Abstract
Background and Scope
In eukaryotes, chromatin remodelling complexes are shown to be responsible for nucleosome mobility, leading to increased accessibility of DNA for DNA binding proteins. Although the existence of such complexes in plants has been surmised mainly at the genetic level from bioinformatics studies and analysis of mutants, the biochemical existence of such complexes has remained unexplored.
Methods
Histone H1-depleted donor chromatin was prepared by micrococcal nuclease digestion of wheat nuclei and fractionation by exclusion chromatography. Nuclear extract was partially purified by cellulose phosphate ion exchange chromatography. Histone octamer trans-transfer activity was analysed using the synthetic nucleosome positioning sequence in the absence and presence of ATP and its analogues. ATPase activity was measured as 32Pi released using liquid scintillation counting.
Key Results
ATP-dependent histone octamer trans-transfer activity, partially purified from wheat nuclei using cellulose phosphate, showed ATP-dependent octamer displacement in trans from the H1-depleted native donor chromatin of wheat to the labelled synthetic nucleosome positioning sequence. It also showed nucleosome-dependent ATPase activity. Substitution of ATP by ATP analogues, namely ATPγS, AMP-PNP and ADP abolished the octamer trans-transfer, indicating the requirement of ATP hydrolysis for this activity.
Conclusions
ATP-dependent histone octamer transfer in trans is a recognized activity of chromatin remodelling complexes required for chromatin structure dynamics in non-plant species. Our results suggested that wheat nuclei also possess a typical chromatin remodelling activity, similar to that in other eukaryotes. This is the first report on chromatin remodelling activity in vitro from plants.
Keywords: ATPase, chromatin remodelling, histone, octamer-transfer, SWI/SNF-like complex, wheat, Triticum aestivum
INTRODUCTION
Eukaryotic chromatin is highly complex and has a dynamic architecture. Biochemical and genetic exploration in recent decades has confirmed the prevailing assumption that the organization of eukaryotic DNA into nucleosomes exerts a general repressive effect on replicative and transcriptional processes (Kornberg and Lorch, 1991; Felsenfeld, 1996), and additionally that it governs the accessibility of DNA for other DNA-centric processes such as repair and recombination. The restricted accessibility of nucleosomal DNA is due to shielding of the DNA helix-facing histone protein surface, resulting in strong binding and kinking of and the electrostatic interaction between the histone surface and terminal tails with DNA backbone. It has proved difficult to understand the principles underlying the organization of eukaryotic genomes in compact chromatin and simultaneously to unravel the genome accessibility to regulatory elements within the nucleus. To overcome this, eukaryotes are armed with sets of proteins that are normally classified as ATP-independent histone-modifying complexes and ATP-dependent chromatin-remodelling complexes (Becker and Hortz, 2002). The histone-modifying complexes modulate the histone code to loosen the contact between DNA and histones by neutralizing the charged groups, thereby producing accessibility to the other regulators, whereas the ATP-dependent remodellers belonging to the typical Saccharomyces cerevisiae Snf2 super family helicases destabilize the contacts between the DNA and histones by physical detachment of histone octamer (Lorch et al., 1999). These two types of complexes can form part of a large super complex and act in concert or independently in choreographing the modulation of DNA-focused events by regulating the temporal and spatial alteration of the chromatin structure in an event-specific manner. A large number of proteins belonging to these categories have been annotated in eukaryotic genomes, of which the ATP-dependent chromatin remodelling enzymes form distinct sub-groups (Flaus et al., 2006).
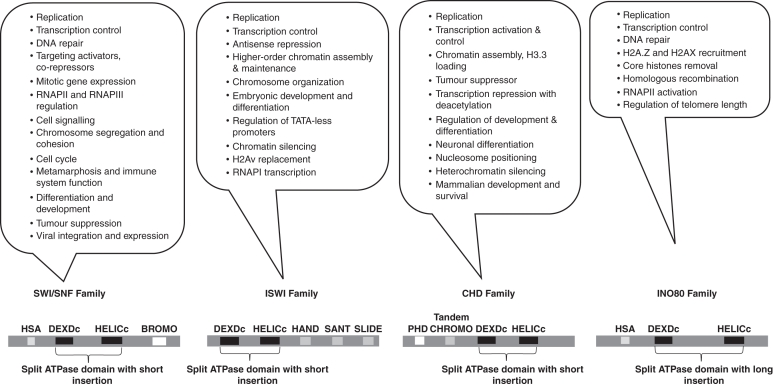
The ATP-dependent chromatin remodelling proteins from yeast, Drosophila and human have characteristic split ATPase domains and are classified into four families, namely SWI/SNF (SWItch/Sucrose Non Fermentable), ISWI (Imitation SWItch), CHD (Chromodomain-Helicase-DNA binding protein) and INO80 (INOsitol requiring 80), depending on the flanking domains. Each family of remodellers has its own mode of operation and although all of them have been implicated in some common vital functions, several events in the eukaryotic life cycle need a specific type of remodeller (as reviewed by Clapier and Cairns, 2009). In vivo functions of SWI/SNF components in development and disease in animal systems based on mutations, gene knockouts and other transgenic approaches have been reviewed by Ko et al. (2008). The domain structures of these chromatin remodelling complexes and the physiological processes affected by them in various eukaryotes are depicted in Fig. 1.
Fig. 1.
The functions and the characteristic domain architecture of the ATPase subunit of chromatin remodelling families from animal systems: chromatin remodellers from yeast and animal systems are classified into four families based on split ATPase domains and the characteristic flanking domains unique for each class. The callout boxes represent the functions that have been assigned to each family of the remodeller by the biochemical and genetic approaches from animal systems and yeast. The bottom panel indicates the domain architecture of each of these families that classifies them into the specific class of the remodeller as described by Clapier and Cairns (2009). Domain detail are described at www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
Models for the signature mechanisms by which each family of remodeller manoeuvre the chromatin have been postulated. The SWI/SNF family remodellers play an important role in nucleosome sliding or eviction while ISWI family remodellers aid in nucleosome assembly except for NURF, an ISWI family member that destabilizes the nucleosomes during transcription. The CHD and INO80 families share activities with SWI/SNF. The CHD complex (NuRD) contains a chromodomain and also has a histone deacetylase (HDAC) associated with it, whereas the INO80 complex has been shown to be important in DNA repair, transcription and H2A.Z-H2B replacement. Moreover, all remodelling complexes, in addition to ATPase-containing subunits, require several accessory subunits, some of which are also conserved in several eukaryotes (Ho and Crabtree, 2010).
Historically, the discovery of ySWI/SNF complexes in the mid-1980s prompted an increase in studies of chromatin remodelling. Genes encoding complex subunits were initially identified in genetic screens for mutants unable to express the HO endonuclease gene required for mating-type switching (SWI, from switch) and for mutants defective in anaerobic fermentation of sucrose (SNF, from sucrose-non-fermenting). As null mutations in any of the genes revealed by these screens produced identical phenotypes, it was assumed that their products, which act as pleiotropic transcriptional activators, must function in a common complex. Based on genetic speculations, yeast whole-cell extracts were fractionated biochemically and five SWI and SNF gene products (SWI1, SWI2/SNF2, SWI3, SNF5 and SNF6) were found to be constituents of a 2-MDa complex named SWI/SNF (Cairns et al., 1994; Peterson et al., 1994). The complex had an ATPase activity that is stimulated by DNA (approx. 30-fold) or by nucleosomes (approx. 40 fold) (Cote et al., 1994). The remodelling complexes BAP and PBAP were subsequently purified from Drosophila embryo extracts which share the BRM ATPase (Mohrmann et al., 2004). In humans two complexes called BAF (Brg1 Associated Factors) and PBAF (Polybromo-associated BAF) were identified.
The ATP-dependent remodelling activity originally detected in Drosophila was later attributed to NURF (Nucleosome Remodeling Factor) that contained an ATPase domain similar to SWI2/SNF2 but with other distinct domains. This led to its classification within another family, known as ISWI. The ISWI family containing remodelling complexes contain fewer subunits as compared with SWI/SNF complexes and three such complexes have been found in yeast (ISW1a, ISW1b, ISW2), Drosophila (NURF, CHRAC, ACF) and humans (hNURF, hCHRAC, hACF) (Corona and Tamkun, 2004). Two more families of chromatin remodellers, CHD and INO80, have also been detected. The CHD1 complex is found in yeast, Drosophila and humans, whereas Mi2 and NuRD complexes belonging to the CHD family are found only in the latter two (Marfella and Imbalzano, 2007). The INO80 family includes ySWR1, dTip60, Pho-dINO80, hINO80, hSCRAP and hTRRAP/hTip60 in all the three organisms and are known to be involved mainly in DNA repair (Bao and Shen, 2007). Over 30 genes encoding ATPase subunits of the remodelling complex have been detected as non-redundant and haplotype-sufficient. Their products show increased nucleosome mobility in vitro in the presence of ATP, which is used as a hallmark assay for ATP-dependent chromatin remodelling activity (Ho and Crabtree, 2010).
In plants, chromatin remodelling is studied mainly in the context of epigenetic switches that occur during reproductive and vegetative development, vernalization, pattern formation, etc., using mutagenesis, transgenics, RNAi techniques and bioinformatics (Goodrich and Tweedie, 2002). Recently, Desvoyes et al. (2010) have reviewed the impact of histone modifications on some of these processes. However, lack of phenotypic traits for all the mutants creates a hurdle for their detailed analysis. Wagner and Mayerowitz (2002) observed in Arabidopsis that the floral transition regulator factor LEAFY (LFY) is repressed by SPLAYED (SYD) and that remodelling by SYD is required for maintenance of meristem identity. SYD and BRM are considered to be members of the SWI/SNF family with a signature terminal bromodomain sequence. Chromatin assembly factors FAS1 and FAS2 play a role in apical meristem identity by regulating WUSCHEL (WUS) activity (Fransz and Jong, 2002). Yeast snf5 and swi3 mutations were complemented by Arabidopsis genes, AtBSH and AtSWI3B respectively, indicating the overlapping roles of plant and animal proteins (Sarnowski et al., 2002). Arabidopsis brm and syd mutants lacking SWI/SNF family ATPases have shown pleotropic developmental defects (Clapier and Cairns, 2009). Forty Arabidopsis SWI2/SNF2 analogues identified by Shaked et al. (2006) were found to be differentially expressed in DNA repair. Among other genes related to chromatin remodelling in plants, PICKLE a CHD3 analogue not belonging to the SWI/SNF family, was shown to be essential for expression of seed-associated genes during germination (Clapier and Cairns, 2009). Using a transgenic approach Mlynarova et al. (2007) revealed a role for AtCHR12, an SNF2/Brahma-type remodelling protein in growth retardation/dormancy under environmental stress in Arabidopsis. Recently, Jarillo et al. (2009) reviewed the role of chromatin remodelling factors that control gene expression in meristematic tissues in Arabidopsis. In contrast to animal cells, where epigenetic states are established in early embryonic development, plant cells show more flexibility. Epigenetic mechanisms would be important in dictating the behaviour of plant cells, as embryonic stem cells in conducive environments require reprogramming of the chromatin. Hence studies on the association between histone codes and chromatin dynamics require particular attention in plant biology.
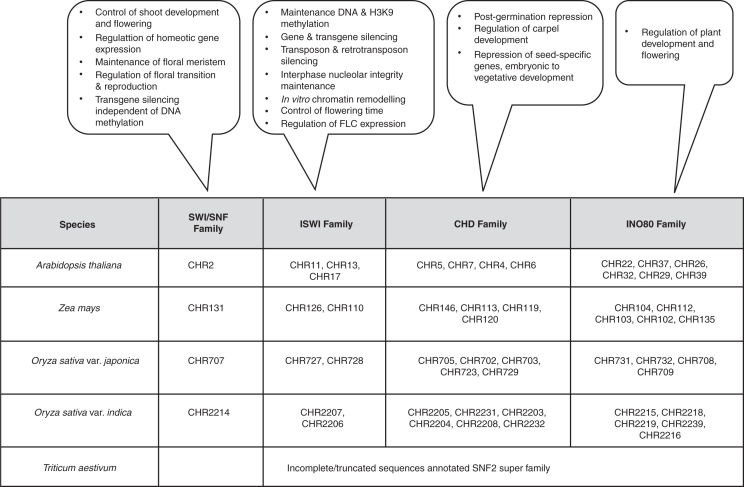
Comparative genome analyses have shown that plants encode a remarkably high number of potential SNF2 family proteins (Verbsky and Richards, 2001; Jerzmanowski, 2007). Figure 2 illustrates the putative chromatin remodelling proteins, detailing the probable functions in plants as described by Hsieh and Fischer (2005). The bottom panel of this figure classifies the SNF2 family proteins of Arabidopsis, rice and maize into four possible remodeller classes on the basis of the SNF2 family split ATPase domain flanked by other remodeller-specific domains. Although bioinformatics tools and genome databases have aided the current grouping of plant SNF2 proteins, their specific roles in different vital processes remains to be explored.
Fig. 2.
The putative functions of the members of the chromatin remodelling families in plant systems: the callout boxes indicate the functions that have been detected mainly by genetic approaches in Arabidopsis as described by Hsieh and Fischer (2005). The table at the bottom indicates the classification of remodellers belonging to the SNF2 superfamily ATPase protein sequences from www.chromdb.org with chromdb i.d. for Arabidopsis, maize and rice on the basis of the domains annotated in the Conserved Domain Database (CDD) www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. The sequences [42 from Arabidopsis, 47 from maize, 42 and 40 from rice (japonica and indica) and 14 from wheat], which were annotated as the SNF2 super family, were used for analysis.
To the best of our knowledge, none of these proteins have been investigated for the hallmark in vitro properties of a typical chromatin remodelling complex. Interestingly, Brzeski and Jerzmanowski (2003) cloned an AtDDM1 gene required for maintenance of methylation in Arabidopsis which showed nucleosome repositioning activity in vitro. In wheat, proteins annotated as belonging to SNF2 class are mostly incomplete and truncated. Here we present data on histone octamer trans-transfer activity of a putative chromatin remodelling complex isolated from wheat nuclei.
MATERIALS AND METHODS
Wheat seeds (Triticum aestivum ‘Tapovan’ NIAW-917) were surface-sterilized with 1 % mercury chloride for 10 min and rinsed three times with distilled water for 5 min each. The seeds were grown by spreading on damp filter paper sheets in plastic trays in the dark for 6 d at 28 °C. Watering was on days 3 and 5.
Nuclei isolation
Nuclei were isolated from wheat seedlings as described by Zhang et al. (1995). All the steps were carried out at 4 °C unless specified otherwise. Etiolated wheat seedlings (approx. 300 g) were harvested and ground in liquid nitrogen. Three volumes of nuclei isolation buffer (NIB) containing 0·5 m sucrose, 10 mm Tris, pH 9·5, 80 mm potassium chloride, 10 mm EDTA, 2 mm spermine and 1 mm spermidine, 0·4 % Triton-X100, 6 mm 2-mercaptoethanol (BME) and Complete EDTA-free protease inhibitor cocktail (Roche, India) containing leupeptin, pafabloc SC, antipain, aprotinin, bestatin, E64, chymostatin, calpain inhibitor I, II, tosyl-l-lysine chloromethyl ketone, phosphoramidon and phenylmethylsulfonyl fluoride (PMSF) was added. The mixture was stirred for 30 min and strained through five layers of cheesecloth. The filtrate containing nuclei was sedimented at 2200g for 10 min and washed twice in 50 mL NIB without Triton-X100 for 5 min. Quantification was done in terms of DNA by lysing a small aliquot in a mixture of NaCl/urea (2 m: 2·5 m). Finally, nuclei were resuspended in NIB without EDTA, spermine and spermidine at a concentration of 5 mg mL−1.
Preparation of donor chromatin
The strategy for preparation of donor chromatin was adapted from that described by Kornberg et al. (1989) with some modifications optimized for plant chromatin. Approximately 0·03 U MNase (Sigma-Aldrich, India) per milligram of DNA was added to the resuspended nuclei and incubated on ice for 2 min with intermittent mixing at an interval of 30 s. Nuclei were centrifuged at 8500g for 5 min and the pellet was resuspended in nuclei digestion buffer containing 2 mm CaCl2 and the suspension was incubated at 37 °C in a water bath with continuous stirring for a further 2 min. EDTA was added at 0·5 m to stop MNase activity. The pellet obtained after centrifugation at 8500g at 4 °C was resuspended in 15 mm Tris/HCl, pH 7·5, with 0·2 mm EDTA and incubated for 30 min on ice. The supernatant containing soluble chromatin was collected and 0·2 mm EDTA was added to prevent any residual MNase activity.
H1 depletion of soluble chromatin
H1-depleted chromatin was prepared as described by Libertini and Small (1980). Briefly, 2 m NaCl was added to the above supernatant to a final concentration of 0·6 m and the soluble chromatin was concentrated using Centricon YM10 filters (Millipore, India). The concentrated soluble chromatin was then loaded on a 45 × 1-cm Sephacryl S-400 (Sigma-Aldrich, India) column equilibrated with H1 depletion buffer (15 mm Tris, pH 7·5, 0·5 m NaCl, 1 mm EDTA, 0·2 mm PMSF, 15 mm BME and protease inhibitors). Fractions of 1·5 mL were collected and elution was monitored at A260nm for the DNA content. DNA from the aliquots of the fractions was precipitated with ethanol and analysed on a 1 % agarose gel. The fractions with DNA size equivalent to approx. 750 bp or more were pooled, concentrated and used as donor chromatin.
Preparation of the acceptor DNA
The DNA fragment containing centrally located synthetic nucleosome positioning sequence was cloned in pGEM plasmid. (This plasmid was obtained from Dr Orjan Wrange, Karolinska Institute, Stokholm by Prof. Madan Mohan Chaturvedi, Department of Zoology, University of Delhi, and renamed as p5TG.). A 210-bp fragment, referred to hereafter as NPS, was amplified with primers for the SP6-T7 promoter as described by Panigrahi et al. (2003). It was labelled with α32P-dATP during PCR amplification for use in remodelling assays.
Reconstitution of nucleosomes in vitro
Nucleosome assembly was carried out by salt jump or octamer exchange as described by Panigrahi et al. (2003). H1-depleted donor chromatin (5 µg) was mixed with 1·0 m NaCl in 50 mm Tris/HCl (pH 7·5) and 5 µg mL−1 bovine serum albumin (BSA). The mixture was gently mixed with pipetting at 2-min intervals for 10 min. Then, 25–50 ng 32P-labelled NPS was added and further incubation was done for 5 min. This reconstitution mixture was gradually diluted with 0·9, 0·8, 0·7, 0·6, 0·5, 0·4, 0·3 or 0·2 m NaCl with nucleosome reconstitution buffer 1 (NRB1: 20 mm Tris/HCl, pH 7·5, 1 mm EDTA, 1 mm dithiothreitol, 0·1 mg mL−1 BSA) followed by incubation at room temperature for 30 min after each dilution. Finally, NRB2 (NRB1 containing 20 % glycerol and 0·1 % NP40) was added to dilute NaCl to a final concentration of 0·1 m. The mobility shift was analysed by autoradiography after resolving the sample on 5 % native PAGE. The reconstituted nucleosomes were purified from donor chromatin and free DNA by size exclusion through Sepharose CL-6B as described by Panigrahi et al. (2003) and stored at 4 °C.
Partial purification of histone octamer trans-transfer activity from wheat nuclei
The nuclear extract was partially purified as described by Kwon et al. (1994). All the buffers were supplemented with protease inhibitors. The nuclei obtained from 500 g of wheat seedlings were extracted by resuspending in 25 ml SWI/SNF buffer (SSB) containing 15 mm Tris/HCl, pH 7·5, 10 % glycerol, 0·1 mm EDTA, 0·1 % Tween 20, 1 mm BME and 0·5 m KCl (hereafter SSB-0·5) with intermittent pippeting for 1 h. The nuclear extract in SSB-0·5 was collected by centrifugation at 19 200g for 30 min. This was diluted with SSB-0 (without KCl) to reduce the concentration of KCl to approx. 0·2 m and mixed with cellulose phosphate equilibrated with SSB-0·2 (SSB plus 0·2 m KCl). Binding to cellulose phosphate was allowed for 2 h at 4 °C on a rocker. The matrix was washed with SSB-0·2 and batch elution was done with 10 mL each of SSB-0·2, SSB-0·35, SSB-0·5 and SSB-0·75, the suffix indicating the KCl concentration. These eluted fractions were desalted and analysed for ATP-dependent histone octamer trans-transfer activity.
Histone octamer transfer assay
The histone octamer trans-transfer activity was monitored as described by Lorch and Kornberg (2004). Donor chromatin (5 ng) was incubated with labelled NPS (50 000 c.p.m./approx. 0·1 ng) and partially purified chromatin remodelling complex (CRC) in the assay buffer containing 15 mm Tris/HCl, pH 7·5, 50 mm KCl, 5 mm MgCl2, 3 mm Mg2+-ATP, 7·5 % glycerol, 0·3 mm EDTA, 100 µg mL−1 BSA and 0·05 % NP-40 at 37 °C for 15–60 min (see figure legends). The reaction was stopped by adding unlabelled plasmid DNA and further incubating for 10 min. Activity was monitored by analysing the mobility shift of NPS bound to nucleosomes by autoradiography after resolving the sample on 5 % native PAGE in 0·5× Tris-borate-EDTA buffer. This activity is referred to as CRC activity. To monitor ATP dependence, 3 mm ATP was substituted with 3 mm ADP, ATPγS and AMP-PNP in assay mixtures as mentioned in the figures.
ATPase assay
Partially purified CRC was added to assay buffer containing unlabelled NPS, donor chromatin and γ-32P ATP (0·25 µCi per reaction mixture) and incubated at 37 °C for 30 min. The 32Pi released during the reaction was quantified as described by Neufeld and Levy (1969). Briefly, the reaction was stopped with 20 % trichloroacetic acid and subsequently 0·1 mm NaH2PO4 was added. The supernatant was collected by centrifugation at 12 000g for 1 min. Twenty per cent acid molybdate was added followed by 10 % reducing solution (0·3 m sodium metabisulfite, 15 mm sodium sulfite, 4 mm 1-amino-2-naphthol-4-sulfonic acid) and an equal volume of isobutanol. The solution was vortexed for 30 s and allowed to stand until two phases were separated. The upper phase containing 32Pi was counted in the liquid scintillation counter.
RESULTS
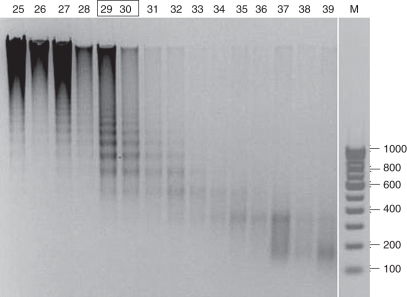
The biochemical activities of CRCs isolated from yeast, Drosophila and animals have been well documented (Becker and Horz, 2002). The main activities include ATP-dependant histone octamer displacement and nucleosome-dependent ATPase activity. The assay mixture requires donor chromatin and nucleosome positioning sequence in addition to ATP and chromatin remodelling complex. Wheat nuclei isolated from 6-d-old seedlings were used for donor chromatin preparation as described above. Treatment with 0·6 m NaCl aided in the release of the linker histones from the soluble chromatin. Figure 3 shows the profile of DNA isolated from the soluble chromatin prepared from wheat nuclei after MNase treatment and fractionation over Sephacryl S-400. The oligonucleosomes started eluting immediately after the void volume as analysed by agarose gel electrophoresis. The oligonucleosome fractions containing DNA of approx. 750 bp or more (fractions between 29 and 30 as shown in the box in Fig. 3) were used as donor chromatin (Supplementary Data Fig. S1, available online, shows the separation of histones on SDS–PAGE from H1-containing and H1-depleted chromatin). Salt treatment resulted in depletion of H1 from the soluble chromatin.
Fig. 3.
Profile of DNA isolated from the soluble chromatin: DNA was extracted from fractionated nucleosomes obtained from chromatin derived from MNase-treated wheat nuclei and analysed on 1 % agarose gel. The lane numbers (25–39) indicate fraction numbers. Lane M: 100-bp DNA ladder. Chromatin from fractions 29 and 30 (boxed) was pooled, concentrated and used as ‘donor chromatin’ for the remodelling assay.
To generate a positive control and substrate for octamer transfer assay, a labelled DNA fragment containing synthetic positioning sequence was amplified from p5TG plasmid. This fragment, referred to here as NPS, acted as acceptor DNA. It was noted that histone octamers released from native wheat oligonucleosomes during salt jump could assemble as nucleosomes on NPS, suggesting their resemblance to animal counterparts (Fig. 4, lane 1). When NPS and donor chromatin were incubated with wheat nuclear extract in the presence of ATP (described in details below), the position of the mononucleosome assembled by the remodelling activity from wheat nuclei was identical to that of the mononucleosomes assembled by salt jump (lane 2). Free NPS had highest mobility (lane 3). The higher molecular weight nucleo-histone, or nucleoprotein or DNA–DNA aggregate was found in all samples. However, the amount was greater in lane 1 where the nucleosomes were assembled by the salt jump method, and this could be attributed to the formation of metastable nucleosomes (Yager et al., 1989) indicating controlled assembly of nucleosome by CRC-mediated enzymatic process as in lane 2.
Fig. 4.
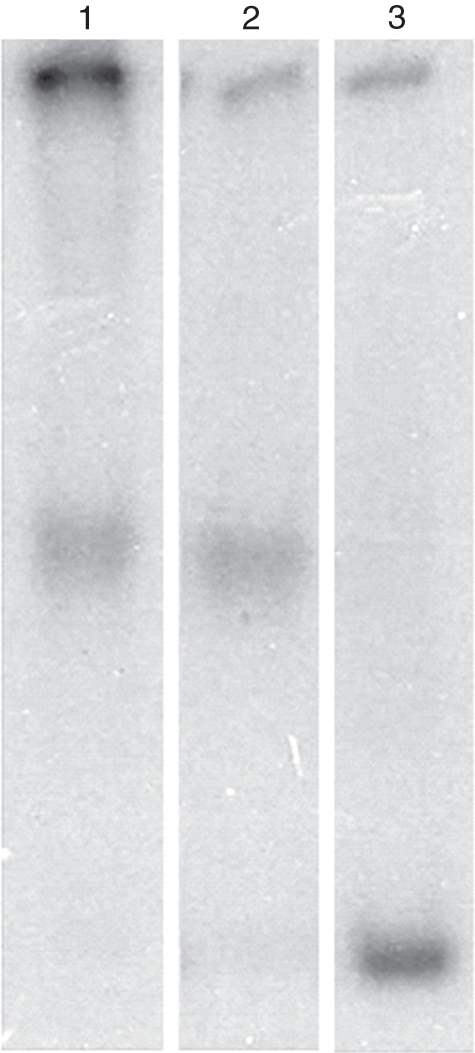
Nucleosome reconstitution in vitro: mononucleosomes were assembled in vitro both non-enzymatically and enzymatically as described in the Material and methods. Lane 1, mobility shift of NPS due to binding of histone octamer by the salt jump method. Lane 2, mobility shift of NPS due to ATP-dependent displacement of histone octamer in trans; Lane 3, free NPS.
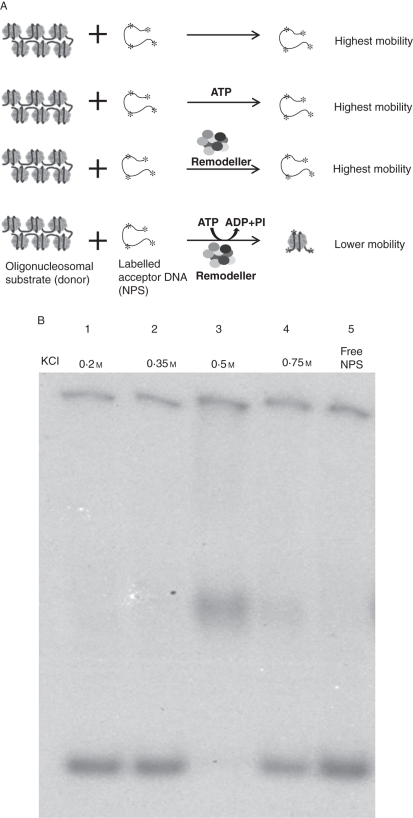
The nuclear extract was adsorbed on cellulose phosphate and eluted with different concentrations of KCl. The histone octamer trans-transfer activity was monitored by electrophoretic mobility shift of NPS. A schematic representation of reaction is shown in Fig. 5A. As shown here, the octamer trans-transfer is expected on labelled NPS only in the presence of ATP and CRC. Octamer transfer was not detected in the fractions eluted with SSB-0·2 and SSB-0·35 (Fig. 5B, lanes 1 and 2) as no mobility shift of labelled NPS was observed (cf. Fig. 5B, lane 5). The NPS mobility shift due to nucleosome formation was maximum in the fraction eluted with SSB-0·5 (lane 3). Residual activity was also found to be eluted with SSB-0·75 (lane 4). We observed interference due to non-specific nucleases during the assay. An increase in EDTA concentration in the assay mixture to 0·3 mm along with ATP (3 mm) prevented this nuclease activity (data not given).
Fig. 5.
(A) Schematic representation of histone octamer trans-transfer activity: the cartoon shows that the labelled NPS assembles into a mononucleosome in the presence of the chromatin remodeler and ATP, resulting in reduction in its mobility on PAGE. In the absence of ATP and/or remodeller the trans-transfer would not occur and hence there will be no reduction in the mobility of the labelled NPS. (B) Histone octamer trans-transfer activity of the elutes from cellulose phosphate: lane 1, activity eluted with SSB-0·2; lane 2, SSB-0·35; lane 3, SSB-0·5; lane 4, SSB-0·75; lane 5, free NPS.
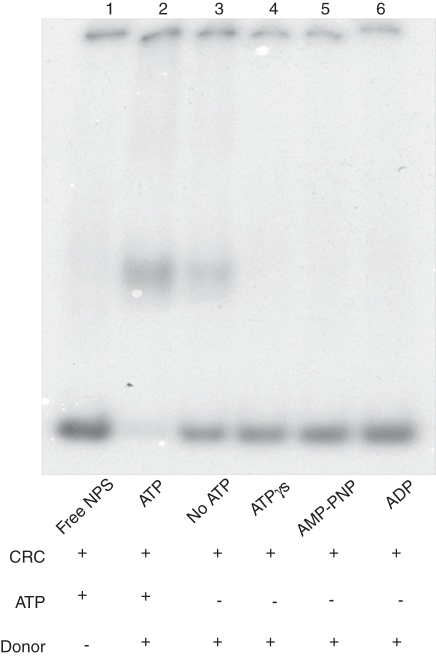
The time kinetics of histone octamer trans-transfer activity is shown in Fig. 6. The observations indicated that the histone octamer transfer was completed in 30 min (lanes 1 and 2). Incubation of the mixture for 45 and 60 min (lanes 3 and 4) did not enhance octamer transfer significantly but caused diffusion of the mononucleosome band, probably due to cis-mobility of the nucleosomes on the labelled NPS after central positioning. We also attempted to monitor cis mobility of the octamer using the mononucleosomes assembled by salt jump on the labelled NPS. Octamer sliding movement in the presence of ATP and the remodeller was observed occasionally (revealing a band with slightly higher mobility as compared with assembled mononucleosomes), indicating terminally positioned octamer (Supplementary Data Fig. S2, available online). As the trans-transfer activity of wheat CRC could be consistently monitored, further experiments were done using this assay. The effect of ATP on chromatin remodelling activity was also explored. Figure 7, lane 1, shows that in the absence of donor chromatin, CRC did not cause a band shift of NPS even in the presence of ATP. ATP enhanced octamer trans-transfer activity of CRC (lane 2) although some amount of activity was seen even in the absence of ATP (lanes 3). Substitution of ATP by ATP analogues, namely ATPγS, AMP-PNP and ADP, completely abolished the nucleosome trans-transfer activity (lanes 4–6) suggesting that energy required for this activity was provided by ATP hydrolysis. These results not only showed the specificity of the remodelling complex but also demonstrated that the remodeller does not bind to free NPS to produce a band with mobility equivalent to that of the assembled mononucleosome. These observations are consistent with the idea that chromatin remodelling activity from wheat is also ATP-dependent.
Fig. 6.
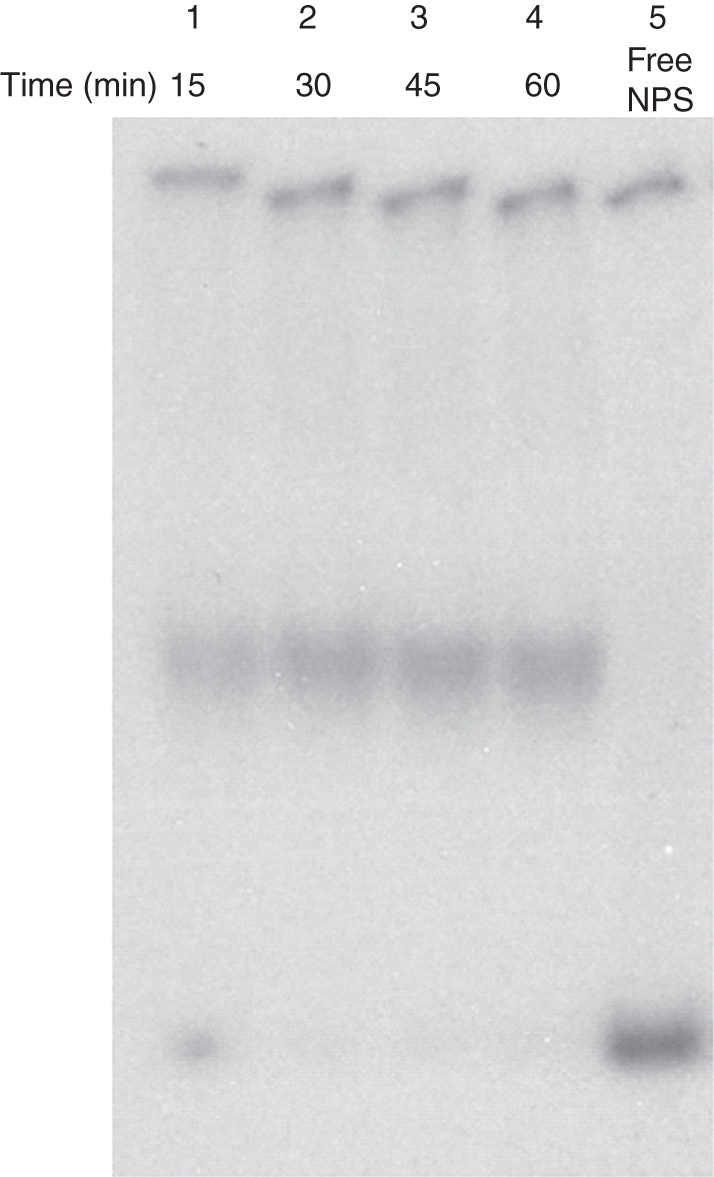
Time course of chromatin remodelling activity: lanes 1–4, 15, 30, 45 and 60 min incubation, respectively, of the assay mixture. Lane 5, free NPS.
Fig. 7.
Chromatin remodelling activity in the presence of ATP and ATP analogues: lane 1, free NPS; lane 2, with ATP; lane 3, without ATP; lane 4, with ATPγS; lane 5, with AMP-PNP; lane 6, with ADP as mentioned at the bottom of each lane.
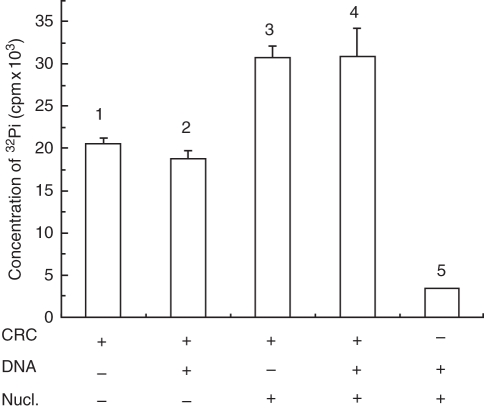
The ATPase activity of the partially purified extracts was monitored by release of Pi (Fig. 8). The nuclear extract showed ATPase activity in the absence of DNA or nucleosomes (lane 1). ATPase activity was slightly reduced when free DNA (unlabelled NPS) was added to the reaction mixture (lane 2). However, there was stimulation of ATPase in the presence of nucleosomes, which is a characteristic feature of chromatin remodelling factors (lane 3). ATPase activity was nearly the same when free DNA was added along with nucleosomes to the assay mixture (lane 4), indicating that the nucleosomes may provide the appropriate stimulation for the ATPase activity of the remodeller in comparison with DNA alone. ATPase activity was not seen in the absence of chromatin remodeller (lane 5).
Fig. 8.
ATPase assay of the chromatin remodelling complex: the x-axis depicts the assay conditions and y-axis shows the concentration of 32Pi released from hydrolysis of ATP in terms of counts (cpm). Nucl., donor chromatin as oligo-nucleosomal DNA; CRC, chromatin remodelling complexes.
DISCUSSION
Chromatin remodelling causes stable yet reversible changes in chromatin structure, resulting in discernible alterations in DNA–histone contacts. Chromatin remodellers are large, multiprotein complexes that utilize the energy of ATP hydrolysis to mobilize and restructure nucleosomes. All remodellers act as directional ATP-dependent DNA translocases capable of pumping DNA by binding to the nucleosome around the histone–octamer surface (Saha et al., 2006). This enables movement of the nucleosome along the DNA, thus permitting exposure of the DNA to regulatory factors. Chromatin remodellers from yeast, Drosophila and animal systems have been investigated in detail at the genetic, epigenetic and biochemical level in the last two decades and have been found to be compositionally and functionally diverse.
In contrast, information about chromatin remodelling largely exits at the genetic level in plants. Recent genome sequencing projects have revealed the presence of analogues of many genes involved in chromatin remodelling activity in plants (Shaked et al., 2006). In addition, analogues of several core components of CRCs such as SWI3-type proteins, SNF5-type proteins, SWP73-type proteins and other non-core subunits such as plant actin, OSA (oncogene suppressor activity), polybromo-type proteins are detected mainly in Arabidopsis (Jerzmanowski, 2007). In spite of extensive breeding programmes, no biochemical insight of the epigenetic aspects in crop plants exists. With advances in the understanding of chromatin structure and its dynamics, it is imperative that extensive studies in this area are undertaken for crop plants such as wheat.
Chromatin remodelling activity is generally measured in vitro by monitoring the mobility of nucleosomes (Owen-Hughes and Workman, 1996; Längst and Becker, 2001). In these assays, cloned, unmodified histones are frequently used for preparing donor chromatin (Duband-Goulet et al., 2004). Remodelling activity is monitored by electrophoretic mobility shift of the acceptor DNA, which is labelled. We monitored chromatin remodelling activity in wheat nuclear extracts. The assay mixture contained H1-depleted donor chromatin, chromatin remodeller, labelled acceptor DNA with NPS and ATP in the presence of 100 mm salt (KCl). We prepared donor chromatin from wheat nuclei by MNase digestion which was used in enzymatic and non-enzymatic assembly of nucleosomes on NPS. As histones are positively charged molecules, their simple association in vitro with DNA results in the formation of nucleo-histone aggregates. The non-covalent and electrostatic interactions between DNA and histones in the donor chromatin can be disrupted by increasing salt concentrations. Gradual elimination of salt by dialysis from an initial high-salt mixture of core histones and DNA allows the histones and DNA to reassociate in a controlled fashion, resulting in the formation of nucleo-histone complexes non-enzymatically. Such complexes are known to resemble native nucleosomal structures (Rhodes and Laskey, 1989).
In the present study, the nucleosome reconstitution method was optimized to utilize H1-depleted donor chromatin from wheat. In the presence of 1 m NaCl, the donor chromatin released histone octamers. Serial dilution of the reaction mix in salt-free buffer resulted in the reassociation of the octamer with available free DNA along with labelled 210-bp NPS in a defined translational and rotational setting with more than 90 % efficiency in 3–4 h. These non-enzymatically assembled mononucleosomes showed one band on PAGE with very high molecular weight band representing non-specific biding of nucleosomes to NPS. Although histone octamers can bind virtually any genomic DNA sequence, the octamer preference to a well-defined position on TG motifs, used in the current study, is well established (Shrader and Crothers, 1989). Reconstitution of wheat mononucleosomes on TG motif DNA suggested that plant mononucleosomes may be similar to that of other organisms such as animals and yeast, although there may be some differences in the structure of plant chromatin (Smith et al., 1995). The results showed, for the first time, the reconstitution of wheat histone octamers on a synthetic positioning motif into a mononucleosome.
As the wheat octamers were able to bind NPS, the chromatin remodelling assay was done using NPS and the donor chromatin isolated from wheat nuclei. Use of wheat donor chromatin for nucleosome reconstitution was aimed at mimicking the native chromatin substrate, including post-translational modifications of histones reported in wheat (Raut and Sainis, 2011). This is in contrast to earlier studies that have utilized chicken histones, mouse histones, derivatized Xenopus histones or recombinant histones to reconstitute nucleosome core for chromatin remodelling assays (Rhodes and Laskey, 1989; Stein, 1989; Duband-Goulet et al., 2004). Along with very few previous studies (e.g. Panigrahi et al., 2003), the present study utilized donor chromatin and remodelling complex, both obtained from the same species, in an in vitro remodelling assay.
Previous studies on SWI/SNF-like complexes from yeast, fly and mammalian systems have shown that 8000–10 000-fold purification is required over several steps (Wang et al., 1996). Dissociation of subunits upon dilution during multiple chromatographic purification steps and its low abundance in the cell nucleus further complicates the difficulties in purification. In the present work, enrichment of the remodelling activity in the nuclear extract was performed using cellulose phosphate ion exchange resin in one step. It was noted that the elution profile of histone octamer trans-transfer activity followed the characteristic pattern reported for SWI/SNF-like complexes in yeast and the avian system (Whitehouse et al., 1999; Panigrahi et al., 2003). The bulk of activity was eluted with 0·5 m KCl from the cellulose phosphate column, whereas the rest (about 10 % of total activity) required 0·75 m KCl to elute. This observation was consistent with the chromatographic behaviour reported for human and chicken SWI/SNF (Wang et al., 1996; Panigrahi et al., 2003). The lower activity in the 0·75 m KCl fraction could be due to either near total elution of activity by 0·5 m KCl wash prior to 0·75 m KCl wash or dissociation of the multiprotein CRC. To simplify the fractionation procedure, the histone octamer trans-transfer activity was routinely eluted from cellulose phosphate with 0·5 m KCl and diluted to 0·1 m KCl before use in the remodelling assay.
The time kinetics of the remodelling assay revealed that activity was saturated in 15–30 min. This was consistent with the established idea that the kinetics of enzymes from different systems are similar. Furthermore, it was observed that octamer transfer and the formation of mononcleosomes was ATP-dependent. However, complete loss of octamer trans-transfer activity upon addition of ATP analogues agrees with earlier reports showing octamer transfer in trans as an ATP-dependent event (Whitehouse et al., 1999; Imbalzano and Xiao, 2004), and also shows that the trans-transfer activity is not an artefact. Chromatin remodellers are known to act as DNA translocating machines fuelling transport of DNA across nucleosomes by ATP hydrolysis (Saha et al., 2006). As even without ATP some trans-transfer was seen, it is conjectured that ATP might enhance the rate of DNA translocation across nucleosomes by chromatin remodellers. Alternatively, there may be some histone-interacting proteins causing nucleosome displacement in an ATP-independent manner as described by Chen et al. (1994) in the case of nucleoplasmin. However, the presence of ATP significantly enhanced the octamer trans-transfer activity. The use of non-hydrolysable ATP analogues demonstrated the requirement not just for ATP but for actual ATP hydrolysis for chromatin remodelling. A conformational change in the quaternary structure of the entire complex during the motorized action driven by ATP hydrolysis may be inhibited due binding of ATP analogues. They might lock the remodellers in a conformation that can no longer act as a DNA-translocating machine or alternatively release the complex from nucleosomes or abolish capacity of its binding domain. The exact mechanism involved needs further study. Another line of evidence that the remodelling activity was ATP-dependent came from the observation that its ATPase activity was stimulated by 47 % in the presence nucleosomes, consistent with similar observations on other SWI/SNF complexes (Duband-Goulet et al., 2004).
This is the first report on detection chromatin remodelling activity in vitro from wheat in particular and plant systems in general. It revealed the hallmark properties of a typical remodeller, such as ATP-dependent nucleosome mobility in cis and trans as well as nucleosome-dependent ATPase activity, suggesting its affinity for nucleosomes beyond DNA. As the assay was done with native histones present on donor chromatin, the chromatin remodeller would have the necessary components to recognize the histone modifications. The nucleosome-dependent ATPase activity would serve as a DNA-translocating motor to break DNA–nucleosome contacts as described by Clapier and Cairns (2009). Inhibition of the remodelling activity by ATP analogues provides support for the requirement of ATP binding domains for regulation of activity by the remodeller. As nucleosome trans-transfer activity is the characteristic property of the yeast RSC (chromatin structure remodelling) (Lorch et al., 1998) and SWI/SNF complex of human (Phelan et al., 2000), the wheat chromatin remodeller can be classified as an SWI/SNF complex. Moreover, we observed that this factor cross-reacted with anti-Brg1 antibody and eluted as a high-molecular-weight fraction on sepharose CL 6B (data not shown), supporting the above assumption. However, the presence of another group of remodelling complexes, such as ISWI and CHD, cannot be completely ruled out because an enriched fraction possessing octamer trans-transfer activity was not affinity purified. Furthermore, confirmation regarding the classification of the wheat chromatin remodeller requires biochemical and proteomic characterization of its subunits and domains.
This report, demonstrating chromatin remodelling activity in wheat nuclei, opens a new vista in research on the chromatin dynamics of the complex genome of this crop plant. The polyploidy of wheat could offer advantages in chromatin research and also poses new challenges in the field of epigenetics.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dr Orjan Wrange, Karolinska Institute, Stokholm, and Prof. Madan Mohan Chaturvedi, Department of Zoology, University of Delhi, India, for the kind gift p5TG plasmid. This work was funded by the Department of Atomic Energy, India.
LITERATURE CITED
- Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutation Research. 2007;618:18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P, Hortz W. ATP-dependent nucleosome remodeling. Annual Review of Biochemistry. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Brzeski J, Jerzmanowski A. Deficient in DNA Methylation 1 (DDM1) defines a novel family of chromatin remodeling factors. The Journal of Biological Chemistry. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proceedings of the National Academy of Sciences USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li B, Workman JL. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO Journal. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C, Cairns B. The biology of chromatin remodeling complexes. Annual Review of Biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Corona DFV, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochemica et Biophisica Acta. 2004;1677:113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the Yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Sanchez MP, Ramirez-Parra E, Gutierrez C. Impact of nucleosome dynamics and histone modifications on cell proliferation during Arabidopsis development. Heredity. 2010;105:80–91. doi: 10.1038/hdy.2010.50. [DOI] [PubMed] [Google Scholar]
- Duband-Goulet I, Ouararhni K, Hamiche A. Methods for chromatin assembly and remodeling. Methods. 2004;33:12–17. doi: 10.1016/j.ymeth.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Research. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, Jong J. Chromatin dynamics in plants. Current Opinion in Plant Biology. 2002;5:560–567. doi: 10.1016/s1369-5266(02)00298-4. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Tweedie S. Remembrance of things past: chromatin remodeling in plant development. Annual Review of Cell and Developmental Biology. 2002;18:707–746. doi: 10.1146/annurev.cellbio.18.040202.114836. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodeling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Fischer RL. Biology of Chromatin dynamics. Annual Review of Plant Biology. 2005;56:327–351. doi: 10.1146/annurev.arplant.56.032604.144118. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Xiao H. Functional properties of ATP-dependent chromatin remodeling complexes. Advances in Protein Chemistry. 2004;67:157–179. doi: 10.1016/S0065-3233(04)67006-9. [DOI] [PubMed] [Google Scholar]
- Jarillo J, Pinerio M, Cubas P, Martinez-Zapater J. Chromatin remodeling in plant development. International Journal of Developmental Biology. 2009;53:1581–1596. doi: 10.1387/ijdb.072460jj. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A. SWI/SNF chromatin remodeling and linker histones in plants. Biochemica et Biophysica Acta. 2007;1769:330–345. doi: 10.1016/j.bbaexp.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Ko M, Sohn DH, Chung H, Seong RH. Chromatin remodeling, development and disease. Mutation Research. 2008;647:59–67. doi: 10.1016/j.mrfmmm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Irresistible force meets immovable object; transcription and the nucleosome. Cell. 1991;67:833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- Kornberg R, LaPoint J, Lorch Y. Preparation of nucleosomes and chromatin. In: Wassarman PM, Kornberg RD, editors. Methods in enzymology. Vol. 170. San Diego: Academic Press; 1989. pp. 3–14. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano A, Khavari P, Kingston R, Green M. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Längst G, Becker P. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. Journal of Cell Science. 2001;114:2561–2568. doi: 10.1242/jcs.114.14.2561. [DOI] [PubMed] [Google Scholar]
- Libertini L, Small E. Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Research. 1980;8:3517–3534. doi: 10.1093/nar/8.16.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Kornberg R. Isolation and assay of the RSC chromatin remodeling complex from Saccharomyces cerevisae. In: Allis DC, Wu C, editors. Methods in enzymology. Vol. 377. Amsterdam: Elsevier Academic Press; 2004. pp. 316–322. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Cairns B, Zhang M, Kornberg R. Activated RSC–nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg R. Histone octamer transfer by a chromatin remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- Marfella GAC, Imbalzano AN. The Chd family of chromatin remodelers. Mutation Research. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova L, Nap J-P, Bisseling T. The SWISNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. The Plant Journal. 2007;51:874–885. doi: 10.1111/j.1365-313X.2007.03185.x. [DOI] [PubMed] [Google Scholar]
- Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Molecular Cell Biology. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld A, Levy H. A second ouabain-sensitive sodium-dependent adenosine triphosphatase in brain. The Journal of Biological Chemistry. 1969;244:6493–6497. [PubMed] [Google Scholar]
- Owen-Hughes T, Workman J. Remodeling the chromatin structure of a nucleosome array by transcription factor-targetted trans-displacement of histones. EMBO Journal. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A, Tomar R, Chaturvedi M. A SWI/SNF-like factor from chicken liver that disrupts nucleosomes and transfers histone octamer in cis and trans. Archives of Biochemistry and Biophysics. 2003;414:24–33. doi: 10.1016/s0003-9861(03)00175-9. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Dingwal A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional activation. Proceedings of the National Academy of Sciences USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan M, Schnitzler G, Kingston R. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Molecular and Cellular Biology. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raut VV, Sainis JK. 60Co-γ radiation induces differential acetylation and phosphorylation of histones H3 and H4 in wheat. Plant Biology. 2011 doi: 10.1111/j.1438-8677.2011.00463.x. doi:10.1111/j.1438-8677.2011.00463.x. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Laskey R. Assembly of nucleosomes and chromatin in vitro. In: Wassarman PM, Kornberg RD, editors. Methods in enzymology. Vol. 170. San Diego: Academic Press; 1989. pp. 575–585. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling: the industrial revolution of DNA around histones. Nature Reviews on Molecular Cell Biology. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Sarnowski T, Swiezewski S, Pawlikowska K, Kaczanowski S, Jerzmanowski A. AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Research. 2002;30:3412–3421. doi: 10.1093/nar/gkf458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Avivi-Ragolski N, Levy A. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;17:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T, Crothers D. Artificial nucleosome positioning sequences. Proceedings of the National Academy of Sciences USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Hill R, Baldwin J. Plant chromatin structure and post-translational modifications. Current Reviews in Plant Sciences. 1995;14:299–328. [Google Scholar]
- Stein A. Reconstitution of chromatin from purified components. In: Wassarman PM, Kornberg RD, editors. Methods in enzymology. Vol. 170. San Diego: Academic Press; 1989. pp. 585–603. [DOI] [PubMed] [Google Scholar]
- Verbsky ML, Richards EJ. Chromatin remodeling in plants. Current Opinion in Plant Biology. 2001;4:494–500. doi: 10.1016/s1369-5266(00)00206-5. [DOI] [PubMed] [Google Scholar]
- Wagner D, Meyerowitz E. SPLAYED a novel SWI-SNF ATPase homolog controls reproductive development in Arabidopsis. Current Biology. 2002;12:85–94. doi: 10.1016/s0960-9822(01)00651-0. [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, et al. Purification and biochemical heterogeneity of the mammalian swi-snf complex. EMBO Journal. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Catalytic nucleosome mobilisation mediated by the SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Yager T, McMurray C, van Holde K. Salt-induced release of DNA from nucleosome core particles. Biochemistry. 1989;28:2271–2281. doi: 10.1021/bi00431a045. [DOI] [PubMed] [Google Scholar]
- Zhang H-B, Xao X, Ding X, Peterson A, Wing A. Preparation of megabase size DNA from plant nuclei. The Plant Journal. 1995;7:175–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.