Abstract
Intracellular membrane fusion requires R-SNAREs and Q-SNAREs to assemble into a four-helical parallel coiled-coil, with their hydrophobic anchors spanning the two apposed membranes. Based on the fusion properties of chemically defined SNARE- proteoliposomes, it has been proposed that the assembly of this helical bundle transduces force through the entire bilayer via the transmembrane SNARE anchor domains to drive fusion. However, an R-SNARE, Nyv1p, with a genetically engineered lipid anchor that spans half of the bilayer suffices for the fusion of isolated vacuoles, although this organelle has other R-SNAREs. To demonstrate unequivocally the fusion activity of lipid-anchored Nyv1p, we reconstituted proteoliposomes with purified lipid-anchored Nyv1p as the only protein. When these proteoliposomes were incubated with those bearing cognate Q-SNAREs, there was trans-SNARE complex assembly but, in accord with prior studies of the neuronal SNAREs, little lipid mixing. However, the addition of physiological fusion accessory proteins (HOPS, Sec17p, and Sec18p) allows lipid-anchored Nyv1p to support fusion, suggesting that trans-SNARE complex function is not limited to force transduction across the bilayers through the transmembrane domains.
Membrane fusion occurs throughout the secretory and endocytic pathways in eukaryotic cells. Although fusion events at certain organelles require unique regulators (e.g., synaptotagmin I for neuronal exocytosis), the general fusion machinery, conserved from yeast to humans, includes a Rab GTPase, one or more tethering factors, SNAREs [soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (SNAP) receptors], SNARE disassembly chaperones [Sec17p/α-SNAP and Sec18p/NSF], and Sec1p/Munc18 (SM) proteins (1). Although each of these components plays a critical role, SNAREs are thought to provide the driving force that merges two apposed membranes (2, 3).
SNAREs are a family of membrane-anchored proteins with 25 members in Saccharomyces cerevisiae, 36 in Homo sapiens, and 54 in Arabidopsis thaliana (1). Although most SNAREs are type II membrane proteins, some associate with the membrane via a covalently linked lipid moiety (e.g., SNAP-25, Ykt6p) or lipid-binding protein domain (e.g., Vam7p) (2). All SNAREs have in common a SNARE motif, 60–70 amino acyl residues that contain α-helix–forming heptad repeats. Cognate SNAREs from tethered membranes use their SNARE motifs to form a four-helical coiled-coil bundle (trans-SNARE complex), bringing membranes into close apposition. Buried at the center of this coiled-coil are the side chains of three glutamyl (Q) residues and 1 arginyl (R) residue, forming the ionic 0 layer. Based on whether they contribute a Q or R to the 0 layer, SNAREs are classified as Q-SNAREs or R-SNAREs (2, 4). A functional, fusogenic trans-SNARE complex is typically composed of three Q-SNAREs from one membrane and one R-SNARE from the other (3). For example, brain synaptic transmission requires VAMP2 (R-SNARE) on the synaptic vesicle to interact with syntaxin1A (Q-SNARE) and SNAP-25 (a Q-SNARE that provides 2 SNARE motifs) on the presynaptic membrane (5). In yeast, homotypic vacuole fusion requires trans-SNARE complexes formed by Nyv1p (R-SNARE) bound to one vacuolar membrane and Vam3p (Qa-SNARE), Vti1p (Qb-SNARE), and Vam7p (Qc-SNARE) on the other (6).
It is unclear how the formation of the trans-SNARE complex triggers membrane fusion. Reconstitution studies using neuronal SNAREs have shown that lipid mixing depends on a short linker that connects the trans-SNARE complex to the transmembrane domains (7). Proteoliposome bilayers held together by trans-SNARE complexes will undergo lipid mixing when they are bound to membranes by peptidic or lipidic anchors that span both leaflets but not when they bear phospholipid anchors spanning only the outer leaflet (8). These observations agree with studies of lipid-anchored Sso2p and Snc1p in yeast secretion (9), and they are in accord with the proposal (10) that the “zippering” of the trans-SNARE complex from the distal end (away from the membranes) to the proximal end transduces force to the membrane anchors to cause the merger of lipid bilayers. The four-helical coiled-coil bundle might even extend beyond the cytoplasmic SNARE motif to the transmembrane domains, as suggested by structural studies of the neuronal SNAREs (11). The important role of the transmembrane domain and its linkage to the coiled-coil SNARE bundle is further supported by the observation that an R-SNARE, Snc2p, with a partial transmembrane domain can prevent the transition from hemifusion to full fusion in a proteoliposomal fusion system (12).
Can SNAREs with lipid anchors spanning only the proximal, outer leaflet mediate fusion? SNAP-25–like proteins are anchored to membranes via palmitoylation or lipid-binding domains, but they function along with other transmembrane domain-bearing Q-SNAREs (i.e., syntaxins) from the same membranes. This partnership might eliminate the need for SNAP-25–like proteins to have their own transmembrane domains. Ykt6p, on the other hand, is an R-SNARE that is involved in multiple trafficking steps in vivo. It has a CCIIM motif at its C terminus, allowing palmitoylation (via the first cysteine) (13) and farnesylation (via the second cysteine) (14), both of which are required for stable membrane association (13). It is not clear whether the lipid-anchored Ykt6p forms fusogenic trans-SNARE complexes through interaction with three Q SNAREs from the opposite membrane in the same fashion as some of the better characterized R-SNAREs (e.g., Nyv1p, VAMP2). Reconstitution studies have implicated Ykt6p in both typical and atypical trans-SNARE complexes (15, 16).
We have used yeast vacuoles, which fuse in several well-delineated stages (6, 17), to determine whether a lipid-anchored Nyv1p (R-SNARE) is fusogenic in a typical trans-SNARE complex (i.e., R-SNARE from one membrane, 3 Q-SNAREs from the other). In priming, SNARE complexes on the same vacuolar membrane (cis-SNARE complexes) are disassembled by the Sec18p ATPase (NSF homolog) and its ligand Sec17p (α-SNAP homolog). Primed vacuoles tether to one another via HOPS, a six-subunit protein that has affinity for multiple membrane constituents, including the Rab GTPase Ypt7p (18), acidic lipids (19), Q-SNAREs Vam3p (20) and Vam7p (19), and the SNARE complex (21). After tethering, Nyv1p from one vacuole can form a trans-SNARE complex with three Q-SNAREs (Vam3p, Vti1p, and Vam7p) from the apposed vacuole, leading to fusion. When the transmembrane domain of Nyv1p was genetically replaced with the CCIIM motif from Ykt6p (Fig. 1A), allowing lipid modification, in vitro vacuole fusion was significantly impaired (22). However, fusion could be restored by the addition of extra Sec18p and Vam7p, suggesting that the remodeling of trans-SNARE complexes is as important for fusion as their steady-state level and that the requirement for a transmembrane domain can be bypassed in vacuole fusion.
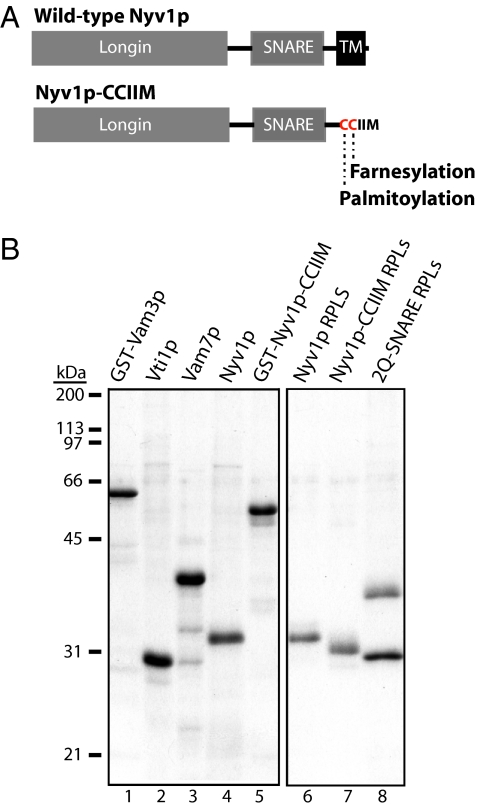
Fig. 1.
SNAREs and RPLs. (A) Domain structures of Nyv1p and Nyv1p-CCIIM. Note that Nyv1p-CCIIM does not have the transmembrane domain (TM). The two cysteines are for palmitoylation and farnesylation, respectively. (B) SDS-PAGE and colloidal blue staining of 250 ng of recombinant proteins (lanes 1–5) and RPLs (lanes 6–8) bearing either Nyv1p, Nyv1p-CCIIM, or the two Q-SNAREs (Vam3p and Vti1p). RPLs were prepared from protein-, lipid-, and detergent-mixed micellar solutions with an initial ratio of each SNARE to lipid of 1:1,000. Although equimolar Vam3p and Vti1p were used to prepare 2Q-SNARE RPLs, their ratio in harvested RPLs varied from 0.65 to 0.84, without effect on their lipid-mixing properties.
To facilitate a direct comparison of the functions of SNARE membrane anchors in model liposomal membranes with that in the intact organelle, we have now reconstituted Nyv1p-CCIIM–dependent fusion using pure proteins and lipids. Proteoliposome membranes bearing lipid-anchored Nyv1p (spanning only the outer leaflet) can fuse with membranes bearing the three Q-SNAREs in the presence of SNARE assembly and disassembly chaperones, indicating that force transduction across the entire bilayer through the transmembrane domains of the SNAREs is not essential for fusion.
Results
The transmembrane domain of Nyv1p (Fig. 1A) can be functionally replaced by short lipid anchors during vacuole fusion (22), although it remained possible that Nyv1p was bound to another integral membrane protein on the vacuole that provided a functional transmembrane anchor. To assess the function of the Nyv1p membrane anchor in a chemically defined setting, we have reconstituted purified lipid-anchored Nyv1p in liposomes of a lipid composition that mimics yeast vacuoles. GST-TEV cleavage site (TCS)-Nyv1p-CCIIM was expressed in yeast to allow proper lipid modification and then purified by glutathione affinity chromatography (Fig. 1B, lane 5). The GST tag was removed by TEV protease before the lipid-modified Nyv1p was mixed with vacuole-mimic lipids to generate proteoliposomes (lane 7). WT Nyv1p (lane 4) was incorporated into separate proteoliposomes in parallel (lane 6). Also shown in Fig. 1B are purified Q-SNAREs used in this study (lanes 1–3). The 2Q-SNARE proteoliposomes (Fig. 1B, lane 8) were made of Vam3p, Vti1p, and lipids.
Lipid-Anchored Nyv1p Participates in Trans-SNARE Complex Formation.
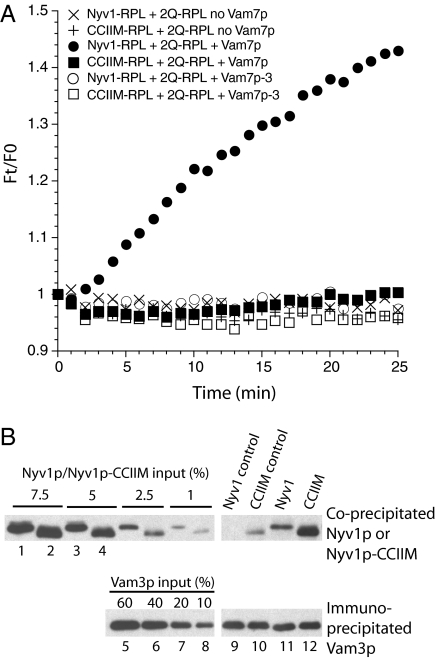
In a SNARE-only reaction, R-SNARE proteoliposomes were mixed with Q-SNARE proteoliposomes and the soluble Q-SNARE Vam7p. Lipid mixing was monitored by the dequenching of the N-(7-nitro-2-1,3-benzoxadiazole-4-yl)-phosphatidylethanolamine (NBD-PE)/rhodamine-PE FRET pair (23) on R-SNARE proteoliposomes (Materials and Methods). Soluble Vam7p was added at a concentration (6 μM) that supports robust lipid mixing of reconstituted proteoliposomes (RPLs) that bear WT Nyv1p (Fig. 2A, ●). As previously observed with purified vacuoles (22), lipid-anchored Nyv1p is ineffective in the absence of physiological accessory proteins (Fig. 2A, ■), whereas WT Nyv1p supports lipid mixing (Fig. 2A, ●). To examine whether the defect in lipid mixing is attributable to an inability of the lipid-anchored Nyv1p to form trans-SNARE complex, we replaced the WT Vam7p with a partially truncated Vam7p, Vam7p-3Δ, which arrests fusion after the formation of trans-SNARE complex (24), preventing postfusion cis-SNARE complexes from complicating the analysis (25, 26). Trans-SNARE complex formation during the fusion reaction was assayed as the Nyv1p that coimmunoprecipitated with Vam3p (Materials and Methods). About 2.5% of the WT Nyv1p coprecipitates with Vam3p (Fig. 2B, lane 11), with very little Nyv1p-Vam3p interaction in the detergent lysates (lane 9). Approximately 5% of the Nyv1p-CCIIM had associated with Vam3p (compare lanes 12 and 4), largely before detergent extraction (compare lanes 10 and 6). Because equal amounts of Vam3p were immunoprecipitated and there had been equal inputs of Nyv1p and Nyv1p-CCIIM, we conclude that the lipid-anchored Nyv1p can effectively form trans-SNARE complexes. These trans-SNARE complexes alone cannot support lipid mixing, in accord with other reports that lipid-anchored SNAREs can form nonfunctional or potentially inhibitory SNARE complexes (9, 22, 27).
Fig. 2.
Nyv1p-CCIIM forms nonfunctional trans-SNARE complexes. (A) R-SNARE RPLs, bearing Nyv1p (circles and X) or Nyv1-CCIIM (squares and +), and 2Q-SNARE RPLs were mixed with either 6 μM Vam7p (closed symbols), 6 μM Vam7p-3Δ (24) to permit trans-SNARE complex assembly while blocking fusion (open symbols), or buffer (X and +). Lipid mixing was measured at 27 °C for 25 min. HOPS is not required for fusion at this level of Vam7p (39). Reaction mixtures without WT Vam7p were transferred to ice and collected for trans-SNARE complex assay. F0, fluorescent signal at 0 min; Ft, fluorescent signal at a given time. (B) Analysis of trans-SNARE complexes. Immediately before the addition of detergent, 6 μM Vam7p-3Δ was added to “control” samples that had not received Vam7p-3Δ (lanes 9 and 10) during the reaction to control for any SNARE complex formation in detergent lysates. Immunoprecipitation was performed with antibodies to Vam3p. Nyv1p that had coimmunoprecipitated with Vam3p was examined by Western blotting. For each primary antibody, regions of the same blot from one gel are shown.
Lipid-Anchored Nyv1p Supports Lipid Mixing in the Presence of Accessory Proteins.
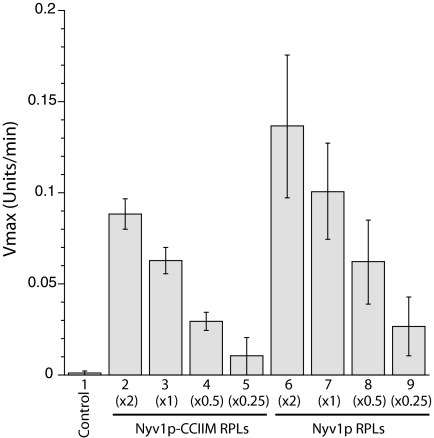
Yeast vacuoles bearing Nyv1p-CCIIM were able to fuse efficiently when provided with additional Sec18p and Vam7p, presumably to drive cycles of SNARE complex disassembly and reassembly for the remodeling of trans-SNARE complexes (22). We therefore assayed whether the addition of the SNARE disassembly chaperones (Sec17p/Sec18p) and the assembly chaperone (HOPS) to proteoliposomes would have a similar stimulatory effect. In the presence of Sec17p/Sec18p, HOPS, and 0.6 μM Vam7p, proteoliposomes bearing lipid-anchored Nyv1p were indeed able to merge with those bearing Q-SNAREs (Fig. 3, bars 2–5, and Fig. S1A, ○). On a molar basis, the lipid-mixing activity of lipid-anchored Nyv1p is ∼1/2 that of WT Nyv1p (Fig. 3, compare lanes 2 and 7 and lanes 3 and 8). Nevertheless, both on vacuoles (22) and proteoliposomes, lipid-anchored Nyv1p is activated by SNARE chaperones to support lipid mixing. This fusion reaction exploits the same pathway as that mediated by the WT Nyv1p, because both reactions require the same components (Fig. 4).
Fig. 3.
Nyv1p-CCIIM is active in chaperone-mediated fusion reactions. Reactions contained 1 mM MgCl2, an ATP-regenerating system, RPLs bearing Nyv1p-CCIIM (×2: protein/lipid ratio = 2:1,000; ×1: 1:1,000; ×0.5: 0.5:1,000; ×0.25: 0.25:1,000) or Nyv1p at corresponding levels, 2Q-SNARE RPLs (protein/lipid ratio = 1:1,000), Vam7p (600 nM), HOPS (40 nM), Sec17p (0.68 μM), and Sec18p (0.24 μM). In a control reaction, the 2Q-SNARE RPLs were omitted. The maximal rates of lipid mixing were calculated based on four independent experiments (Materials and Methods). Error bars represent SDs. A typical experiment is shown in Fig. S1.
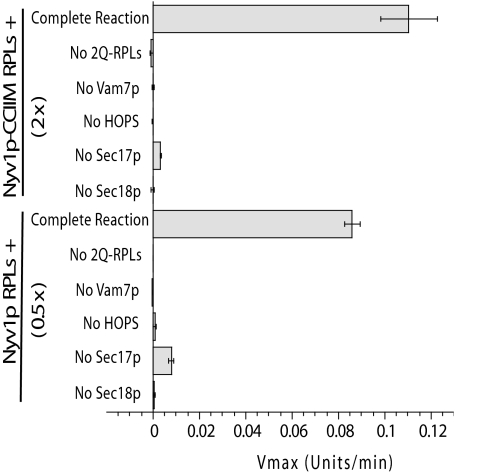
Fig. 4.
Protein requirements for Nyv1p-CCIIM–mediated lipid mixing. RPLs bearing Nyv1p-CCIIM (protein/lipid ratio = 2:1,000) or Nyv1p (protein/lipid ratio = 0.5:1,000) were assayed for lipid mixing as in Fig. 3. As previously reported (39) and as shown in Fig. 2, vacuolar R-SNARE RPLs will undergo lipid mixing with 2Q-SNARE RPLs with 6 μM Vam7p, whereas with only 0.6 μM Vam7p, HOPS, Sec17p, and Sec18p are required. Where specified, a single component was replaced with its buffer. Fusion rates were the mean of three independent experiments (Materials and Methods). Error bars represent SDs. A typical experiment is shown in Fig. S2.
Nyv1p-CCIIM and WT Nyv1p Catalyze Content Mixing and Lysis in the Same Proportion.
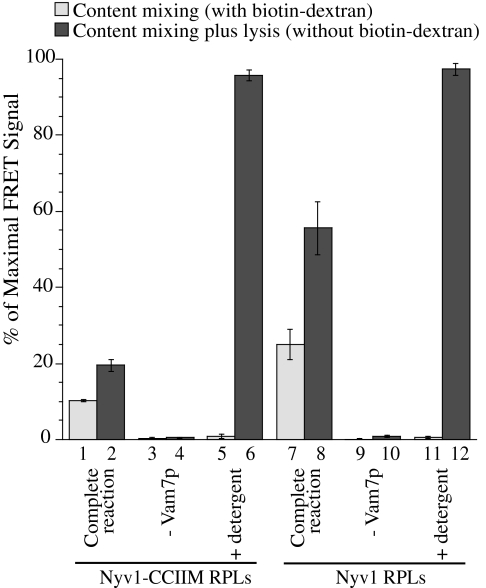
The fusion of proteoliposomes is routinely monitored by lipid mixing, yet SNARE-driven lysis (28) and the ensuing membrane reannealing (29) are also detected by this assay. To determine whether lipid-anchored Nyv1p mediates content mixing and/or lysis, we turned to a recently developed content-mixing assay (30) that employs streptavidin-Cy5 entrapped within R-SNARE proteoliposomes and biotin-R-phycoerythrin entrapped in Q-SNARE proteoliposomes. Full fusion leads to the almost irreversible association of streptavidin with biotin, bringing their bound R-phycoerythrin and Cy5 into close proximity and generating a robust FRET signal. Because a large molar excess of nonfluorescent biotin-dextran effectively blocks streptavidin and biotin association, and hence the FRET signal (Fig. 5, compare bars 5 and 6 and bars 11 and 12), we added it to the buffer surrounding the proteoliposomes in the fusion reaction (bars with odd numbers) to prevent any signal derived from proteoliposome lysis. We also measured the overall FRET signal from content mixing plus lysis in parallel reactions without biotin-dextran (bars with even numbers). When all the fusion components were present, both lipid-anchored Nyv1p and WT Nyv1p supported content mixing (lanes 1 and 7). Omission of the Qc-SNARE Vam7p abolished the content mixing signal (bars 3 and 9), showing that the reaction is SNARE-dependent. The fusion activity of lipid-anchored Nyv1p is ∼40% of that of WT Nyv1p, in accord with lipid-mixing assays (Fig. 3). Lysis accompanied fusion for both Nyv1p proteoliposomes and Nyv1p-CCIIM proteoliposomes, but the ratio of lysis to content mixing was unchanged (compare bars 1 and 2 vs. 7 and 8), suggesting that the lipid-anchored Nyv1p-CCIIM did not cause more lysis than WT Nyv1p. Together, these data show that lipid-anchored Nyv1p suffices as the sole SNARE on one membrane to mediate crucial steps leading to content mixing.
Fig. 5.
Content mixing of RPLs bearing Nyv1p-CCIIM. In a complete fusion reaction with all the SNAREs and chaperones, content mixing was monitored by the FRET signal between streptavidin-Cy5 (from R-SNARE RPLs) and biotin-R-phycoerythrin (from Q-SNARE RPLs) with a large molar excess of biotin-dextran in the reaction (light gray columns). Content mixing plus lysis was monitored in a parallel reaction without biotin-dextran (dark columns). Where indicated, Vam7p was replaced with buffer or detergent was added. The maximal FRET signal in the absence of biotin-dextran was determined at t = 60 min. The FRET signals at 45 min as a percentage of the maximal signal from three independent experiments were averaged and used to compare the content mixing or lysis under different fusion conditions. Error bars represent SDs.
Discussion
SNARE proteins form trans-complexes that are anchored in apposed membranes to cause membrane fusion. There are several nonexclusive mechanistic possibilities for their action. SNAREs might bring apposed bilayers so close that they can fuse spontaneously; however, even liposomes that are sedimented to a pellet under enormous pressure do not fuse. SNAREs might provide a platform that can spatially enrich other fusion proteins (31), although these other proteins alone cannot mediate bilayer rearrangement. SNAREs can also regulate the enrichment of inherently fusogenic lipids, such as diacylglycerol, at docking microdomains (32), although it is unclear whether this enrichment alone would support fusion. The tilted membrane anchor domain of syntaxin may itself be fusogenic (33). SNAREs in trans-complex might join their α-helical SNARE domains to their α-helical transbilayer anchor domains, forming long, continuous, and parallel helices that exert a destabilizing stress on docked membranes (11, 34). However, trans-SNARE complexes anchored by isoprenoid chains, which can span each docked bilayer but cannot join in an α-helix, will mediate the fusion of SNARE proteoliposomes, whereas shorter membrane anchors embedded in only one monolayer do not (8). These studies have suggested that the completely “zippered” SNARE complex may drive fusion by using the transmembrane anchor of each SNARE to exert a pull across the entire bilayer of each membrane.
In accord with our earlier organelle-based studies, we find that the fusion of proteoliposomes bearing the R-SNARE Nyv1p with Q-SNARE proteoliposomes, in the absence of any other proteins, strongly depends on the Nyv1p transmembrane domain and that a lipid-anchored Nyv1p engages in normal SNARE complex assembly but will not suffice for fusion. However, when HOPS and the SNARE disassembly chaperones Sec17p and Sec18p are present, substantial fusion is restored to proteoliposomes with lipid-anchored Nyv1p. Because these proteins are absolutely required for fusion in vivo and in cell-free incubations with the intact organelle (6), we conclude that a fully transbilayer anchor is not essential for fusion.
What are the functions of HOPS, Sec17p, and Sec18p in this reaction? Sec17p and Sec18p work together as an ATP-driven machine to disassemble SNARE complexes. HOPS is a tethering factor (19, 35) and protects functional trans-SNARE complexes from disassembly by Sec17p/Sec18p (25). Together, HOPS, Sec17p, and Sec18p may promote membrane fusion by converting nonfunctional trans-SNARE complexes into functional ones. The dependence on Sec17p/Sec18p for the fusion of liposomes bearing lipid-anchored Nyv1p suggests that it may be more prone than WT Nyv1p to assemble into nonfunctional trans-SNARE complexes (e.g., antiparallel coiled-coil), although direct evidence for this is lacking. Because the SM homolog Vps33p is a subunit of HOPS, the HOPS complex may also have a direct role in promoting membrane merger, as suggested for other SM proteins (31).
It will be important to investigate whether other lipid-anchored SNAREs, particularly R-SNAREs, can support fusion reactions in the presence of physiological fusion factors. Overexpression of the exocytic yeast SNARE Snc1p (R-SNARE) or Sso2p (Qa-SNARE) with its transmembrane domain replaced by a geranylgeranyl anchor had a dominant negative effect on cell growth. These cells exhibited ∼1/5 the normal secretion rate (9), whereas we see ∼1/2 the rate of fusion with lipid-anchored Nyv1p compared with the WT. However, direct comparison of the study by Grote et al. (9) and our current findings is difficult because they differ in lipid anchors (geranylgeranyl vs. paltmitoyl and farnesyl anchors) and in the context of the study (in vivo vs. RPLs). In vacuole fusion, the R-SNARE Ykt6p, with its native lipid anchor, can functionally replace Nyv1p under certain conditions (36). Synaptic transmission requires the lipid-anchored SNAP-25 in complex with VAMP2 and syntaxin1, each with a transmembrane anchor of apolar amino acyl residues; it will be of great interest to determine whether these neuronal SNARE anchors, if rendered lipidic, could also function when provided NSF, α-SNAP, and the Exocyst complex.
Assays of liposome lipid mixing have suggested that SNAREs are the engine of membrane fusion, directly transferring the energy of four-helical bundle assembly to their transmembrane domains to overcome the energy barrier to fusion. However, elevated levels of SNAREs can also drive lysis (28, 29), and other proteins, such as the Sec1p/Munc18 family members, are as essential to fusion as the SNAREs. Lysis-free fusion at low SNARE levels may be a multiple-step process: first, tethering mediated by a Rab family GTPase and its large effector complex, such as HOPS, and then SNARE complex assembly. SNARE assembly can be promiscuous and nonfunctional, but complexes like HOPS, which includes the SM factor (HOPS subunit Vps33p), may protect only the authentic and functional SNARE complex from disassembly by Sec18p/NSF and Sec17p/α-SNAP (25, 37). The energy barrier to bilayer rearrangement for fusion may then be lowered by some combination of SNARE-mediated local enrichment of fusogenic lipids (32), bilayer disruption via tilted SNARE transmembrane anchor (33), and force transmission from SNARE domains to transmembrane domains (7, 8); the absence of one or another of these factors may still allow fusion to occur.
Materials and Methods
Strain Construction.
To prepare Nyv1p-CCIIM from yeast cells, cDNA encoding GST-fused Nyv1p-CCIIM was constructed by overlap extension PCR. Briefly, the DNA fragment encoding GST, followed by a TCS, was PCR-amplified from pGST-Parallel1 (38) using primer-1 and primer-2, and the DNA fragment for Nyv1p-CCIIM was PCR-amplified from pRS406-NYV1-CCIIM (22) using primer-3 and primer-4. The DNA fragment encoding GST-TCS-NVY1-CCIIM was generated and amplified by overlap extension PCR using primer-1 and primer-4. The resulting DNA fragment was digested with HindIII and XhoI, and it was inserted into the HindIII/XhoI sites of the pYES2-NT-C plasmid vector (Invitrogen), generating pYES2-GST-TCS-NYV1-CCIIM. This construct contains a 4-aa-long linker sequence (TVDA) between the TCS and NYV1P-CCIIM regions for efficient cleavage by TEV:
Primer-1: ctg AAG CTT acc atg tcc cct ata cta ggt tat tgg
Primer-2: gcg ttt cat ggc gtc aac ggt gcc ctg aaa ata cag gtt ttc
Primer-3: cag ggc acc gtt gac gcc atg aaa cgc ttt aat gta agt tat g
Primer-4: cgc CTC GAG cta cat gat gat gca aca att ttt g
Proteins and Antibody Preparation.
To purify GST-Nyv1p-CCIIM, the galactose (GAL)-positive and vacuolar protease-deficient strain BJ5459 was transformed with pYES2-GST-TCS-NYV1-CCIIM and incubated on complete synthetic media lacking uracil at 30 °C for 2 d. Yeast cells from a single colony were grown at 30 °C to OD600 = 0.4 in 10 L of CSM-URA (0.77 g/L; Q-Biogene) minimal medium containing 0.17% yeast nitrogen base (DIFCO), 0.5% ammonium sulfate, 2% (wt/vol) d-(+)-raffinose pentahydrate, and 0.2% d-sucrose (pH 6.5). After addition of 530 mL of 40% (wt/vol) GAL (final = 2%), growth was continued for 24 h before harvesting by centrifugation [5,000 rpm, 5 min, room temperature, JLA-10.500 rotor (Beckman) in a Beckman centrifuge]. The pellet was resuspended in 400 mL of ice-cold wash buffer [20 mM Hepes⋅NaOH (pH 7.4), 500 mM NaCl, 1 mM MgCl2, and 10% (vol/vol) glycerol] and centrifuged as before. The pellet was resuspended in wash buffer at a quarter of the weight of the pellet (i.e., 10 mL for 40 g of pellet), followed by addition of protease inhibitors [1 mM PMSF, 0.62 μg/mL leupeptin, 4 μg/mL pepstatin A, and 24.4 μg/mL Pefabloc-SC (Roche)]. The mixture was flash-frozen as droplets in liquid N2, blended in a Waring blender in the presence of liquid N2, and stored at −80 °C.
To extract GST-Nyv1p-CCIIM, the blended yeast cells were diluted with 150 mL of wash buffer containing protease inhibitors (above) and then centrifuged [30,000 rpm, 1 h, 4 °C, 45Ti rotor (Beckman) in a Beckman centrifuge]. The pellet was resuspended in 160 mL of wash buffer, homogenized using a dounce homogenizer, and centrifuged (45Ti, 4 °C, 1 h). The pellet was resuspended and homogenized in 160 mL of extraction buffer [20 mM Hepes⋅NaOH (pH 7.4), 500 mM NaCl, 1 mM MgCl2, 10% (vol/vol) glycerol, and 0.5% Thesit (Sigma)], and it was then incubated at 4 °C for 2 h with nutation. The mixture was centrifuged (15,000 rpm, 45Ti, 4 °C, 10 min), and the supernatant was transferred to prechilled 45Ti tubes for a 35-min centrifugation (30,000 rpm, 4 °C). The supernatants were pooled and mixed at 4 °C overnight with 10 mL of glutathione agarose resin (General Electric) pre-equilibrated with extraction buffer.
In the cold room, the suspension was poured into a chromatography column (2.5 × 20 cm). After draining, the resin was washed with two 50-mL portions of ice-cold extraction buffer and then with 50 mL of preelution buffer [20 mM Hepes⋅NaOH (pH 7.4), 150 mM NaCl, 10% glycerol, 1 mM MgCl2, and 1% (wt/vol) n-Octyl-β-D-Glucopyranoside (β-OG)]. Bound proteins were eluted with 50 mL of elution buffer [20 mM Hepes⋅NaOH (pH 7.4), 150 mM NaCl, 10% (vol/vol) glycerol, 1 mM MgCl2, 1% β-OG, and 20 mM glutathione], and fractions (1.2 mL) enriched with proteins (Bradford) were pooled, concentrated with a Millipore centrifugal filter (30-kDa cutoff), flash-frozen in liquid N2 in small aliquots, and stored at −80 °C.
All other proteins and antibodies used in this study were prepared as described elsewhere (26).
RPLs and Lipid-Mixing Assay.
The 2Q-SNARE proteoliposomes with vacuole-mimic lipids were prepared as described (39). R-SNARE RPLs bearing Nyv1p-CCIIM use the same lipid film as above, except that the lipid film was resuspended in 545 μL of 20 mM Hepes⋅NaOH (pH 7.4), 150 mM NaCl, 10% (vol/vol) glycerol, 1 mM MgCl2, and 1% β-OG before the addition of 455 μL of Nyv1p-CCIIM. For R-SNARE RPLs with the typical 1:1,000 SNARE/lipid ratio, 223 μg of GST-TEV-Nyv1p-CCIIM was incubated with 65 μg of MBP-TEV at 4 °C overnight in 520 μL of cleavage buffer [20 mM Hepes⋅NaOH (pH 7.4), 150 mM NaCl, 10% (vol/vol) glycerol, 1 mM MgCl2, and 1% β-OG] with nutation. Following centrifugation (5,000 × g for 5 min), 455 μL of supernatant was collected for RPL preparation. R-SNARE RPLs bearing WT Nyv1p were prepared in parallel.
Lipid-mixing assays, including R-SNARE RPLs (50 μM) and Q-SNARE RPLs (400 μM), were performed as described (25). Fusion is calculated as the increased fluorescence (attributable to lipid mixing) at any time divided by the fluorescence at the first minute [(Ft − F0)/F0]. An increase of 1 in this parameter is defined as one unit of fusion. To compare fusion reactions, the maximal rate of fusion was calculated. Error bars are SDs from three or more experiments.
Content-Mixing Assay.
Content-mixing assays were performed as described (30). In brief, liposomes bearing two Q-SNAREs and R-SNARE proteoliposomes were prepared as above with some modifications [1-Palmitoyl-2-oleoyl-3-sn-phosphatidylcholine (43.6% mol/mol or 46.6% mol/mol) for donor or acceptor], no cardiolipin, addition of Ypt7p at a lipid/protein ratio of 10,000:1 (mol/mol), and fluorescent lipids only for donors [1.5% (mol/mol) Marina Blue-PE and 1.5% (mol/mol) NBD-PE]. During proteoliposome preparation, the following probes were entrapped: R-SNARE donor (8 μM Cy5-labeled streptavidin; KPL, Inc.) and 2Q-SNARE acceptors (4 μM biotinylated R-phycoerythrin; Invitrogen).
To measure content mixing, R-SNARE donor and two Q-SNARE acceptor proteoliposomes (0.25 mM lipid each) were mixed with Sec17p (32.5 nM), Sec18p (0.15 μM), HOPS (40 nM), ATP (1 mM), Vam7p (1 μM), and dextran-biotin (Mr = 70,000, 0.5 μM). Reaction mixtures were incubated in 384-well plates at 27 °C in a fluorescence plate reader for 45 min, and FRET signals between R-phycoerythrin and Cy5 were recorded (Excitation: 565 nm; Emission: 670 nm; cutoff: 630 nm).
Trans-SNARE Complex Assay.
Reaction mixtures (450 μM lipids, 20 μL) were chilled (ice, 5 min), with each receiving 2.2 μg of GST-Nyv1(ΔTM) and, after an additional 10 min, 800 μL of 25 mM Tris⋅Cl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40-Alternative (Calbiochem), 1% deoxycholate, 0.1% SDS, 10 mM EDTA, and protease inhibitors (above). GST-Nyv1(ΔTM) was added to block potential binding of native Nyv1p to Vam3p in detergent. Samples were nutated at 4 °C for 20 min and then centrifuged (16,000 × g, 4 °C, 5 min). Supernatant (700 μL) was used for immunoprecipitation with immobilized αVam3p as described elsewhere (25).
Supplementary Material
Acknowledgments
We thank Amy Orr and Holly Jakubowski for excellent technical support. This work was supported by National Institutes of Health Grant GM23377. M.Z. was supported by a Research Fellowship (ZI 1339/1-1) through the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113888108/-/DCSupplemental.
References
- 1.Malsam J, Kreye S, Söllner TH. Membrane fusion: SNAREs and regulation. Cell Mol Life Sci. 2008;65:2814–2832. doi: 10.1007/s00018-008-8352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs—Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNew JA. Regulation of SNARE-mediated membrane fusion during exocytosis. Chem Rev. 2008;108:1669–1686. doi: 10.1021/cr0782325. [DOI] [PubMed] [Google Scholar]
- 6.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 7.McNew JA, Weber T, Engelman DM, Söllner TH, Rothman JE. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 8.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grote E, Baba M, Ohsumi Y, Novick PJ. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J Cell Biol. 2000;151:453–466. doi: 10.1083/jcb.151.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 11.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Zhang F, Su Z, McNew JA, Shin YK. Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 13.Dietrich LE, et al. The SNARE Ykt6 is released from yeast vacuoles during an early stage of fusion. EMBO Rep. 2005;6:245–250. doi: 10.1038/sj.embor.7400350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNew JA, et al. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- 15.Parlati F, et al. Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc Natl Acad Sci USA. 2002;99:5424–5429. doi: 10.1073/pnas.082100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 17.Wickner W. Yeast vacuoles and membrane fusion pathways. EMBO J. 2002;21:1241–1247. doi: 10.1093/emboj/21.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulubova I, Yamaguchi T, Wang Y, Südhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- 21.Kramer L, Ungermann C. HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites. Mol Biol Cell. 2011;22:2601–2611. doi: 10.1091/mbc.E11-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun Y, Xu H, Thorngren N, Wickner W. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. EMBO J. 2007;26:4935–4945. doi: 10.1038/sj.emboj.7601915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz ML, Merz AJ. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol. 2009;185:535–549. doi: 10.1083/jcb.200811082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, Jun Y, Thompson J, Yates J, Wickner W. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 2010;29:1948–1960. doi: 10.1038/emboj.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Wickner W. Phosphoinositides function asymmetrically for membrane fusion, promoting tethering and 3Q-SNARE subcomplex assembly. J Biol Chem. 2010;285:39359–39365. doi: 10.1074/jbc.M110.183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde J, Dietrich L, Langosch D, Ungermann C. The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J Biol Chem. 2003;278:1656–1662. doi: 10.1074/jbc.M209522200. [DOI] [PubMed] [Google Scholar]
- 28.Starai VJ, Jun Y, Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci USA. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennison SM, Bowen ME, Brunger AT, Lentz BR. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zucchi PC, Zick M. Membrane fusion catalyzed by a Rab, SNAREs, and SNARE chaperones is accompanied by enhanced permeability to smal molecules and by lysis. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-08-0680. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizo J, Chen X, Araç D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann MW, et al. Self-interaction of a SNARE transmembrane domain promotes the hemifusion-to-fusion transition. J Mol Biol. 2006;364:1048–1060. doi: 10.1016/j.jmb.2006.09.077. [DOI] [PubMed] [Google Scholar]
- 34.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 35.Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21:2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorngren N, Collins KM, Fratti RA, Wickner W, Merz AJ. A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J. 2004;23:2765–2776. doi: 10.1038/sj.emboj.7600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starai VJ, Hickey CM, Wickner W. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell. 2008;19:2500–2508. doi: 10.1091/mbc.E08-01-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 39.Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.