Abstract
Viruses have evolved as combinations of genes whose products interact with cellular components to produce progeny virus throughout the plants. Some viral genes, particularly those that are involved in replication and assembly, tend to be relatively conserved, whereas other genes that have evolved for interactions with the specific host for movement and to counter host–defense systems tend to be less conserved. Closteroviridae encode 1–5 nonconserved ORFs. Citrus tristeza virus (CTV), a Closterovirus, possesses nonconserved p33, p18, and p13 genes that are expendable for systemic infection of the two laboratory hosts, Citrus macrophylla and Mexican lime. In this study, we show that the extended host range of CTV requires these nonconserved genes. The p33 gene was required to systemically infect sour orange and lemon trees, whereas either the p33 or the p18 gene was sufficient for systemic infection of grapefruit trees and the p33 or the p13 gene was sufficient for systemic infection of calamondin plants. Thus, these three genes are required for systemic infection of the full host range of CTV, but different genes were specific for different hosts. Remarkably, either of two genes was sufficient for infection of some citrus hybrids. These findings suggest that CTV acquired multiple nonconserved genes (p33, p18, and p13) and, as a result, gained the ability to interact with multiple hosts, thus extending its host range during the course of evolution. These results greatly extend the complexity of known virus–plant interactions.
Keywords: gene addition, host-specific activity, long-distance transport
Viruses have evolved as combinations of genes whose products interact with cellular components to produce progeny throughout the plants and, in most cases, to interact with a vector to be moved to other plants. These processes require several layers of precise interactions with the host. Some viral genes tend to be relatively conserved, whereas other genes appear to be completely unrelated to any gene in otherwise similar viruses or, in some cases, to any known gene. In general, the more conserved genes tend to be those whose products interact within the cell to replicate the viral genome. This process appears to be somewhat generic because many viruses have been found to be capable of multiplying in individual cells (protoplasts) but cannot move throughout the intact plant. The less conserved genes tend to be those that have evolved for interactions with the specific host for movement and to counter host–defense systems (1, 2).
The ability of the virus to move from the initially infected cell throughout the plant appears to be one of the major selective forces for the evolution of plant viruses. Successful systemic infection of plant viruses results from replication in initially infected cells, followed by two distinct processes: cell-to-cell and long-distance movement (2–9). Cell-to-cell movement is a process that allows the virus to pass to adjacent cells by successful interactions between virus-encoded movement proteins and host factors (2, 7, 9). Long-distance movement is a multistep process that allows the virus to enter the sieve element from an adjacent cell, followed by passive movement of virus through the phloem to a distal region of the plant by exiting into a cell adjacent to the phloem (2, 3). Further cell-to-cell movement from the phloem-associated cells allows the virus to invade most of the cells at a distal region of the plant. Viral proteins and host factors that are involved in cell-to-cell movement of plant viruses have been widely examined (7, 9). However, the host factors that are involved in long-distance transport of plant viruses and the mechanisms of long-distance movement such as factors that are involved in virus entry into phloem tissue and virus exit at a distal region of the plant are less well understood. Additionally, plants have host–defense mechanisms including RNA silencing that must be overcome by the virus for effective movement within the plant. Viruses have evolved gene products to suppress these defense mechanisms (1).
Citrus tristeza virus (CTV), one of the most economically important viruses, is a member of the genus Closterovirus in the family Closteroviridae (10, 11). CTV has a 19.3-kb single-stranded positive-sense RNA genome (12, 13). The genomic RNA of CTV is organized into 12 ORFs, which potentially encode at least 19 final proteins (14). Several of the ORFs are conserved among all of the Closteroviridae, including ORFs 1a and 1b, which are expressed from the genomic RNA as two polyproteins that contain two papain-like proteases, methyltransferase, helicase, and RNA-dependent RNA polymerase-like domains, and a set of five signature ORFs encoding two coat proteins, a HSP70 homolog, and ≈60-kDa and ≈6-kDa proteins (12, 13). Four of the signature ORFs are involved in formation of the long flexuous virions (2,000 nm × 10–12 nm) that are encapsidated by the minor coat protein on the 5′ 630 nucleotides of the genomic RNA with the major coat protein (CP) completing the rest of the genomic RNA (15, 16). The HSP70h and 61-kDa proteins work in concert to enhance assembly (15, 17, 18). The 10 internal genes, which are dispensable for replication at the single-cell level, are expressed through a nested set of 3′ coterminal subgenomic mRNAs (19, 20). The function of the other signature protein (p6) is unknown but was shown to be needed for systemic infection of citrus trees (21). The p20 and p23 proteins and CP have been shown to counter the host–defense system by suppressing host RNA silencing (22).
Additionally, CTV has three nonconserved genes, p33, p18, and p13, with no significant homology with reported GenBank sequences. Remarkably, these nonconserved genes can be deleted without affecting the ability of the virus to systemically infect the more susceptible citrus trees (21). Because CTV is able to move in these hosts by both cell-to-cell and long-distance movement, it is expected that the virus has other genes that function as a minimal set of movement genes. In this study, we further examined the roles of these expendable genes (p33, p18, and p13) in a wider range of citrus species and relatives and found that they are involved in systemic infection of some of the hosts. However, different genes were required for systemic infection of different hosts. The p33 gene was required for systemic infection of sour orange (C. aurantium L.) and lemon [C. limon (L.) Burm.f.], either the p33 or the p13 was sufficient for systemic infection of calamondin (C. mitis Blanco), whereas either the p33 or the p18 gene was sufficient to systemically infect grapefruit (C. paradisi Macf.). These results demonstrate that CTV has evolved combinations of these genes and the other movement genes to define its host range. These findings greatly extend the complexity of known virus–plant interactions.
Results
Host Range of CTV Deletion Mutants.
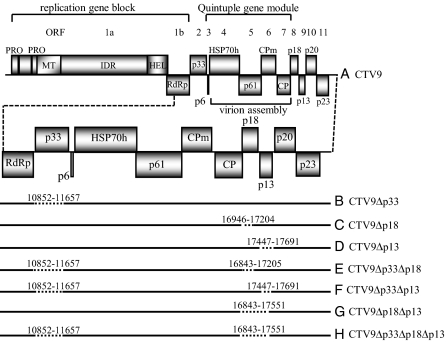
We reported that CTV (T36) with a deletion of most of the p33, p18, or p13; double deletions in p33 and p18, p33 and p13, or p18 and p13; or triple deletions in p33, p18, and p13 ORFs systemically infected C. macrophylla and Mexican lime [Citrus aurantifolia (Christm.) Swing.] plants (Fig. 1; ref. 21). Why would CTV have genes that it does not need? Because wild-type CTV infects several citrus species, we wondered whether the genes that are dispensable for infection of C. macrophylla and Mexican lime were required for systemic infection of other citrus species. The ability of the mutants with deletions in a single gene (CTV9Δp33, CTV9Δp18, and CTV9Δp13), two genes (CTV9Δp33Δp18, CTV9Δp33Δp13, and CTV9Δp18Δp13), or three genes (CTV9Δp33Δp18Δp13) to infect a range of citrus species was examined by graft inoculation from C. macrophylla- or Mexican lime-infected plants (Table 1 and Table S1). The inoculated plants were incubated in a greenhouse and examined for systemic infection at 4 and 8 wk after inoculation (wpi) by double antibody sandwich indirect ELISA (DAS-I-ELISA) by using CTV-specific antiserum (23).
Fig. 1.
(A) Schematic diagram of the genomic organization of CTV (CTV9) with ORFs (open boxes) showing the putative papain-like proteases (PRO), large interdomain region (IDR), and the methyl transferase-like (MT), helicase-like (HEL), and RNA-dependent RNA polymerase-like (RdRp) domains. The ORFs with numbers and corresponding translation products are indicated. HSP 70h, homolog of heat shock protein 70; CPm, minor coat protein; CP, major coat protein. ORFs corresponding to replication gene block, quintuple gene module, and virion assembly are indicated. The enlarged view of the 3′ end ORFs showed below the genome organization. CTV deletion mutants are shown in B–H with deleted sequences shown as dotted lines, and nucleotide coordinates of deletions are indicated.
Table 1.
Biological indexing of CTV deletion mutants on selected varieties of citrus that are differentially infected
| Mutant | Plant no. | Sour orange | Lemon | Grapefruit | Calamondin |
| CTV9 | 1 | 1.37 ± 0.00 | 1.46 ± 0.02 | 3.21 ± 0.08 | 0.80 ± 0.01 |
| 2 | 3.15 ± 0.01 | 1.75 ± 0.08 | 3.42 ± 0.09 | 0.52 ± 0.02 | |
| 3 | 3.05 ± 0.01 | 0.88 ± 0.07 | 2.22 ± 0.10 | 0.46 ± 0.00 | |
| 4 | — | — | 3.15 ± 0.03 | — | |
| CTV9Δp33 | 1 | 0.09 ± 0.00 | 0.09 ± 0.00 | 2.25 ± 0.02 | 0.92 ± 0.02 |
| 2 | 0.07 ± 0.00 | 0.10 ± 0.00 | 1.85 ± 0.08 | 0.41 ± 0.02 | |
| 3 | 0.19 ± 0.01 | 0.07 ± 0.00 | 1.15 ± 0.06 | 0.95 ± 0.02 | |
| 4 | — | — | 2.27 ± 0.08 | — | |
| CTV9Δp18 | 1 | 2.79 ± 0.01 | 1.10 ± 0.04 | 1.89 ± 0.32 | 0.35 ± 0.01 |
| 2 | 2.13 ± 0.04 | 1.76 ± 0.02 | 2.84 ± 0.07 | 0.27 ± 0.00 | |
| 3 | 2.90 ± 0.02 | 1.84 ± 0.00 | 2.83 ± 0.04 | 0.39 ± 0.00 | |
| 4 | 2.26 ± 0.04 | — | 3.44 ± 0.02 | — | |
| CTV9Δp13 | 1 | 3.22 ± 0.01 | 0.35 ± 0.00 | 2.40 ± 0.67 | 2.38 ± 0.00 |
| 2 | 2.94 ± 0.03 | 0.45 ± 0.02 | 1.89 ± 0.06 | 0.82 ± 0.01 | |
| 3 | 3.02 ± 0.03 | 1.11 ± 0.02 | 3.29 ± 0.06 | 0.72 ± 0.03 | |
| 4 | 3.14 ± 0.03 | — | 1.79 ± 0.18 | — | |
| CTV9Δp33Δp18 | 1 | 0.06 ± 0.00 | 0.06 ± 0.00 | 0.06 ± 0.03 | 1.29 ± 0.00 |
| 2 | 0.07 ± 0.00 | 0.06 ± 0.00 | 0.08 ± 0.00 | 1.00 ± 0.01 | |
| 3 | 0.12 ± 0.00 | 0.07 ± 0.00 | 0.09 ± 0.00 | 1.75 ± 0.04 | |
| 4 | 0.08 ± 0.00 | — | — | — | |
| CTV9Δp33Δp13 | 1 | 0.11 ± 0.01 | 0.06 ± 0.00 | 0.65 ± 0.02 | 0.07 ± 0.00 |
| 2 | 0.08 ± 0.00 | 0.06 ± 0.00 | 1.45 ± 0.05 | 0.11 ± 0.00 | |
| 3 | 0.08 ± 0.00 | 0.06 ± 0.00 | 1.59 ± 0.02 | 0.07 ± 0.00 | |
| 4 | 0.07 ± 0.00 | — | — | — | |
| CTV9Δp18Δp13 | 1 | 0.71 ± 0.00 | 0.60 ± 0.01 | 1.15 ± 0.12 | 2.65 ± 0.02 |
| 2 | 1.96 ± 0.23 | 0.78 ± 0.00 | 2.29 ± 0.19 | 0.81 ± 0.01 | |
| 3 | 1.00 ± 0.21 | 1.00 ± 0.13 | 2.69 ± 0.06 | — | |
| 4 | 0.47 ± 0.004 | — | 3.23 ± 0.15 | — | |
| CTV9Δp33Δp13Δp18 | 1 | 0.08 ± 0.00 | 0.07 ± 0.00 | 0.12 ± 0.02 | 0.09 ± 0.00 |
| 2 | 0.11 ± 0.01 | 0.06 ± 0.00 | 0.07 ± 0.00 | 0.09 ± 0.00 | |
| 3 | 0.08 ± 0.00 | 0.06 ± 0.00 | 0.10 ± 0.00 | 0.06 ± 0.00 | |
| 4 | 0.08 ± 0.00 | — | 0.12 ± 0.03 | — | |
| Healthy | 1 | 0.07 ± 0.00 | 0.07 ± 0.00 | 0.10 ± 0.00 | 0.09 ± 0.00 |
| 2 | 0.08 ± 0.00 | 0.07 ± 0.00 | 0.12 ± 0.00 | 0.07 ± 0.00 | |
| 3 | 0.08 ± 0.01 | 0.07 ± 0.00 | 0.08 ± 0.00 | 0.09 ± 0.00 | |
| 4 | 0.08 ± 0.01 | — | 0.08 ± 0.00 | — |
Citrus plants inoculated with CTV9 and its deletion mutants were assayed at 8 wk after inoculation by double antibody sandwich indirect ELISA. ELISA values (A405) are averages for three wells ± SDs. Bold letters represent mutants failed to infect.
CTV with deletions in p33, p18, and p13 ORFs in all possible combinations were found to have infected the following citrus cultivars at 8 wpi: C. macrophylla, Mexican lime, Madam vinous sweet orange (C. sinensis L.), C. indica, C. hystrix DC, C. micrantha, Persian lime (C. latifolia Tan), and citron (C. medica L.) (Table S1). The full-length virus (CTV9) and the deletion mutants induced visible foliar symptoms only on C. macrophylla and Mexican lime plants with mild to moderate veinal chlorosis and leaf cupping symptoms, and all produced mild to symptomless infection on Madam vinous, C. indica, C. hystrix, C. micrantha, Persian lime, and citron plants. These data demonstrated that none of these genes (p33, p18, and p13) were needed for systemic infection of these citrus trees by CTV (Table S1).
p33 ORF Is Required for Systemic Infection of Sour Orange and Lemon Plants.
Sour orange and Eureka lemon plants were graft-inoculated with CTV deletion mutants by using bark patches/budwood from C. macrophylla- or Mexican lime-infected trees as the source of inoculum. Sour orange and lemon plants became systemically infected with CTV9, CTV9Δp18, CTV9Δp13, and CTV9Δp18Δp13 (Table 1), and sour orange trees developed strong yellowing symptoms, referred to as the seedling yellows reaction, as is typical of the T36 and many other CTV isolates. However, all mutants with a deletion in the p33 ORF (CTV9Δp33, CTV9Δp33Δp18, CTV9Δp33Δp13, and CTV9Δp33Δp18Δp13) failed to systemically infect sour orange or lemon plants (Table 1). These data demonstrated that the p33 ORF was required for systemic infection of sour orange and lemon plants.
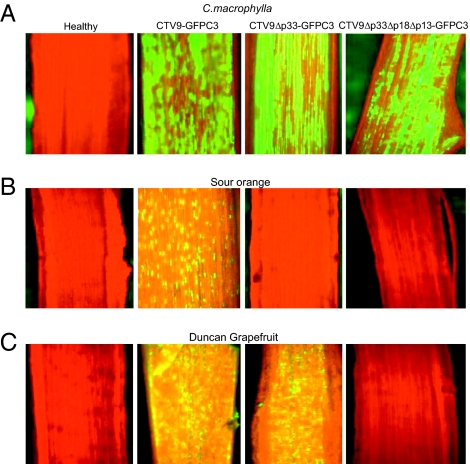
We tagged the full-length virus and deletion mutants CTV9Δp33 and CTV9Δp33Δp18Δp13 with GFP to allow visualization of the distribution of the virus in infected citrus trees (21). Sour orange plants were graft-inoculated with CTV9Δp33-GFPC3 and CTV9Δp33Δp18Δp13-GFPC3 from infected C. macrophylla. The full-length virus (CTV9-GFPC3) was used as a positive control in sour orange and C. macrophylla plants. The phloem of bark patches from sour orange and C. macrophylla plants was examined for the presence of GFP fluorescence by stereo-fluorescence microscopy at 4–12 wpi. As expected, the GFP-tagged full-length virus and the deletion mutants infected C. macrophylla plants and expressed abundant levels of GFP in large stretches of cells throughout the bark (Fig. 2A). The GFP fluorescence was observed in sour orange plants inoculated with CTV9-GFPC3 as small stretches of cells with fewer phloem cells expressing GFP fluorescence (Fig. 2B), as has been shown (24). However, no detectable fluorescence was observed from sour orange plants grafted with CTV9Δp33-GFPC3 or CTV9Δp33Δp18Δp13-GFPC3 (Fig. 2B). These results were confirmed by ELISAs of virions that found no virus of the deletion mutants in the sour orange plants, again demonstrating that the p33 ORF is required for systemic infection of sour orange.
Fig. 2.
Detection of GFP fluorescence in Citrus macrophylla (A), sour orange (B), and grapefruit (C) plants inoculated with CTV9-GFPC3, CTV9Δp33-GFPC3, or CTV9Δp33Δp18Δp13-GFPC3. The internal side of the bark patches of citrus plants was observed at 12 wk after inoculation under Zeiss Stemi SV 11 UV-fluorescence dissecting microscope with an attached Olympus Q-color 5 camera.
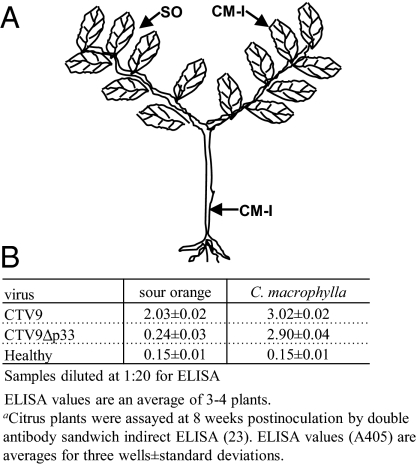
To assess the stringency of the p33 requirement for infection and spread, we modified conditions to increase the inoculum pressure. The first approach was to use “duplex” plants of sour orange and C. macrophylla (Fig. 3A). Healthy sour orange budwood was grafted onto CTV9Δp33-infected C. macrophylla plants, and the new shoots of sour orange and C. macrophylla were forced to grow in parallel from the same infected C. macrophylla rootstock (Fig. 3A). In this assay C. macrophylla, a susceptible host, is a source of abundant levels of inoculum for sour orange shoots. The sour orange and C. macrophylla shoots from “duplex” plants were examined by DAS-I-ELISA at 10–12 wpi. The result was that the sour orange branches remained free from CTV9Δp33 infection, whereas high levels of virus were detected in the parallel C. macrophylla branches (Fig. 3B). In contrast, CTV9 infected both sour orange and C. macrophylla shoots (Fig. 3B).
Fig. 3.
Schematic diagram of duplex plant of CTV9Δp33-infected C. macrophylla and sour orange. (A) Sour orange budwood was grafted onto CTV9Δp33-infected C. macrophylla plant, and the branches of C. macrophylla were pruned to force the shoots of sour orange (SO) and C. macrophylla (CM-I) simultaneously. (B) Analysis of sour orange and C. macrophylla branches from duplex plants for virus infection by DAS-I-ELISA.
The second approach was to substitute small rectangles of bark patches from healthy sour orange plants into C. macrophylla plants infected with CTV9Δp33-GFPC3 or CTV9Δp33Δp18Δp13-GFPC3 and allowed substituted sour orange bark patches to establish vascular connections (Fig. 4Aa). In reciprocal experiments, bark patches from CTV9Δp33-GFPC3- or CTV9Δp33Δp18Δp13-GFPC3-infected C. macrophylla plants were excised and replaced with the same sized bark patches onto sour orange plants (Fig. 4Bb). CTV9-GFPC3 was used as a positive control in the bark patch experiments. The junctions of sour orange and C. macrophylla bark patches were examined at 8 wpi for the presence of GFP under a stereo-fluorescence microscope. The sour orange bark patches on CTV9Δp33-GFPC3- or CTV9Δp33Δp18Δp13-GFPC3-infected C. macrophylla plants remained virus free as no GFP fluorescence was detected in the sour orange bark patches, whereas abundant levels of GFP fluorescence was observed in C. macrophylla bark patches (Fig. 4 Ac and Ad). In reciprocal bark patch experiments, CTV9Δp33-GFPC3- or CTV9Δp33Δp18Δp13-GFPC3-infected C. macrophylla bark patches on sour orange plants, GFP fluorescence was detected only in C. macrophylla bark patches, but not on sour orange bark patches (Fig. 4 Bc and Bd). In control experiments, CTV9-GFPC3 infected-C. macrophylla bark patches on sour orange or vice versa, infection foci were detected in sour orange as well as C. macrophylla bark patches (Fig. 4 Ab and Bb). These data demonstrated that failure to infect sour orange plants by CTV with a deletion in p33 ORF was not related to the quantity of virus inoculum and further confirmed that the p33 ORF of CTV is required for systemic infection of sour orange.
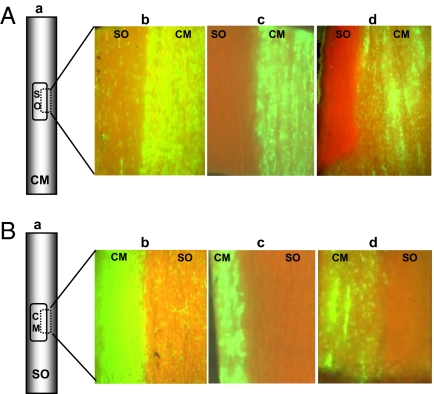
Fig. 4.
CTV9Δp33 failed to infect sour orange in bark patch experiment. (A) Schematic representation of bark patch from healthy sour orange onto C. macrophylla plants infected with GFP-tagged CTV deletion mutants (a). A small piece of healthy sour orange (SO) bark patch was graft inoculated onto C. macrophylla (CM) plants infected with CTV9-GFPC3 (b), CTV9Δp33-GFPC3 (c), and CTV9Δp33Δp18Δp13-GFPC3 (d). (B) Schematic diagram of C. macrophylla (CM) bark patch from GFP-tagged deletion mutants onto healthy sour orange (SO) plants (a). A small piece of C. macrophylla bark patch from CTV9-GFPC3– (b), CTV9Δp33-GFPC3– (c), CTV9Δp33Δp18Δp13-GFPC3–infected (d) plants were graft inoculated onto healthy sour orange plants. The substituted bark patches were allowed to establish vascular connections, and a small piece of bark patch junction of C. macrophylla and sour orange was excised and observed under Zeiss Stemi SV 11 UV-fluorescence dissecting microscope. Note that GFP fluorescence was not observed at detectable levels in sour orange bark patches in Ac, Ad, Bc, and Bd.
Deletion of the p33 and p18 ORFs Prevents Systemic Infection of Grapefruit Trees.
Grapefruit is an intermediately susceptible host for CTV, less than C. macrophylla and Mexican lime hosts, but more than sour orange (24). The ability of the deletion mutants to infect grapefruit was examined by graft inoculating young grapefruit trees with budwood pieces from C. macrophylla- or Mexican lime-infected plants. Full-length virus CTV9 and the deletion mutants CTV9Δp33, CTV9Δp18, CTV9Δp13, CTV9Δp33Δp13, and CTV9Δp18Δp13 systemically infected the grapefruit plants (Table 1). However, CTV9Δp33Δp18 and CTV9Δp33Δp18Δp13 failed to infect grapefruit plants at 8–12 wpi (Table 1). The CTV9Δp33Δp18 mutant was equivocal. Although there was no infection at 8–12 wpi, after 6 mo, a small proportion of plants exhibited a few infected cells. In contrast, grapefruit plants were not infected with CTV9Δp33Δp18Δp13 even after a prolonged incubation of 12 mo or more after inoculation. These data suggested that the p33 gene or the p18 gene was sufficient for systemic infection of grapefruit trees, but the p13 gene alone appears to allow a minimal amount that resulted in a very slow and limited systemic infection.
We next used GFP-tagged CTV9Δp33 and CTV9Δp33Δp18Δp13 mutants to further examine the roles of the p33, p18, and p13 ORFs in systemic infection of grapefruit plants. Budwood pieces from C. macrophylla-infected plants with CTV9-GFPC3, CTV9Δp33-GFPC3, or CTV9Δp33Δp18Δp13-GFPC3 were grafted into grapefruit plants. CTV9-GFPC3 and CTV9Δp33-GFPC3 produced infection foci in grapefruit at 8 wpi as expected (Fig. 2C). In contrast, no infection foci were observed in grapefruit plants inoculated with CTV9Δp33Δp18Δp13-GFPC3 (Fig. 2C). These data further confirmed that the p33 could be dispensable for systemic infection of grapefruit plants, but deletion of all three genes prevented systemic infection of grapefruit.
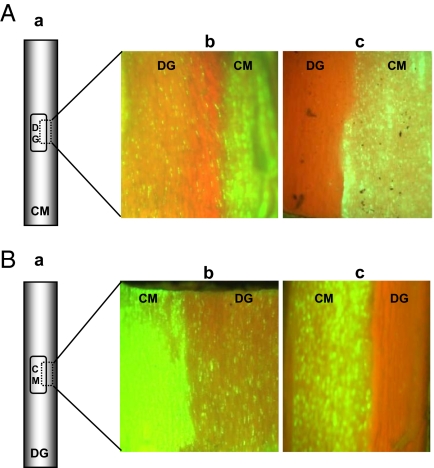
We also examined the ability of the mutants to infect grapefruit by replacing small rectangle bark patches from CTV9Δp33Δp18Δp13-GFPC3–infected C. macrophylla with those of healthy grapefruit and allowing the substituted bark patches to establish a vascular connection. In reciprocal experiments, small rectangle bark patches from CTV9Δp33Δp18Δp13-GFPC3–infected C. macrophylla plants were grafted onto grapefruit plants. The junctions of bark patches were examined under a stereo-fluorescence microscope at 8 wpi and the result was that grapefruit bark patches were free from GFP fluorescence (Fig. 5 Ac and Bc), demonstrating that failure to infect grapefruit by the triple gene deletion mutant was not due to limitation of virus inoculum. As expected, in a control experiment, CTV9-GFPC3 exhibited fluorescent foci in the grapefruit bark patches (Fig. 5 Ab and Bb).
Fig. 5.
CTV9Δp33Δp18Δp13 failed to infect grapefruit in bark patch experiment. (A) Schematic representation of bark patch from healthy grapefruit (DG) onto C. macrophylla (CM) plants infected with GFP-tagged CTV deletion mutant (a). A small piece of healthy grapefruit bark patch was graft inoculated onto C. macrophylla plants infected with CTV9-GFPC3 (b), and CTV9Δp33Δp18Δp13-GFPC3 (c). (B) Schematic diagram of C. macrophylla (CM) bark patch from GFP-tagged deletion mutant onto healthy grapefruit (DG) plants (a). A small piece of C. macrophylla bark patch from CTV9-GFPC3– (b), and CTV9Δp33Δp18Δp13-GFPC3–infected (c) plants was graft inoculated onto healthy grapefruit plants. The substituted bark patches were allowed to establish vascular connections, and a small piece of bark patch junction of C. macrophylla and grapefruit was excised and observed for GFP fluorescence. GFP fluorescence was not observed at detectable levels in grapefruit bark patches in Ac and Bc.
p33 or the 13 ORFs Were Required for Systemic Infection of Calamondin Plants.
Calamondin plants were graft inoculated with budwood and/or leaf pieces from C. macrophylla plants infected with the deletion mutants and examined for systemic infection at 8 wpi. The full-length virus and CTV with deletions in p33, p18 or p13; p33 and p18; and p18 and p13 systemically infected calamondin plants (Table 1). However, CTV with deletions in p33 and p13; and p33, p18, and p13 ORFs failed to infect calamondin plants at 8 wpi (Table 1). These data suggested that either the p33 or p13 gene was sufficient for systemic infection of CTV in calamondin plants.
Discussion
The members of Closteroviridae family encode conserved signature gene modules across the Closterovirus, Crinivirus and Ampelovirus genera with mono-, bi-, or tripartite genomes. These conserved gene products are involved primarily in replication and virion assembly. Additionally, members within a genus possess 1–5 genes, which are unique with no sequence identity with other members of the family, whose products are thought to interact with their specific hosts (14, 25). CTV encodes three nonconserved genes, p33, p18, and p13, that are dispensable for systemic infection of the more sensitive hosts (21). In this study, we found several other hosts in which none of these genes were needed, but that CTV needed these genes for systemic infection of some of its citrus hosts. However, we have not found a host in which all three genes were needed by CTV.
The bark patch substitution assay allowed a side-by-side examination of the ability of the virus mutants to move into different host tissues within the same plant. It works for examining both cell-to-cell and long-distance movement of the virus. Because the inoculum source from infected areas of the mother plant is the same for both tissues, this approach minimizes any variation of inoculum pressure. This procedure is a useful complement to scion-rootstock grafts in examining virus interactions with different hosts, particularly when the virus is visually tagged.
Systemic infection is the final result of a series of reactions that include initiation of infection, replication, cell-to-cell movement, long-distance movement, and mitigation of host–defense systems. Although with CTV infections, cell-to-cell movement may not be a requisite in some hosts (24). A defect in any of these processes can result in the lack of systemic infection. Thus, we cannot conclude that the specific function of the p33, p18, or p13 protein in certain hosts is to facilitate the virus crossing a cell wall and membrane. However, these proteins are not required for replication (20) or assembly (17) and appear not to be involved in mitigating RNA silencing (22). The bark-patch experiments with GFP-tagged deletion mutants suggest that one likely possibility is that the CTV deletion mutants are defective in entering and/or long-distance transport of the virus from initially infected cells.
CTV with a deletion in the p33 ORF failed to infect sour orange and lemon plants. CTV movement throughout sour orange by the wild-type virus is unusual in that it spreads by long-distance movement with essentially no cell-to-cell movement (24). This observation is in contrast to both processes occurring in susceptible C. macrophylla and Mexican lime plants. Experiments with bark patches from CTV9Δp33-GFPC3–infected C. macrophylla in sour orange plants and “duplex” plants with sour orange branches on CTV9Δp33-infected C. macrophylla plants demonstrated that failure to infect sour orange plants by CTV with a deletion in p33 ORF was not because of low concentration of virus inoculum. It would appear that the p33 is involved in interactions with host proteins of sour orange and lemon for successful long-distance transport of CTV.
CTV with deletions in both the p33 and p18 ORFs failed to systemically infect grapefruit trees. However, the presence of either p33 or p18 allowed movement in grapefruit. These results suggest that the p33 and p18 genes provided similar or redundant functions for CTV in this host. However, in some experiments, after a prolonged incubation (6 mo to 2 yr), CTV with deletions in p33 and p18 ORFs was able to infect a limited number of cells of the new growth of grapefruit plants, suggesting a slight amount of activity by the p13 alone. However, the mutant with deletions in all three genes (p33, p18, and p13) failed to infect grapefruit plants even after prolonged incubations of >12 mo. The function of different genes in grapefruit and the possible redundancy of the genes might be due to the complex genetics of grapefruit, because it is known to be a hybrid of several citrus species. One gene could function with one haploid genome and the other gene with the other haploid genome.
The presence of either the p33 or the p13 gene was sufficient for systemic infection of calamondin plants. Deletions in either p33 or p13 ORF did not affect the systemic infection of calamondin plants, but deletions in both p33 and p13 ORFs prevented the virus from systemically infecting calamondin plants. These results suggest that either p33 or p13 was sufficient for systemic infection by CTV in this plant. It should be noted that calamondin also is a hybrid—a hybrid of mandarin orange and kumquat, which come from two different genera.
Several viral proteins have been shown to be differential for specific hosts. For example, the papain-like leader proteases of Grapevine leafroll-associated virus-2, and -3a protein of Cowpea chlorotic mottle virus have been reported to fail to function in some hosts but not others (26, 27). Host-specific infections by potyvirid species have been studied extensively, and P1, P3, 6K2, VPg, NIa-Pro, and CP have been found to be differential in certain hosts (28–33). In general, different versions of these viral gene products function in specific hosts. However, the situation with CTV is quite different. In contrast to other viruses, acquisitions that changed hosts of CTV did not cause the loss of other hosts. CTV has genes that are needed in some hosts but are expendable in others. Thus, CTV was able to invade new hosts when additional genes had been incorporated into its genome.
The relationship between different strains of CTV is unusual. Although the 3′ portions of the genomes are within expected levels of sequence variation for members of the same virus group, the sequence similarity progressively decreases toward the 5′ termini to levels expected for unrelated viruses. Based on this level of differences, one possibility is that the different virus groups (strains) evolved in different hosts, perhaps thousands of years ago. An observation that could support this hypothesis is that most isolates of CTV are symptomless in most of their hosts. However, they often cause severe disease in a subset of their host range, supporting the argument that these hosts are different from that which the isolate evolved. However, from the results presented here, one possibility is that CTV progressively acquired genes for interacting with multiple hosts. Although viruses are known to have acquired nonconserved genes/domains (25, 34–36), the involvement of these genes to extend the virus host range had not been demonstrated. CTV has multiple genes required for systemic infection of different citrus hosts including three that are only needed for a subset of its hosts. If the virus evolved primarily in one citrus host, it would not be expected to have all of the expendable genes. However, no isolate has been found without all three genes. Approximately 20 isolates of CTV have been sequenced with no deletions found in any of these genes. In addition, Harper et al. (37) found that the percentage of negatively selected codons in p33, p18, and p13 was similar to that of all other CTV ORFs except for ORF 1b, which had a majority of codons apparently under purifying selection. Although the p33, p18, and p13 genes can be deleted with no defects being obvious in some hosts, perhaps these genes provide other functions that increase the fitness of the virus in these hosts.
Materials and Methods
CTV Deletion Mutants and Inoculation of Citrus Plants.
The wild-type cloned virus CTV9; deletion mutants CTV9Δp33, CTV9Δp18, CTV9Δp13, CTV9Δp33Δp18, CTVΔp33Δp13, CTV9Δp18Δp13, and CTV9Δp33Δp18Δp13; GFP-tagged wild-type virus CTV9-GFPC3; and GFP-tagged deletion mutants CTV9Δp33-GFPC3 and CTV9Δp33Δp18Δp13-GFPC3 have been maintained in C. macrophylla and/or Mexican lime plants under greenhouse conditions (21, 38). The bark tissue, budwood, and/or leaf pieces from C. macrophylla- or Mexican lime-infected plants were used as the source of inoculum to graft inoculate different citrus species used in host range studies (Table 1 and Table S1). A minimum of 3–4 test plants per citrus species per mutant were inoculated in host range experiments, and host range experiment was repeated at least 3 times. The grafted plants were pruned at 1 wpi, which resulted in the appearance of a new flush of leaves. Young stems were cut into small pieces, and extracts were analyzed by DAS-I-ELISA.
Serological Assays.
DAS-I-ELISA of tissue extracts was performed as described by using antibodies specific to CTV virions (23) to confirm infection in inoculated plants. Purified IgG from rabbit polyclonal antibody CTV-908 (1 μg/mL) was used as coating antibody. ECTV172, a broadly reactive CTV monoclonal antibody, was used as the detecting antibody. Citrus plants inoculated with deletion mutants were examined for infection by DAS-I-ELISA at different times beginning at 4 wpi.
Examination of Fluorescence in Citrus Plants Infected with GFP-Tagged Viruses.
Citrus plants inoculated with GFP-tagged variants of CTV and bark patch inoculation of sour orange or Duncan grapefruit/C. macrophylla with GFP-tagged deletion mutants were examined for fluorescence at different times beginning at 4 wpi by using a Zeiss Stemi SV 11 UV fluorescence dissecting microscope (Carl Zeiss Jena) with an attached Olympus Q-color 5 camera (Olympus America).
Supplementary Material
Acknowledgments
We thank Roy French for critical reading of the manuscript. This research was supported in part by an endowment from the J. R. and Addie S. Graves family and grants from the Florida Citrus Production Research Advisory Board and the National Research Initiative for the US Department of Agriculture Cooperative State Research, Education, and Extension Service.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113227108/-/DCSupplemental.
References
- 1.Li F, Ding SW. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucas WJ. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology. 2006;344:169–184. doi: 10.1016/j.virol.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Lucas WJ, Gilbertson RL. Plasmodesmata in relation to viral movement within leaf tissue. Annu Rev Phytopathol. 1994;32:387–411. [Google Scholar]
- 4.Carrington JC, Kasschau KD, Mahajan SK, Schaad MC. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang HL, Wang Y, Giesman-Cookmeyer D, Lommel SA, Lucas WJ. Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system. Virology. 1998;245:75–89. doi: 10.1006/viro.1998.9154. [DOI] [PubMed] [Google Scholar]
- 6.Xoconostle-Cázares B, Ruiz-Medrano R, Lucas WJ. Proteolytic processing of CmPP36, a protein from the cytochrome b(5) reductase family, is required for entry into the phloem translocation pathway. Plant J. 2000;24:735–747. doi: 10.1046/j.1365-313x.2000.00916.x. [DOI] [PubMed] [Google Scholar]
- 7.Waigmann E, Ueki S, Trutnyeva K, Citovsky V. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit Rev Plant Sci. 2004;23:195–250. [Google Scholar]
- 8.Harries PA, Schoelz JE, Nelson RS. Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol Plant Microbe Interact. 2010;23:1381–1393. doi: 10.1094/MPMI-05-10-0121. [DOI] [PubMed] [Google Scholar]
- 9.Harries P, Ding B. Cellular factors in plant virus movement: At the leading edge of macromolecular trafficking in plants. Virology. 2011;411:237–243. doi: 10.1016/j.virol.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Bar-Joseph M, Garnsey SM, Gonsalves D. The closteroviruses: A distinct group of elongated plant viruses. Adv Virus Res. 1979;25:93–168. doi: 10.1016/s0065-3527(08)60569-2. [DOI] [PubMed] [Google Scholar]
- 11.Moreno P, Ambrós S, Albiach-Martí MR, Guerri J, Peña L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol Plant Pathol. 2008;9:251–268. doi: 10.1111/j.1364-3703.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappu HR, et al. Nucleotide sequence and organization of eight 3′ open reading frames of the citrus tristeza closterovirus genome. Virology. 1994;199:35–46. doi: 10.1006/viro.1994.1095. [DOI] [PubMed] [Google Scholar]
- 13.Karasev AV, et al. Complete sequence of the citrus tristeza virus RNA genome. Virology. 1995;208:511–520. doi: 10.1006/viro.1995.1182. [DOI] [PubMed] [Google Scholar]
- 14.Karasev AV. Genetic diversity and evolution of closteroviruses. Annu Rev Phytopathol. 2000;38:293–324. doi: 10.1146/annurev.phyto.38.1.293. [DOI] [PubMed] [Google Scholar]
- 15.Satyanarayana T, Gowda S, Ayllón MA, Dawson WO. Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc Natl Acad Sci USA. 2004;101:799–804. doi: 10.1073/pnas.0307747100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowda S, Tatineni S, Folimonova SY, Hilf ME, Dawson WO. Accumulation of a 5′ proximal subgenomic RNA of Citrus tristeza virus is correlated with encapsidation by the minor coat protein. Virology. 2009;389:122–131. doi: 10.1016/j.virol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Satyanarayana T, et al. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology. 2000;278:253–265. doi: 10.1006/viro.2000.0638. [DOI] [PubMed] [Google Scholar]
- 18.Tatineni S, Gowda S, Dawson WO. Heterologous minor coat proteins of Citrus tristeza virus strains affect encapsidation, but the coexpression of HSP70h and p61 restores encapsidation to wild-type levels. Virology. 2010;402:262–270. doi: 10.1016/j.virol.2010.03.042. [DOI] [PubMed] [Google Scholar]
- 19.Hilf ME, et al. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology. 1995;208:576–582. doi: 10.1006/viro.1995.1188. [DOI] [PubMed] [Google Scholar]
- 20.Satyanarayana T, et al. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc Natl Acad Sci USA. 1999;96:7433–7438. doi: 10.1073/pnas.96.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatineni S, et al. Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology. 2008;376:297–307. doi: 10.1016/j.virol.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 22.Lu R, et al. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci USA. 2004;101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnsey SM, Cambra M. Enzyme-linked immunosorbent assay (ELISA) for citrus pathogens. In: Roistacher CN, editor. Graft Sixteenth IOCV Conference, 2005-Citrus Tristeza Virus Transmissible Diseases of Citrus. Handbook for Detection and Diagnosis. Rome: Food and Agric Org; 1991. pp. 193–216. [Google Scholar]
- 24.Folimonova SY, Folimonov AS, Tatineni S, Dawson WO. Citrus tristeza virus: Survival at the edge of the movement continuum. J Virol. 2008;82:6546–6556. doi: 10.1128/JVI.00515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolja VV, Kreuze JF, Valkonen JP. Comparative and functional genomics of closteroviruses. Virus Res. 2006;117:38–51. doi: 10.1016/j.virusres.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita Y, et al. Bromovirus movement protein conditions for the host specificity of virus movement through the vascular system and affects pathogenicity in cowpea. Mol Plant Microbe Interact. 2000;13:1195–1203. doi: 10.1094/MPMI.2000.13.11.1195. [DOI] [PubMed] [Google Scholar]
- 27.Liu YP, Peremyslov VV, Medina V, Dolja VV. Tandem leader proteases of Grapevine leafroll-associated virus-2: Host-specific functions in the infection cycle. Virology. 2009;383:291–299. doi: 10.1016/j.virol.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaad MC, Lellis AD, Carrington JC. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J Virol. 1997;71:8624–8631. doi: 10.1128/jvi.71.11.8624-8631.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajamäki ML, Valkonen JPT. The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides. Mol Plant Microbe Interact. 1999;12:1074–1081. doi: 10.1094/MPMI.1999.12.12.1074. [DOI] [PubMed] [Google Scholar]
- 30.Salvador B, Delgadillo MO, Sáenz P, García JA, Simón-Mateo C. Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol Plant Microbe Interact. 2008;21:20–29. doi: 10.1094/MPMI-21-1-0020. [DOI] [PubMed] [Google Scholar]
- 31.Chen KC, et al. A single amino acid of niapro of papaya ringspot virus determines host specificity for infection of papaya. Mol Plant Microbe Interact. 2008;21:1046–1057. doi: 10.1094/MPMI-21-8-1046. [DOI] [PubMed] [Google Scholar]
- 32.Decroocq V, et al. The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N-terminal region of the coat protein. Mol Plant Microbe Interact. 2009;22:1302–1311. doi: 10.1094/MPMI-22-10-1302. [DOI] [PubMed] [Google Scholar]
- 33.Tatineni S, Van Winkle DH, French R. The N-terminal region of wheat streak mosaic virus coat protein is a host- and strain-specific long-distance transport factor. J Virol. 2011;85:1718–1731. doi: 10.1128/JVI.02044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 35.Susaimuthu J, Tzanetakis IE, Gergerich RC, Martin RR. A member of a new genus in the Potyviridae infects Rubus. Virus Res. 2008;131:145–151. doi: 10.1016/j.virusres.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Mbanzibwa DR, Tian Y, Mukasa SB, Valkonen JP. Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC-Pro. J Virol. 2009;83:6934–6940. doi: 10.1128/JVI.00537-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harper SJ, Dawson TE, Pearson MN. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol. 2010;155:471–480. doi: 10.1007/s00705-010-0604-5. [DOI] [PubMed] [Google Scholar]
- 38.Satyanarayana T, et al. Amplification of Citrus tristeza virus from a cDNA clone and infection of citrus trees. Virology. 2001;280:87–96. doi: 10.1006/viro.2000.0759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.