational change involving both CBD1 and CBD2 (Wu et al., 2010). However, no structure of the complete Ca2+ regulatory CBD domain (CBD12) from any member of the exchanger family is available to date, and no structural interaction between CBDs has been deduced from individual CBD domain structures ([Hilge et al., 2006], [Besserer et al., 2007], [Wu et al., 2010], [Wu et al., 2010] and [Nicoll et al., 2006]). In addition, our CALX CBD1 structures demonstrate three consecutive Ca2+ binding states with cumulative binding of two Ca2+ pairs (primary and secondary) in each stage (Wu et al., 2010). Whether successive Ca2+ binding signals are converted to responsive domain motion within the intracellular region requires the structural information of CBD12.
The Ca2+ regulatory property of NCXs is also fine-tuned by alternative splicing ([Dyck et al., 1999] and [Quednau et al., 1997]). Two splicing variants of CALX 1.1 and 1.2, which only differ in five residues, exhibit remarkably distinct Ca2+ regulatory properties (Omelchenko et al., 1998). CALX 1.1 shows prominently greater inhibition by regulatory Ca2+. In contrast, CALX 1.2 exhibits higher affinity for Ca2+ but much smaller inhibitory effect. The splicing region is located in a distal loop region of CBD2 (Wu et al., 2009). How the alternative residues on CBD2 affect the Ca2+ regulatory properties of CALX, which in fact are determined in CBD1, remains unknown.
To elucidate any domain interaction between CBD1 and CBD2 and to gain structural insight into the Ca2+ regulatory mechanism of CALX and its modification by splicing, we determined the crystal structures of CBD12 containing both CBD1 and CBD2 from each of the two CALX splice variants and characterized their Ca2+ binding properties by isothermal titration calorimetry (ITC). Our data strongly suggested that the Ca2+ regulatory property of CALX and its modification by alternative splicing are mediated by interdomain conformational changes between CBD1 and CBD2.
Results
The crystal structure of the Ca2+-bound CBD12 from CALX 1.1 was first determined at 2.35 Å resolution (Table 1). In the CBD12 structure, both CBD1 and CBD2 exhibit similar immunoglobulin-like conformations with each consisting of seven β strands as seen in the individual domain structure (Figure 2A) ([Wu et al., 2010] and [Wu et al., 2009]). Four Ca2+ ions were found in the Ca2+ binding site of CBD1 in a conformation nearly identical to that previously observed in the individual CBD1 structure (Figure 3A) (Wu et al., 2010). However, CBD2 undergoes disorder at its distal region including the inactive Ca2+ binding site in the CBD12 structure. i.e. the 2E-2F loop and an additional carboxyl-terminal helix CH1 are not visible in the electron density map.
Table 1.
Data collection and structure refinement statistics
| CALX1.1-CBD12 | CALX1.2-CBD12 | |
|---|---|---|
| Data collection | ||
| Space group | P 43 | R 3 |
| Cell dimensions | ||
| a, b, c (Å) | 62.8, 62.8, 226.3 | 107.5, 107.5, 358.7 |
| α, β, γ (o) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 40.0-2.35 (2.39-2.35)* | 46.2-2.9 (2.96-2.9)* |
| Rsym (%) | 7.8 (50.2) | 12.3 (40.1) |
| I /σ(I) | 7.7 (2.2) | 5.1 (1.5) |
| Completeness (%) | 99.7 (99.8) | 91.2 (93.0) |
| Redundancy | 4.6 (4.5) | 2.7 (2.5) |
| Refinement | ||
| Resolution (Å) | 40.0-2.35 | 46.2-2.9 |
| No. of unique reflections | 33954 | 34762 |
| Rwork / Rfree (%) | 21.9/26.0 | 23.0/27.8 |
| No. atoms/a.s.u. | ||
| Protein | 3877 | 7903 |
| Ca2+ | 8 | 16 |
| Water | 42 | 38 |
| B-factors | ||
| Protein | 51.7 | 63.3 |
| Ca2+ | 41.4 | 55.1 |
| Water | 29.6 | 40.6 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.008 |
| Bond angles (o) | 1.191 | 1.255 |
One crystal for each structure; values in parentheses are for highest-resolution shell.
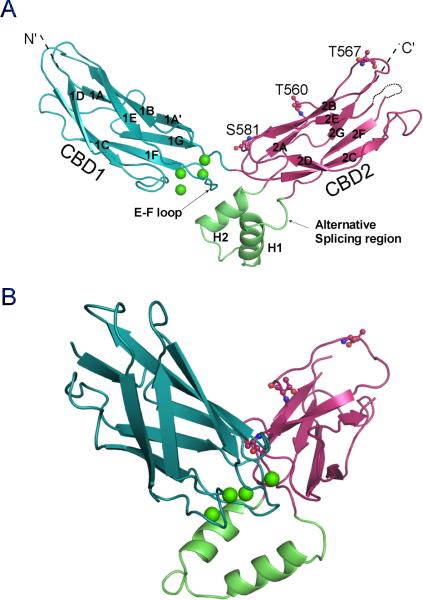
Figure 2.
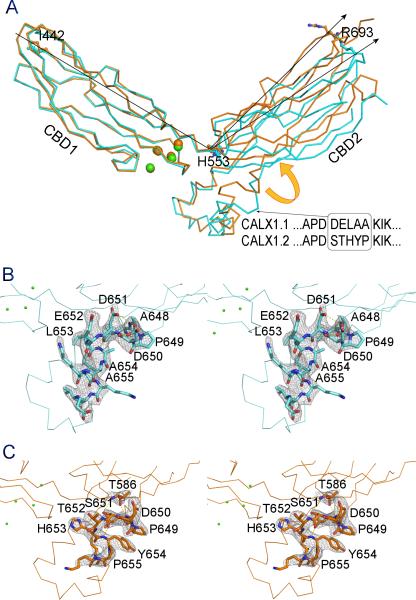
Overall structure of the Ca2+ regulatory domain CBD12 of CALX 1.1. A. The front view of the CBD12 structure in cartoon. CBD1 or CBD2 are depicted in blue or red with β strands labeled from 1A-2G or 2A-2G. The missing 2E-2F loop of CBD2 is marked as a dashed line. The 2F-2G loop of CBD2 is colored in green and the splicing alternative region is highlighted. The four Ca2+ ions bound at the hinge region are shown as green spheres. The three residue positions for the LRET labeling are shown as stick-balls. B. Side view of the CBD12 structure. See also Figures S1 and S2.
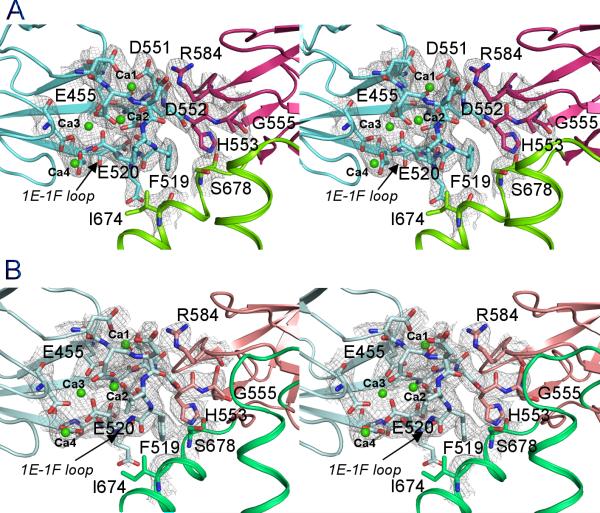
Figure 3.
Stereo views of the domain interface between CBD1 and CBD2. A. CALX 1.1; B. CALX 1.2. CBD1, CBD2 or 2F-2G loop are depicted as cartoon in blue, red or green. The residues on the domain interface are shown as stick-balls. Four Ca2+ ions (Ca1-Ca4) are drawn as green spheres. The electron density maps contoured at 1.5σ are drawn as grey meshes. See also Figure S1.
The most remarkable feature of the CBD12 structure is the domain orientation of CBD1 and CBD2. These two rigid domains are arranged in a V-shaped conformation at a 115° angle from each other around an axis at H553 (Figure 2A). The overall conformation of CBD12 is reminiscent of a “soaring eagle” with CBD1 and CBD2 forming its two open “wings”. As a result, I442 at the amino terminus of CBD1 is separated from R693 at the carboxyl terminus of CBD2 by a large distance of 70 Å. The 2F-2G loop of CBD2, where alternative splicing occurs, forms two helices (H1 and H2) nearly perpendicular to the plane of the two “wings” and lies under a hinge region as the “eagle” body (Figure 2B).
One conserved short linker sequence was predicted between CBD1 and CBD2 in NCXs. In fact, there is no obvious spacer between CBD1 and CBD2 in the CBD12 structure. While D552 is involved in Ca2+ binding at CBD1, its neighboring residue H553 is considered to be part of CBD2 (Figure 3A). G555, the residue that is mutated to proline abolishes Ca2+ regulation of CALX (Dyck et al., 1998), is located on the β-strand 2A of CBD2. The proximity of these residues consequently facilitates direct interaction between the two CBD domains with the Ca2+ binding site of CBD1 positioned in the central domain interface.
In addition to the linker between CBD1 and CBD2, the domain interface is predominantly contributed by the Ca2+ binding site, particularly the 1E-1F loop of CBD1, which is packed in the central hinge region of CBD12 (Figure 3A). The CBD12 structure reveals the extensive interactions of this loop with other structural components. On one side, four Ca2+ ions, aligned in line in the Ca2+ binding site of CBD1, stabilize the 1E-1F loop and the physical linker region with a compact Ca2+ coordination network formed by nine carboxylate residues. On another side, the 1E-1F loop is structurally supported by the perpendicular H2 helix. F519 on the outer arc of the 1E-1F loop directly interacts with I674 and S678 of the H2 helix. On the top of the interface, R584 from CBD2 lays on the hinge axis to form two salt-bridges with D551 and D552 and also two hydrogen bonds with D517 from the 1E-1F loop of CBD1. Four Ca2+ ions appear at different positions at the domain interface. The primary Ca2+ pair (Ca1 and Ca2) is closer to the hinge region than the secondary Ca2+ pair (Ca3 and Ca4). The former aids the assembly of the 1E-1F loop with the physical linker on the interface.
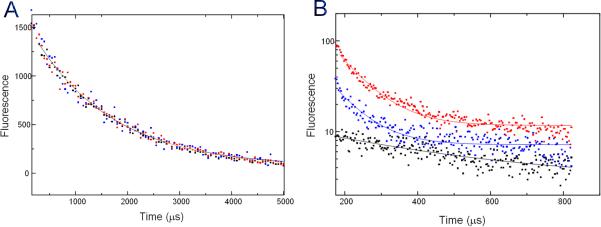
To confirm that the observed open V-shaped conformation exists also in solution, we used a luminescence resonance energy transfer (LRET)-based assay to investigate the protein conformation without crystallization constraints. The N-terminal His tag-associated Cy3 derivative of nitrilotriaacetic acid chelate of nickel ((Ni-NTA)2Cy3) of the protein served as the acceptor and three individual cysteine mutations representing positions S581, T560 and T567 (Figure 2A) were made in CBD2 to allow the introduction of a terbium chelate donor fluorophore to probe relative distances using lifetime LRET measurements. There was no difference in fluorescence signal lifetime among the three cysteine mutants without the acceptor probe (Figure 4A), indicating no significant perturbation of the structure from the cysteine mutations. Therefore lifetimes indicating the extent of energy transfer at different donor positions in the donor-acceptor labeled proteins can be used to estimate relative donor distances from the acceptor (Figure 4B, Table 2). These measurements confirmed that the distance from the approximate N-terminal His-tag to the C-terminus (T567) is significantly longer than to the hinge region (S581) and to the middle point (T560) of CBD2, as predicted from the CBD12 structure. Notably, the distance differences between LRET measurements and structural observations can be attributed to the extra N-terminal spacer sequence before the acceptor. We conclude that the open V-shaped conformation observed in our crystal structures exists also in solution, and therefore probably also represents the physiological condition of CALX on the cell membrane.
Figure 4.
LRET measurements. A. Donor-only life collected at 545 nm. B. The LRET lifetimes as measured by the sensitized emission of the acceptor at 575 nm for the mutant proteins containing an N-terminal His-tag and cysteines located at positions 581 (red), 560 (blue), and 567 (black) on CBD2.
Table 2.
LRET lifetimes and calculated distances
| Donor position | 581 | 560 | 567 |
|---|---|---|---|
| Donor lifetime (μs) | 1607 ± 55 | 1607 ± 69 | 1612 ± 38 |
| Sensitized emission lifetime (μs) | 81 ± 2 | 77 ± 4 | 380 ± 32 |
| Distance (LRET) (Å) | 42 ± 2 | 41 ± 2 | 56 ± 5 |
| Distance in crystal structure *(Å) | 44 | 53 | 70 |
Distances are measured between the Ca atoms between the N-terminal I422 and respective donor residues.
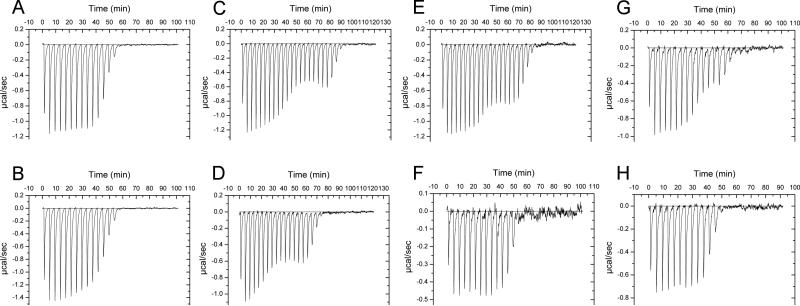
The two CALX splicing variants 1.1 and 1.2 have five residue differences within the region (651-655) (Figure 5A). These five residues are located within a loop between the H1 helix and the β strand 2F of CBD2 (Figure 1B). Despite the clearly resolved conformation of this region, this splicing region has no direct contact with the Ca2+ binding site of CBD1 (Figure 5B). The molecular mechanism for alternative splicing modification of CALX could not be deduced from the CALX 1.1 structure. In addition, both CBD12 proteins from the two isoforms exhibit similar monophasic Ca2+ binding profiles with similar binding affinities (160±35 nM for CALX 1.1, 210±38 nM for CALX 1.2), determined by ITC measurements (Figure 6A & 6B), indicating no direct impact of splicing alternation on the Ca2+ binding property of CBD12.
Figure 5.
Conformational difference between two CBD12 structures from alternative splicing variants CALX 1.1 and 1.2. A. Superimposition of the CBD12 ribbon structures from CALX 1.1 and 1.2 colored in blue and in orange. The arrows indicate the rotating angles of CBD2 from CBD1 measured between Cα atoms of terminal residues I442 and R693 with Cα atom of H553 as the axis. The alternative splicing residues are highlighted. B & C are the stereo views of the alternative regions of CALX 1.1 and 1.2 structures. Five alternative residues and other interactive residues nearby are depicted as stick-balls. H-bonds are showed in yellow dashed lines. The electron density maps contoured at 1.5σ are drawn as grey meshes. See also Figure S1.
Figure 6.
Ca2+ binding assays of CBD12 and mutants from CALX by ITC. A: CALX 1.1 CBD12 wild type; B: CALX 1.2 CBD12 wild type; C: F519A; D: I674Y; E: S678Y; F: H553P; G: G555P; H: R584A.
To demonstrate the structural modification of the CBD12 conformation by alternative splicing, we determined the crystal structure of CBD12 from the splicing variant CALX 1.2 at 2.9 Å resolution (Table 1). The CBD12 structure of CALX 1.2 shows a similar V-shaped conformation as seen in the CALX 1.1 structure. CBD1 and CBD2 are also identical to those in CALX 1.1 and four Ca2+ ions were bound at the same Ca2+ binding site as seen in CALX 1.1 (Figure 3B). However, the angle between the CBD1 and CBD2 “wings” undergoes a considerable change. By superimposing the invariant CBD1 of the two isoform structures, the CBD2 of CALX 1.2 rotates along the hinge axis of H553 upward by 9° and results in a movement of the C-terminal R693 by 6.6 Å (Figure 5A). This movement is clearly attributable to the alternative splicing region. In CALX 1.1, the five alternative residues constitute a 310 end of H1 (Figure 5B). The carbonyl group of D651 forms a hydrogen bond with the amide group of A654 to terminate the H1 helix. Meanwhile its carboxyl group interacts with D649 to stabilize the P648-induced turn conformation. In sharp contrast, the alternative residues extend the H1 helix by one additional turn toward the 2B-2C loop of CBD2 in CALX 1.2 (Figure 5C). The interaction is reinforced by two hydrogen bond formations of A648 and S651 with T586 on the 2B-2C loop of CBD2. The interface between H2 and the 1E-1F loop of CBD1 remains unaffected. Thus, the conformational changes occurring on the loop between the H1 helix and the β-strand body of CBD2 eventually lead the rigid CBD2 domain to undergo a closure rotation around the hinge. The splicing region in CALX 1.2 is closer to the Ca2+ binding site; one of the alternative residues H653 interacts with the main chain of D517 on the 1E-1F loop of CBD1. Notably, none of the splicing region residues is involved in crystal contacts (Figure S1).
The carboxylate residues in the Ca2+ binding site of CBD1 have been extensively studied ([Matsuoda et al., 1995], [Wu et al., 2010] and [Dyck et al., 1998]). In the CBD12 structures, the Ca2+-bound 1E-1F loop is a key component of the domain interface conformation (Figure 3). To investigate cross-talk between Ca2+ binding and the interface, we mutated these residues on the domain interface and evaluated the Ca2+ binding properties of the mutant proteins by ITC. Despite the fact that these residues are not directly involved in Ca2+ binding, perturbations of the 1E-1F loop interface remarkably alters the Ca2+ binding profile. F519A exhibits a biphasic Ca2+ binding curve instead of the monophasic one characteristic of wild type. In addition, saturation of Ca2+ binding was shifted to a higher [Ca2+] (Figure 6C). We also mutated I674 or S678, the interface partners of F519 on the H2 helix of CBD2, to bulky tyrosine residues. Both mutants exhibited a similar biphasic curve as that of F519A (Figure 6D & 6E), suggesting the interface is important for the conformation of the Ca2+ binding site.
To explore the physical linker region between CBD1 and CBD2, we mutated the hinge residue H553 to proline. Although no notable difference from the wild type was observed (Figure 6F), the trans configuration of proline at the hinge region may favor the V-shaped conformation. Interestingly, the G555P mutant showed a near-biphasic titration curve with 10-fold reduction of Ca2+ affinity (Figure 6G). This mutation, at least 10 Å from the Ca2+ binding site, might also disrupt the domain interface by altering domain orientation. However, mutant R584A did not affect the conformation of the domain interface (Figure 6H). The above results conclusively show that the 1E-1F loop interface, perhaps together with the linker, is important for maintaining the integrity of the Ca2+ binding property of the CBD12 domain.
Discussion
Ca2+ regulatory mechanism of CALX
Ca2+ efflux mediated by NCXs is crucial for maintaining the cellular Ca2+ homeostasis cycle. The Ca2+ regulatory mechanism permits NCXs and CALX to properly conduct Na+/Ca2+ exchange activity in response to cellular Ca2+ signaling needs. In Drosophila CALX, CBD1 has been recognized as the only Ca2+ binding region that can sense elevation of intracellular Ca2+ concentration to down-regulate Na+/Ca2+ flux ([Wu et al., 2010] and [Wu et al., 2009]). However, CBD2 is required to maintain the Ca2+ regulatory property of CALX (Dyck et al., 1998). In this study, we determined the atomic structures of CBD12 from the two known CALX splicing variants. Both crystal structures feature an open V-shaped conformation, which was further confirmed in solution by LRET measurements. The characteristic short linker between CBD1 and CBD2 enables close assembly of two adjacent CBD1 and CBD2 domains, geometrically positioning the Ca2+ binding site in the center of the CBD interdomain interface. This structural feature explains the requirement for both CBD1 and CBD2 domains to maintain the negative Ca2+ regulatory property of CALX.
The CBD12 structures demonstrate the major contribution of the 1E-1F loop of CBD1 to the interdomain interface, which enables communication between the Ca2+ binding site of CBD1 with CBD2. The strong Ca2+ interaction network coordinated by nine carboxylate residues maximizes the strength of the 1E-1F loop and the interdomain conformation. Any mutation in the Ca2+ binding site of CBD1 causes reduction of Ca2+ regulation ([Wu et al., 2010] and [Dyck et al., 1998]). These findings demonstrate the critical importance of the integrity and rigidity of the Ca2+ interaction network for the interdomain conformation. Our previous studies have shown that decalcification of CBD1, in particular removing the primary Ca2+ pair, results in disorder of the entire 1E-1F loop instead of the overall structure of rigid CBD1 (Wu et al., 2010). Based on the CBD12 structures, it is unambiguous that destabilization of the 1E-1F loop disrupts the interface conformation, leading to a rigid body movement of CBD1.
Our ITC results indicate that Ca2+ binding at CBD1 can also be influenced by the CBD domain interface. Any disturbance of the interface between the 1E-1F loop and H2 helix of CBD2 influences Ca2+ binding, implying that the interfacial residues maintain cooperativity of binding of the four Ca2+ ions rather than acting directly on Ca2+ binding affinity. Our previous structural and electrophysiological analyses suggested that these four Ca2+ ions interact with the Ca2+ binding site in a progressive two-step manner stimulated by two Ca2+ pairs (Wu et al., 2010). The two Ca2+ pairs are coupled by direct interaction between Ca2 and Ca3, as reflected by the monophasic Ca2+ binding property. Three interfacial residues F519, I674 and S678 are located between the primary and secondary Ca2+ pair. Mutations at this region perhaps disturb the coupling of the binding of the two Ca2+ pairs. This decoupling may cause delay of the secondary Ca2+ binding step as evidenced by the diphasic Ca2+ binding property of the three mutants. These findings lend further support to the two-step Ca2+ binding hypothesis for CBD1.
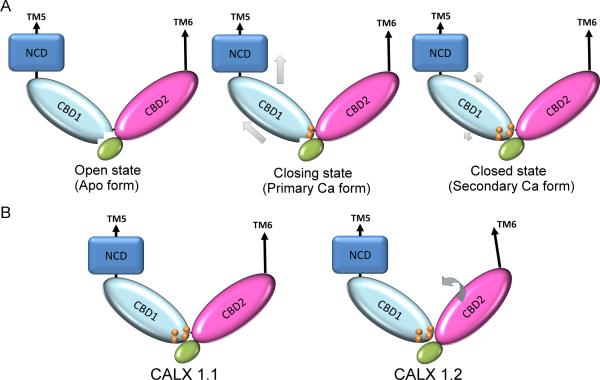
The above observations support interdomain movement as a key structural feature responsible for CBD1-mediated Ca2+ regulation of CALX and form the basis of a three-step model (Figure 7A): 1) CALX remains active while its Ca2+ binding site at CBD1 is disrupted in the absence of regulatory Ca2+ binding; 2) The primary Ca2+ pair first accesses the Ca2+ binding site of CBD1 to stabilize the collapsed conformation of the 1E-1F loop and the linker region, generating protein motion along CBD1 towards the upstream TM domain; 3) Following the primary Ca2+ pair, the secondary Ca2+ pair further enhances binding of the primary Ca2+ pair, leading to stabilization of the 1E-1F loop via the Ca2+ coordination network to completely close the Na+/Ca2+ exchange pathway.
Figure 7.
hypothetical models of Ca2+ regulatory mechanism (A) and its alternative splicing modification (B) for the Drosophila Na+/Ca2+ exchanger CALX. The grey arrows indicate hypothetic protein conformational changes.
Differences between CALX and NCXs
In contrast to the negative Ca2+ regulatory property of CALX, all of the characterized mammalian NCXs exhibit a positive effect of Ca2+i. These opposing regulatory properties have been exclusively attributed to their intracellular regulatory domains (Dyck et al., 1998). However, both CALX and canine NCX1 share identical Ca2+ binding conformations at CBD1 ([Wu et al., 2010] and [Nicoll et al., 2006]). Furthermore, exchanging CBD1s between CALX and canine NCX1 resulted in no significant change in their respective Ca2+ regulatory properties (Personal communication by Larry V. Hryshko, University of Manitoba, Canada). These common structural and functional features strongly indicate an analogous role of CBD1 in the Ca2+ regulatory property of the NCX family. Although no CBD12 structure from any mammalian NCXs is available, mammalian NCXs share a nearly identical sequence of the 1E-1F loop of CBD1 and the linker with CALX (Figure 1B). A very similar V-shaped CBD12 orientation model was recently proposed for NCX1 by other research groups ([Giladi et al., 2010] and [John et al., 2011]). An NMR study of canine CBD1 has revealed a similar Ca2+ binding mechanism to that seen for CALX. Also, removal of Ca2+ from the Ca2+ binding site also only resulted in disruption of the structure of the Ca2+ binding sites, mainly the 1E-1F loop, while the overall structure of CBD1 remained unchanged (Johnson et al., 2008). Their similar Ca2+ binding mechanism and their predicted similar interdomain interface together imply that the Ca2+-mediated interdomain conformational change at CBD1 may be generally applicable to the larger NCX family. These features also exclude CBD1 as the basis of the negative Ca2+ regulatory properties. Instead, CBD1 may serve as the common Ca2+i sensor of the NCX family.
Although CALX has a conformation of the Ca2+ binding site region of CBD2 distinct from that of NCX1, the inactivation of CBD2 might not be the cause (at least not the complete reason) of the negative Ca2+ regulatory effect of CALX since mutations abolishing the Ca2+ binding site of CBD2 did not lead to reversal of the positive Ca2+ regulatory effect of NCX1 (Besserer et al., 2007). Instead, the reversal can be accomplished (partially if not all) by swapping a fragment (amino acid sequence 398-583) of CALX with the counterpart sequence of NCX1, which mainly includes CBD1 and an upstream fragment (Dyck et al., 1998) (Figure 1B). Given the clear lack of involvement of CBD1, the enigmatic opposite Ca2+ regulatory effects might be caused by other conformational differences within the intracellular regulatory region rather than the CBD12 domain.
CBD2 is essential for maintaining the Ca2+ regulatory property of cardiac NCX1 (Besserer et al., 2007). However, the CBD2-mediated Ca2+ regulatory mechanism is unclear since Ca2+ binding does not result in any significant conformational change in CBD2. In the NCX1-CBD2 structure, the 2E-2F loop exhibits an equivalent conformation for Ca2+ binding to that of the 1E-1F loop in CBD1 (Besserer et al., 2007). In the two CALX CBD12 structures, the 2E-2F loop and a carbonyl helical peptide CH1 (696-730) are disordered. In contrast, the missing regions are clearly resolved in our previous CALX1.1-CBD2 structure, in which the carbonyl helix (CH1) lies toward the pseudo- Ca2+ binding region of CBD2 and F698 rides on the 2E-2F loop of CBD2 to stabilize the pseudo- Ca2+ binding sites (Wu et al., 2009) (Figure S2). Despite the possibility that these conformational differences may be caused by different crystal packing, they suggest that the conformation of the Ca2+ binding sites of CBD2 can be altered by an adjacent structural component and argue that NCX1-CBD2 shares a similar Ca2+-mediated inter-domain conformational change mechanism to that of CBD1. Notably, the CH1 helix is rather conserved between CALX and mammalian NCXs (Figure 1B). The residue F698 located at the hinge region between CBD2 and CH1 appears in every exchanger protein. The conformational change illustrated for CALX-CBD2 may shed light on the CBD2-involved regulatory mechanism of mammalian NCXs.
Alternative splicing modification of Ca2+ regulatory properties
Alternative splicing occurring within the 2F-2G loop of CBD2 offers tailored modulation of the regulatory property of Na+/Ca2+ exchangers in different tissues (Hryshko, 2002). The two CALX splicing variant CBD12 structures clearly demonstrate that altered residues in the loop between the H1 helix and the β strand 2G of CBD2 induce a change in the orientation angle of CBD12. The resulting ~7 Å movement between the two termini of CBD12 as shown in Figure 5A may substantially influence regulatory Ca2+ binding signal transmission to the adjacent TM domain. Additionally, this rigid domain movement occurring between CBD1 and CBD2 also provides direct evidence in support of our model for the CBD1-mediated Ca2+ regulatory mechanism.
Our ITC results taken together with structural observations indicate that alternative splicing of CALX does not cause direct modification of the Ca2+ binding site of CBD1 despite previous findings that spliced residues modified regulatory Ca2+ affinity (Omlchenko et al., 1998). However, this modification (enhancement) was assessed by measuring inward Ca2+ current changes at different [Ca2+i], which actually recorded final output of the exchanger TM domain instead of direct measurement of regulatory Ca2+ binding affinity at CBD1. NCXs and CALX proteins consist of many structural components, including CBD12 for regulatory Ca2+ binding, the Na+ regulatory region, and the binding site for Ca2+ transport residing at the intracellular side of the proteins (Figure 1A). The Ca2+ current output is a summation of all regulatory and transport events. It is also changed during Na+/Ca2+ exchanging activity since both Ca2+ and Na+ are both transport substrates and regulatory species. These complicated structural and dynamic regulatory features are reflected in two different regulatory Ca2+ affinities, 400 nM for the peak current and 130 nM for the steady state current for CALX 1.1, regardless of the presence of only one regulatory Ca2+ binding site in CBD1 of CALX (Wu et al., 2010).
Our previous study showed that different mutations in the Ca2+ binding site of CBD1 uniformly lead to reductions in the apparent Ca2+i affinities of both peak and steady state currents at similar levels (Wu et al., 2010). In contrast, splicing alteration of CALX 1.2 generates Ca2+ regulatory changes limited to the peak current (Omlchenko et al., 1998). This difference suggests that alternative splicing may not directly modify the Ca2+ binding site of CBD1, in agreement with our structural observations and our hypothetical model that alternative splicing induces a rotating domain conformational change that is distinct from that mediated by Ca2+ binding (Figure 7B). Possibly, the Ca2+ current change of CALX 1.2 is a manifestation of conformational change between the TM domain and CBD12 to adjust regulatory Ca2+ binding signal transmission, even though Ca2+ binding at CBD12 is unaffected.
Although alternative splicing only occurs at the 2F-2G loop of CBD2, it triggers multiple regulatory changes, in particular Na+ inactivation, which is predictably controlled by a region other than CBD12 within the intracellular loop. CALX 1.2 exhibits a stronger inhibition by Na+i and a higher recovery rate from the Na+-dependent inactive state (Omlchenko et al., 1998). Multiple regulatory responses by simple alternative splicing reflect the interplay of various regulatory components. Indeed, many regulatory features of NCXs including CALX, such as Ca2+ and Na+ regulatory properties, are coupled and mutually responsive (Hryshko et al., 1996). These interrelated modifications are as yet not all revealed by the CBD12 structures. Instead, the interdomain orientation angle change between two CBDs is suggestive of similar interactions between different domains within the intracellular region of Na+/Ca2+ exchangers. The CBD12 structures of the two CALX splicing variants may serve as a starting point to examine different regulatory features as an integrated functional unit in Na+/Ca2+ exchangers.
Experimental Procedures
Protein expression and purification
The gene fragments (amino acid sequences 433-730 (Figure 1B)) encoding the CBD12 domain were cloned into the vector pET28a (EMD Biochemicals, NJ) from cDNAs of CALX 1.1 or CALX 1.2 ([Hryshko et al., 1996] and [Omlchenko et al., 1998]). The resulted CBD12 proteins have an N-terminal His-tag spaced by a thrombin cleavage site. All proteins were expressed and purified using similar approaches as follows: Proteins were expressed in E.coli BL21(DE3) in LB medium after overnight induction with 1mM IPTG. Cells were disrupted by a microfluidizer, and then soluble CBD12 proteins were purified using Ni2+-NTA resin (GE-Healthcare, PA). After His-tag cleavage by thrombin protease (GE-Healthcare, PA), the CBD12 proteins were further purified by size-exclusion chromatography using a Superdex 75 10/300 GL column (GE-Healthcare, PA) and concentrated to 10 mg/ml for crystallization.
Crystallization and structural determination
The crystallization experiments were performed with the sitting-drop vapor diffusion method at 18°C. Both CALX 1.1 and 1.2 crystals were obtained at a condition of 0.1 M Bis-Tris, pH 5.5, 0.4 M ammonium sulfate and 16% Polyethylene glycol 3350. All crystals were flash-cooled to 100 K with additional 22% 2-methyl-2,4-pentanediol as cryo-protectant. Diffraction data were collected at the Advanced Light Sources beamline 4.2.2 (Berkeley, CA). Data processing, merging and reduction were carried out with the programs D*TREK (Pflugrath, 1999). Both crystals are twinned with similar twin factors of 43.8%, assessed by the program phenix.xtriage (Adam et al., 2002). Both structures were solved with the molecular replacement method by the program Phaser (McCoy et al., 2007) and using CBD1 or CBD2 domain from CALX ([Wu et al., 2010] and [Wu et al., 2009]) as search models. The model building was performed using COOT (Emsley et al., 2004) and refined using the program PHENIX with respective twin laws (Adam et al., 2002). All amino acid residues are in the favorable and allowed region in Ramachandran plot. Crystallographic data and the model refinement statistics are given in Table 1. The structure coordinates and structural factors of CBD12 structures from CALX 1.1 and 1.2 have been deposited in Protein Data Bank (PDB code: 3RB5 and 3RB7)
Fluorescence labeling
Prior to LRET experiments, two endogenous cysteines (C456 & C523) exposed on the molecular surface of CBD12 from CALX 1.1 were mutated to serines. Cysteine was introduced at S581, T560 or T567 position, respectively. The protein expression and purification were performed with a similar procedure as described above except for the His-tag cleavage step. Each mutant protein with a His-tag at the N-terminus of CBD (0.5 μM) was labeled with donor, (0.75 μM) maleimide terbium chelate, and dialyzed against TBS buffer to remove excess fluorophore. The labeled protein was then used for donor-only measurements or the acceptor, 2 μM (Ni-NTA)2-Cy3, was added for donor:acceptor measurements. (Ni-NTA)2-Cy3 was synthesized as described (Kapanidis et al., 2001) and the triethylenetetraaminehexaacetic acid chelate (TTHA-Tb) of terbium was purchased from Invitrogen.
Fluorescence measurements
The fluorescence measurements were carried out using a cuvette-based fluorescence lifetime spectrometer, TimeMaster™ Model TM-3/2003 (Photon Technology International, NJ). A nitrogen/dye laser coupled to the sample compartment by a fiber optic served as the energy source. The emitted light was collected and passed through a 200-mm monochromator to a stroboscopic detector. Data were collected with Felix 32 software (Photon Technology International, NJ) and analyzed with Origin (OriginLab Corp. MA). The lifetimes presented are an average of three sets of data; however, each single set of data for a given mutant was examined to ensure that each exhibited a similar trend. LRET lifetimes were obtained as the sensitized emission at 575 nm for Cy3, and the donor-only lifetimes were collected at 545 nm. The distance between the terbium chelate and (Ni-NTA)2-Cy3 fluorophores was measured using Förster's theory for energy transfer. Time constants were obtained for the donor fluorescence decay in the absence of the acceptor and the sensitized emission of the acceptor due to energy transfer from the donor. R0 was obtained as previously described using the overlap integral (Ramanoudjame et al., 2006).
Ca2+ binding assay by isothermal titration calorimetry
Ca2+ binding assays were performed with a similar ITC protocol previously reported (Wu et al., 2009). Briefly, prior to ITC titration, protein samples were treated with 10 mM EDTA and then extensively dialyzed against a buffer containing 20 mM HEPES pH 7.4, 20mM β-mercaptoethanol and 150 mM KCl to remove endogenous Ca2+. ITC experiments were carried out using a VP-ITC device (Microcal, NJ) with 5 μM of each protein sample. All ITC measurements were performed with the same program: the system was thermally equilibrated at 25°C; after an initial delay of 120 seconds, 26 serial injections (10 μl each) were done with a spacing time of 240 seconds at 307 rpm stirring speed. Each measurement was corrected with a background titration in which Ca2+ was titrated into the buffer solution. Corrected titration curves were fit using the Origin software .
Supplementary Material
Highlights.
▶ Crystal structures of the complete Ca2+ regulatory domain of the exchanger CALX
▶ Ca2+ binding at the domain interface between CBD1 and CBD2
▶ Alternative splicing induces CBD domain orientation angle change
▶ Implication of the Ca2+ regulatory mechanism of the Na+/Ca2+ exchanger family
Acknowledgements
This work was supported by American Heart Association grant 0830353N (LZ) and postdoctoral fellowship 2260265 (MW), National Institutes of Health Grants R01GM097290 (LZ), R01GM073102 (VJ), NRSA-GM082023-01 (JG), R37GM27750 (JLS), and the Robert A. Welch Foundation (JLS). We thank Dr. Larry Hryshko for kindly providing cDNAs of CALX 1.1 and 1.2 and personal communication, Jay Nix from the Molecular Biology Consortium ALS beamline 4.2.2 for crystal data collections, and Julia E. Lever for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bers DM. Excitation-contraction coupling and cardiac contractile force. Kluwer Academic Publications; Boston, USA: 2001. pp. 71–92. [Google Scholar]
- 2.Blaustein MP, Fontana G, Rogowski RS. The Na+-Ca2+ exchanger in rat brain synaptosomes. Kinetics and regulation. Ann. N. Y. Acad. Sci. 1996;779:300–317. doi: 10.1111/j.1749-6632.1996.tb44803.x. [DOI] [PubMed] [Google Scholar]
- 3.Hryshko LV, Matsuoka S, Nicoll DA, Weiss JN, Schwarz EM, Benzer S, Philipson KD. Anomalous regulation of the Drosophila Na+-Ca2+ exchanger by Ca2+. J. Gen. Physiol. 1996;108:67–74. doi: 10.1085/jgp.108.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz EM, Benzer S. Calx, a Na-Ca exchanger gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1997;94:10249–10254. doi: 10.1073/pnas.94.19.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Xu H, Oberwinkler J, Gu Y, Hardie RC, Montell C. Light activation, adaptation, and cell survival functions of the Na+/Ca2+ exchanger CalX. Neuron. 2005;45:367–378. doi: 10.1016/j.neuron.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Nicoll DA, Ottolia M, Philipson KD. Toward a topological model of the NCX1 exchanger. Ann. N. Y. Acad. Sci. 2002;976:11–18. doi: 10.1111/j.1749-6632.2002.tb04709.x. [DOI] [PubMed] [Google Scholar]
- 7.Hilgemann DW. Regulation and deregulation of cardiac Na+-Ca2+ exchange in giant excised sarcolemmal membrane patches. Nature. 1990;344:242–245. doi: 10.1038/344242a0. [DOI] [PubMed] [Google Scholar]
- 8.Hilge M, Aelen J, Vuister GW. Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors. Mol. Cell. 2006;22:15–25. doi: 10.1016/j.molcel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka S, Nicoll DA, Hryshko LV, Levitsky DO, Weiss JN, Philipson KD. Regulation of the cardiac Na+-Ca2+exchanger by Ca2+. Mutational analysis of the Ca2+-binding domain. J. Gen. Physiol. 1995;105:403–420. doi: 10.1085/jgp.105.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besserer GM, Ottolia M, Nicoll DA, Chaptal V, Cascio D, Philipson KD, Abramson J. The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis. Proc. Natl. Acad. Sci. USA. 2007;104:18467–18472. doi: 10.1073/pnas.0707417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu M, Le HD, Wang M, Yurkov V, Omelchenko A, Hnatowich M, Nix J, Hryshko LV, Zheng L. Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor CBD1 from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions. J. Biol. Chem. 2010;285:2554–2561. doi: 10.1074/jbc.M109.059162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, Wang M, Nix J, Hryshko LV, Zheng L. Crystal structure of CBD2 from the Drosophila Na+/Ca2+ exchanger: diversity of Ca2+ regulation and its alternative splicing modification. J. Mol. Biol. 2009;387:104–112. doi: 10.1016/j.jmb.2009.01.045. [DOI] [PubMed] [Google Scholar]
- 13.Nicoll DA, Sawaya MR, Kwon S, Cascio D, Philipson KD, Abramson J. The crystal structure of the primary Ca2+ sensor of the Na+/Ca2+ exchanger reveals a novel Ca2+ binding motif. J. Biol. Chem. 2006;281:21577–21581. doi: 10.1074/jbc.C600117200. [DOI] [PubMed] [Google Scholar]
- 14.Johnson E, Bruschweiler-Li L, Showalter SA, Vuister GW, Zhang F, Brüschweiler R. Structure and dynamics of Ca2+-binding domain 1 of the Na+/Ca2+ exchanger in the presence and in the absence of Ca2+. J. Mol. Biol. 2008;377:945–955. doi: 10.1016/j.jmb.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyck C, Maxwell K, Buchko J, Trac M, Omelchenko A, Hnatowich M, Hryshko LV. Structure-function analysis of CALX1.1, a Na+-Ca2+ exchanger from Drosophila. Mutagenesis of ionic regulatory sites. J. Biol. Chem. 1998;273:12981–12987. doi: 10.1074/jbc.273.21.12981. [DOI] [PubMed] [Google Scholar]
- 16.Dyck C, Omelchenko A, Elias CL, Quednau BD, Philipson KD, Hnatowich M, Hryshko LV. Ionic regulatory properties of brain and kidney splice variants of the NCX1 Na+-Ca2+ exchanger. J. Gen. Physiol. 1999;114:701–711. doi: 10.1085/jgp.114.5.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quednau BD, Nicoll DA, Philipson KD. Tissue specificity and alternative splicing of the Na+/Ca2+ exchanger isoforms NCX1, NCX2 and NCX3 in rat. Am. J. Physiol. 1997;272:C1250–C1261. doi: 10.1152/ajpcell.1997.272.4.C1250. [DOI] [PubMed] [Google Scholar]
- 18.Omelchenko A, Dyck C, Hnatowich M, Buchko J, Nicoll DA, Philipson KD, Hryshko LV. Functional differences in ionic regulation between alternatively spliced isoforms of the Na+-Ca2+ exchanger from Drosophila melanogaster. J. Gen. Physiol. 1998;111:691–702. doi: 10.1085/jgp.111.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giladi M, Boyman L, Mikhasenko H, Hiller R, Khananshvili D. Essential role of the CBD1-CBD2 linker in slow dissociation of Ca2+ from the regulatory two-domain tandem of NCX1. J. Biol. Chem. 2010;285:28117–28125. doi: 10.1074/jbc.M110.127001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John SA, Ribalet B, Weiss JN, Philipson KD, Ottolia M. Ca2+-dependent structural rearrangements within Na+-Ca2+ exchanger dimers. Proc. Natl. Acad. Sci. USA. 2011;108:1699–1704. doi: 10.1073/pnas.1016114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hryshko LV. Tissue-specific modes of Na/Ca exchanger regulation. Ann. N. Y. Acad. Sci. 2002;976:166–175. doi: 10.1111/j.1749-6632.2002.tb04738.x. [DOI] [PubMed] [Google Scholar]
- 22.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Cryst. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta. Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta. Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Kapanidis AN, Ebright YW, Ebright RH. Site-specific incorporation of fluorescent probes into protein: hexahistidine-tag-mediated fluorescent labeling with Ni2+:nitrilotriacetic Acid (n)-fluorochrome conjugates. J. Am. Chem. Soc. 2001;123:12123–12125. doi: 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- 27.Ramanoudjame G, Du M, Mankiewicz KA, Jayaraman V. Allosteric mechanism in AMPA receptors: A FRET-based investigation of conformational changes. Proc. Natl. Acad. Sci.USA. 2006;103:10473–10478. doi: 10.1073/pnas.0603225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.