Abstract
Peroxidation of cellular membrane lipids, rich in polyunsaturated fatty acids, generates electrophilic, α,β-unsaturated aldehydes such as 4-hydroxy-2-nonenal (HNE). HNE is a highly reactive and cytotoxic molecule that can react with the nucleophilic sites in proteins causing posttranslational modification. The identification of protein targets is an important first step; however, quantitative profiling of site-specific modifications is necessary to understand the biological impact of HNE-induced carbonylation. We report a method that uses light (H12CHO) and heavy (D13CDO) isotopic variant of formaldehyde to differentially label primary amines (N-termini and ε-amino group of lysines) in peptides through reductive methylation and, combined with selective enrichment of modified peptides, permits comparison of the extent of carbonylation in two samples after mixing for simultaneous liquid chromatography–mass spectrometry. Specifically, dimethyl-labeled peptide carbonyls were fractionated from unmodified peptides using solid-phase hydrazide chemistry to immobilize them to porous glass beads and, after removing the unmodified peptides by thoroughly washing the beads, subsequently recover them by acid-catalyzed hydrolysis. The method was developed using HNE-modified synthetic peptides and also showing enrichment from a complex matrix of digested human plasma proteins. Applicability was confirmed using apomyoglobin as an analyte, implicating thereby its potential value to proteome-wide identification and relative quantification of posttranslational protein carbonylation with residue-specific information. Because HNE attachment may not necessarily cause change in protein abundance, this modification-focused quantification should facilitate the characterization of accompanied changes in protein function and, also, provide important insights into molecular signaling mechanisms and a better understanding of cellular processes associated with oxidative stress.
Keywords: protein carbonylation, 4-hydroxy-2-nonenal, LC–MS/MS, stable-isotope tagging, solid-phase hydrazide chemistry, differential quantitative proteomics
Introduction
Polyunsaturated fatty acids present in membrane lipids are susceptible to free radical-initiated peroxidation.[1] 4-Hydroxy-2-nonenal (HNE), a major product of lipid peroxidation (LPO), is regarded as a highly toxic aldehyde formed during peroxidation of polyunsaturated (n−6) fatty acids such as linoleic acid and arachidonic acid.[1,2] This aldehyde acts as a toxic second messenger of oxidative stress by disseminating and augmenting initial free-radical events. HNE has been implicated in numerous pathologies, such as atherosclerosis, cancer, neurodegenerative disorders (Alzheimer’s disease, Parkinson’s disease and Amyotrophic lateral sclerosis), ethanol-associated liver injury, and injury from ischemia/reperfusion.[3–5]
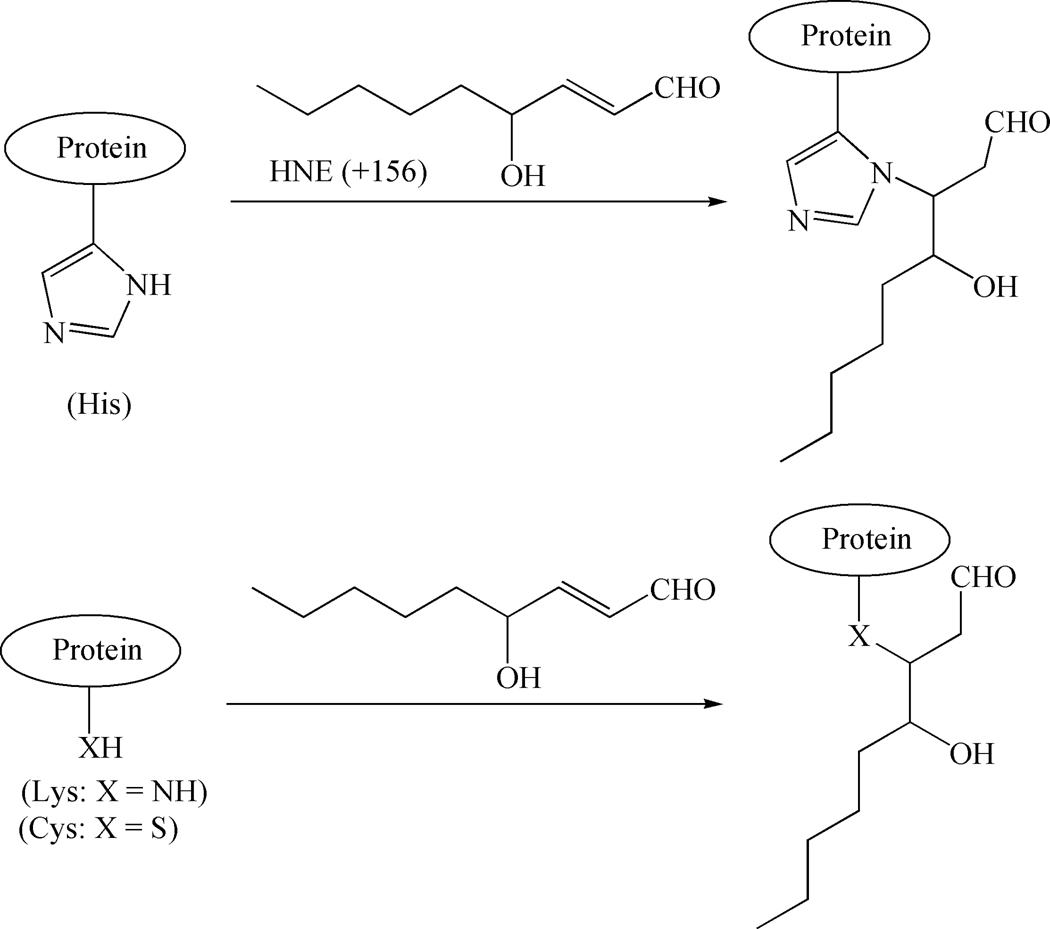
HNE, a bifunctional aldehyde, is susceptible to nucleophilic addition at both the double bond (C3) and the carbonyl moiety (C1). The presence of electron-withdrawing carbonyl group at C1 position in HNE makes the double bond at the C3 position highly electrophilic and, hence, can covalently bind to proteins via Michael addition (MA) to the sulfhydryl group of cysteine (Cys, C), imidazole group of histidine (His, H), and ε-amino group of lysine (Lys, K) residues resulting in mass increase of 156 Da (Fig. 1).[6] The reactivity of these amino acids to HNE occurs in the following order: Cys >> His > Lys >> Arg.[7] Although the Michael adducts of HNE with Cys and His are stable, Michael adducts to Lys ε-amino groups may form reversibly.[8] The C1 carbonyl moiety of HNE can also yield Schiff-base adducts with the ε-NH2 group of Lys and a concomitant loss of water produces a gain of 138 Da in molecular mass.[9] Less abundant Schiff base formation with Lys groups results in some late stage stable adducts such as the 2-pentylpyrroles. The bifunctionality of HNE also allows it to induce protein cross-links such as the 2-hydroxy-2-pentyl-1,2-dihydropyrrol-3-one iminium that are fluorescent four-electron oxidation products.[10]
Figure 1.
Primary reaction chemistry observed for 4-hydroxy-2-nonenal (HNE) modification of proteins. The α,β-unsaturated aldehyde is susceptible to Michael addition by a variety of nucleophiles including the histidine (His) imidazole group, lysine (Lys) ε-amino group, or thiol group of cysteine (Cys).
Quantification or comparison of relative abundance of proteins expressed in cells or tissues during normal or diseased conditions is essential to identify the putative target proteins correlated with the diseased states. However, not all proteins undergo change in their relative expression profile; instead, they may undergo change in the levels of their posttranslational modification. A wide array of functions is controlled by HNE-mediated protein carbonylation. Several studies have shown that change in protein carbonylation in various diseases occurs without any concomitant variation in the protein expression levels;[11–15] however, most of these studies have used gel electrophoresis for validation. In these studies, changes in protein carbonylation are determined by comparing the spot intensities upon labeling with antibodies specific for HNE or prior labeling of protein carbonyl groups with digoxigenin-hydrazide[13] or 2,4-dinitrophenylhydrazine[14] and detecting by anti-digoxigenin or anti-2,4-dinitrophenol antibodies in immunoblots obtained from different samples. An additional method involves biotinylation of oxidized proteins and detecting them in two-dimensional (2D-) gels using avidin-fluorescein isothiocyanate staining.[15] The protein identity in the spot or band is then determined by excising it from the gel, performing enzymatic digestion, and analyzing by mass spectrometry.[16] The 2D-gel-based method is labor-intensive and time-consuming which confines high-throughput proteome analysis. The low specificity of anti-HNE antibodies and the lack of site-specific HNE antibodies that recognize a particular carbonylation site within a protein sequence make this type of quantification challenging. These limitations have accentuated the need for a technique that can quantify changes in the relative carbonylation states in proteins along with identification of the modification sites.
Mass spectrometry has an unprecedented advantage in protein identification and determination of posttranslational modifications. Several strategies have also been reported to quantitate differential protein expression in tissues.[17] Extending beyond our previous studies that involve the development of methods for the determination of HNE modification,[18–20] we describe here an approach that incorporates stable-isotope dimethyl labeling of N-terminus and ε-amino side chains of lysine residues of the peptides for differential quantification of HNE-modified protein. The applicability of stable-isotope dimethyl labeling for quantitative protein profiling has been reported previously.[21,22] The corresponding stable-isotope labeling allows samples to be analyzed and compared in a single analytical experiment, and does not require the use of labeled peptide standards such as the so-called absolute quantification approach.[23] The peptide amines are differentially labeled via reductive alkylation with isotopically coded formaldehydes (d0,12C-formaldehyde or d2,13C-formaldehyde). Following reductive methylation of primary amino groups, each isotopic peptide pair differs in mass by n-times 6 Da, where n is the number of primary amino groups in the peptide. This provides sufficient spacing by m/z without overlap of the isotope envelopes, facilitating the measurement of relative abundances of peptides by LC–MS. For carbonylated peptides that are normally present in extremely minute amounts, enrichment techniques to minimize interference, such as ion suppression effects caused by large amounts of non-carbonylated peptides,[24] are normally necessary. Hence, we have incorporated a chemoprecipitation method to selectively isolate HNE-modified peptides and thereby minimize such ion suppression effects from unmodified peptides. The enriched fraction is then subjected to LC–MS and the quantification between the relative abundance of isotopically labeled populations of peptide carbonyls can be computed from comparative area measurements using extracted ion chromatograms (XICs) of light- and heavy-isotopic dimethyl-labeled peptides, respectively. The implementation of a quantification method for protein carbonyls in a biological system is expected to further our understanding of disease mechanisms associated with oxidative stress and, thus, assist the discovery of novel therapeutics for these conditions.[5,25]
Experimental
Chemicals and reagents
4-Hydroxy-2-nonenal (HNE, 10 mg/mL in ethanol) was obtained from Cayman Chemical Company (Ann Arbor, MI). Water, acetonitrile and methanol were of high-performance liquid chromatography (HPLC) grade and purchased from Honeywell Burdick and Jackson (Morristown, NJ, USA). Trypsin (TPCK treated) was obtained from Applied Biosystems (Foster City, CA). Formaldehyde (“d0,12C-,“ 37% solution in H2O), apomyoglobin from equine skeletal muscle, and all other chemicals were provided by Sigma-Aldrich (St. Louis, MO). Stable-isotope labeled d2,13C-formaldehyde (20% solution in D2O) was purchased from Isotec (Miamisburg, OH). The peptides, angiotensin I (DRVYIHPFHL) and II (DRVYIHPF) were purchased from AnaSpec (Fremont, CA), and oxidized insulin β-chain was obtained from Sigma-Aldrich (St. Louis, MO). ATP synthase subunit beta peptide, LVLEVAQHLGESTVR, aconitate hydratase peptide, IVYGHLDDPANQEIER, and vimentin peptide, QVQSLTCEVDALK (shown to be targets for HNE modification[18,26]) were custom-synthesized by Peptide 2.0, Inc. (Chantilly, VA) and used without further purification. Pooled normal human plasma was obtained from Innovative Research, Inc. (Novi, MI).
4-Hydroxy-2-nonenal modification of synthetic peptides and apomyoglobin
HNE adducts of synthetic peptides (DRVYIHPFHL, DRVYIHPF, LVLEVAQHLGESTVR and IVYGHLDDPANQEIER, QVQSLTCEVDALK, 1 mg/mL) were prepared by reaction with 2 mM HNE in 0.1 M phosphate buffer, pH 7.4, at 37 °C for 2 h. Excess HNE was removed by extracting the solution three times with ethyl acetate. The resultant aqueous (stock) solutions of HNE-modified peptides were used for differential labeling with light and heavy stable isotopes of formaldehyde without further purification in the experiments detailed below.
Apomyoglobin (1 mg/mL) was incubated with 2 mM HNE in phosphate buffer (50 mM pH 7.4) at 37 °C for 2 h to obtain carbonylated protein. Then, the HNE-modified protein was precipitated by adding 4 volumes of ice-cold acetone and keeping the mixture at −20 °C for 2 h. The sample was centrifuged at 13000 rpm for 10 min at 4 °C. The protein pellet was resuspended in 50 mM triethylammonium bicarbonate and subjected to proteolytic digestion at 37 °C for 18 h by adding trypsin at a ratio of 1:50. Following digestion, proteolytic activity was terminated by acidifying the reaction mixture with acetic acid to pH <3.0 and lyophilized subsequently.
Dimethyl-labeling of HNE-modified peptides by reductive alkylation
Reductive methylation, with either light or heavy formaldehyde, of HNE-modified synthetic peptides and apomyoglobin tryptic peptides were accomplished as described previously[21,22] with some modifications. The synthetic peptides, diluted in sodium acetate buffer (100 mM, pH 5–6), were mixed with freshly prepared sodium cyanoborohydride (1 M, 20 µL), vortexed, and divided into two aliquots, each containing 0.1 mg, and separately labeled via reductive methylation with excess of either d0,12C-formaldehyde (HCOH, 4% in water, 20 µL) or d2,13C-formaldehyde (D13COD, 4% in water, 20 µL). Following addition of formaldehyde, the mixtures were vortexed and allowed to react for 2 h at room temperature and the reaction was stopped by adding ammonium hydroxide (4% in water, 20 µL) to consume the excess aldehyde. After labeling, the solution in each vial was acidified to pH 3.0 by adding 1% TFA.
The lyophilized apomyoglobin tryptic digest was reconstituted with sodium acetate buffer (100 mM, pH 5–6), mixed with freshly prepared 1 M NaBH3CN (20 µL) and vortexed. The tryptic peptide solution was split into two aliquots and dimethylated, as desribed above, by reacting one sample with light formaldehyde and the other with heavy formaldehyde; i.e., d0,12C-formaldehyde (4% in water, 25 µL) and d2,13C-formaldehyde (4% in water, 25 µL), respectively. The reaction mixture was then incubated for 2 h at room temperature. Excess formaldehyde was consumed by addition of 20 µL of ammonium hydroxide. After labeling, the solution in each vial was acidified to pH 3.0 by adding 1% TFA.
The isotopic dimethyl-labeled HNE-modified synthetic peptides and apomyoglobin tryptic peptide samples were combined at abundance ratios (light/heavy) of 0.33, 1.0 and 3.0, respectively, desalted using a Sep-Pak C18 solid-phase extraction cartridge (Waters, Milford, MA) and completely dried in a speed vacuum microfuge. The residues were reconstituted in reaction buffer [0.2% (v/v) acetic acid, 10% (v/v) acetonitrile in 89.8% (v/v) water, (pH 3.6)] and subjected to solid-phase enrichment using hydrazide-coated glass beads[27] to selectively capture (as hydrazones) the peptide carbonyls and release from the beads in their original forms, following cleavage of acid labile hydrazone linkage.
Preparation of tryptic digest of human plasma protein containing light and heavy dimethyl-labeled HNE-modified Angiotensin I
Proteins were precipitated from 250 µl (~10 mg proteins) of human plasma by adding 4 volumes of ice-cold acetone and keeping the mixture at −20 °C for 2 h. The sample was centrifuged at 12000 rpm for 10 min at 4 °C and the supernatant was decanted and properly disposed. The residual acetone was evaporated by incubating the pellet at room temperature. For the in-solution digestion, the pellet was solubilized by the addition of 250 µl 8 M urea, and the mixture was vortexed thoroughly. The disulfide bonds of proteins were reduced with freshly prepared dithiothreitol (DTT) and the free thiol groups were then alkylated by reaction with iodoacetamide. The urea concentration in denatured, reduced, and carbamidomethylated plasma protein solution was lowered to <2 M by diluting the sample with 50 mM ammonium bicarbonate and trypsin was added to a final protease:protein ratio of 1:100 (w/w) and allowed to digest ~16 h at 37 °C. Following digestion, proteolytic activity was terminated by acidifying the reaction mixture with acetic acid to pH <3.0 and subjected to solid-phase extraction using Oasis® HLB (hydrophilic-lipophilic balanced) extraction cartridges (Waters Coroporation, Milford, MA). The eluate from the cartridge was evaporated to dryness in an Eppendorf (Westbury, NY) Vacufuge concentrator and resuspended in the reaction buffer. Six hundred micrograms of plasma protein tryptic digests in 200 µl reaction buffer was aliquoted and spiked with 0.8 µg each of light and heavy or 0.8 µg of light and 2.4 µg of heavy or 2.4 µg of light and 0.8 µg of heavy dimethyl-labeled HNE-modified angiotensin I (DRVYIHPFH*L and DRVYIH*PFH*L, where H* indicates HNE-modification of His as Michael adduct), resulting in carbonylated peptide abundance ratios (light/heavy) of 1:1, 1:3 or 3:1, respectively. The sample was then subjected to solid-phase enrichment using hydrazide-coated glass beads as described below.
Solid-phase enrichment of HNE-modified peptides
For capturing isotope-coded peptide carbonyls from the synthetic peptides, apomyoglobin tryptic digests or plasma protein tryptic digests spiked with a dimethyl-labeled modified peptides DRVYIHPFH*L and DRVYIH*PFH*L, 4–5 mg of hydrazide-coated glass beads[27] were added to each of them and the resulting mixture was rotated end-over-end overnight at room temperature. The carbonylated peptides are selectively captured by the hydrazide group in the beads. After centrifugation to settle the beads, the supernatant (labeled as “flowthrough”) was collected, dried by lyophilization, and saved for LC–MS/MS analysis. The beads were subsequently washed four times each with 400 µL of reaction buffer followed by 1 M NaCl, distilled water, 80% (v/v) acetonitrile, and a second round of distilled water to remove the non-carbonylated tryptic peptides. Following stringent washing, the peptide carbonyls (captured as hydrazones) were released from the beads in their original forms with 200 µL of trifluoroacetic acid (90%, v/v) at room temperature for 30 min. This step was repeated one more time and the “eluate” containing the peptide carbonyls were combined and lyophilized. The residues in the flowthrough and eluate fraction were reconstituted in 15–20 µL of loading solvent containing 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in 94.9% (v/v) water. Five µL aliquots were used for LC–ESI-MS/MS analyses.
LC–ESI-MS/MS
LC–ESI–MS/MS analyses were performed using a hybrid linear ion trap-Fourier transform ion cyclotron resonance (7-T) mass spectrometer (LTQ-FT, Thermo, San Jose, CA) equipped with a nano-electrospray ionization source and operated with Xcalibur (version 2.2) and Tune Plus (version 2.2) data acquisition software. Online reversed phase-high performance liquid chromatography (RPLC) was performed with an Eksigent nano-LC-2D (Eksigent, Dublin, CA) system. An amount of 5 µL of the eluate or flowthrough fraction was automatically loaded onto the IntegraFrit™ sample trap (2.5 cm × 75 µm) (New Objective, Woburn, MA), for sample concentration and desalting, at a flow rate of 1.5 µl/min in a loading solvent containing 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in 94.9% (v/v) water prior to injection onto a reverse-phase column (NAN75-15-03-C18-PM; 75 µm i.d. × 15 cm, LC Packings, Sunnyvale, CA) packed with C18 beads (3 µm, 100 Å pore size, PepMap). Mobile-phase buffer A consisted of 0.1% (v/v) acetic acid and 99.9% (v/v) water, and mobile-phase B consisted of 0.1% (v/v) acetic acid and 99.9% (v/v) acetonitrile. Following desalting and injection onto the analytical column, peptides were separated using the following gradient conditions: (1) 5 min in 95.2% solvent A for equilibration; (2) linear gradient to 40% solvent B over 90 min and holding at 40% solvent B for isocratic elution for 5 min; (3) increasing the gradient to 90% solvent B and maintaining for 5 min; and finally (4) 95.2% solvent A in the next 20 min. The flow rate through the column was 250 nL/min. Peptides eluted through a Picotip emitter (internal diameter 10 ± 1 µm; New Objective) were directly supplied into the nano-electrospray source of the mass spectrometer. Spray voltage and capillary temperature during the gradient run were maintained at 2.0 kV and 250 °C. Conventional data-dependent mode of acquisition was utilized in which an accurate m/z survey scan was performed in the FTICR cell followed by parallel MS/MS linear ion trap analysis of the top five most intense precursor ions. FTICR full-scan mass spectra were acquired at 50000 mass resolving power (m/z 400) from m/z 350 to 1500 using the automatic gain control mode of ion trapping. Peptide fragmentation was performed by collision-induced dissociation (CID) in the linear ion trap using a 3.0-Th isolation width and 35% normalized collision energy with helium as the target gas. The precursor ion that had been selected for CID was dynamically excluded from further MS/MS analysis for 60 s.
The ratios of differentially labeled peptides were calculated by manually extracting total ion chromatograms (XICs) of each ion pair and determining ratios of the total area under curve of the first three isotopic clusters of the tagged species observed in the MS survey scan using the Xcalibur software. In all cases, ratios are reported as area under the curve for d0,12C-tagged peptide ion divided by area under the curve for d2,13C-tagged peptide ion. Retention times of the differentially dimethyl-tagged peptide pairs were compared for the corresponding XIC pairs. CID-MS/MS spectra were manually interpreted to increase the confidence in identification of HNE-modified peptides.
The efficiency of the enrichment of stable isotope-labeled HNE-modified angiotensin I from plasma protein tryptic digest by the use of hydrazide-coated glass beads was estimated by the calculation of enrichment factors. This was done by obtaining first peak areas from XICs for three contaminating peptide ions (m/z 671.82, 682.71 and 992.46) along with the combined peak areas of dimethyl-labeled HNE-modified angiotensin I [(M+3H)3+: m/z 494.28 and m/z 496.29] in the samples before enrichment (‘pre-enrichment’) and in the ‘eluate’ fraction. The enrichment factor (f) was calculated for each mixing ratio and each chosen contaminating peptide as follows:
| (1) |
The average of the values was considered an estimation of the (‘fold’) enrichment efficiency for the carbonylated peptide from a complex biological matrix of digested proteins.
Results and Discussion
The strategy of combining stable-isotope labeling with LC–MS/MS is widely being used in quantitative proteomic studies.[17,28,29] The significance of various enrichment techniques for reducing sample complexity and allowing selective isolation and detection of low abundance HNE-modified peptides/proteins have been demonstrated.[27,30–32] Our quantification strategy is based on labeling peptides by reductive dimethylation with isotopomers of formaldehyde, combining the labeled samples, selective fractionation of HNE-peptides using carbonyl-specific hydrazide-reagent immobilized on glass beads[27], and performing LC–MS/MS analysis. For quantification, isotope-coded dimethyl labeling of the peptides was performed before hydrazide chemistry-based enrichment. Unlike the AQUA approach as a possible alternative,[23] the chosen method does not need stable isotope-labeled HNE-modified peptides standards whose reliable synthesis has not been reported.
Principle of quantification
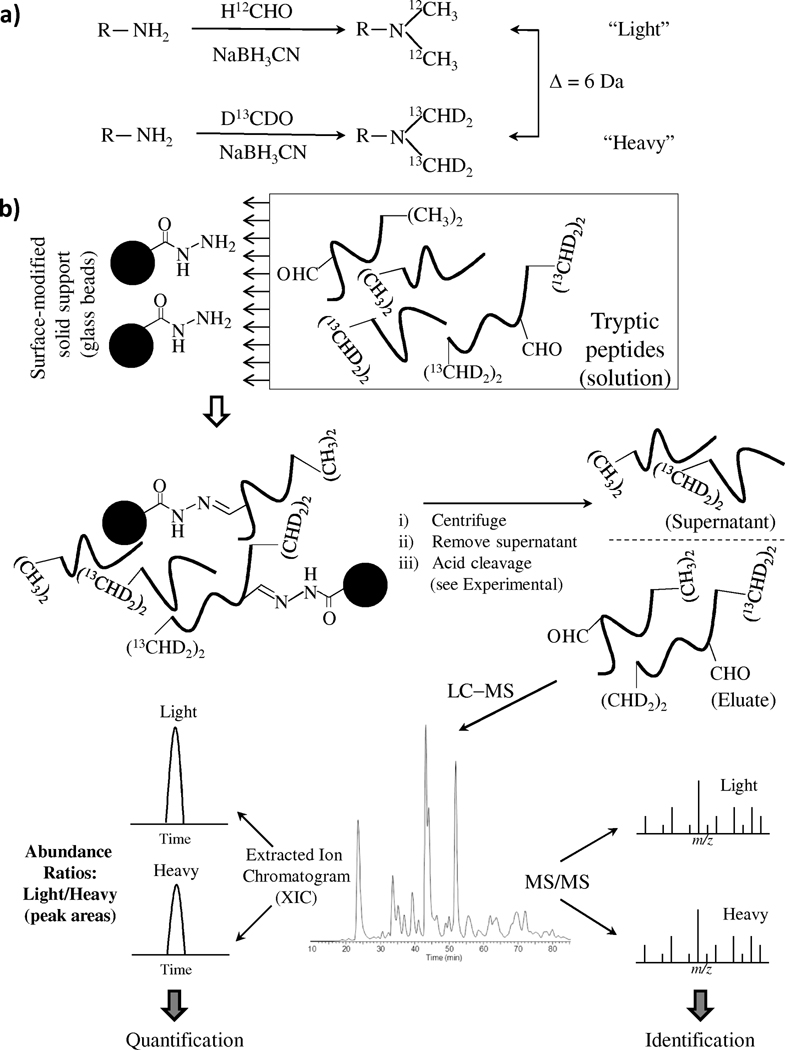
The strategy developed for the quantitative analysis of HNE-carbonylated peptides (and, hence, the corresponding proteins) consists of the steps summarized in Fig 2. First, differential dimethyl labeling of peptide amine groups (N-terminus and ε-amino group of a lysine residues) using either d0,12C- or d2,13C-formaldehyde isotopic variants was performed. Fig. 2(a) shows the reaction scheme of stable isotope dimethyl labeling, where peptide primary amine reacts with formaldehyde to generate a Schiff-base that is rapidly reduced with the addition of cyanoborohydride to the mixture. The labeling adds 28 or 34 Da to each peptide corresponding to the formation of light or heavy dimethylamines and, thus, produces a mass difference of 6 Da per labeling site between the isotopically labeled peptides. However, the cyclic structure of proline (Pro, P) results in the formation of monomethylamine and hence an increase of only 14 Da and 17 Da, respectively, occurs with peptides harboring proline at the N-terminus.[33] In case of tryptic peptides having arginine (Arg, R) at their carboxy-terminus, label is introduced only at the N-terminus and, hence, the mass difference will be 6 Da (3 Da with N-terminal Pro), which is equivalent to the mass difference between the light and heavy isotopic versions of the methyl groups introduced. On the other hand, lysine-terminating (Lys, K) peptides will have two labels, one at the N-terminus and the other at the ε-amino group of the Lys residue. Furthermore, the number of labels will increase with each missed cleavage at this residue. Hence, the peptide ions with more than one potential site of labeling will generally (except for those with N-terminal Pro) have a mass difference (Δ) of n × δ, where n is the number of dimethyl labels in the corresponding peptide fragment and δ is the mass difference between the (CH3)2- and (13CHD2)2-group; i.e., 6 Da. After differential labeling, the light and heavy isotope-labeled peptide samples are combined and the excess labeling reagents were removed by solid-phase extraction. Then, the dimethylated HNE-modified peptides are covalently conjugated to a solid support via hydrazide chemistry in which covalent hydrazone bonds are formed between aldehyde groups in the peptides with hydrazide groups immobilized on a solid support (Fig. 2(b)). Non-carbonylated peptides are washed away, whereas the HNE-modified peptides remain on the solid support and are subsequently released by acid cleavage. Finally, mass spectrometric analysis of the recovered HNE-modified peptide isotopomers for determining the peptide sequence information and location of the modification site from CID-MS/MS data along with pairwise quantification with the XICs of the modified peptides labeled with isotopic tags from full-scan MS spectra are performed.
Figure 2.
Schematic representation of the steps involved in the isotope-coded dimethyl labeling for differential quantification of posttranslational protein modification by 4-hydroxy-2-nonenal (HNE) (a) Reductive methylation reaction scheme of d0,12C-formaldehyde and d2,13C-formaldehyde, corresponding to light and heavy isotopic versions of formaldehyde, with a primary amine group in a peptide. The isotopic mass shift between the isotopically labeled peptide is 6 Da. (b) Solid-phase hydrazide-based enrichment and quantification of isotopic dimethyl-labeled HNE-modified peptides. The relative abundance between the peptide carbonyls can be determined by integrating the peak areas of the extracted ion chromatograms (XIC, lower left) for each isotope-coded peptide; see text for details.
Method development through quantification of isotopically-labeled HNE-modified synthetic peptides labeled in various ratios
We first developed the method and explored the accuracy and reproducibility of dimethyl labeling strategy using an isotopic variant of formaldehyde for quantitative analysis using a commercially available Glu–C fragment of the oxidized insulin β-chain, FVNQHLC#GSHLVE (where # represents cysteine residue oxidized to cysteine-sulfonic acid, Cys-SO3H). Two identical solutions of the peptide were dimethylated using light (H12CHO) or heavy (D13CDO) isotopes of formaldehyde, respectively. Upon labeling, peptides were combined in various molar ratios (1:1, 1:5, 5:1, 1:10 and 10:1), desalted and analyzed by LC–MS for relative quantification of the isotopic pairs from XICs of the doublet ions at m/z 779.87 (2+) and 782.89 (2+), respectively, which corresponds to the light and heavy dimethyl-labeled FVNQHLC#GSHLVE peptide. The peptide was labeled only at its N-terminus as it contained a single amine group and, hence, Δ was 6 Da (3 Th for the doubly charged ions), which was indeed sufficient for obtaining baseline-resolved isotopic envelopes as shown in Supplemental Fig. S1 (see Supporting Information online). Additionally, the dimethylation reaction was complete with no traces of the starting material. Quantification was performed by comparing peak areas of the differentially labeled peptides in XICs based on the monoisotopic peak and the first two 13C-containing isotope peaks. Supplemental Fig. S1 (a) shows that the calculated coefficient of determination (R2) between the mixing ratio and the observed ratio of the XICs of the isotopic pair is greater than 0.99, indicating a practically perfect linear correlation between the expected and observed ratios and substantiating the accuracy of this quantification method.
The influence of dimethyl labeling on CID-based fragmentation of HNE-modified peptides, when compared with the unlabeled peptide, was then studied. The CID-MS/MS spectra of doubly charged unlabeled (m/z 904.02), light (m/z 918.03) and heavy (m/z 921.04) dimethyl-labeled HNE-modified peptide fragment of ATP synthase β-subunit (LVLEVAQH*LGESTVR, from residues 21 to 34 in the protein) is shown in the Supporting Information online (Supplemental Fig. S2). The m/z shifts correspond to mass increments of 28 and 34 Da, respectively, upon labeling at the only available N-terminal site in the peptide. The spectra resulted in nearly identical fragmentation patterns with a characteristic high intensity product ion peak at m/z 78 Th (156 Da for singly-charged peptide) lower than the precursor ion, corresponding to the neutral loss of HNE moiety. Few b-type ions, however, were more pronounced in dimethyl-labeled peptides compared to the unlabeled peptide. For the peptide LVLEVAQH*LGESTVR, the dimethylation at the N-terminal primary amine, resulted in the values of m/z of the labeled b-ions 28 mass units higher for the light-isotope dimethylated and 34 mass units higher for the heavy-isotope dimethylated relative to those obtained for the unlabeled b ions; however, no shift in mass was observed for y-ions. Moreover, it is apparent that CID of dimethylated HNE-modified peptides did not show the evidence of fragmenting the dimethyl group during MS/MS. The presence of [M+2H-HNE]2+ product ion serves as a signature for the existence of the HNE group and the fragment ions y7 (m/z 761), y8 (m/z 1054) and b11 (m/z 1046 for unlabeled, m/z 1074 for light dimethyl-labeled and m/z 1080 for heavy dimethyl-labeled). This indicated the HNE modification to be localized at the histidine (His-8) residue in the peptide. Even though a1 ion signal is enhanced upon dimethylation of peptides due to the formation of quaternary immonium ions that can be used to validate the N-terminal amino acid of the assigned sequence,[34] we did not observe them. This is due to the operating principle of the ion trap instruments, where product ions below a lower limit of the fragment mass range (below about a quarter of the parent ion mass) cannot be detected.[35]
The reliability of dimethylation strategy in quantification of HNE-modified peptides upon enrichment with hydrazide chemistry-based reagents was assessed by using synthetic peptides. Table 1 shows the theoretical and observed peak area ratios of the quantitative labeling of several HNE-modified synthetic peptides. In each of the mixed combinations, the observed relative abundance ratios of the isotopic variants of peptide pairs was very close to the theoretical ratios, indicating the reliability of the method to provide accurate quantitative information.
Table 1.
Observed ratios of dimethyl stable isotope labeling and enrichment of HNE-modified synthetic peptides.Δ
| Peptides | Ratio (Expected 0.33) | Ratio (Expected 1.0) | Ratio (Expected 3.0) |
|---|---|---|---|
| @FVNQH*LC#GSHLVE | 0.31 | 1.00 | 3.09 |
| @DRVYIH*PF | 0.33 | 1.02 | 3.34 |
| @LGFLGSNTPHVNHHMPPH¶ | 0.35 | 1.05 | 3.75 |
| @LVLEVAQH*LGESTVR | 0.33 | 0.99 | 3.12 |
| @IVYGH*LDDPANQEIER | 0.34 | 1.02 | 3.78 |
| @QVQSLTC*EVDALK@ | 0.31 | 0.96 | 2.86 |
| 0.33±0.02 | 1.0±0.03 | 3.32±0.37 |
Dimethyl label,
HNE-modified amino acid residue,
Oxidized cysteine residue
Ratios were given as averages from triplicate measurements.
The neutral loss of HNE from the fragment ions during CID-MS/MS precluded the localization of the site of HNE modification.
A challenging problem is to detect and quantitate carbonylated peptides in a proteome due to their low abundance compared to unmodified analogs. In a proof-of-principle experiment to show that the relative abundance of HNE-modified peptide is maintained when enriched from a complex matrix, different relative concentrations of dimethyl labeled HNE-modified angiotensin I was mixed in a ratio of 1:3, 1:1 or 3:1 and added into a contrived mixture of plasma protein tryptic digest and reisolated from the complex matrix by hydrazide-based chemoprecipitation strategy. A representative base-peak chromatogram and averaged full-scan mass spectrum of plasma protein tryptic digest spiked with differentially dimethyl-labled HNE-modified angiotensin I in a ratio of 3:1 (without employing ennrichment) acquired in the 0–90 min retention time range during LC–ESI-MS analysis is shown in Supplemental Fig. S3 of the Supporting Information online. The mass spectrum displays ions mostly corresponding to peptides from plasma protein, and the relative abundance of dimethyl-labeled HNE-modified angiotensin I is about 2% of that of the base peak. A notable difference was obtained after selective enrichment of HNE-modified peptides via hydrazide-based chemistry, as shown in Supplemental Fig. S4 of the Supporting Information online. An acid-catalyzed hydrolysis of the captured hydrazones (‘eluate’ fraction) and subsequent LC–ESI-MS analysis resulted in the dimethyl-labeled HNE-modified angiotensin I giving the two most intense ions in the mass spectrum obtained by averaging over the entire elution period. The average f calculated for dimethyl-labeled DRVYIHPFH*L according to equation (1) indicated about 4000-fold enrichment (with ±2000 standard error considering three abundant tryptic peptides of plasma proteins in the samples as contaminating peptides), which indicated a high degree of enrichment for the carbonylated peptide.
XICs of DRVYIHPFH*L dimethyl-labeled on the N-terminus with d0,12C-formaldehyde or d2,13C-formaldehyde and modified with the attachment of one ([M+3H]3+: m/z 494.28 and m/z 496.29) or two ([M+3H]3+: m/z 546.32 and m/z 548.33) HNE and obtained upon enrichment are shown in Supplemental Figs. S5 (a) and (b), respectively, of the Supporting Information online. Ratios of the triply charged precursor ions are determined by the total area under these traces and that the observed ratios of the [M+3H]3+ peptide pairs were in close agreement to the expected ratios of 1:3, 1:1 and 3:1. The reduction in the complexity of the sample by the removal of high-abundance unmodified peptides and selective capture of subpopulation of carbonylated peptides with chemoprecipitation by solid-phase hydrazide produce a highly enriched sample containing carbonylated peptides, which is very valuable to the detection and identification of carbonylated peptides. In addition, differential dimethylation with isotopic variant of formaldehyde prior to the capture of carbonylated peptides by hydrazide-modified glass beads allows the quantitation of their relative concentrations. Supplemental Fig. S6 of the Supporting Information online demonstrates the applicability of the method involving solid-phase hydrazide enrichment followed by isotope-coded dimethyl tagging for differential quantification of QVQSLTC*EVDALK, a tryptic peptide carbonylated by HNE on its Cys (C) residue. In general, the strategy could be applicable to any posttranslational modification that introduces a carbonyl group to proteins and, hence, commonly referred to as protein carbonylation.[36]
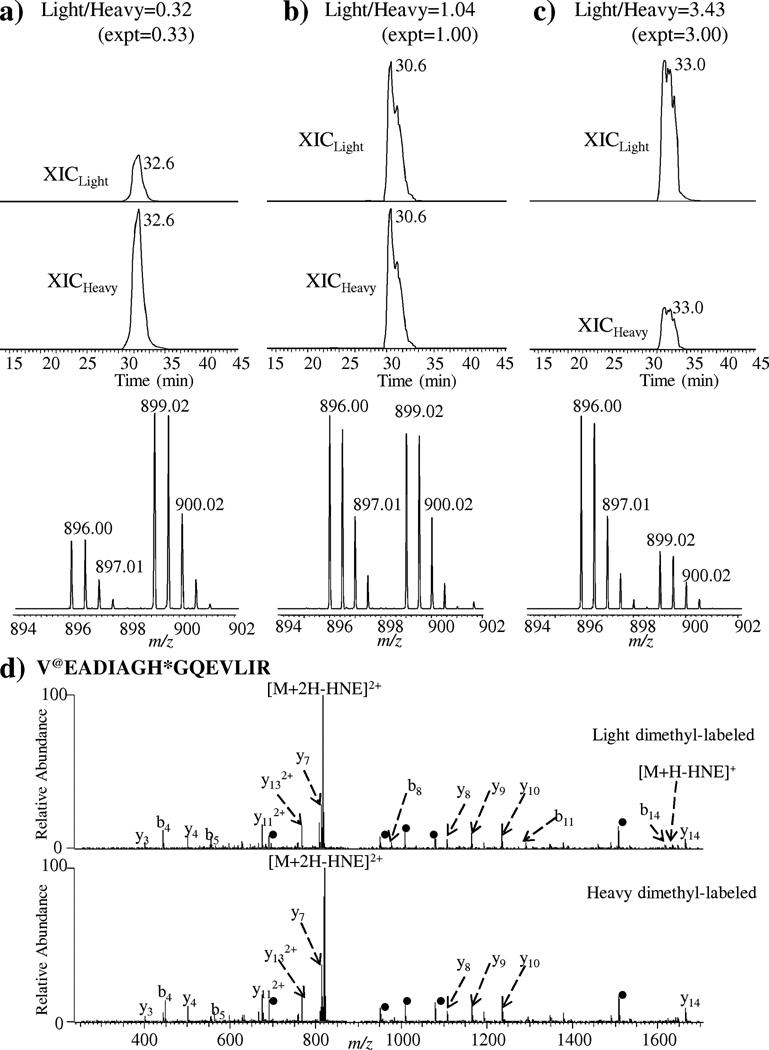
Finally, the feasibility of method summarized in Fig. 2 to permit quantitative analysis of protein carbonylation was demonstrated by following the change in abundance ratios of the tryptic peptide, VEADIAGH*GQEVLIR, derived from HNE-modified apomyoglobin obtained from horse skeletal muscle (Fig. 3). The focus of this study was not to identify the maximum number of peptides in the sample but rather to ensure that the method was quantitatively accurate and reproducible for carbonylated peptides. The susceptibility of His residue in the peptide VEADIAGH*GQEVLIR to Michael addition reaction by HNE has been shown by our group[24] and others.[10,37] The HNE-modified apomyoglobin was digested with trypsin and aliquots of the digest were reacted separately with light and heavy dimethyl isotopomers. Upon labeling, the two aliquots were combined in light to heavy stable isotopic ratios of 1:3, 1:1 and 3:1, and the HNE-modified peptides were fractionated by chemoprecipitation with hydrazide-coated glass beads that can substantially reduce the complexity of the peptide mixture. The eluate fraction, apparently containing enriched dimethyl-labeled HNE-modified apomyoglobin tryptic peptides, was subjected to LC–MS analysis. Figs. 3(a)–3(c) depicts the XICs and mass spectra of doubly charged precursor ions of the HNE-modified apomyoglobin tryptic peptide VEADIAGH*GQEVLIR, labeled at N-terminus with d0,12C-formaldehyde and d2,13C-formaldehyde. The observed ratios of the [M+2H]2+ peptide pairs were in congruence with the theoretical ratios of 1:3, 1:1 and 3:1. The experiment was performed three times to ensure repeatability. Fig. 3(d) shows the CID-MS/MS spectra of [M+2H]2+ ions of light and heavy dimethylated VEADIAGH*GQEVLIR peptide with HNE modification on its His residue (indicated by asterisk). As this peptide has only one site that can be dimethyl labeled, the m/z difference between the isotopically labeled doubly charged species is 3.0. Compared to light isotopes, the b-ions containing the heavy labeled isotopes were 6 mass units (for +1 ion series) higher while the y-ions not containing the label have the same m/z value in the two spectra. The detection of b8 ion at m/z 977.5 (m/z 983.5 for heavy isotope labeled) and the m/z difference between the y7 (at m/z 814.5) and the y8 ion (at m/z 1107.7) localizes the attachment of the HNE moiety to the His-8 residue in the peptide. These results illustrate that the differential dimethyl labeling combined with solid phase enrichment and LC–MS can provide accurate information on stoichiometry of protein carbonylation by enriching and quantifying the relative abundances of HNE-modified isotopic peptide pairs present in the two samples.
Figure 3.
XICs and mass spectra of doubly-charged precursor ions at m/z 896.00 (light dimethyl label) and m/z 899.01 (heavy dimethyl label) mixed in a ratio of (a) 1:3, (b) 1:1, and (c) 3:1 of the HNE-modified peptide, @VEADIAGH*GQEVLIR, derived from trypsin digestion of apomyoglobin obtained from horse skeletal muscle (where, @ = dimethyl label and * = HNE-modified residue). The dimethylation reaction with d0,12C-formaldehyde and d2,13C-formaldehyde occurs on the N-terminal amino-group of this peptide. The ratio of the calculated areas was used to determine the relative concentrations of the carbonylated peptide upon LC–MS analysis. (d) CID-MS/MS spectra of [M+2H]2+ ions of light- and heavy-dimethylated HNE-modified peptide, @VEADIAGH*GQEVLIR. The dot symbol (•) corresponds to the product ions with neutral loss of HNE moiety.
Examination of potential isotope effect on chromatographic separation
For accurate quantification, it is important that the isotopically labeled peptide pairs co-elute from the reversed-phase column. The peptide isotopomers labeled with deuterium isotope tags are frequently resolved in typical RPLC analyses. The stronger nature of the hydrogen bonding compared to deuterium bonding may result in partial resolution of light- and heavy-labeled small- to intermediate-sized peptides (<2500 Da) during RPLC separation.[38–40] The ionization efficiency could vary between the peptide isotopomers when they are chromatographically resolved causing error in calculating relative concentration of isoforms based on integrated peak areas.[38] The isotopic shift, however, is not observed in ions labeled with 12C/13C stable isotopes.[38] In our case, the heavy isotope of formaldehyde for dimethyl labeling includes deuterium and 13C; hence, the XICs of several methylated peptides were examined for difference in retention times between each of the isotopic peptide pairs. The XICs of light- versus heavy-methylated HNE-modified synthetic peptides and tryptic peptides of HNE-modified apomyoglobin (examples shown, along with corresponding MS/MS spectra, in Supplemental Figs. S7–S11 of the Supplementary Information online) indicated no noticeable differences in the elution times of the differentially labeled peptides. This substantiates the study by Ji et al. for quantification of the proteome upon differential dimethyl labeling of the N-termini of peptides by d0,12C- and d2,13C-formaldehyde, performed after guanidinylation to block the labeling at lysine side chains.[22] Regardless whether the peptides incorporated a single dimethyl-tag or multiple dimethyl-tags and, hence, have large mass difference (data not shown), the isotopically labeled peptide pairs had identical retention times during separation in the 15 cm × 75 µm i.d. nano-RPLC column packed with 3-µm C18 beads. The absence or insignificant isotope effect may be due to the proximity of deuterium atoms to the hydrophilic amine groups in the peptides that do not interact strongly with the nonpolar stationary phase and, also, due to the deuterium atoms contributing only 4 Da in a mass shift of 6 Da between a single dimethyl-labeled peptide pair.[22]
Several classes of labeling reagents that modify the N-terminus and ε-amino groups of Lys residues have been described, such as acetic anhydride, N-acetoxysuccinimide or urea, for the quantification of global protein expression. However, acylation of basic amino groups changes the ionic states of peptides and may reduce the ionization efficiency of tryptic digests containing C-terminal lysine residues.[21,41] Hence, we used formaldehyde to universally label the N-terminus and ε-amino groups of lysine residues via reductive amination, as described previously.[21] This labeling reagent preserves the charge state of the modified peptides. Moreover, the mass difference between each light- and heavy-labeled isotopic pair is 6 mass units, which can be resolved even in the triply charged isotopic pair with one labeled site in which the isotopic pair will have a difference in m/z of 2 Th. A minimum mass difference of >4 Da is needed to surpass the interferences from natural isotopes, mostly originating from 13C.[42] A mass difference of 6 Da between light and heavy isotopically labeled peptide pairs eliminates overlap of the isotope envelopes even for peptide pairs with a molecular mass of 3000 Da.[22] Although no apparent distortion of the employed mixing ratios indicating discrimination between the light and heavy formaldehyde reagents to label peptides was seen during the development of the procedure (Fig. 3, Supplemental Fig. S1, and S5–S11), we nevertheless recommend both forward and, then, reverse labeling with samples obtained from an independent experiment to control potential variability upon application of the method.[43]
Few other mass spectrometry-based methods for quantifying HNE-carbonylated proteins, such as the hydrazide-functionalized isotope-coded affinity tag (HICAT)[31] and stable isotopes of Girard’s P reagent (GPR),[44] have been reported. These approaches are based on incorporating a stable isotope-containing affinity reagent in the HNE group that enables enrichment as well as its quantification. The drawback of HICAT-based quantification is that the tag is covalently bound to HNE, making the conjugate bulkier and hence the MS/MS spectrum is complicated by side-chain fragment ions from the HICAT moiety. The presence of nonpeptide ions in the fragment ion spectrum can adversely affect the identity scores of database search algorithms by providing scores that are often lower than those considered to be acceptable for conclusive identification of a peptide. In GPR-based enrichment techniques, basic peptides are also enriched along with HNE-modified peptides. The method is based on enrichment of tagged peptides by strong cation-exchange chromatography due to the presence of a quaternary amine group in the reagent. In our work described above, the HNE group is left intact; instead, the peptide’s primary amine groups are labeled. As the label is not linked with an HNE group, we believe the isotopic dimethyl labeling to be a better quantitative tag for quantification of protein carbonylation, because the corresponding MS/MS spectra do not contain any decomposition product of the dimethyl tag and the intact HNE group can easily be captured reversibly by the solid surface-bound hydrazide. Since dimethylation does not have any effect per se on the CID-based fragmentation pattern of HNE-modified peptides and the intact HNE group in the peptide can be recovered after immobilization on the solid support, this differential quantification strategy permits the use of our previously reported techniques involving neutral loss-driven MS3 acquisition[18] and neutral loss-driven electron capture dissociation[20] that are designed to specifically detect HNE-modified peptides in complex mixtures. Considering HNE modification is substoichiometric, quantification aside, even the detection of modified proteins is challenging. However, implementation of the strategy involving selective isolation of HNE-modified peptides with solid-phase hydrazide chemistry is useful in enhancing the identification of such modified peptides.[20,27] Also, prior labeling of peptides with stable isotopes of dimethyl tags enables the reliable quantification of the differences in the relative carbonylation state of protein obtained from two distinct sources. The necessity of selective enrichment of HNE-modified peptides from native peptides for its detection by MS has been reported.[24] We have shown that only three HNE-modified peptides were detected without enrichment; however, nine HNE-modification sites in apomyoglobin tryptic digest were reported after enrichment by the use of LC–CID-MS/MS and LC–ECD-MS/MS strategy.
The described binary dimethyl labeling strategy has the potential to provide direct comparison about the degree of specific protein carbonylation between two samples (e.g., diseased versus healthy control). This method can also be used for simultaneous differential MS quantification of protein carbonylation in different stages of disease progression by multiplexing to three- or four-sets of isotopic labeling by utilizing isotopomers of formaldehyde (HCHO, DCDO, D13CDO) and reducing agents, sodium cyanoborohydride (NaBH3CN) and sodium cyanoborodeuteride (NaBD3CN).[33,45,46] The variability in mass shift due to Arg-terminating or misscleaved peptides (at Lys residues) may, however, complicate data interpretation in a complex proteome sample. This may be circumvented by protein digestion utilizing endoproteinase Lys–C that can result in at least two labeling sites per peptide, providing sufficient resolution and mass difference for quantitative analysis. Although this study has been focused on quantification of carbonylation by HNE, the strategy should also be generally applicable to protein carbonylation.
Conclusions
In this study, we have designed a strategy to quantify differences in the relative abundance of HNE-carbonylated synthetic peptides by using isotopic variants of formaldehyde to label the N-terminus and ε-NH2 group of Lys residues followed by chemoprecipitation via hydrazide chemistry-based enrichment technique. The accuracy and precision of the technique in LC–MS mode were further evaluated through the analysis of a differentially labeled HNE-modified apomyoglobin. The reductive methylation protocol is simple, fast and completely labels all the primary amines in a peptide. Additionally, the formaldehyde isotopes are commercially available and inexpensive. Since the reaction dimethylates the N-terminus as well as the Lys residues, the tryptic peptides with C-terminal Arg can still be tagged and quantitated. Also, the isotopically labeled peptides co-elute upon RPLC and the mass difference between the heavy and light dimethyl tag (6 Da per label) are wide apart to provide baseline separation and accurate determination of the relative abundance of each form. The results show the feasibility of stable-isotope tagging followed by enrichment of HNE-modified peptides by solid-phase hydrazide chemistry, along with subsequent LC–MS analysis, for quantitative profiling of protein carbonyls in a complex proteome. Furthermore, the method is based on post-isolation stable-isotope labeling of peptides obtained by proteolytic digestion of proteins and is, therefore, not limited to cells and tissues that permit metabolic labeling to afford quantification.
Supplementary Material
Acknowledgements
This research has been supported by grant AG025384 from the National Institutes of Health and by the Robert A. Welch Foundation (endowment BK-0031). We thank Dr. Katalin Prokai-Tatrai for guidance in the synthesis and use of the solid-phase hydrazide beads.
Footnotes
Supporting information
Supporting information may be found in the online version of this article.
References
- 1.Catala A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem. Phys. Lipids. 2009;157:1. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Schneider C, Porter NA, Brash AR. Routes to 4-hydroxynonenal: Fundamental issues in the mechanisms of lipid peroxidation. J. Biol. Chem. 2008;283:15539. doi: 10.1074/jbc.R800001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog. Lipid Res. 2003;42:318. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 4.Carini M, Aldini G, Facino RM. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrom. Rev. 2004;23:281. doi: 10.1002/mas.10076. [DOI] [PubMed] [Google Scholar]
- 5.Aldini G, Dalle-Donne I, Facino RM, Milzani A, Carini M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Med. Res. Rev. 2007;27:817. doi: 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]
- 6.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 7.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: Focus on the HNE and ONE. Drug Metab. Rev. 2006;38:651. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 8.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemsitry of 4-hydroxynonenal. Chem. Res. Toxicol. 1995;8:284. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 9.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem. Res. Toxicol. 2009;22:1499. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem. Res. Toxicol. 2003;16:901. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 11.Butterfield DA, Bader Lange ML, Sultana R. Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer's disease. Biochim. Biophys. Acta. 2010;1801:924. doi: 10.1016/j.bbalip.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed TT, Pierce WM, Markesbery WR, Butterfield DA. Proteomic identification of HNE-bound proteins in early Alzheimer disease: Insights into the role of lipid peroxidation in the progression of AD. Brain Res. 2009;1274:66. doi: 10.1016/j.brainres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Bautista J, Mateos-Nevado MD. Immunological detection and quantification of oxidized proteins by labelling with digoxigenin. Biosci. Biotechnol. Biochem. 1998;62:419. doi: 10.1271/bbb.62.419. [DOI] [PubMed] [Google Scholar]
- 14.Robinson CE, Keshavarzian A, Pasco DS, Frommel TO, Winship DH, Holmes EW. Determination of protein carbonyl groups by immunoblotting. Anal. Biochem. 1999;266:48. doi: 10.1006/abio.1998.2932. [DOI] [PubMed] [Google Scholar]
- 15.Yoo BS, Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis. 2004;25:1334. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 16.Prokai L, Yan LJ, Vera-Serrano JL, Stevens SM, Forster MJ. Mass spectrometry-based survey of age-associated protein carbonylation in rat brain mitochondria. J. Mass Spectrom. 2007;42:1583. doi: 10.1002/jms.1345. [DOI] [PubMed] [Google Scholar]
- 17.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 18.Stevens SM, Rauniyar N, Prokai L. Rapid characterization of covalent modifications to rat brain mitochondrial proteins after ex vivo exposure to 4-hydroxy-2-nonenal by liquid chromatography-tandem mass spectrometry using data-dependent and neutral loss-driven MS3 acquisition. J. Mass Spectrom. 2007;42:1599. doi: 10.1002/jms.1349. [DOI] [PubMed] [Google Scholar]
- 19.Rauniyar N, Prokai L. Detection and identification of 4-hydroxy-2-nonenal Schiff-base adducts along with products of Michael addition using data-dependent neutral loss-driven MS3 acquisition: Method evaluation through an in vitro study on cytochrome c oxidase modifications. Proteomics. 2009;9:5188. doi: 10.1002/pmic.200900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal. Chem. 2009;81:782. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu JL, Huang SY, Chow NH, Chen SH. Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 2003;75:6843. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 22.Ji C, Guo N, Li L. Differential dimethyl labeling of N-termini of peptides after guanidination for proteome analysis. J. Proteome Res. 2005;4:2099. doi: 10.1021/pr050215d. [DOI] [PubMed] [Google Scholar]
- 23.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Nat. Acad. Sci. U. S. A. 2003;100:6940. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rauniyar N, Prokai-Tatrai K, Prokai L. Identification of carbonylation sites in apomyoglobin after exposure to 4-hydroxy-2-nonenal by solid-phase enrichment and liquid chromatography–electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2010;45:398. doi: 10.1002/jms.1725. [DOI] [PubMed] [Google Scholar]
- 25.Aldini G, Vistoli G, Regazzoni L, Benfatto MC, Bettinelli I, Carini M. Water-soluble alpha,beta-unsaturated aldehydes of cigarette smoke induce carbonylation of human serum albumin. Antioxid. Redox Signal. 2010;12:381. doi: 10.1089/ars.2009.2806. [DOI] [PubMed] [Google Scholar]
- 26.Chavez J, Chung W, Miranda CL, Singhal M, Stevens JF, Maier CS. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: Protein carbonylation is diminished by ascorbic acid. Chem. Res. Toxicol. 2010;23:37. doi: 10.1021/tx9002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal. Chem. 2007;79:3747. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 28.Julka S, Regnier FE. Recent advancements in differential proteomics based on stable isotope coding. Brief. Funct. Genom. Proteom. 2005;4:158. doi: 10.1093/bfgp/4.2.158. [DOI] [PubMed] [Google Scholar]
- 29.Regnier FE, Julka S. Primary amine coding as a path to comparative proteomics. Proteomics. 2006;6:3968. doi: 10.1002/pmic.200500553. [DOI] [PubMed] [Google Scholar]
- 30.Kim HY, Tallman KA, Liebler DC, Porter NA. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell. Proteomics. 2009;8:2080. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal. Chem. 2007;79:3342. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 32.Mirzaei H, Regnier F. Enhancing electrospray ionization efficiency of peptides by derivatization. Anal. Chem. 2006;78:770. doi: 10.1021/ac0602266. [DOI] [PubMed] [Google Scholar]
- 33.Hsu JL, Huang SY, Chen SH. Dimethyl multiplexed labeling combined with microcolumn separation and MS analysis for time course study in proteomics. Electrophoresis. 2006;27:3652. doi: 10.1002/elps.200600147. [DOI] [PubMed] [Google Scholar]
- 34.Hsu JL, Huang SY, Shiea JT, Huang WY, Chen SH. Beyond quantitative proteomics: Signal enhancement of the a(1) ion as a mass tag for peptide sequencing using dimethyl labeling. J. Proteome Res. 2005;4:101. doi: 10.1021/pr049837+. [DOI] [PubMed] [Google Scholar]
- 35.Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem. 2001;70:437. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 36.Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, Colombo R, Rossi R, Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: The contribution of redox proteomics. Mass Spectrom. Rev. 2005;24:55. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 37.Fenaille F, Tabet JC, Guy PA. Identification of 4-hydroxy-2-nonenal-modified peptides within unfractionated digests using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2002;74:6298. doi: 10.1021/ac0303822. [DOI] [PubMed] [Google Scholar]
- 38.Zhang R, Regnier FE. Minimizing resolution of isotopically coded peptides in comparative proteomics. J. Proteome Res. 2002;1:139. doi: 10.1021/pr015516b. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE. Controlling deuterium isotope effects in comparative proteomics. Anal. Chem. 2002;74:3662. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- 40.Zhang R, Sioma CS, Wang S, Regnier FE. Fractionation of isotopically labeled peptides in quantitative proteomics. Anal. Chem. 2001;73:5142. doi: 10.1021/ac010583a. [DOI] [PubMed] [Google Scholar]
- 41.Regnier FE, Riggs L, Zhang R, Xiong L, Liu P, Chakraborty A, Seeley E, Sioma C, Thompson RA. Comparative proteomics based on stable isotope labeling and affinity selection. J. Mass Spectrom. 2002;37:133. doi: 10.1002/jms.290. [DOI] [PubMed] [Google Scholar]
- 42.Lill J. Proteomic tools for quantitation by mass spectrometry. Mass Spectrom. Rev. 2003;22:182. doi: 10.1002/mas.10048. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, He J, Ji S, Wang Q, Pu H, Jiang T, Meng L, Yang X, Ji J. Comparative studies of early liver dysfunction in senescence-accelerated mouse using mitochondrial proteomics approaches. Mol. Cell. Proteomics. 2008;7:1737. doi: 10.1074/mcp.M800109-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirzaei H, Regnier F. Identification and quantification of protein carbonylation using light and heavy isotope labeled Girard's P reagent. J. Chromatogr. A. 2006;1134:122. doi: 10.1016/j.chroma.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 45.Boersema PJ, Aye TT, van Veen TA, Heck AJ, Mohammed S. Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics. 2008;8:4624. doi: 10.1002/pmic.200800297. [DOI] [PubMed] [Google Scholar]
- 46.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.