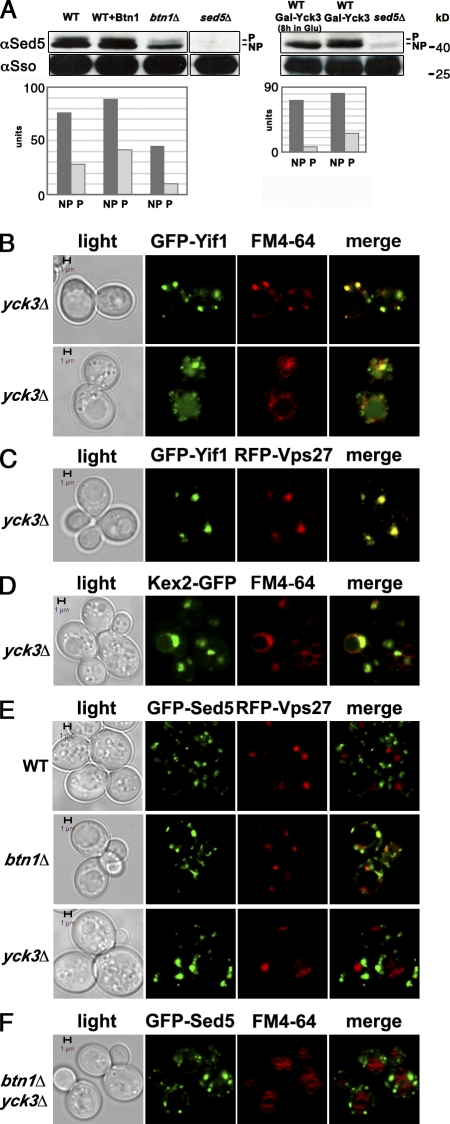
Btn1 controls endosome–Golgi retrograde sorting by regulating SNARE phosphorylation and assembly.
Abstract
The human Batten disease gene CLN3 and yeast orthologue BTN1 encode proteins of unclear function. We show that the loss of BTN1 phenocopies that of BTN2, which encodes a retromer accessory protein involved in the retrieval of specific cargo from late endosomes (LEs) to the Golgi. However, Btn1 localizes to Golgi and regulates soluble N-ethyl-maleimide sensitive fusion protein attachment protein receptor (SNARE) function to control retrograde transport. Specifically, BTN1 overexpression and deletion have opposing effects on phosphorylation of the Sed5 target membrane SNARE, on Golgi SNARE assembly, and on Golgi integrity. Although Btn1 does not interact physically with SNAREs, it regulates Sed5 phosphorylation by modulating Yck3, a palmitoylated endosomal kinase. This may involve modification of the Yck3 lipid anchor, as substitution with a transmembrane domain suppresses the deletion of BTN1 and restores trafficking. Correspondingly, deletion of YCK3 mimics that of BTN1 or BTN2 with respect to LE–Golgi retrieval. Thus, Btn1 controls retrograde sorting by regulating SNARE phosphorylation and assembly, a process that may be adversely affected in Batten Disease patients.
Introduction
Neuronal ceroid lipofuscinoses (NCLs) are autosomal recessive disorders that lead to progressive neurodegeneration and early death in humans (Mole et al., 2005; Kyttälä et al., 2006; Siintola et al., 2006). NCLs are typified by the accumulation of autofluorescent material in the lysosome followed by subsequent neuron loss and result from mutations in genes encoding lysosomal enzymes or transmembrane domain (TMD)–containing proteins of unknown function possibly connected to protein palmitoylation (Phillips et al., 2005; Kyttälä et al., 2006; Siintola et al., 2006; Rakheja et al., 2008; Getty and Pearce, 2011). For example, CLN1, which is involved in infantile NCL, encodes a palmitoyl protein thioesterase (Vesa et al., 1995; Kyttälä et al., 2006). More recently, CLN3, which is involved in juvenile-onset NCL or Batten disease, was suggested to encode a palmitoyl protein Δ-9 desaturase predicted to convert palmitoylated protein substrates into palmitoylated derivatives (Narayan et al., 2006; Rakheja et al., 2008). How defects in these genes lead to neurodegeneration is unknown, and the connection between NCL-related proteins, protein palmitoylation, and identity of the palmitoylated substrates is obscure.
Some genes associated with NCLs are conserved. The best example is CLN3, which has a yeast orthologue called BTN1 (see Table S9 for name description; Pearce and Sherman, 1997; Croopnick et al., 1998). The deletion of BTN1 in Saccharomyces cerevisiae yields defects in vacuolar pH homeostasis, amino acid uptake to the vacuole, vacuolar-type H+–ATPase (V-ATPase)–dependent H+ transport, and nitric oxide signaling and leads to the up-regulation of BTN2, a gene possibly involved in cellular adaptation to the loss of BTN1 (Pearce et al., 1999; Chattopadhyay et al., 2000; Chattopadhyay and Pearce, 2002; Padilla-López and Pearce, 2006; Osório et al., 2007). Although Btn1 is proposed to localize to and act at the level of the vacuole (Croopnick et al., 1998; Pearce and Sherman, 1998, 1999; Pearce et al., 1999; Gachet et al., 2005; Kim et al., 2005; Wolfe et al., 2011), many of these studies used gene overexpression, which often results in protein mislocalization. Recent studies suggest that Btn1 might localize to the Golgi in S. cerevisiae (Vitiello et al., 2010) and Schizosaccharomyces pombe (Codlin and Mole, 2009).
We previously demonstrated that Btn2, which has similarity to the Hook family of coiled-coil proteins associated with endosomal processes (Krämer and Phistry, 1996; Sunio et al., 1999; Walenta et al., 2001; Richardson et al., 2004), localizes to late endosomes (LEs) and mediates the retrieval of specific cargo proteins from LEs to the Golgi (Kama et al., 2007). Btn2 binds to components involved in LE–Golgi protein retrieval, such as elements of the retromer complex (e.g., Vps26) and the Snc1/2 exo/endocytic vesicle SNAREs. Moreover, it forms a complex between a retrieved cargo protein (e.g., Yif1), the endocytic SNARE complex (e.g., Snc1/2, Vti1, Tlg1, and Tlg2), retromer (e.g., Vps5, 17, 26, 29, and 35), and a sorting nexin (e.g., Snx4; Kama et al., 2007). This complex is specific to a subset of LE–Golgi-retrieved proteins because Vps10, the carboxypeptidase Y (CPY) receptor, cannot be coprecipitated with Btn2 and is not mislocalized in btn2Δ cells. This contrasts to mutants defective in retromer, Snx4, or Ypt6 (a Rab involved in LE–Golgi transport) that mislocalize both Vps10 and CPY. Thus, multiple routes may be involved in LE–Golgi protein retrieval, and Btn2 may be specific to one (Kama et al., 2007). Interestingly, Btn2 also facilitates prion curing, indicating a potential role in the targeting of misfolded protein aggregates for degradation (Kryndushkin et al., 2008). Recently, we demonstrated that Btn2 can localize to the insoluble protein deposit compartment (Bagola and Sommer, 2008; Kaganovich et al., 2008) thought to be involved in prion sequestration and identified Btn3 as a negative regulator of Btn2-mediated LE–Golgi transport and prion curing (Kanneganti et al., 2011).
Given the connection between Btn2 and LE–Golgi transport, we examined whether Btn1 acts upon protein retrieval. We find that BTN1 encodes a Golgi protein involved in the regulation of SNARE phosphorylation and assembly and controls LE–Golgi and intra-Golgi protein sorting. The deletion of BTN1 phenocopies the loss of BTN2, resulting in the mislocalization of Yif1 and Kex2, and can be complemented by the heterologous expression of human CLN3. This suggests that the biochemical function of Btn1/Cln3 is conserved. Specifically, BTN1 overexpression or deletion has opposing effects upon SNARE assembly and Golgi morphology, which are mediated through Sed5, a SNARE whose control of retrograde protein trafficking and Golgi morphology is sensitive to phosphorylation (Weinberger et al., 2005). Btn1 appears to regulate the phosphorylation of Sed5 via Yck3, a palmitoylated kinase involved in Golgi–vacuole transport (Sun et al., 2004) and whose deletion phenocopies btn1Δ or btn2Δ cells. The ability of Btn1 to regulate kinase function may involve modulation of the palmitate lipid anchor, as substitution of the cysteine acceptor region in Yck3 with a TMD suppresses the loss of BTN1. Thus, Btn1 controls a membrane-anchored kinase that functions in Golgi–endosome transport.
Results
The deletion of BTN1 results in the mislocalization of Yif1 and Kex2
The loss of BTN2 results in the mislocalization of a trans-Golgi protein, Yif1, to the vacuole (Chattopadhyay et al., 2003; Kama et al., 2007), where it is degraded (Kama et al., 2007). As BTN2 is up-regulated in the absence of BTN1 and because Btn2 acts upon LE–Golgi sorting, we determined whether Btn1 is involved in transport. We examined the localization of GFP-tagged Yif1 in wild-type (WT) cells or cells lacking either BTN1 or BTN2 (Fig. 1 A). In WT cells, GFP-Yif1 labeled small punctate structures that correspond to Golgi (Matern et al., 2000) and did not colabel with FM4-64 (Fig. 1 A and Table S1), a dye that labels endosomes and then vacuoles. In contrast, GFP-Yif1 mislocalized to vacuoles in btn2Δ cells, as previously shown (Chattopadhyay et al., 2003; Kama et al., 2007), and to FM4-64–labeled compartments situated adjacent to the vacuole (along with limited vacuolar labeling) in btn1Δ cells (Fig. 1 A and Table S1). The nonvacuolar compartments labeled by GFP-Yif1 in btn1Δ cells are likely to be LEs, as they colabeled with RFP-tagged Vps27 (Fig. 1 B), a protein involved in the multivesicular body (MVB) pathway (Piper et al., 1995). Thus, the deletion of BTN1 affects Yif1 localization in a manner similar to the deletion of BTN2. Importantly, Yif1 localization to the Golgi was almost fully restored to btn1Δ cells by expression of the human CLN3 gene (Fig. 1 A [bottom] and Table S1). Thus, the function of these orthologues may be conserved.
Figure 1.
The deletion of BTN1 results in defects in LE–Golgi sorting. (A) Yif1 is mislocalized to LEs in cells lacking BTN1. (top) WT (BY4741), btn1Δ, and btn2Δ cells expressing GFP-Yif1 from a single-copy plasmid were labeled with FM4-64 and visualized, as described in Materials and methods. Merge indicates merger of the GFP and FM4-64 (i.e., RFP) panels. Light indicates the DIC panel. The third row illustrates endosomal labeling seen with Yif1 in the majority of btn1Δ cells, whereas the fourth row illustrates vacuolar labeling. (bottom) btn1Δ cells expressing GFP-Yif1 from a single-copy plasmid and Cln3 from a multicopy plasmid; cells were labeled as previously described. (B) Yif1 localizes to Vps27-labeled endosomes in btn1Δ cells. WT and btn1Δ cells expressing GFP-Yif1 from a single-copy plasmid and RFP-Vps27 from a multicopy plasmid are shown. (C) Kex2 is mislocalized to LEs in cells lacking BTN1. (top) WT, btn1Δ, and btn2Δ cells expressing Kex2-GFP from a single-copy plasmid are shown. (bottom) btn1Δ cells expressing Kex2-GFP from a single-copy plasmid and Cln3 from a multicopy plasmid are shown. Bars, 1 µm.
Next, we examined whether GFP-tagged Kex2, a protease retrieved to the trans-Golgi via LEs in WT cells (Brickner and Fuller, 1997), is mislocalized in btn1Δ cells (Fig. 1 C). Kex2 accumulates at LEs in btn2Δ cells, and its mislocalization results in a loss in the secretion of active α-mating factor (Kanneganti et al., 2011). We found that Kex2-GFP accumulates in large LE-like structures that label with FM4-64 and are situated adjacent to the vacuole in both btn1Δ and btn2Δ cells (Fig. 1 C and Table S2). This indicates that Kex2 is not efficiently retrieved to the Golgi in the absence of either Btn1 or Btn2. Interestingly, Kex2 also relocalized to the Golgi when the CLN3 was overexpressed in btn1Δ cells (Fig. 1 C and Table S2). Together, these results suggest that both Btn1 and Btn2 act upon LE–Golgi protein sorting.
To show specificity, we examined the localization of other proteins that undergo sorting on the secretory pathway in cells lacking BTN1 (Fig. S1 A). These proteins (see Table S9) included GFP-tagged Snx4 (Hettema et al., 2003), Vps27, Vps10 (Deloche and Schekman, 2002), Tlg1 (Coe et al., 1999), Tlg2 (Abeliovich et al., 1998; Holthuis et al., 1998), Snc1 (Protopopov et al., 1993; Gurunathan et al., 2000), Fur4 (Bugnicourt et al., 2004), Ste2 (Stefan and Blumer, 1999), and CPY. However, we did not observe significant changes in their localization when expressed (from single-copy plasmids) in btn1Δ cells in comparison with WT cells (Fig. S1 A). This result was identical to that observed for btn2Δ cells (Kama et al., 2007). We also examined CPY secretion, which occurs as a result of defects in vacuolar protein sorting. Although CPY was detected on filters where cells lacking VPS10 or VPS26 (which encode retromer components; Seaman et al., 1998) were grown, it was not detected where either WT or btn1Δ cells were grown (Fig. S1 B). Thus, in contrast to Yif1 or Kex2, neither CPY sorting nor the localization of other trafficked markers was altered in the absence of BTN1.
To verify the imaging data, we examined properties of these proteins that might support the idea that Yif1 and Kex2 are mislocalized. First, we examined the stability of GFP-Yif1 by Western analysis (Fig. S1 C), as the robust accumulation of free GFP was observed in cells lacking BTN2 but not in WT cells (Kama et al., 2007). Upon quantification, we found that the level of free GFP in btn1Δ cells was twofold that of WT cells but only half as much as that seen in control btn2Δ cells (unpublished data). Thus, Yif1 does not undergo the same level of degradation in btn1Δ cells as in btn2Δ cells, but this result parallels the microscopy data showing that 16% of btn1Δ cells have vacuolar labeling, whereas WT and control btn2Δ cells show 7 and 32% vacuolar labeling, respectively (Table S1). Next, we examined Kex2 function in α-factor maturation by scoring halo formation on lawns of sst2Δ tester cells. We found that the halo size (i.e., which delimits the area of mature and active secreted α-factor) was significantly reduced in the presence of either btn1Δ cells or btn2Δ cells in contrast to WT cells (e.g., the halo diameter was 7.5 mm for the spotted btn1Δ and btn2Δ cells vs. 10 mm for WT cells; n = 2; Fig. S1 D). Thus, Kex2 function is reduced in btn1Δ cells, suggesting that it localizes to a compartment that is unable to access immature α-factor.
Btn1 localizes to the Golgi and is retrieved from LEs in a Btn2-dependent manner
The intracellular localization of Btn1 is unclear; GFP-tagged Btn1 localizes to the vacuole upon overexpression (i.e., from multicopy plasmids; Croopnick et al., 1998; Pearce and Sherman, 1998, 1999; Pearce et al., 1999; Gachet et al., 2005; Kim et al., 2005) or to the Golgi in S. cerevisiae (Vitiello et al., 2010) and S. pombe (Codlin and Mole, 2009) at lower levels of expression. We examined the localization of Btn1 tagged at the amino terminus with GFP to avoid masking a potential ER export sequence present at the carboxy terminus (position 353–357; LNILE) and expressed it from the genome or single-copy plasmids. GFP-Btn1 expressed from the chromosome under a GAL promoter, constitutively from a single-copy plasmid via an ADH1 promoter, or endogenously under its own promoter localized to numerous small punctate structures that did not colocalize with FM4-64 (Fig. 2 A, top, middle, and bottom rows, respectively). These structures are likely to be trans-Golgi, as GFP-Btn1 colocalized with either RFP-tagged Yif1 or Sec7 (Fig. 2 B), which are both trans-Golgi markers. In contrast, GFP-Btn1 did not overlap with Btn2-RFP (Fig. 2 C), which localizes to LEs (Kama et al., 2007). Thus, in contrast to experiments that show Btn1 to be vacuolar, we observe Btn1 as a Golgi protein. Moreover, GFP-Btn1 localization to the Golgi appears to be mediated by LE–Golgi protein retrieval, as GFP-Btn1 was mislocalized to the vacuole in cells lacking BTN2 (Fig. 2 D), as with Yif1 or Kex2 (Fig. 1, A and C). However, we note that GFP-BTN1 overexpression from multicopy plasmids indeed leads to labeling of the vacuole membrane (unpublished data).
Figure 2.
Btn1 localizes to the Golgi in a manner dependent on Btn2. (A) Btn1 does not colocalize with FM4-64–labeled compartments. (top) WT cells expressing GFP-tagged BTN1 from its genomic locus under the control of a GAL promoter (GAL integrated [GAL int]) were grown on galactose-containing medium and were labeled with FM4-64. (middle) WT cells expressing GFP-BTN1 constitutively from a single-copy plasmid (CEN plasmid) were grown on glucose-containing medium and were labeled with FM4-64. (bottom) WT cells expressing GFP-BTN1 from its genomic locus and under the control of the BTN1 promoter were grown on glucose-containing medium and were labeled with FM4-64. (B) Btn1 colocalizes with Yif1. WT cells expressing GFP-BTN1 from its genomic locus under the control of a GAL promoter and either RFP-YIF1 from a single-copy plasmid (top row) or SEC7-DsRed from a chromosomal integration (bottom row) were grown on galactose-containing medium and visualized. (C) Btn1 and Btn2 do not colocalize. WT cells expressing GFP-BTN1 from its chromosomal locus under the control of a GAL promoter and BTN2-RFP from a single-copy plasmid are shown; cells were grown on galactose-containing medium. (D) Btn1 is mislocalized to the vacuole in btn2Δ cells. btn2Δ cells expressing GFP-BTN1 from a single-copy plasmid were grown on glucose-containing medium and were labeled with FM4-64. Bars, 1 µm.
BTN1 overexpression or deletion affects the growth of SNARE mutants
Previously, we demonstrated that BTN2 overexpression ameliorates the growth of vti1-11 cells (Kama et al., 2007; Kanneganti et al., 2011), which are defective in Golgi to LE/MVB transport at elevated temperatures (Fischer von Mollard and Stevens, 1999). We examined the effect of BTN1 overexpression or deletion in mutants of the secretory pathway and found that overexpression inhibited the growth of cells bearing temperature-sensitive alleles of VTI1, YKT6, and SED5 (Fig. S2, A–C), which encode SNAREs involved in endosome–Golgi and intra-Golgi protein sorting. This is specific, as BTN1 overexpression had little to no effect on the growth of cells bearing a temperature-sensitive allele of SEC21, which encodes a COPI component involved in retrograde Golgi–ER transport; SEC22, which encodes an ER–Golgi SNARE; and either SEC1 or SEC9, which encode an exocytic SNARE regulator and SNARE, respectively (Fig. S2 D). Thus, the deleterious effects of BTN1 overexpression are limited to cells that bear mutations in genes involved in endosome–Golgi and intra-Golgi transport. Correspondingly, the deletion of BTN1 ameliorated growth defects seen in ykt6-1 cells at elevated temperatures (Fig. S2 B). No effect by either the overexpression or deletion of BTN1 was observed in WT cells (Fig. S2 E).
BTN1 overexpression and deletion affect the assembly of Golgi SNAREs
As Btn1 overproduction impedes the functioning of SNAREs specific to Golgi transport steps (e.g., Ykt6 and Sed5; Fig. S2), we examined SNARE assembly in cells either overexpressing or lacking BTN1. First, we immunoprecipitated an myc epitope–tagged temperature-sensitive form of Ykt6 (e.g., Ykt6-1; see Materials and methods) from WT and either BTN1-overexpressing or btn1Δ cells. In WT cells grown at permissive temperatures, we found that Ykt6-1 readily formed complexes with Sed5 and other SNAREs involved in ER–Golgi transport (e.g., Bet1 and Bos1), intra-Golgi transport (e.g., Sft1 and Gos1), and endosome–Golgi transport (e.g., Tlg1, Tlg2, and Vti1) but not with Sso1/2 or Snc1/2, which act primarily upon exocytosis (Fig. 3 A). In contrast, BTN1 overexpression greatly reduced Ykt6-1 binding to Sed5 as well as to Vti1 and Tlg1 (Fig. 3 A). This indicates that SNARE partners involved in endosome–Golgi transport were less able to form complexes with Ykt6 upon BTN1 up-regulation.
Figure 3.
Overexpression or deletion of BTN1 has opposing effects on Golgi SNARE assembly. (A) BTN1 overexpression or deletion affects SNARE partnering with Ykt6. WT and btn1Δ cells expressing myc-tagged Ykt6-1 from a single-copy plasmid and WT cells expressing both myc-tagged Ykt6-1 from a single-copy plasmid and HA-tagged Btn1 from a multicopy plasmid were processed for IP with anti-myc antibodies. Samples of the TCL were separated by SDS-PAGE in parallel to the IP samples. Proteins were detected using antibodies to SNAREs as well as the HA and myc epitopes to detect Btn1 and Ykt6-1, respectively. Note that Ykt6 did not precipitate Sso, Snc, or Btn1. (B) BTN1 overexpression or deletion affects SNARE partnering with Sed5. Control WT and btn1Δ cells expressing HA-Sed5 from a single-copy plasmid and WT cells overexpressing myc-Btn1 from a multicopy plasmid were grown and processed for IP. Samples of the TCL and the IP precipitates were electrophoresed and detected in Western blots, as previously described. Note that Sed5 did not precipitate Tlg2, Sso1/2, or Btn1.
As Sed5 is an essential SNARE involved in Golgi transport (Banfield et al., 1994) and whose binding to Ykt6 was influenced by Btn1 (Fig. 3 A), we examined the effect of BTN1 overexpression or deletion on the ability of Sed5 to assemble into SNARE complexes. We immunoprecipitated an HA-tagged form of Sed5 from WT and either BTN1-overexpressing or btn1Δ cells. In WT cells, Sed5 bound weakly to the Vti1 and Tlg1 endosome–Golgi transport SNAREs, the Ykt6, Gos1, and Sft1 intra-Golgi transport SNAREs, and the Bos1 ER–Golgi SNARE (Fig. 3 B). In contrast, BTN1 overexpression greatly reduced, if not eliminated, SNARE binding to Sed5, whereas, correspondingly, the deletion of BTN1 greatly enhanced binding (Fig. 3 B). This result is specific to Vti1, Tlg1, Ykt6, Gos1, Sft1, and Bos1, as Sed5 did not bind to the Tlg2, Sso1/2, or Snc1/2 SNAREs under any condition. Thus, the levels of Btn1 appear to control Sed5 assembly into specific SNARE complexes.
BTN1 overexpression or deletion modulates Golgi clustering
Previous work revealed that Sed5 undergoes phosphorylation at serine 317, which is adjacent to the TMD (Weinberger et al., 2005). A constitutive nonphosphorylated (NP) form of Sed5 (e.g., Sed5317A) conferred normal Golgi function in terms of protein export and retrograde sorting events and readily entered into COPI vesicles but greatly enhanced Golgi clustering (Weinberger et al., 2005). In contrast, a phosphomimicking mutant (e.g., Sed5317D) inhibited retrograde protein trafficking within the Golgi (i.e., leading to Kar2 secretion, defects in Gas1 processing, and Sec22 mislocalization) and cell growth and induced Golgi vesiculation (Weinberger et al., 2005). Because Sed5 phosphorylation regulates Golgi morphology and function, we examined whether BTN1 overexpression or deletion has similar effects.
First, we examined the effect of BTN1 overexpression or deletion in WT cells expressing GFP-tagged Sed5, Sed5317A, or Sed5317D from single-copy plasmids (Fig. 4; see Table S3 for the percentage of cells having Sed5 in dispersed/clustered Golgi). In cells expressing GFP-tagged Sed5, which yielded numerous variably sized puncta (Fig. 4 A, top row; Weinberger et al., 2005), we observed that the deletion of BTN1 led to an enhancement of their size (i.e., increased clustering; Fig. 4 A, bottom row). In cells expressing Sed5317A (Fig. 4 B, top row), which were previously shown to have greatly enlarged Golgi puncta (Weinberger et al., 2005), the deletion of BTN1 yielded even larger (although typically fewer) puncta (i.e., superclustering; Fig. 4 B, bottom row). Similar results were observed in btn1Δ cells expressing Sed5317D (Fig. 4 C, bottom row), although these puncta were smaller than in the other cells lacking BTN1 (Fig. 4, A and B [bottom rows]), probably as a result of the enhanced vesiculation exerted by Sed5317D (Weinberger et al., 2005). Conversely, BTN1 overexpression led to smaller and more dispersed puncta in all cell types (Fig. 4, A–C [middle rows]). Thus, Btn1 levels greatly modulate Golgi clustering. The notable changes in size and number of Golgi puncta likely result from an effect that Btn1 has upon native Sed5 that is present in the WT background (and expressed from the plasmid in Fig. 4 A).
Figure 4.
BTN1 overexpression or deletion has opposing effects on Golgi morphology. (A) Deletion of BTN1 enhances Golgi clustering in cells expressing GFP-Sed5. WT, btn1Δ, and WT cells overexpressing BTN1 from a multicopy plasmid (BTN1) and all expressing GFP-Sed5 from a single-copy plasmid are shown. Note the large puncta in btn1Δ cells. (B) Deletion of BTN1 enhances Golgi clustering in cells expressing GFP-Sed5317A, whereas BTN1 overexpression enhances fragmentation (same as in A, except all strains express GFP-Sed5317A). Note the large puncta in WT and btn1Δ cells. (C) Golgi clustering is reduced in btn1Δ cells expressing GFP-Sed5317D (same as in A, except all strains express GFP-Sed5317D). (D) Deletion of BTN1 enhances Golgi clustering in cells expressing GFP-Sed5 from its genomic locus (GFP-Sed5 integrated [int.]). WT, btn1Δ, and WT cells overexpressing BTN1 from a multicopy plasmid (BTN1) and all expressing GFP-SED5 from its genomic locus are shown. Note the larger puncta in btn1Δ cells. (E) BTN1 overexpression reduces Golgi clustering in cells expressing GFP-Sed5317A from its genomic locus (same as in D, except all cells express GFP-SED5317A). Note the reduction in puncta size in cells overproducing Btn1. (F) Deletion of BTN1 enhances Golgi clustering in cells expressing GFP-Sed5317D from its genomic locus (same as in D, except all cells express GFP-SED5317D). Note the enhancement in puncta size in cells lacking BTN1. Bars, 1 µm.
Second, we examined the effect of BTN1 overexpression or deletion in WT cells expressing GFP-tagged Sed5, Sed5317A, or Sed5317D from their genomic loci (i.e., as the sole copy of SED5; Fig. 4, D–F). We observed similar results as those seen with the plasmid-expressed forms of Sed5 (Fig. 4, A–C), although we note that the size of the Golgi clusters formed in the presence of Sed5317A (Fig. 4 E) and/or in the absence of BTN1 (Fig. 4, D–F [bottom rows]) was not as dramatic as that observed with plasmid-based SED5 overexpression (Fig. 4, A–C). This connection between SED5 expression levels and Golgi size was previously observed (Weinberger et al., 2005).
Btn1 regulates the phosphorylation state of Sed5
Because the deletion of BTN1 enhances Golgi size (Fig. 4), which corresponds to the NP form of Sed5 (Weinberger et al., 2005), and BTN1 overexpression has an opposite effect that corresponds with phosphorylated (P) Sed5 (Weinberger et al., 2005), we examined Sed5 phosphorylation in cells lacking or overexpressing BTN1 by Western blotting with anti-Sed5 antibodies. Cell extracts from WT, BTN1-overexpressing, and btn1Δ cells were resolved on 11.5% acrylamide gels and were probed with anti-Sed5 antibodies to reveal the lower (NP) and higher (P) molecular mass forms (Weinberger et al., 2005). As previously seen, Sed5 can exist in the P form in WT cells (Weinberger et al., 2005), and this signal appeared somewhat stronger (≥15%) in WT cells overexpressing BTN1 than in WT cells (see representative experiment shown in Fig. 5 A, top and bottom left). More strikingly, the P form of Sed5 was greatly reduced in btn1Δ cells, and quantification revealed that its levels were three- to fourfold lower than in WT cells after normalization for loading (Fig. 5 A, bottom left), although a small reduction in the levels of Sed5 protein was also apparent. The NP/P ratio for Sed5 in WT cells, WT cells overexpressing BTN1, and btn1Δ cells was 2.7:1, 2.2:1, and 5:1 in this representative experiment (NP/P ratio for btn1Δ cells was 7.2 ± 1.7:1 in four experiments; see Figs. 6 (C and F) and S5 A for similar results). Thus, excess Btn1 appears to enhance Sed5 phosphorylation, whereas its absence greatly enhances the NP state. These results could account for the changes in the aforementioned Golgi morphology observed (Fig. 4).
Figure 5.
BTN1 overexpression and deletion have opposing effects on Sed5 phosphorylation. (A) Sed5 is underphosphorylated in cells lacking BTN1 or YCK3. (left) WT cells, WT cells overexpressing BTN1 from a multicopy plasmid (BTN1), and btn1Δ cells were grown on glucose-containing medium. In parallel, cells expressing SED5 under a galactose-inducible promoter (GAL-SED5) were grown on galactose-containing medium and were shifted to glucose-containing medium for 16 h (to yield the sed5Δ condition). Cells were processed for Western analysis and probed with anti-Sso antibodies to detect Sso1/2 (as a loading control), whereas anti-Sed5 antibodies were used to detect Sed5. NP indicates the lower molecular mass/NP form of Sed5, whereas P indicates the higher molecular mass/P form. The histogram shows quantification (in arbitrary units) of the upper (P) and lower bands (NP) of Sed5 after normalization for Sso loading. (right) Cells expressing YCK3 under a GAL promoter (GAL-YCK3) were grown on galactose-containing medium and were shifted to glucose-containing medium for 8 h (to yield the yck3Δ condition) or were maintained on galactose-containing medium as a control. In parallel, cells expressing SED5 under a galactose-inducible promoter (GAL-SED5) were grown on galactose-containing medium and were shifted to glucose-containing medium for 16 h (to yield the sed5Δ condition). Cells were processed for Western analysis, and both Sed5 and Sso were detected as previously described. Note the presence of only the NP form of Sed5 (NP) in btn1Δ cells and in GAL-YCK3–expressing cells grown on glucose-containing medium. Also, note lack of Sed5 in GAL-SED5 cells grown on glucose-containing medium. The histogram shows quantification of the upper (P) and lower bands (NP) after normalization. The data shown are representative of multiple replicates of the experiment (n = 4). (B) Yif1 is mislocalized in the absence of Yck3. Yeast lacking YCK3 (yck3Δ) and expressing GFP-Yif1 from a single-copy plasmid were labeled with FM4-64. The top row depicts an endosomal pattern of labeling, whereas the bottom row depicts endosomal and vacuolar labeling. (C) Yif1 is localized to LEs in cells lacking YCK3. yck3Δ cells expressing GFP-Yif1 and RFP-Vps27 from single-copy plasmids are shown. (D) Kex2 is mislocalized in the absence of Yck3. yck3Δ yeast expressing Kex2-GFP from a single-copy plasmid and labeled with FM4-64 are shown. (E) The deletion of YCK3 enhances Golgi size. WT, btn1Δ, and yck3Δ cells expressing GFP-Sed5 and RFP-Vps27 from single-copy plasmids are shown. Note that the GFP-Sed5 and RFP-Vps27 signals do not overlap. (F) The double deletion of BTN1 and YCK3 does not further enhance Golgi size. btn1Δyck3Δ cells expressing GFP-Sed5 from a single-copy plasmid are shown. Bars, 1 µm.
Figure 6.
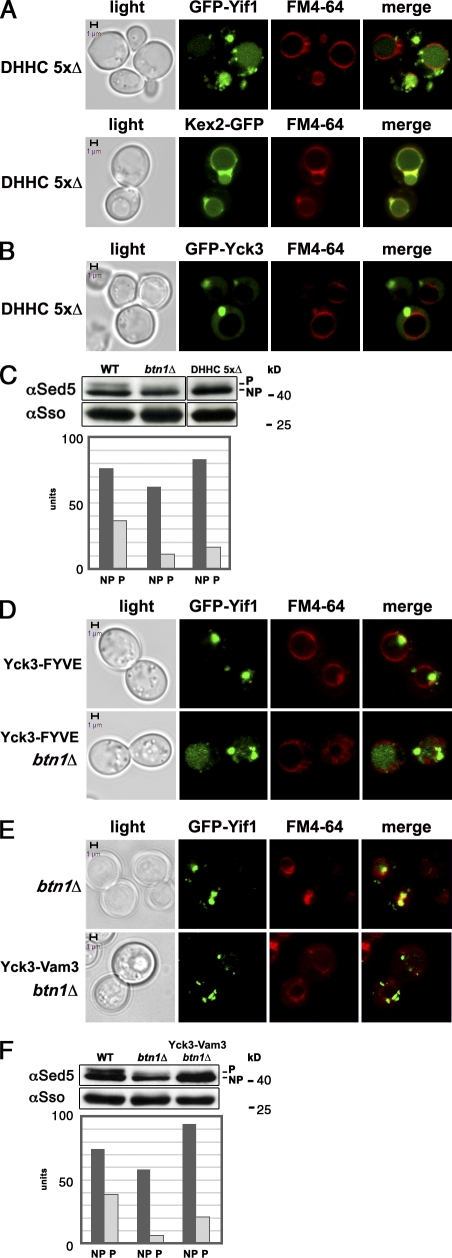
Yck3 palmitoylation is required for LE–Golgi sorting but can be obviated upon substitution with a TMD. (A) Palmitoyltransferase activity is required for LE–Golgi retrieval. Yeast lacking five DHHC palmitoyltransferases (DHHC 5×Δ) and expressing either GFP-YIF1 (top) or KEX2-GFP (bottom) are shown. Note the localization of Yif1 to LE and vacuole and Kex2 to the vacuole membrane. (B) Yck3 is mislocalized to LEs in cells depleted of palmitoyltransferases. DHHC 5×Δ yeast expressing GFP-YCK3 are shown. (C) Depletion of DHHC palmitoyl transferases reduces Sed5 phosphorylation. WT, btn1Δ, and DHHC 5×Δ cells were processed for Western analysis with anti-Sed5 and -Sso antibodies, as described in Fig. 5 A. NP and P indicate the NP and P form of Sed5, respectively. The histogram shows quantification of the Sed5 bands after normalization. The data shown are representative of multiple replicates of the experiment (n = 3). (D) Yif1 is mislocalized to LEs in yck3Δ cells expressing a Yck3-FYVE fusion protein. WT and btn1Δ cells expressing both YCK3-FYVE from the YCK3 locus and GFP-YIF1 from a single-copy plasmid are shown. (E) Expression of Yck3 fused to a TMD bypasses the Btn1 requirement for Yif1 localization to the Golgi. btn1Δ cells and btn1Δ cells expressing YCK3-VAM3 from the YCK3 locus were transformed with a plasmid expressing GFP-Yif1, labeled with FM4-64, and examined. Note the localization of Yif1 to LEs in btn1Δ cells and to smaller puncta (that do not colabel with FM4-64) in cells expressing Yck3-Vam3. (F) Expression of Yck3-Vam3 partially restores Sed phosphorylation. WT, btn1Δ, and btn1Δ cells expressing YCK3-VAM3 were processed for Western analysis and Sed5 quantification, as described in Fig. 5 A. The data shown are representative of multiple replicates of the experiment (n = 2). Bars, 1 µm.
Loss of Yck3, a palmitoylated protein kinase, leads to defects in LE–Golgi retrieval and Golgi clustering
Btn1 encodes a multipass membrane-spanning protein that is not a kinase; thus, we determined how it could influence Sed5 phosphorylation and changes in Golgi morphology. Because human CLN3 was proposed, but not independently verified, to encode a palmitoyl protein Δ-9 desaturase of unknown consequence (Narayan et al., 2006, 2008), we hypothesized that it might act upon palmitoylated proteins that control Sed5 function. Although several palmitoylated proteins function at the Golgi–endosome in yeast, Yck3 and Hrr25 constitute an essential pair of palmitoylated endomembrane-associated kinases (Wang et al., 1996) that could potentially regulate Sed5 phosphorylation. Yck3 localizes to vacuolar membranes and regulates vacuole biogenesis by phosphorylating proteins like Vps41, a homotypic vacuole fusion and vacuole protein sorting complex subunit that localizes to LEs when NP (LaGrassa and Ungermann, 2005; Cabrera et al., 2009), and the Vam3 t-SNARE that mediates vacuolar fusion (Brett et al., 2008). YCK3 overexpression inhibits the fusion of fragmented vacuoles (LaGrassa and Ungermann, 2005), and palmitoylation is required for both function and AP-3–dependent protein sorting to the vacuole (Sun et al., 2004). Recently, Yck3 fused to a FYVE domain was shown to be sufficient to target the kinase to LEs and to locally activate Vps41 (Cabrera et al., 2010).
To determine whether Yck3 acts downstream of Btn1, we examined whether Yif1 localization is affected by the deletion of YCK3. As in btn1Δ cells (Fig. 1 A), Yif1 mislocalized to large puncta and vacuoles in yck3Δ cells (Fig. 5 B [top and bottom, respectively] and Table S1). These puncta are probably LEs because Yif1 colocalized with RFP-tagged Vps27 in yck3Δ cells (Fig. 5 C), as previously shown for btn1Δ cells (Fig. 1 B). We also examined whether Kex2 localization is affected by the deletion of YCK3 and found that Kex2 was mislocalized to large LE-like structures that label with FM4-64 and that are situated adjacent to the vacuole (Fig. 5 D and Table S2), as seen in btn1Δ cells (Fig. 1 C). As Kex2 mislocalization leads to reduced secretion of active α-mating factor in cells that lack either BTN1 (Fig. S1 D) or BTN2 (Kanneganti et al., 2011), we examined whether a similar effect was apparent in yck3Δ cells. We found that halo formation induced by the yck3Δ cells was also reduced relative to WT cells (Fig. S1 D). Overall, the results suggest that the deletion of YCK3 phenocopies that of either BTN1 or BTN2. We also examined Yif1 and Kex2 localization in btn1Δyck3Δ cells but did not discern significant differences in relation to the single mutants, nor were defects in growth observed (unpublished data). This suggests that BTN1 and YCK3 are probably epistatic.
As Yck3 is involved in protein uptake to the vacuole via the AP-3 pathway and not via the clathrin- and endosome-dependent AP-2 route that Yif1 likely takes, we examined whether the AP-3 pathway is affected in btn1Δ cells. To do this, we expressed a GFP-tagged Nyv1-Snc1 fusion protein that localizes to the vacuole-limiting membrane in WT cells but is partly mislocalized to the plasma membrane in cells lacking an AP-3 component (e.g., apl5Δ). We examined GFP–Nyv1-Snc1 in btn1Δ cells and found no mislocalization, unlike in apl5Δ or yck3Δ cells (Fig. S3 A). Thus, the AP-3 pathway appears intact in cells lacking BTN1. In addition, we examined localization of GFP-tagged Apl5 expressed from its chromosomal locus in cells lacking BTN1 but saw no difference with WT cells (Fig. S3 B).
Because the deletion of BTN1 leads to enhanced Golgi clustering (Fig. 4), we examined the effect of the deletion of YCK3 in WT cells and btn1Δ cells expressing GFP-Sed5 (Fig. 5, E and F). In both yck3Δ (Fig. 5 E and Table S4) and yck3Δbtn1Δ cells, Sed5 labeled large puncta that did not colabel with RFP-Vps27 or FM4-64 (Fig. 5 F), indicating that these are not endosomal compartments. Thus, the loss of Btn1 (Figs. 4 and 5 E and Table S4), Yck3 (Fig. 5 E and Table S4), or both (Fig. 5 F) results in enlargement of the Golgi.
Btn1 may control Yck3 phosphorylation of Sed5
With the exception of its necessity for AP-3–dependent protein sorting, the deletion of YCK3 parallels that of BTN1. Thus, Yck3 could act downstream of Btn1 and phosphorylate Sed5. To test this hypothesis, we examined Sed5 phosphorylation in cells lacking YCK3; however, no significant change in Sed5 phosphorylation was observed therein (unpublished data). Thus, either Yck3 does not phosphorylate Sed5 or, because Yck3 and Hrr25 form an essential pair (Wang et al., 1996), then Hrr25 (or the other Yck kinases, e.g., Yck1 and Yck2) substitutes for Yck3 in the constitutive yck3Δ deletion background. To test the latter possibility, we created an inducible form of YCK3 whose expression from its chromosomal locus is under the control of the GAL1 promoter. When cells were grown continually in the presence of galactose, both the NP and P forms of Sed5 were observed in Western blots (NP/P ratio = ∼3:1; Fig. 5 A, right). However, only the NP form of Sed5 was apparent in cells shifted to glucose for 8 h (NP/P ratio = ∼7:1; Fig. 5 A, top and bottom right). Thus, the turn-off of YCK3 results in a significant decrease in Sed5 phosphorylation. This suggests that either Yck3 phosphorylates Sed5 directly or that its activation is necessary for Sed5 phosphorylation by another kinase.
One means by which Btn1 might regulate Yck3 is by controlling its localization. Yck3 is mainly found on the vacuolar membrane, although endosome- and plasma membrane–localized Yck3 is functional (Sun et al., 2004; Cabrera et al., 2010), and the protein likely undergoes palmitoylation early in the secretory pathway (e.g., ER and Golgi) where the DHHC class of palmitoyl transferases is found (Ohno et al., 2006). We examined the localization of GFP-tagged Yck3 expressed from its chromosomal locus in cells lacking BTN1 and found that it labeled the limiting membrane of the vacuole and plasma membrane in both WT and btn1Δ cells (Fig. S4 A). Thus, Btn1 does not appear to regulate Yck3 localization.
Substitution of the Yck3 lipid anchor with a TMD bypasses the requirement for palmitoylation or Btn1 in LE–Golgi retrieval
Because Yck3 palmitoylation is essential for its function, we examined whether mutations in the DHHC palmitoyl transferases affect LE–Golgi sorting (Fig. 6, A and B). The deletion of individual palmitoyltransferase genes, such as AKR1, a Golgi protein that palmitoylates Yck kinases (Sun et al., 2004), or PFA3, a vacuolar transferase that palmitoylates Vac8 and other substrates (Hou et al., 2005, 2009), had no effect on Yif or Kex2 localization (unpublished data). However, we found that both Yif1 and Kex2 were completely mislocalized in cells lacking five of the seven yeast DHHC transferases (i.e., DHHC 5×Δ cells; Fig. 6 A and Tables S5 and S2, respectively), which are known to mislocalize Yck3 (Hou et al., 2009). Moreover, we could verify that GFP-Yck3 did not show vacuolar, but rather endosome-like labeling, in these cells (Fig. 6 B). Thus, selective LE–Golgi protein retrieval is sensitive to changes in DHHC levels, indicating that the palmitoylation of substrates, such as Yck3, is likely to be involved. It is noteworthy to add that the overexpression of BTN1 did not restore Yif1 or Kex2 Golgi localization in DHHC 5×Δ cells (unpublished data), which suggests that Btn1 probably does not enhance Yck3 palmitoylation via the remaining acyltransferases. Lastly, we examined whether Sed5 undergoes phosphorylation in the DHHC 5×Δ cells by Western analysis (Fig. 6 C, top and bottom). We found that levels of the P form of Sed5 were greatly reduced in btn1Δ cells (NP/P ratio = 6:1 in btn1Δ cells vs. 2:1 in WT cells), as shown in Fig. 5 A. In the DHHC 5×Δ cells, the NP/P ratio remained at ∼6:1 (5.8 ± 0.3; n = 2), although the levels of Sed5 protein were similar to those seen in WT cells. Thus, removal of the DHHC transferases inhibits both selective LE–Golgi retrieval and Sed5 phosphorylation.
If Btn1 is a palmitoyl protein desaturase and thus a potential regulator of palmitoylated proteins as suggested for CLN3 (Narayan et al., 2006, 2008) and LE–Golgi retrieval as suggested here, then substitution of the cysteine-rich palmitoylated domain in Yck3 with either an endosome localization sequence (e.g., FYVE domain) or a TMD could render it functional and restore LE–Golgi retrieval in cells lacking Btn1. Therefore, we deleted BTN1 in cells expressing Yck3-FYVE (Cabrera et al., 2010) or Yck3 bearing the Vam3 TMD (which fully supports Vps41 phosphorylation and vacuolar protein sorting; unpublished data) and examined the localization of GFP-Yif1 therein (Fig. 6, D and E). We found that Yif1 mislocalized to LEs in the btn1Δ control and btn1Δ YCK3-FYVE cells but not in btn1Δ YCK3-VAM3 cells (Fig. 6 [D and E] and Table S5). Yif1 localization in the btn1Δ YCK3-VAM3 cells did not colabel with FM4-64 and appeared Golgi-like. This indicates that addition of a TMD (which confers transport through the secretory pathway) to Yck3 is sufficient to bypass (at least in part) the requirement for Btn1 in selective LE–Golgi sorting. Furthermore, this result implies that Btn1 may affect the lipid anchor of Yck3 in some manner and thus regulate protein function.
We then examined whether Sed5 phosphorylation is restored in btn1Δ YCK3-VAM3 cells and found that the ratio of NP to P Sed5 was 4.5 ± 0.2:1 (n = 2; see representative experiment in Fig. 6 F, top and bottom). This result was intermediate to that of WT cells, which gave an average ± SD ratio of 2.1 ± 0.1:1 (n = 4), and btn1Δ cells, which had an average of 7.2 ± 1.7:1 (n = 4). Thus, substitution of the Yck3 acyl anchor with a TMD not only appears to restore Yif1 trafficking but may confer the ability of Sed5 to undergo modification.
Sed5 phosphorylation and Golgi morphology are affected in part by the deletion of BTN2
Because the deletion of BTN1 largely phenocopies that of BTN2 with respect to protein trafficking (i.e., Yif1 and Kex2 mislocalization and normal Vps10 and CPY localization; Figs. 1 [A and C] and S1 [A, B, and D]; Kama et al., 2007), we examined whether the deletion of BTN2 has parallel effects upon Sed5 phosphorylation and Golgi morphology as that of BTN1. First, we examined Sed5 phosphorylation in WT, btn1Δ, and btn2Δ cells and observed a robust reduction in the P form relative to the NP form (e.g., NP/P ratio = 6.5:1) in cells lacking BTN2 (Fig. S5 A). This ratio was 2.7:1 in WT control cells and ∼10:1 in the btn1Δ cells (in this representative experiment), and no significant effect upon the overall levels of Sed5 protein was observed in the btn2Δ cells. Next, we examined Golgi morphology in WT, btn1Δ, and btn2Δ cells expressing either GFP-Sed5 or GFP-Sed5317A. We found that the deletion of BTN2 had similar effects as the deletion of BTN1; namely, an increase in Golgi clustering was observed in btn2Δ cells expressing GFP-Sed5 (Fig. S5 B and Table S6). In contrast, BTN2 overexpression or deletion had little effect on Golgi clusters formed in the presence of GFP-Sed5317A (Fig. S5 C and Table S6). This indicates that Btn2 overproduction does not block Golgi clustering, which is in contrast to BTN1 overexpression (Fig. 4 B). Overall, the removal of BTN2 results in changes in Sed5 phosphorylation and Golgi morphology that are similar to the deletion of BTN1. Although this could indicate that Btn1 and Btn2 confer like functions, it is more probable that the removal of BTN2 results in Btn1 mislocalization to the vacuole (as shown in Fig. 2 D), hence resulting in a loss of function.
Discussion
We previously demonstrated that BTN2, a gene up-regulated in the absence of BTN1, encodes a component of a transport complex that retrieves specific proteins back to the Golgi (Kama et al., 2007). We now show that in the absence of BTN1, Golgi proteins like Yif1 and TGN-EE proteins like Kex2 fail to retrieve to the Golgi and accumulate in late compartments of the endosomal pathway (Fig. 1 and Tables S1 and S2). This is indicated by the colocalization of Yif1 and Kex2 with FM4-64 and/or Vps27 in cells lacking BTN1 (Fig. 1). This phenotype shared between btn1Δ and btn2Δ cells implies common defects in the sorting of material away from the vacuole to the Golgi. Thus, defects in LE–Golgi protein recycling may contribute to the mechanism underlying Batten Disease/NCL pathogenesis.
Yeast Btn1 and mammalian CLN3 are membrane proteins of unclear function, and, although a wide range of localization patterns and actions have been attributed to them, recent studies from S. cerevisiae (Vitiello et al., 2010), S. pombe (Codlin and Mole, 2009), and mammalian cells (Metcalf et al., 2008) imply that Golgi localization and/or function may be involved. We find that S. cerevisiae Btn1 expressed from its chromosomal locus or from single-copy plasmids localizes to Golgi structures (Fig. 2 A) that colabel with either Yif1 or Sec7 (Fig. 2 B). Thus, although Btn1 and Btn2 have distinct patterns of localization (i.e., Btn2 localizes to LEs; Kama et al., 2007), both contribute to the same transport process. However, Btn1 probably acts in a different manner than Btn2, which binds to retrieval factors like retromer, endosomal SNAREs, Snx4, and even to Yif1 (Kama et al., 2007). Studies made with cultured mammalian cells or fission yeast suggest that CLN3 and btn1 act in an unknown fashion upon protein export from the Golgi and mislocalize the mannose-6-phosphate receptor or its S. pombe equivalent, vps10, respectively, to varying degrees (Metcalf et al., 2008; Codlin and Mole, 2009). In contrast, Vps10 and CPY trafficking is normal in btn1Δ cells (Fig. S1, A and B), as observed for btn2Δ cells (Kama et al., 2007; Kanneganti et al., 2011). This suggests that the Vps10–CPY sorting pathway of S. cerevisiae differs from that of S. pombe. This difference may relate to the finding that both Btn2 and Vps10 colocalize with Vps27 but do not colocalize with each other (Kama et al., 2007), indicating that there may be different LE populations in budding yeast.
Despite differences in Vps10–CPY trafficking, our results suggest that S. cerevisiae Btn1 controls protein trafficking within the Golgi. BTN1 overexpression inhibited Ykt6, an R-SNARE that confers intra-Golgi protein sorting and protein trafficking into and out of the Golgi, from assembling into a canonical 1R–3Q complex with an essential Golgi Q-SNARE, Sed5, and two additional Q-SNAREs implicated in endosome–Golgi trafficking, Vti1 and Tlg1 (Fig. 3 A). Likewise, the overexpression or deletion of BTN1 had opposing effects on the ability of Sed5 to assemble into multiple SNARE complexes (Fig. 3 B) as well as to maintain Golgi morphology (Fig. 4 and Tables S3 and S4). Importantly, Btn1 regulates the phosphorylation state of Sed5, which was shown to control retrograde protein trafficking from the Golgi as well as Golgi morphology (Weinberger et al., 2005). In the absence of BTN1, Sed5 is in an underphosphorylated state (Figs. 5 A, 6 (C and F), and S5 A) that mimics the NP form and results in an enhancement of Golgi clustering (Fig. 4 (A–F) and Table S3) and an increase in SNARE assembly (Fig. 3 B). In contrast, BTN1 overexpression mimicked the constitutively P form of Sed5 by dispersing Golgi clusters (e.g., those formed by Sed5317A; Figs. 4 (B and E) and Table S3), reducing SNARE assembly (Fig. 3 B), and inhibiting the growth of cells bearing mutant SNAREs involved in Golgi trafficking (Fig. S2). Finally, BTN1 overexpression may even enhance Sed5 modification slightly (Fig. 5 A). Thus, it appears that Btn1 regulates Sed5 phosphorylation and, therefore, function. Importantly, we noted that the deletion of BTN2 also had effects on Sed5 phosphorylation and Golgi morphology (Fig. S5); however, these could result from the mislocalization of Btn1 that occurs in btn2Δ cells (Fig. 2 D).
Because Btn1 is not a kinase, it must be indirectly involved in Sed5 phosphorylation. Two pieces of evidence suggested that Yck3 might be involved. First, mammalian CLN3 was suggested to function as a palmitoyl protein desaturase (Narayan et al., 2006, 2008). Second, Yck3 is a palmitoylated endosome- and vacuole-associated casein kinase involved in protein sorting to the vacuole (Sun et al., 2004; LaGrassa and Ungermann, 2005). Moreover, although Yck3 shares essential functions with a paralog, Hrr25 (Wang et al., 1996), and is able to suppress deletions in homologues that function primarily at the plasma membrane (e.g., Yck1 and Yck2; Sun et al., 2004), it also phosphorylates proteins involved in vacuole protein transport, such as Vps41 and Vam3 (LaGrassa and Ungermann, 2005; Brett et al., 2008). We examined Sed5 phosphorylation in cells expressing an inducible form of YCK3 and found that this t-SNARE was underphosphorylated after the turn-off of expression (Fig. 5 A, right). Thus, Yck3 is a candidate kinase for Sed5 phosphorylation. Moreover, the deletion of YCK3 resulted in strong defects in LE–Golgi sorting (Fig. 5, B–D), which parallel those seen in btn1Δ and btn2Δ cells (Fig. 1 and Tables S1 and S2). This strengthens the idea that Yck3 is involved with Btn1 function, although Sed5 may not be the only substrate involved in the regulation of LE–Golgi sorting. A model for the control of Sed5 and LE–Golgi sorting by Btn1 and Yck3 is shown in Fig. 7.
Figure 7.
A model for the control of Sed5 phosphorylation and LE–Golgi transport by Btn1 and Yck3. Yif1 is in the trans-Golgi in WT cells but delocalizes to the LE in cells lacking either BTN1 or YCK3. Thus, Btn1 and Yck3 mediate Yif1 retrieval. Sed5 is a Golgi SNARE that undergoes a phosphorylation/dephosphorylation cycle important for its localization and function (Weinberger et al., 2005). Btn1 regulates Yck3 kinase function, resulting in Sed5 phosphorylation either directly by Yck3 (as shown) or indirectly through another kinase (not depicted). Sed5 phosphorylation and dephosphorylation alter SNARE assembly in a manner that regulates retrograde transport, resulting in the retrieval of Yif1 to the Golgi. The phosphatase that dephosphorylates Sed5 is unknown but whose function must be coordinated with Yck3 to control SNARE assembly.
How Btn1 actually regulates Yck3 function is unclear; however, it might occur via regulation of the lipid anchor moiety. This conjecture is supported by the fact that substitution of the palmitate acceptor region in Yck3 with a TMD allows yeast to bypass the Btn1 requirement in LE–Golgi sorting (Fig. 6 E and Table S5) and partially restores Sed5 phosphorylation (Fig. 6 F). Yck3 palmitoylation and membrane anchoring are required for normal vacuolar fusion (Hou et al., 2009) as well as its function in LE–Golgi sorting, the latter being indicated by the fact that Yif1 and Kex2 are missorted in cells lacking five of the DHHC class of palmitoyl transferases (Fig. 6 A and Tables S2 and S5) and that Sed5 is underphosphorylated therein (Fig. 6 C). Although we have not proven whether Btn1 (or CLN3) is a palmitoyl protein desaturase, it does show that modulation of the palmitate anchor could play an important role in kinase function in yeast and, potentially, could form the basis for disease onset in postmitotic cells (e.g., neurons) lacking an inherent ability to either control or turn over palmitoylated proteins in a proper fashion. In this regard, it is highly interesting and probably not coincidental that the infantile form of NCL occurs as a result of mutation in a lysosomal palmitoyl thioesterase, although the target substrates for this enzyme have yet to be identified (Vesa et al., 1995; Mole et al., 2005; Siintola et al., 2006; Getty and Pearce, 2011). If, as in yeast, mammalian CLN3 controls a secretory kinase, screens for regulators of kinase function may identify therapeutic agents of benefit to Batten disease patients.
Materials and methods
Media, DNA, and genetic manipulations
Yeast were grown to midlog phase at 26°C in standard growth media containing either 2% glucose or 3.5% galactose. Synthetic complete and dropout media were prepared as previously described (Haim-Vilmovsky and Gerst, 2009), whereas rich growth medium (yeast extract peptone dextrose) and an amino acid–enriched synthetic complete medium were prepared according to Rose et al. (1990). Standard methods were used for the introduction of DNA into yeast and the preparation of genomic DNA (Rose et al., 1990). The tagging of genes at their genomic loci with GFP or deletion of genes was performed by homologous recombination using PCR products with flanking regions to the appropriate genomic sequences generated using specific primers to genome-tagging plasmids used as templates (Longtine et al., 1998).
Growth tests
For growth tests on plates, yeast were grown to log phase, normalized for optical density (OD600), diluted serially (i.e., 10-fold), and plated by drops onto solid medium preincubated at different temperatures.
Halo assays
Halo assays for measuring the production of biologically active α-factor were performed by spotting MAT-α yeast strains onto a lawn of MATa sst2Δ indicator yeast, as previously performed (Kanneganti et al., 2011). Assays were performed in triplicate.
Yeast strains and plasmids
The yeast strains used are listed in Table S7. T. Lagrassa (University of Osnabrück, Osnabrück, Germany) provided the the YCK3-VAM3 strain. The plasmids used in this study are listed in Table S8. To create a plasmid expressing HA-tagged Ykt6-1 (including its 3′ untranslated region [UTR], which could be used as a temperature-sensitive tool for use in immunoprecipitation [IP] assays), the ykt6-1 gene (Ykt6 bearing substitutions S35G, L125G, D149G, S182R, and M185T; a gift from D. Banfield, Hong Kong University of Science and Technology, Hong Kong, China) was amplified by PCR using the isolated balancer plasmid recovered from the SARY 166 strain as a template and primers encoding SalI and SacI sites for amplification at the 5′ and 3′ ends, respectively. The SalI–SacI fragment was inserted in-frame and downstream to the myc epitope encoded in vector pRS313 (CEN and URA3). To create a yeast expression plasmid for CLN3, the human CLN3 gene was amplified by PCR using cDNA derived from HeLa cells as a template and primers encoding SalI and SacI sites for amplification at the 5′ and 3′ ends, respectively. The SalI–SacI fragment was inserted in-frame and downstream to the HA epitope encoded by vector pAD54 (2μ and LEU2). A construct for the integration of GFP-tagged BTN1 at its genomic locus was created by subcloning an SalI and BglII fragment bearing GFP-BTN1-3′UTR from plasmid pUG46-GFP-BTN1(+3′ UTR) into plasmid pFA6a–natMX4, which encodes a PCR amplification module for integration at target genes using selection for resistance to nourseothricin via the SalI and BglII sites. After verification by sequencing, an integration cassette containing GFP-BTN1-3′UTR-natMX-BTN1 was amplified using a 60-bp forward chimeric oligonucleotide corresponding to the 5′ UTR of BTN1 and GFP (which follows directly downstream) and a 60-bp reverse chimeric oligonucleotide corresponding to the pFA6a-nat-MX4 plasmid (after the nat gene sequence) and a region of the 3′ UTR of BTN1 beginning 210 bp after the stop codon. Fragments obtained by PCR amplification were used to transform WT yeast and were selected for medium containing clonNAT (nourseothricin; CAS no. 96736-11-7; WERNER BioAgents). Integration at the BTN1 locus was verified by PCR.
Microscopy
GFP and RFP fluorescence in strains expressing the appropriate GFP- and RFP-tagged fusion proteins/FM4-64 fluorescence, respectively, was visualized by confocal microscopy. Unless mentioned, cells were grown to midlog phase at 26°C in synthetic selective medium before visualization. For vacuolar staining, cells were pulsed with 5.4 µM FM4-64 (Invitrogen) for 30 min in the dark at 26°C. After the pulse, a chase of 20 min in the same medium lacking FM4-64 was performed. In the figures, merge indicates either merger of the GFP and FM4-64 panels or GFP and RFP panels where appropriate. Light indicates the differential interference contrast (DIC) panel. All bars equal 1 µm. Images were captured with a confocal imaging system (LSM 710; Carl Zeiss) mounted on an inverted microscope (AxioObserver; Carl Zeiss) using a Plan Apochromat 63×/1.40 N.A. objective. Image acquisition was accomplished at 26°C using the detection system and accompanying software provided by the manufacturer. Photoshop (Adobe) was used to assemble and size the images for figure preparation.
IP and Western analysis
Cell lysates for use in IP experiments and immunoblots were prepared using glass beads to break intact cells. Cells were grown to midlog phase at 26°C before harvesting, washed with TE (10 mM Tris-Hcl, pH 7.5, and 1 mM EDTA), and resuspended in 250–300 µl of lysis buffer consisting of TE containing 1% NP-40 (volume/volume), 10 µg/ml aprotinin, 10 µg/ml leupeptin, 10 µg/ml soybean trypsin inhibitor, 10 µg/ml pepstatin, and 100 µM PMSF. For the detection of P Sed5 in Western blots, the lysis buffer was supplemented with the following phosphatase inhibitors: 10 mM NaF, 20 mM NaPPi, 25 mM β-glycerophosphate, and 0.5 mM sodium vanadate. For IP experiments, 15 OD600 units of cells were used for each pull-down. Cell extracts were prepared by adding an equal volume of washed glass beads (e.g., 0.25 ml) to the cells followed by prolonged shaking at 2,200 rpm in a Vibrax shaker (IKA Laboratories Ltd.) at 4°C for 45 min. Extracts were clarified by centrifugation at 12,000 g for 10 min to yield total cell lysates (TCLs). Protein concentration was determined using the Pierce MicroBCA protein assay kit (Thermo Fisher Scientific). For each IP reaction, 0.5 mg of TCL was diluted with ice-cold IP buffer (1% NP-40 in TE) to a final volume of 0.5 ml and was incubated with the appropriate IP antibody (see below). IP was performed at 4°C with constant rotation for 16 h, and precipitation of the complexes was performed by adding a 30-µl bed volume of protein G–agarose (Santa Cruz Biotechnology, Inc.) prewashed in IP buffer to the IP mixture. After 2 h of incubation with constant rotation, the complexes were spun down and washed three times with ice-cold IP wash buffer (TE containing 1% NP-40 and 150 mM NaCl). Precipitated proteins were resuspended in 30 µl SDS-PAGE sample buffer and boiled (3 min) before electrophoresis on 10 and 12.5% polyacrylamide gels (i.e., low molecular mass proteins <60 kD were resolved on 12.5% gels, whereas high molecular mass proteins >60 kD were resolved in parallel on 10% gels). Where relevant, samples of TCLs (e.g., 30 µg of protein per lane) were electrophoresed in parallel to the IP samples. For immunoblotting, gels were electroblotted onto pure nitrocellulose blotting membranes (BioTrace; Pall Corporation). Blots were blocked with 5% nonfat milk in TBS (PBS solution containing 0.05% Tween 20) and then probed with the appropriate antibody in TBS containing 5% BSA and 3 mM NaN3. Protein expression was detected by an ECL detection kit (GE Healthcare).
Affinity-purified IP antibodies included monoclonal anti-myc (9E10, 3 µl per IP; Santa Cruz Biotechnology, Inc.) and anti-HA antibodies (16B12, 2.5 µl per IP; Covance). Antibodies for protein detection in Western blots or on nitrocellulose filters included polyclonal antibodies against Bet1 (diluted 1:1,000; provided by C. Barlowe, Dartmouth College, Hanover, NH); CPY (a gift from S. Emr, University of California, San Diego, La Jolla, CA); Gos1, Sft1, and Ykt6 (diluted 1:1,000; gifts from D. Banfield); Sed5 (diluted 1:3,000; a gift from H. Pelham, Medical Research Council, Cambridge, England, UK); Snc1 (diluted 1:500; Protopopov et al., 1993); Sso1/2 (diluted 1:1,000; a gift from S. Keranen, Technical Research Centre of Finland, Espoo, Finland); Tlg1 (diluted 1:3,000; a gift from H. Pelham); Tlg2 (diluted 1:750; a gift from H. Abeliovich, Hebrew University of Jerusalem, Rehovot, Israel); Vti1 (diluted 1:1,000; a gift from G. Fischer von Mollard, University of Göttingen, Göttingen, Germany); and monoclonal antibodies to the HA and myc epitopes (diluted 1:1,000).
To distinguish between the higher and lower molecular mass forms of Sed5 (i.e., upper and lower bands), TCL samples were subjected to SDS-PAGE on 11.5% acrylamide gels. Quantification of the upper (i.e., P) and lower (i.e., NP) bands of Sed5 in Western blots by density measurement was performed using ImageJ software (National Institutes of Health). Molecular mass markers are listed in kilodaltons for all Western blots.
Online supplemental material
Fig. S1 demonstrates that the deletion or overexpression of BTN1 does not alter localization of a wide variety of endosome-trafficked proteins. Fig. S2 shows that the deletion and overexpression of BTN1 have opposing effects on the growth of temperature-sensitive mutants of Golgi SNAREs. Fig. S3 shows that the deletion of BTN1 does not affect AP3-mediated protein sorting or localization of an AP-3 component. Fig. S4 shows that Yck3 is not mislocalized in btn1Δ cells, although YCK3 overexpression inhibits the growth of Golgi SNARE mutants. Fig. S5 shows that the deletion of BTN2 also affects Sed5 phosphorylation and Golgi morphology. Table S1 provides statistics for the localization of GFP-Yif1 in btn1Δ and yck3Δ cells. Table S2 provides statistics for the localization of Kex2-GFP in btn1Δ, yck3Δ, and DHHC 5×Δ cells. Table S3 provides statistics for the localization of GFP-Sed5 in WT cells lacking or overexpressing BTN1. Table S4 provides statistics for the localization of GFP-Sed5 in yck3Δ cells. Table S5 provides statistics for the localization of GFP-Yif1 in cells expressing mutant Yck3 proteins or lacking DHHC palmitoyl transferases. Table S6 provides statistics for the localization of GFP-Sed5 in cells lacking or overexpressing BTN2. Table S7 includes the yeast strains used in this study, and Table S8 includes the plasmids used for this study. Finally, Table S9 provides a glossary describing the genes used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201102115/DC1.
Supplementary Material
Acknowledgments
We thank H. Abeliovich, D. Banfield, G. Fischer von Mollard, S. Keranen, H. Pelham, and T. LaGrassa for reagents.
This study was supported by grants to J.E. Gerst from the Israel Science Foundation (188/07 and 358/10), Y. Leon Benoziyo Institute for Molecular Medicine, Irving B. Harris Foundation, and Weizmann Institute of Science and to V. Kanneganti and J.E. Gerst from the National Contest for Life Foundation. C. Ungermann is supported by grants from the Deutsche Forschungsgemeinschaft (SFB 944 and 431) and Hans-Mühlenhoff Foundation. J.E. Gerst holds the Besen-Brender Chair of Microbiology and Parasitology.
Footnotes
Abbreviations used in this paper:
- CPY
- carboxypeptidase Y
- DIC
- differential interference contrast
- IP
- immunoprecipitation
- LE
- late endosome
- MVB
- multivesicular body
- NCL
- neuronal ceroid lipofuscinose
- NP
- nonphosphorylated
- P
- phosphorylated
- TCL
- total cell lysate
- TMD
- transmembrane domain
- UTR
- untranslated region
- WT
- wild type
References
- Abeliovich H., Grote E., Novick P., Ferro-Novick S. 1998. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 273:11719–11727 10.1074/jbc.273.19.11719 [DOI] [PubMed] [Google Scholar]
- Bagola K., Sommer T. 2008. Protein quality control: On IPODs and other JUNQ. Curr. Biol. 18:R1019–R1021 10.1016/j.cub.2008.09.036 [DOI] [PubMed] [Google Scholar]
- Banfield D.K., Lewis M.J., Rabouille C., Warren G., Pelham H.R. 1994. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J. Cell Biol. 127:357–371 10.1083/jcb.127.2.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett C.L., Plemel R.L., Lobinger B.T., Vignali M., Fields S., Merz A.J. 2008. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J. Cell Biol. 182:1141–1151 10.1083/jcb.200801001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J.H., Fuller R.S. 1997. SOI1 encodes a novel, conserved protein that promotes TGN–endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23–36 10.1083/jcb.139.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnicourt A., Froissard M., Sereti K., Ulrich H.D., Haguenauer-Tsapis R., Galan J.M. 2004. Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins. Mol. Biol. Cell. 15:4203–4214 10.1091/mbc.E04-05-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., Ostrowicz C.W., Mari M., LaGrassa T.J., Reggiori F., Ungermann C. 2009. Vps41 phosphorylation and the Rab Ypt7 control the targeting of the HOPS complex to endosome-vacuole fusion sites. Mol. Biol. Cell. 20:1937–1948 10.1091/mbc.E08-09-0943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H.J., et al. 2010. Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cell Biol. 191:845–859 10.1083/jcb.201004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Pearce D.A. 2002. Interaction with Btn2p is required for localization of Rsglp: Btn2p-mediated changes in arginine uptake in Saccharomyces cerevisiae. Eukaryot. Cell. 1:606–612 10.1128/EC.1.4.606-612.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Muzaffar N.E., Sherman F., Pearce D.A. 2000. The yeast model for batten disease: Mutations in BTN1, BTN2, and HSP30 alter pH homeostasis. J. Bacteriol. 182:6418–6423 10.1128/JB.182.22.6418-6423.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Roberts P.M., Pearce D.A. 2003. The yeast model for Batten disease: A role for Btn2p in the trafficking of the Golgi-associated vesicular targeting protein, Yif1p. Biochem. Biophys. Res. Commun. 302:534–538 10.1016/S0006-291X(03)00209-2 [DOI] [PubMed] [Google Scholar]
- Codlin S., Mole S.E. 2009. S. pombe btn1, the orthologue of the Batten disease gene CLN3, is required for vacuole protein sorting of Cpy1p and Golgi exit of Vps10p. J. Cell Sci. 122:1163–1173 10.1242/jcs.038323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe J.G., Lim A.C., Xu J., Hong W. 1999. A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2407–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croopnick J.B., Choi H.C., Mueller D.M. 1998. The subcellular location of the yeast Saccharomyces cerevisiae homologue of the protein defective in the juvenile form of Batten disease. Biochem. Biophys. Res. Commun. 250:335–341 10.1006/bbrc.1998.9272 [DOI] [PubMed] [Google Scholar]
- Deloche O., Schekman R.W. 2002. Vps10p cycles between the TGN and the late endosome via the plasma membrane in clathrin mutants. Mol. Biol. Cell. 13:4296–4307 10.1091/mbc.02-07-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G., Stevens T.H. 1999. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 10:1719–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Codlin S., Hyams J.S., Mole S.E. 2005. btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell Sci. 118:5525–5536 10.1242/jcs.02656 [DOI] [PubMed] [Google Scholar]
- Getty A.L., Pearce D.A. 2011. Interactions of the proteins of neuronal ceroid lipofuscinosis: Clues to function. Cell. Mol. Life Sci. 68:453–474 10.1007/s00018-010-0468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S., Chapman-Shimshoni D., Trajkovic S., Gerst J.E. 2000. Yeast exocytic v-SNAREs confer endocytosis. Mol. Biol. Cell. 11:3629–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim-Vilmovsky L., Gerst J.E. 2009. m-TAG: a PCR-based genomic integration method to visualize the localization of specific endogenous mRNAs in vivo in yeast. Nat. Protoc. 4:1274–1284 10.1038/nprot.2009.115 [DOI] [PubMed] [Google Scholar]
- Hettema E.H., Lewis M.J., Black M.W., Pelham H.R. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22:548–557 10.1093/emboj/cdg062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C., Nichols B.J., Dhruvakumar S., Pelham H.R. 1998. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 17:113–126 10.1093/emboj/17.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Subramanian K., LaGrassa T.J., Markgraf D., Dietrich L.E., Urban J., Decker N., Ungermann C. 2005. The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc. Natl. Acad. Sci. USA. 102:17366–17371 10.1073/pnas.0508885102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., John Peter A.T., Meiringer C., Subramanian K., Ungermann C. 2009. Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic. 10:1061–1073 10.1111/j.1600-0854.2009.00925.x [DOI] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R., Robinson M., Gerst J.E. 2007. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol. 27:605–621 10.1128/MCB.00699-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V., Kama R., Gerst J.E. 2011. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol. Biol. Cell. 22:1648–1663 10.1091/mbc.E10-11-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Chattopadhyay S., Locke S., Pearce D.A. 2005. Interaction among Btn1p, Btn2p, and Ist2p reveals potential interplay among the vacuole, amino acid levels, and ion homeostasis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 4:281–288 10.1128/EC.4.2.281-288.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer H., Phistry M. 1996. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J. Cell Biol. 133:1205–1215 10.1083/jcb.133.6.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D.S., Shewmaker F., Wickner R.B. 2008. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 27:2725–2735 10.1038/emboj.2008.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttälä A., Lahtinen U., Braulke T., Hofmann S.L. 2006. Functional biology of the neuronal ceroid lipofuscinoses (NCL) proteins. Biochim. Biophys. Acta. 1762:920–933 [DOI] [PubMed] [Google Scholar]
- LaGrassa T.J., Ungermann C. 2005. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J. Cell Biol. 168:401–414 10.1083/jcb.200407141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Matern H., Yang X., Andrulis E., Sternglanz R., Trepte H.H., Gallwitz D. 2000. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 19:4485–4492 10.1093/emboj/19.17.4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D.J., Calvi A.A., Seaman M.Nj., Mitchison H.M., Cutler D.F. 2008. Loss of the Batten disease gene CLN3 prevents exit from the TGN of the mannose 6-phosphate receptor. Traffic. 9:1905–1914 10.1111/j.1600-0854.2008.00807.x [DOI] [PubMed] [Google Scholar]
- Mole S.E., Williams R.E., Goebel H.H. 2005. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 6:107–126 10.1007/s10048-005-0218-3 [DOI] [PubMed] [Google Scholar]
- Narayan S.B., Rakheja D., Tan L., Pastor J.V., Bennett M.J. 2006. CLN3P, the Batten’s disease protein, is a novel palmitoyl-protein Δ-9 desaturase. Ann. Neurol. 60:570–577 10.1002/ana.20975 [DOI] [PubMed] [Google Scholar]
- Narayan S.B., Tan L., Bennett M.J. 2008. Intermediate levels of neuronal palmitoyl-protein Δ-9 desaturase in heterozygotes for murine Batten disease. Mol. Genet. Metab. 93:89–91 10.1016/j.ymgme.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Ohno Y., Kihara A., Sano T., Igarashi Y. 2006. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta. 1761:474–483 [DOI] [PubMed] [Google Scholar]
- Osório N.S., Carvalho A., Almeida A.J., Padilla-Lopez S., Leão C., Laranjinha J., Ludovico P., Pearce D.A., Rodrigues F. 2007. Nitric oxide signaling is disrupted in the yeast model for Batten disease. Mol. Biol. Cell. 18:2755–2767 10.1091/mbc.E06-11-1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-López S., Pearce D.A. 2006. Saccharomyces cerevisiae lacking Btn1p modulate vacuolar ATPase activity to regulate pH imbalance in the vacuole. J. Biol. Chem. 281:10273–10280 10.1074/jbc.M510625200 [DOI] [PubMed] [Google Scholar]
- Pearce D.A., Sherman F. 1997. BTN1, a yeast gene corresponding to the human gene responsible for Batten’s disease, is not essential for viability, mitochondrial function, or degradation of mitochondrial ATP synthase. Yeast. 13:691–697 [DOI] [PubMed] [Google Scholar]
- Pearce D.A., Sherman F. 1998. A yeast model for the study of Batten disease. Proc. Natl. Acad. Sci. USA. 95:6915–6918 10.1073/pnas.95.12.6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D.A., Sherman F. 1999. Investigation of Batten disease with the yeast Saccharomyces cerevisiae. Mol. Genet. Metab. 66:314–319 10.1006/mgme.1999.2820 [DOI] [PubMed] [Google Scholar]
- Pearce D.A., Ferea T., Nosel S.A., Das B., Sherman F. 1999. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 22:55–58 10.1038/8861 [DOI] [PubMed] [Google Scholar]
- Phillips S.N., Benedict J.W., Weimer J.M., Pearce D.A. 2005. CLN3, the protein associated with batten disease: structure, function and localization. J. Neurosci. Res. 79:573–583 10.1002/jnr.20367 [DOI] [PubMed] [Google Scholar]
- Piper R.C., Cooper A.A., Yang H., Stevens T.H. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603–617 10.1083/jcb.131.3.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V., Govindan B., Novick P., Gerst J.E. 1993. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 74:855–861 10.1016/0092-8674(93)90465-3 [DOI] [PubMed] [Google Scholar]
- Rakheja D., Narayan S.B., Bennett M.J. 2008. The function of CLN3P, the Batten disease protein. Mol. Genet. Metab. 93:269–274 10.1016/j.ymgme.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Richardson S.C., Winistorfer S.C., Poupon V., Luzio J.P., Piper R.C. 2004. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell. 15:1197–1210 10.1091/mbc.E03-06-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston F., Hieter P. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 198 pp [Google Scholar]
- Seaman M.N., McCaffery J.M., Emr S.D. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siintola E., Lehesjoki A.E., Mole S.E. 2006. Molecular genetics of the NCLs — status and perspectives. Biochim. Biophys. Acta. 1762:857–864 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Blumer K.J. 1999. A syntaxin homolog encoded by VAM3 mediates down-regulation of a yeast G protein-coupled receptor. J. Biol. Chem. 274:1835–1841 10.1074/jbc.274.3.1835 [DOI] [PubMed] [Google Scholar]
- Sun B., Chen L., Cao W., Roth A.F., Davis N.G. 2004. The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXPhi adaptin sorting signal. Mol. Biol. Cell. 15:1397–1406 10.1091/mbc.E03-09-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunio A., Metcalf A.B., Krämer H. 1999. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: Hook is required for normal maturation of multivesicular endosomes. Mol. Biol. Cell. 10:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesa J., Hellsten E., Verkruyse L.A., Camp L.A., Rapola J., Santavuori P., Hofmann S.L., Peltonen L. 1995. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 376:584–587 10.1038/376584a0 [DOI] [PubMed] [Google Scholar]
- Vitiello S.P., Benedict J.W., Padilla-Lopez S., Pearce D.A. 2010. Interaction between Sdo1p and Btn1p in the Saccharomyces cerevisiae model for Batten disease. Hum. Mol. Genet. 19:931–942 10.1093/hmg/ddp560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta J.H., Didier A.J., Liu X., Krämer H. 2001. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152:923–934 10.1083/jcb.152.5.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hoekstra M.F., DeMaggio A.J., Dhillon N., Vancura A., Kuret J., Johnston G.C., Singer R.A. 1996. Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 16:5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger A., Kamena F., Kama R., Spang A., Gerst J.E. 2005. Control of Golgi morphology and function by Sed5 t-SNARE phosphorylation. Mol. Biol. Cell. 16:4918–4930 10.1091/mbc.E05-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe D.M., Padilla-Lopez S., Vitiello S.P., Pearce D.A. 2011. pH-dependent localization of Btn1p in the yeast model for Batten disease. Dis Model Mech. 4:120–125 10.1242/dmm.006114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.