Abstract
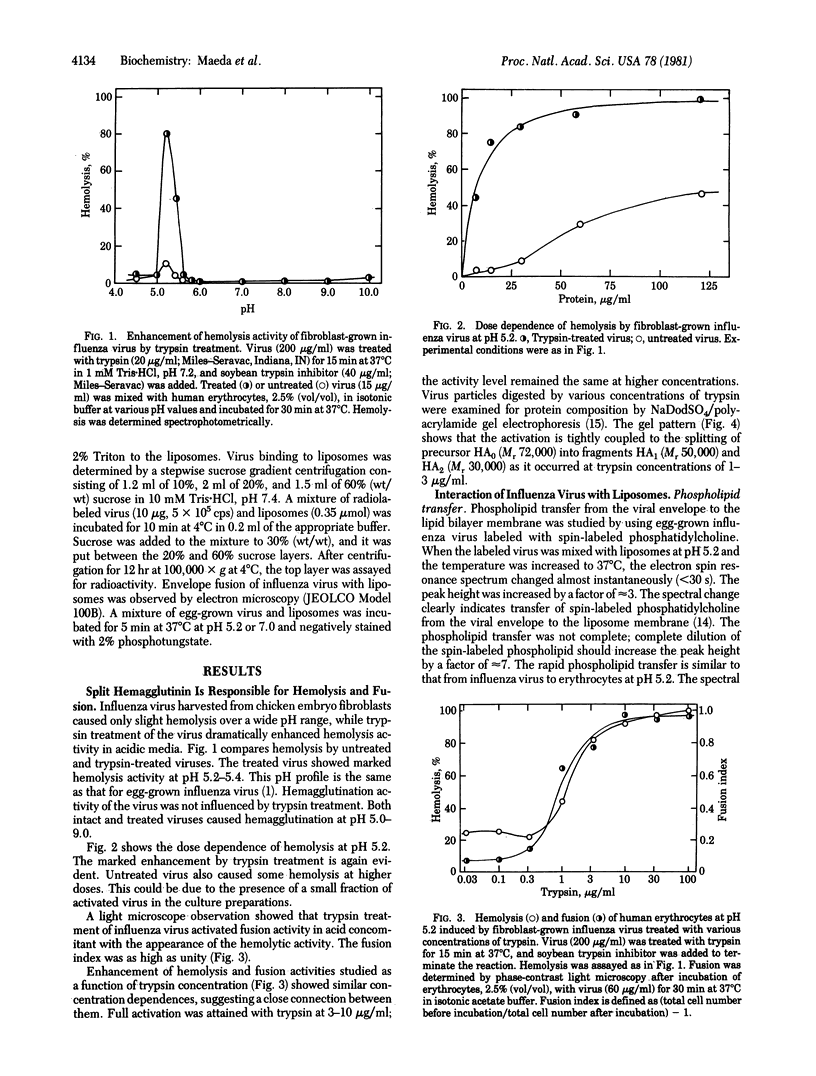
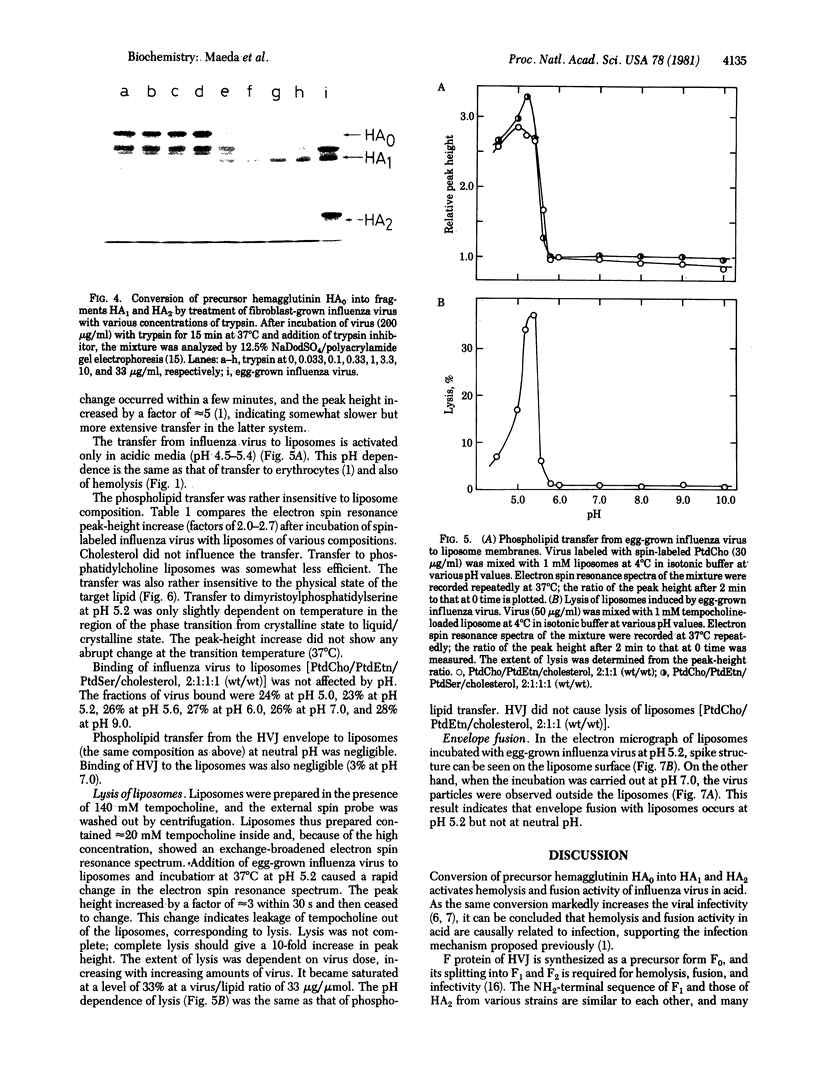
The molecular mechanism of hemolysis and fusion by influenza virus in acidic media was studied. First, the effect of trypsin treatment on the activity of fibroblast-grown influenza virus was studied. The results showed that the split form of viral hemagglutinin, HA1 and HA2, but not the precursor, is responsible for the activity. Second, the interaction of egg-grown influenza virus, which contains the split hemagglutinin, with lipid liposomes was studied by spin labeling and electron microscopy. Phospholipid transfer from the viral envelope to the lipid bilayer membrane occurred within 30 s at pH 4.5-5.4. The transfer is largely independent of the lipid composition and the crystalline vs. liquid/crystalline state of the membrane. Virus-induced lysis of liposomes also took place rapidly in the same pH range. Envelope fusion with liposomes occurred at pH 5.2 but not at pH 7.0. These characteristic interactions were similar to those between influenza virus and erythrocytes reported previously. On the other hand, hemagglutinating virus of Japan did not interact with liposomes at neutral pH. These results suggest that protonation of the NH2-terminal segment of the HA2 form causes interaction of the segment with the lipid core of the target cell membrane, leading to hemolysis and fusion.
Full text
PDF

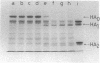
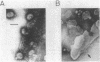
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Comfurius P., Zwaal R. F. The enzymatic synthesis of phosphatidylserine and purification by CM-cellulose column chromatography. Biochim Biophys Acta. 1977 Jul 20;488(1):36–42. doi: 10.1016/0005-2760(77)90120-5. [DOI] [PubMed] [Google Scholar]
- Compans R. W., Klenk H. D., Caliguiri L. A., Choppin P. W. Influenza virus proteins. I. Analysis of polypeptides of the virion and identification of spike glycoproteins. Virology. 1970 Dec;42(4):880–889. doi: 10.1016/0042-6822(70)90337-5. [DOI] [PubMed] [Google Scholar]
- Dourmashkin R. R., Tyrrell D. A. Electron microscopic observations on the entry of influenza virus into susceptible cells. J Gen Virol. 1974 Jul;24(1):129–141. doi: 10.1099/0022-1317-24-1-129. [DOI] [PubMed] [Google Scholar]
- Garoff H., Frischauf A. M., Simons K., Lehrach H., Delius H. Nucleotide sequence of cdna coding for Semliki Forest virus membrane glycoproteins. Nature. 1980 Nov 20;288(5788):236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Bye J., Skehel J., Waterfield M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature. 1980 Sep 25;287(5780):301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- Gething M. J., White J. M., Waterfield M. D. Purification of the fusion protein of Sendai virus: analysis of the NH2-terminal sequence generated during precursor activation. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2737–2740. doi: 10.1073/pnas.75.6.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Kartenbeck J., Simons K., Fries E. On the entry of Semliki forest virus into BHK-21 cells. J Cell Biol. 1980 Feb;84(2):404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Ouchi M. Trypsin action on the growth of Sendai virus in tissue culture cells. 3. Structural difference of Sendai viruses grown in eggs and tissue culture cells. J Virol. 1973 Dec;12(6):1457–1465. doi: 10.1128/jvi.12.6.1457-1465.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R. T., Wahn K., Klenk H. D., Rott R. Fusion between cell membrane and liposomes containing the glycoproteins of influenza virus. Virology. 1980 Jul 30;104(2):294–302. doi: 10.1016/0042-6822(80)90334-7. [DOI] [PubMed] [Google Scholar]
- Kato N., Eggers H. J. Inhibition of uncoating of fowl plague virus by l-adamantanamine hydrochloride. Virology. 1969 Apr;37(4):632–641. doi: 10.1016/0042-6822(69)90281-5. [DOI] [PubMed] [Google Scholar]
- Klenk H. D., Rott R., Orlich M., Blödorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975 Dec;68(2):426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- Koff W. C., Knight V. Effect of rimantadine on influenza virus replication. Proc Soc Exp Biol Med. 1979 Feb;160(2):246–253. doi: 10.3181/00379727-160-40428. [DOI] [PubMed] [Google Scholar]
- Koff W. C., Knight V. Inhibition of influenza virus uncoating by rimantadine hydrochloride. J Virol. 1979 Jul;31(1):261–263. doi: 10.1128/jvi.31.1.261-263.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., McConnell H. M. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971 Mar 30;10(7):1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- Kuroda K., Maeda T., Ohnishi S. Enhancement of phospholipid transfer from Sendai virus to erythrocytes is mediated by target cell membrane. Proc Natl Acad Sci U S A. 1980 Feb;77(2):804–807. doi: 10.1073/pnas.77.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S. G., Choppin P. W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975 Dec;68(2):440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- Maeda T., Asano A., Oki K., Okada Y., Onishi S. A spin-label study on fusion of red blood cells induced by hemagglutinating virus of Japan. Biochemistry. 1975 Aug 26;14(17):3736–3741. doi: 10.1021/bi00688a003. [DOI] [PubMed] [Google Scholar]
- Maeda T., Ohnishi S. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 1980 Dec 29;122(2):283–287. doi: 10.1016/0014-5793(80)80457-1. [DOI] [PubMed] [Google Scholar]
- Ohkuma S., Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGLETON W. S., GRAY M. S., BROWN M. L., WHITE J. L. CHROMATOGRAPHICALLY HOMOGENEOUS LECITHIN FROM EGG PHOSPHOLIPIDS. J Am Oil Chem Soc. 1965 Jan;42:53–56. doi: 10.1007/BF02558256. [DOI] [PubMed] [Google Scholar]
- Sanders H. Preparative isolation of phosphatidyl serine from brain. Biochim Biophys Acta. 1967 Oct 2;144(2):485–487. doi: 10.1016/0005-2760(67)90184-1. [DOI] [PubMed] [Google Scholar]
- Skehel J. J., Hay A. J., Armstrong J. A. On the mechanism of inhibition of influenza virus replication by amantadine hydrochloride. J Gen Virol. 1978 Jan;38(1):97–110. doi: 10.1099/0022-1317-38-1-97. [DOI] [PubMed] [Google Scholar]
- Vänänen P., Käriäinen L. Fusion and haemolysis of erythrocytes caused by three togaviruses: Semliki Forest, Sindbis and rubella. J Gen Virol. 1980 Feb;46(2):467–475. doi: 10.1099/0022-1317-46-2-467. [DOI] [PubMed] [Google Scholar]
- Vänänen P., Käriäinen L. Haemolysis by two alphaviruses: Semliki Forest and Sindbis virus. J Gen Virol. 1979 Jun;43(3):593–601. doi: 10.1099/0022-1317-43-3-593. [DOI] [PubMed] [Google Scholar]
- White J., Helenius A. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]