Abstract
Conventional wisdom holds that microbes support their growth in vertebrate hosts by exploiting a large variety of nutrients. We show here that use of a specific nutrient (ethanolamine) confers a marked growth advantage on Salmonella enterica serovar Typhimurium (S. Typhimurium) in the lumen of the inflamed intestine. In the anaerobic environment of the gut, ethanolamine supports little or no growth by fermentation. However, S. Typhimurium is able to use this carbon source by inducing the gut to produce a respiratory electron acceptor (tetrathionate), which supports anaerobic growth on ethanolamine. The gut normally converts ambient hydrogen sulfide to thiosulfate, which it then oxidizes further to tetrathionate during inflammation. Evidence is provided that S. Typhimurium's growth advantage in an inflamed gut is because of its ability to respire ethanolamine, which is released from host tissue, but is not utilizable by competing bacteria. By inducing intestinal inflammation, S. Typhimurium sidesteps nutritional competition and gains the ability to use an abundant simple substrate, ethanolamine, which is provided by the host.
Keywords: diarrhea, gastroenteritis, metabolism
The pathogen Salmonella enterica serotype Typhimurium (S. Typhimurium) is a common cause of gastroenteritis (1), a diarrheal illness characterized by acute intestinal inflammation and the presence of neutrophils in stool samples (2). The pathogen triggers inflammation in the gut by using its virulence factors, two type III secretion systems (T3SS), termed T3SS-1 and T3SS-2, to invade the intestinal epithelium and survive in tissue phagocytes, respectively (3–6). Inflammation supports growth of S. Typhimurium in the intestinal lumen (7–9), thereby promoting transmission of the pathogen to the next susceptible host (10). However, it remains unknown what nutrients promote growth of S. Typhimurium in the inflamed intestine.
Because of extensive metabolic redundancies and access to a surprisingly diverse range of nutrients in vivo, many metabolic pathways in S. Typhimurium are not essential for growth in tissue (11). Although the array of nutrients is certainly also diverse in the gut lumen, it seems likely that competing microbes limit the usefulness of many of the substrates that can support fermentative growth in this anaerobic environment. Furthermore, inflammatory diarrhea changes this environment dramatically by removing potential nutrients from the gut lumen and reducing the diversity of available nutrients (12). Nutrient sources in the inflamed intestinal lumen include the mucein and enterocytes released from the tips of villi. Phosphatidylethanolamine is the most abundant phospholipid in membranes of released enterocytes (13). Ethanolamine, a nutrient derived from phosphatidylethanolamine, is present in the intestine of healthy calves (14). Here we investigated whether ethanolamine utilization supports growth of S. Typhimurium in an inflamed intestinal lumen.
Results
Ethanolamine Utilization Is Not Required for Virulence in the Mouse Typhoid Model.
The S. Typhimurium eut gene cluster encodes 17 genes required for the degradation and utilization of ethanolamine (eutS, -P, -Q, -T, -D, -M, -N, -E, -J, -G, -H, -A, -B, -C, -L, -K, -R) (15, 16) (Fig. 1A). To investigate the role of ethanolamine utilization in S. Typhimurium, we constructed an eutC mutant (PT100) in S. Typhimurium strain IR715. This mutant lacks ethanolamine ammonia lyase, which catalyzes the first step in ethanolamine utilization. Because ethanolamine has been shown to be present in the mammalian intestine (14), we investigated whether this nutrient supports growth of S. Typhimurium in the murine gut lumen. Mice infected with S. Typhimurium develop a typhoid fever-like disease, but acute neutrophilic inflammation does not develop in the intestine (mouse typhoid model) (Fig. S1A). In this mouse typhoid model, the eutC mutant (PT100) showed full virulence. Equal numbers of S. Typhimurium wild-type (IR715) and the eutC mutant (PT100) were recovered from intestinal contents and organs of mice (C57BL/6) 4 d after inoculation with an equal mixture of both strains (Fig. S1B). This result was consistent with a previous report showing that inactivation of the eut operon does not markedly attenuate S. Typhimurium virulence in the mouse typhoid model (16). Thus, data derived from infection of inbred susceptible mice support the idea that ethanolamine utilization is not essential for systemic infection.
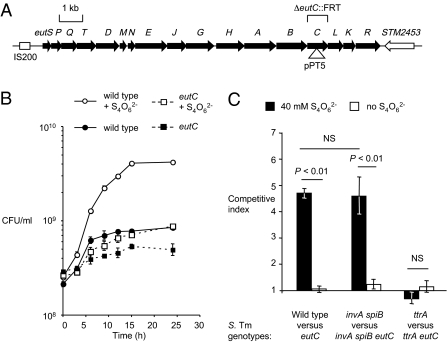
Fig. 1.
Anaerobic growth of S. Typhimurium on ethanolamine as the sole carbon source in vitro. (A) Schematic drawing of the eut locus in the chromosome of the S. Typhimurium strain LT2. The locations, orientations and sizes of genes are indicated by arrows. Mutation in strains PT100 (eutC::pPT5) and TT26355 (ΔeutC::FRT) are indicated. (B) Anaerobic growth curve of the S. Typhimurium wild-type (IR715; circles) and an eutC mutant (PT100; squares) with ethanolamine as the sole carbon source in the presence (open symbols) or absence (filled symbols) of the respiratory electron acceptor tetrathionate (S4O62−). The number of viable bacteria was determined at the indicated time points. (C) Competitive growth of S. Typhimurium strains [wild-type (IR715) vs. eutC mutant (PT100); invA spiB mutant (SPN452) vs. invA spiB eutC mutant (PT102); ttrA mutant (SW661) vs. ttrA eutC mutant (PT106)]. Bacterial numbers were determined 24 h after anaerobic growth with ethanolamine as the sole carbon source in the presence (black bars) or absence (white bars) of tetrathionate. Data represent the geometric mean of three independent experiments ± SE. Statistical significance (P value) is indicated above the bars.
Tetrathionate Respiration Supports Anaerobic Growth on Ethanolamine in Vitro.
We next investigated the ability of S. Typhimurium to grow on ethanolamine anaerobically in vitro. In the absence of an exogenous electron acceptor, ethanolamine only poorly supported anaerobic growth by fermentation (Fig. 1 B and C). However, in the presence of the respiratory electron acceptor tetrathionate (S4O62−), the S. Typhimurium wild-type (IR715), but not the eutC mutant (PT100), was able to anaerobically grow on ethanolamine as the sole carbon source (Fig. 1 B and C). Ethanolamine did not support anaerobic growth of S. Typhimurium in the absence of tetrathionate (Fig. 1 B and C). These data were consistent with previous studies demonstrating that under anaerobic conditions S. Typhimurium can use ethanolamine as a carbon source only in the presence of tetrathionate as a respiratory electron acceptor (17). Tetrathionate respiration does not provide a growth benefit for S. Typhimurium in the mouse typhoid model (8, 18). One limitation of the mouse typhoid model is that mice show no signs of gastroenteritis. This limitation can be overcome if, before S. Typhimurium infection, mice are treated with streptomycin, which results in intestinal inflammation with marked neutrophil transmigration into the lumen (Fig. S1C) (mouse colitis model) (5). In the mouse colitis model, it has recently been shown that neutrophils transmigrating into the intestinal lumen during inflammation increase the relative luminal abundance of S. Typhimurium (7) by oxidizing endogenous sulfur compounds (thiosulfate) to generate tetrathionate (8). These observations suggested that the importance of ethanolamine utilization in vivo should be reevaluated.
eut Gene Cluster Confers a Growth Advantage in the Mouse Colitis Model.
Because S. Typhimurium requires tetrathionate respiration for ethanolamine utilization under anaerobic conditions in vitro (Fig. 1 B and C) (17), we reasoned that this might also be true under conditions of the mouse colitis model. Streptomycin-pretreated mice (C57BL/6) were orally infected with an equal mixture of the S. Typhimurium wild-type strain (IR715) and the eutC mutant (PT100). These mice developed acute cecal inflammation (Fig. 2 A and B and Fig. S2B) and showed increased levels of mRNA from the Kc gene, encoding a neutrophil chemoattractant, and the Nos2 gene, encoding inducible nitric oxide synthase (Fig. 2C). A marked enrichment (P < 0.05) for the S. Typhimurium wild-type strain over the eutC mutant was observed 4 d after infection in the colon contents (Fig. 2D and Fig. S1D). A similar phenotype was observed with a S. Typhimurium mutant (TT26355) carrying an in-frame deletion of the eutC gene (ΔeutC::FRT), which was not polar on expression of the downstream eutLKR genes (Figs. S3 and S4). To determine whether a growth benefit is observed at later time points, genetically resistant mice (129/svJ) were pretreated with streptomycin and infected with an equal mixture of the S. Typhimurium wild-type strain (IR715) and the eutC mutant (PT100). A marked enrichment (P < 0.05) for the S. Typhimurium wild-type strain over the eutC mutant was observed 11, 12, and 15 d after infection in the fecal contents (Fig. S5). Growth of the eutC mutant (PT100) on ethanolamine in vitro and in the mouse colitis model (C57BL/6) could be restored by either bringing back the wild type eutC allele (PT238) or by complementing the strain with an F plasmid carrying the eut operon (F′ 606) (Fig. S3 A and B). Collectively, these data suggested that ethanolamine utilization provided a luminal growth advantage in the mouse colitis model.
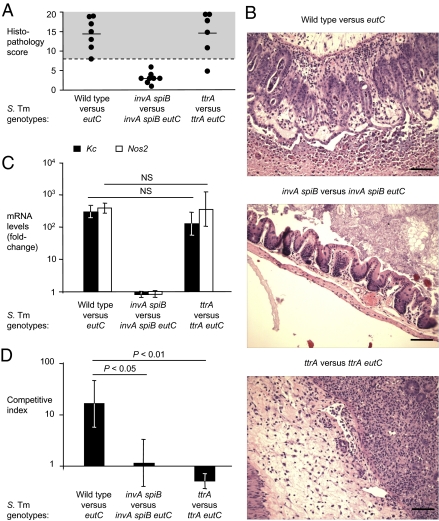
Fig. 2.
Ethanolamine utilization genes confer a growth advantage in the inflamed intestine. (A–D) Streptomycin-pretreated mice (mouse colitis model) were orally inoculated with equal mixtures of S. Typhimurium strains [wild-type (IR715) vs. eutC mutant (PT100); invA spiB mutant (SPN452) vs. invA spiB eutC mutant (PT102); ttrA mutant (SW661) vs. ttrA eutC mutant (PT106)] as indicated below each panel. Samples were collected 4 d after infection. The number of animals is indicated in A. (A) Blinded, combined histopathology score of cecal inflammation showing averages (lines) of individual scores (circles). (B) Representative images of H&E-stained cecal sections. (Scale bars, 100 μm.) (C) Expression of proinflammatory markers in the cecal mucosa. Relative mRNA levels of Kc (black) and Nos2 (white) were determined by quantitative real-time RT-PCR analysis. Data shown are fold-changes over mock-treated control animals. (D) Competitive indices of S. Typhimurium strains recovered from the colon content. Data represent the geometric mean ± SE. Statistical significance (P value) is indicated above the bars.
Ethanolamine Utilization Supports Growth in Inflamed Intestine.
S. Typhimurium‘s ability to invade and survive in host tissues depends on T3SSs (T3SS-1 and T3SS-2) that promote transfer of bacterial proteins into host cells. We inactivated T3SS-1 (through a mutation in invA) and T3SS-2 (through a mutation in spiB), thus rendering S. Typhimurium unable to induce intestinal inflammation in the mouse colitis model (5, 6, 8). The invA and spiB mutations did not reduce the ability of S. Typhimurium to grow anaerobically on ethanolamine in vitro (Fig. 1C). Streptomycin-pretreated mice were inoculated with an equal mixture of an invA spiB mutant (SPN452) and an invA spiB eutC mutant (PT102). Mice infected with this mixture developed no signs of intestinal inflammation (Fig. 2 A–C). Equal numbers of both strains were recovered from colon contents (Fig. 2D and Fig. S1D). This finding demonstrated that in the absence of inflammation, ethanolamine did not promote luminal growth of S. Typhimurium.
Effects of Inflammation on Ethanolamine Levels in the Gut.
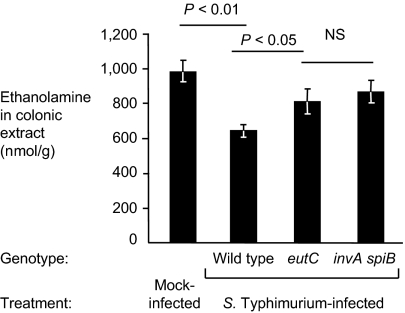
We next investigated whether inflammation induces release of ethanolamine into the gut lumen because epithelial erosion is sometimes observed in the cecum during S. Typhimurium infection. To address this question, streptomycin-pretreated mice were orally inoculated with either the S. Typhimurium wild-type strain (IR715), or an eutC mutant (PT100) or an invA spiB mutant (SPN452). Colonic extracts of whole colons from these mice were analyzed for free ethanolamine by reverse-phase HPLC coupled with mass spectrometry (MS) (Fig. 3 and Fig. S6). Ethanolamine levels were assayed 4 d after infection. Mice infected with the fully virulent eutC mutant (PT100) showed colon ethanolamine levels that were similar to the colon levels in uninfected mice and to mice infected with a strain unable to cause inflammation (invA spiB mutant) (Fig. 3). This finding indicated that altered ethanolamine concentrations during inflammation were not the reason for the inflammation-dependence of the observed ethanolamine utilization phenotype.
Fig. 3.
Quantification of intestinal ethanolamine concentrations in the mouse colitis model. Streptomycin-pretreated mice were orally infected with the S. Typhimurium wild-type (IR715), an eutC mutant (PT100), an invA spiB mutant (SPN452), or mock-treated, as indicated below each panel. Four days after infection, the ethanolamine concentration in colonic extracts was determined by LC-MS/MS. Data represent the geometric mean of four animals ± SE. Statistical significance (P value) is indicated above the bars.
Ethanolamine Utilization Provides No in Vivo Growth Advantage in the Absence of Tetrathionate Respiration.
The ttrA gene encodes tetrathionate reductase subunit A and is essential for growth of S. Typhimurium on ethanolamine in vitro (Fig. 1C). To determine whether growth on ethanolamine provided an advantage in the absence of tetrathionate respiration in vivo, streptomycin-pretreated mice were infected with an equal mixture of a ttrA mutant (SW661) and a ttrA eutC mutant (PT106). Mice infected with this mixture developed acute intestinal inflammation (Fig. 2 A–C and Fig. S2B). Both strains were recovered from colon contents in equal numbers (Fig. 2D), suggesting that even in the presence of inflammation, ethanolamine utilization did not provide a growth benefit without the ability to respire tetrathionate.
Ethanolamine Utilization Increases Numbers of S. Typhimurium Cells Present in Inflamed Intestine.
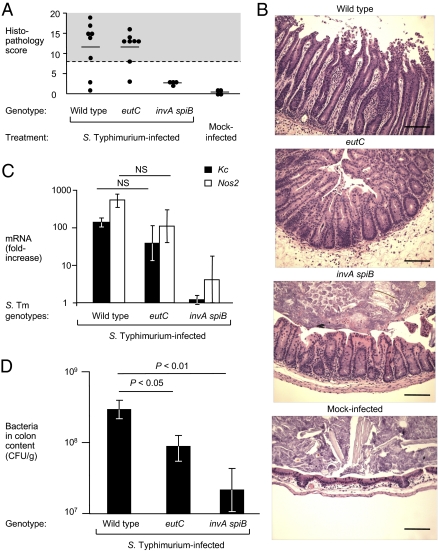
To further investigate whether ethanolamine consumption contributes to intestinal colonization, streptomycin pretreated mice were infected with individual S. Typhimurium strains. Mice were orally inoculated with the S. Typhimurium wild-type (IR715), an eutC mutant (PT100), or an invA spiB mutant (SPN452) and bacteria were recovered 4 d after infection (Fig. 4). Infection with the S. Typhimurium wild-type and the eutC mutant resulted in intestinal inflammation, but no marked inflammatory changes were observed in mice infected with the invA spiB mutant (Fig. 4 A–C). The S. Typhimurium wild-type was recovered in significantly (P < 0.05) higher numbers from colon contents than the eutC mutant or the invA spiB mutant (Fig. 4D). Collectively, these data support the concept that ethanolamine utilization enhances the ability of S. Typhimurium to grow the lumen of the inflamed gut.
Fig. 4.
Contribution of ethanolamine utilization to intestinal colonization in the mouse colitis model. (A–D) Streptomycin-pretreated mice were orally inoculated with the S. Typhimurium wild-type (IR715), an eutC mutant (PT100), an invA spiB mutant (SPN452), or mock-treated, as indicated below each panel. Samples were collected 4 d after infection. The number of animals is indicated in A. (A) Blinded histopathology scoring of cecal inflammation showing averages (lines) of individual scores (circles). (B) Representative images of H&E-stained cecal sections. (Scale bars, 100 μm.) (C) Expression of proinflammatory markers in the cecal mucosa. Relative mRNA expression of Kc (black) and Nos2 (white) was determined by quantitative real-time RT-PCR analysis. Data shown are fold-changes over mock-treated animals. (D) Recovery of S. Typhimurium from the colon content of infected mice. Data represent the geometric mean ± SE. Statistical significance (P value) is indicated above the bars.
Discussion
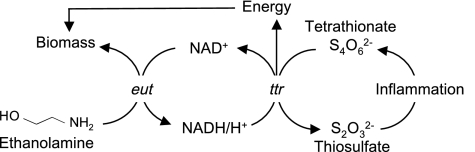
The principal strategy of S. Typhimurium for colonizing and disseminating through the intestinal route is to use its virulence factors (T3SS-1 and T3SS-2) to elicit acute intestinal inflammation (3). This host response stimulates oxidation of endogenous sulfur compounds (i.e., thiosulfate) and generates tetrathionate, which can be used as an electron acceptor that supports anaerobic respiration in S. Typhimurium (8) (Fig. 5). Here we show that tetrathionate respiration in the inflamed intestine enabled S. Typhimurium to use ethanolamine, a nutrient that supports very little anaerobic growth by fermentation. Thus, by inducing intestinal inflammation, a pathogen may gain the ability to consume substrates (by respiration) that cannot be readily fermented. Through this mechanism pathogens can take advantage of the host inflammatory response to foster their own growth in the gut.
Fig. 5.
Schematic of ethanolamine utilization and tetrathionate respiration in the inflamed gut.
The ability to use specific, nonfermentable carbon sources may provide a substantial growth benefit in the inflamed intestine by reducing competition with microbes that rely entirely on a fermentative metabolism. The effectiveness of the strategy is illustrated by the finding that tetrathionate respiration enables S. Typhimurium to outgrow the microbiota in the inflamed intestine (8). Here we show that this growth relies at least in part on the ability to use the nonfermentable carbon source ethanolamine. The resulting outgrowth contributes to the long-term success of S. Typhimurium by enhancing its transmission to the next host by fecal shedding (10). The concept that intestinal inflammation empowers facultative anaerobic bacteria to sidestep nutritional competition in the gut might be applicable to a wide range of other host microbe interactions and to therapeutic strategies.
Materials and Methods
Bacterial Strains.
The S. Typhimurium strains used in this study are listed in Table S1. Unless stated otherwise, bacteria were grown aerobically at 37 °C in Luria-Bertani (LB) broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) or on LB agar (15 g/L agar). When appropriate, antibiotics were added at the following concentrations: carbenicillin (0.1 mg/mL), chloramphenicol (0.03 mg/mL), nalidixic acid (0.05 mg/mL), kanamycin (0.05 mg/mL), and tetracycline (0.01 mg/mL).
Construction of S. Typhimurium Mutants.
Standard cloning techniques were performed as described previously (19). P22 HT105 int-201 generalized phage transduction was performed as describe previously (20). Plasmids and oligonucleotide sequences used in this study are listed in Tables S1 and S2, respectively. An internal fragment of the eutC gene was PCR amplified from the S. Typhimurium wild-type strain IR715 and cloned into pCR2.1 (Invitrogen) to generate pPT3. The “eutC” fragment from pPT3 was cloned into plasmid pEP185.2 to give rise to pPT5. Plasmid pPT5 was introduced into IR715 by conjugation to yield strain PT100. Strain PT100 was transduced to eut+ (PT238), which removed the eutC insertion mutation. For complementation, a his mutation (hisG::Tn10dTc) was added to the eutC mutant (PT100) and a low-copy F′ his+ eut+ plasmid (F′ 606) was introduced by conjugational transfer selecting for his+ (yielding strain TT26356). As a control, the eutC::pPT5 mutation was introduced into plasmid F′ 606 to give rise to F′ pPT5. Plasmid F′ pPT5 was introduced into PT100 by conjugational transfer selecting for his+ to give rise to strain TT26358. A phoN mutation was introduced into TT26356 by transduction (yielding strain PT251) to distinguish the strain from TT26358 during competitive infection experiments. An eutC::FRT deletion was added to the chromosome of strain IR715 by first introducing Δ763(cysA-eutS)::MudA by transduction to yield strain TT26354. Strain TT26354 was then used as recipient in a transduction cross with a cysA+ donor carrying the in-frame deletion mutation eutC::FRT to yield strain TT26355.
In Vitro Anaerobic Growth Assays.
Three milliliters of ethanolamine broth [5 mM ethanolamine (Sigma-Aldrich) in No-carbon-E medium (21) supplemented with trace elements (17) and 40 mM sodium tetrathionate, as indicated] was inoculated with 1 × 108 CFU/mL of each strain and anaerobically incubated at 37 °C (Bactron I anaerobic chamber; Sheldon Manufacturing). At the indicated time points, dilutions of the bacterial cultures were plated on LB agar plates containing the appropriate antibiotics.
Animal Experiments.
All experiments in this study were approved by the Institutional Animal Care and Use Committee at the University of California at Davis.
Mouse typhoid fever model.
Ten- to 12-wk-old female C57BL/6J wild-type mice were obtained from The Jackson Laboratory. Mice were inoculated intragastrically with 5 × 108 CFU of the indicated bacterial strains. Bacterial numbers in the inoculum were determined by spreading serial 10-fold dilutions on agar plates containing the appropriate antibiotics. Animals were killed 4 d after infection. Tissue samples were weighed, homogenized in 5 mL PBS (T-25 homogenizer; IKA Works), and bacterial numbers enumerated by plating serial 10-fold dilutions of homogenates on selective media.
Mouse colitis model.
C57BL/6J female wild-type mice and 129/svJ mice were obtained from The Jackson Laboratory. Ten- to 12-wk-old animals were pretreated with 20 mg streptomycin 24 h before infection. Mice were inoculated intragastrically with either 0.1 mL LB broth or S. Typhimurium strains (5). For competitive infections, mice were inoculated with an equal mixture of 5 × 108 CFU of each strain. The ratio of recovered wild-type to mutant bacteria was normalized by the ratio in the inoculum to determine the competitive index. All animals were killed at the indicated time points and organs were collected for mRNA purification, histopathology analysis and to determine bacterial numbers, as described previously (22).
Quantitative Real-Time RT-PCR Analysis.
Expression analysis was conducted as described previously (22). Briefly, RNA from mouse cecal tissues was isolated using TRI reagent (Molecular Research Center) according to the recommendations of the manufacturer. RNA was reversely transcribed using TaqMan reverse transcription reagents (Applied Biosystems). Then, SYBR-Green (Applied Biosystems) based real-time PCR was performed using the primers listed in Table S2 and analyzed with comparative Ct method (Applied Biosystems). The levels of mRNA expression of target genes (Kc and Nos2) were normalized by the levels of Gapdh mRNA.
Bacterial RNA was isolated using the Aurum Total RNA Kit (Bio-Rad) according to the recommendations of the manufacturer. Afterward, an additional DNase treatment was performed using the Ambion DNA removal and inactivation kit. RT-PCR and real-time PCR was performed as described previously (23) with the following modifications: 0.3 μg total RNA was used for reverse transcription and real-time PCR was performed using an ABI Viia7. The comparative CT method was used to analyze the data. Transcription of eutL, eutK, and eutR in each sample was normalized to the respective levels of guanyl nucleotide kinase mRNA, encoded by the gmk gene.
Histopathology.
Formalin fixed, H&E-stained cecal tissue sections were blindly evaluated by an experienced veterinary pathologist. The scoring scheme is shown in Fig. S2A. Images were produced by an Olympus BX41 microscope.
Measurement of Ethanolamine Concentrations in Intestine.
Ten- to 12-wk-old female wild-type mice (C57BL/6) were pretreated with 20 mg streptomycin for 24 h and orally inoculated with either 0.1 mL LB broth or 1 × 109 CFU of the indicated bacterial strains. Four days after infection, a section of the colon was collected in 1 mL of ultrapure H2O (Invitrogen), weighed, and homogenized (T-25 homogenizer). Ethanolamine in the tissue homogenate was derivatized as described previously (14). Briefly, 1 mL of tissue homogenate was incubated with 100 μL of 40% sulfosalicylic acid for 15 min then centrifuged at 11,000 × g for 15 min at room temperature. One milliliter of the supernatant was collected and 300 μL of 0.5 M NaHCO3, 2 mL of fresh dansyl chloride solution [20 mg/mL of dansyl chloride (Sigma Aldrich) in acetone], and 200 μL of 1 M NaOH was added. The samples were incubated for 20 min in the dark. Next, 200 μL of 25% NH4OH was added to inactivate residual dansyl chloride. The volume was adjusted to 5 mL with acetonitrile. One milliliter of this sample was centrifuged at 11,000 × g for 1 min and the supernatant was used for further analysis. LC-MS/MS analysis of dansyl monoethanolamine was performed using a Surveyor HPLC separation module (ThermoFischer) coupled to a LTQ linear ion trap MS (ThermoFinnigan) equipped with APCI ion source. The sample tray compartment was kept at 15 °C. Ten milliliters of methanolic solution of dansyl monoethanolamine synthesized starting from monoethanolamine was injected onto a Chromolith Monolithic RP-18e C18 analytical column [100 × 3 mm, (Merck)] equipped with a C18 guard cartridge. Mobile phases used were: A, 0.1% formic acid in deionized water and B, pure acetonitrile. Column temperature was 30 °C, and the flow rate was 0.4 mL/min. After a 0.2 min of isocratic run at 5% B, a gradient to 95% B was concluded at 6 min, and then continued at 5% B with to 0.5 min. The columns were equilibrated with the starting buffer for at least 3.5 min. The flow from the HPLC column was directed into the APCI ionization source of a LTQ linear ion trap MS (ThermoFinnigan) controlled with Xcalibur software (V1.4; ThermoFinnigan). Acquisition was performed in positive mode. Nitrogen sheath and auxiliary gas flow was 50 and 5 arbitrary units, respectively. Sweep gas flow rate was set to 5 units. Typical ion gauge pressure was 0.90_10_5. Vaporizer temperature was set to 450 °C and the discharge current was set to 5 mA. The capillary voltage was set to +9 V. Nitrogen sheath and auxiliary gas flow was 60 and 20 arbitrary units, respectively. The ion transfer capillary temperature was set to 275 °C. The AGC setting for selected ion monitoring and MS/MS was set to 1 × 103. Collision energy was set to 35%. A general MS/MS experiment was conducted to determine the fragmentation pattern (Fig. S3A), optimize the collision energy, and to choose the appropriate daughter ions for monitoring. Single ion monitoring scan spectra of parent ions were acquired from 292.5 to 297.5 at unit mass resolution with maximum injection time set to 200 ms in one micro-scan. The range of selected atomic mass units values corresponds to [M+H]+ parent ion of dansyl monoethanolamine (Mi 295). SRM transitions were found 295/280, 295/171, and 295/157 (Fig. S3B). These transitions were used for dansyl monoethanolamine detection. The transition 295/280 generated the most intense daughter ion and was chosen for quantitation. The target was eluted at 1.79 min (Fig. S3C). HPLC grade acetonitrile was purchased from Burdick and Jackson (VWR International). Each lot of organic solvents was investigated by LC/MS infusion. Extra pure formic acid was purchased from EMD. The ultrapure water was supplied by an in-house Millipore system.
Statistical Analysis.
Bacterial numbers, ethanolamine concentrations and changes in mRNA levels were transformed logarithmically before statistical analysis using a Student t test.
Supplementary Material
Acknowledgments
Work in A.J.B.'s laboratory is supported by Public Health Service Grants AI040124, AI044170, AI073120, AI076246, and AI088122. P.T. was supported by a scholarship from the Faculty of Medicine, Chiang Mai University, Thailand.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107857108/-/DCSupplemental.
References
- 1.Rabsch W, Tschäpe H, Bäumler AJ. Non-typhoidal salmonellosis: Emerging problems. Microbes Infect. 2001;3:237–247. doi: 10.1016/s1286-4579(01)01375-2. [DOI] [PubMed] [Google Scholar]
- 2.Harris JC, Dupont HL, Hornick RB. Fecal leukocytes in diarrheal illness. Ann Intern Med. 1972;76:697–703. doi: 10.7326/0003-4819-76-5-697. [DOI] [PubMed] [Google Scholar]
- 3.Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S, et al. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD and SopE2 act in concert to induce diarrhea in calves. Infect Immun. 2002;70:3843–3855. doi: 10.1128/IAI.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun. 2005;73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekirov I, et al. Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes. 2010;1(1):30–41. doi: 10.4161/gmic.1.1.10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stecher B, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley TD, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker D, et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 12.Santos RL, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai K, Fujita M, Nakao M. Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim Biophys Acta. 1974;369:222–233. [PubMed] [Google Scholar]
- 14.Bertin Y, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 15.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stojiljkovic I, Bäumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: Nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 20.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 21.Davis RW, Botstein D, Roth JR. Advanced Bacterial Genetics. New York: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 22.Winter SE, et al. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect Immun. 2009;77:1904–1916. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winter SE, et al. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog. 2010;6:e1001060. doi: 10.1371/journal.ppat.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.