It was known that the RNA binding proteins Nrd1, Nab3, and the RNA helicase Sen1 were involved in terminating Pol II transcription of noncoding RNAs in yeast. Here, in vivo cross-linking was used to identify binding sites of the three proteins. All three proteins are shown to bind to a number of noncoding RNAs in an unanticipated manner. Evidence shows that these proteins play unexpected roles in proper processing of noncoding RNAs including telomerase RNA.
Keywords: transcription termination, noncoding RNA, RNA polymerase II, Nrd1, Nab3, Sen1, Rpb2
Abstract
RNA polymerase II transcribes both coding and noncoding genes, and termination of these different classes of transcripts is facilitated by different sets of termination factors. Pre-mRNAs are terminated through a process that is coupled to the cleavage/polyadenylation machinery, and noncoding RNAs in the yeast Saccharomyces cerevisiae are terminated through a pathway directed by the RNA-binding proteins Nrd1, Nab3, and the RNA helicase Sen1. We have used an in vivo cross-linking approach to map the binding sites of components of the yeast non-poly(A) termination pathway. We show here that Nrd1, Nab3, and Sen1 bind to a number of noncoding RNAs in an unexpected manner. Sen1 shows a preference for H/ACA over box C/D snoRNAs. Nrd1, which binds to snoRNA terminators, also binds to the upstream region of some snoRNA transcripts and to snoRNAs embedded in introns. We present results showing that several RNAs, including the telomerase RNA TLC1, require Nrd1 for proper processing. Binding of Nrd1 to transcripts from tRNA genes is another unexpected observation. We also observe RNA polymerase II binding to transcripts from RNA polymerase III genes, indicating a possible role for the Nrd1 pathway in surveillance of transcripts synthesized by the wrong polymerase. The binding targets of Nrd1 pathway components change in the absence of glucose, with Nrd1 and Nab3 showing a preference for binding to sites in the mature snoRNA and tRNAs. This suggests a novel role for Nrd1 and Nab3 in destruction of ncRNAs in response to nutrient limitation.
INTRODUCTION
The yeast Nrd1-Nab3-Sen1 complex is required to terminate RNA polymerase (Pol) II transcription of noncoding transcripts including small nucleolar RNAs (snoRNAs), cryptic unstable transcripts (CUTs), and 5′ regulatory RNAs (Ursic et al. 1997; Rasmussen and Culbertson 1998; Steinmetz et al. 2001; Arigo et al. 2006a,b; Thiebaut et al. 2006, 2008; Kopcewicz et al. 2007; Jenks et al. 2008; Kuehner and Brow 2008; Marquardt et al. 2011). In this process, Nrd1 and Nab3 act as RNA-binding sensors to detect specific terminator sequences in nascent transcripts (Carroll et al. 2004, 2007).
Nrd1, Nab3, and Sen1 are part of a larger complex that includes Pol II, the nuclear cap binding proteins Cbp20 and Cbp80, Rnt1, Spt5, and the TRAMP and exosome complexes (Vasiljeva and Buratowski 2006). Nrd1 preferentially interacts with the Ser5 phosphorylated form of the CTD present on Pol II near the 5′ end of most genes (Komarnitsky et al. 2000; Gudipati et al. 2008; Vasiljeva et al. 2008a), and this interaction is required for efficient termination of non-poly(A) transcripts (Conrad et al. 2000; Steinmetz et al. 2001).
How the Nrd1-Nab3-Sen1 complex facilitates termination of non-poly(A) transcripts is unknown. The 3′ ends of noncoding RNAs (ncRNAs) could be formed either directly by termination or by cleavage of the transcript as in the poly(A) pathway (Kuehner et al. 2011). Unlike the poly(A) pathway, the Rat1 5′–3′ exonuclease is not required for Nrd1-Nab3-Sen1–directed termination, ruling out a torpedo-like model (Kim et al. 2006). Termination of non-poly(A) transcripts is coupled to RNA degradation by the nuclear exosome (Arigo et al. 2006b; Thiebaut et al. 2006) through the TRAMP complex consisting of the poly(A) Pol Trf4 or Trf5, the RNA-binding proteins Air1 and Air2, and the helicase Mtr4 (Anderson and Wang 2009). TRAMP adds a short oligo(A) tail to the 3′ end of these ncRNA transcripts, and this serves to prime the RNA for 3′–5′ degradation by the nuclear exosome (Wyers et al. 2005; Houseley et al. 2006; Wlotzka et al. 2011).
We have used a newly described in vivo RNA cross-linking protocol (Hafner et al. 2010; Creamer et al. 2011) to map in vivo binding sites for Nrd1, Nab3, Sen1, and Rpb2. In this article, we present an analysis of Nrd1, Nab3, and Sen1 binding to snoRNAs and show that the pattern of binding suggests additional, unanticipated roles for the non-poly(A) termination pathway in processing these RNAs. Nrd1 and Nab3 bind, as expected, to terminator sequences downstream from snoRNAs, but Sen1 binds specifically to box H/ACA snoRNAs. Nrd1 also binds upstream of some snoRNAs, suggesting a role in degrading the products of Rnt1 cleavage. We show that the Nrd1 pathway plays a role in mRNA 3′-end formation at several protein-coding genes and is required for efficient processing of the telomerase RNA TLC1. As recently reported, we find that Nrd1 and Nab3 also bind to transcripts derived from genes normally transcribed by Pol III (Wlotzka et al. 2011). Finally, we show that removing glucose from the growth medium alters the binding of Nrd1 and Nab3 to snoRNAs and transfer RNAs (tRNAs), suggesting a role for these proteins in directing destruction of these ncRNAs under nutrient limitation.
RESULTS
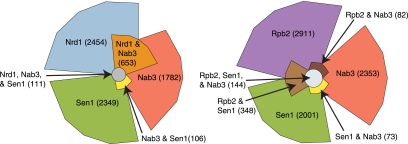
The photoactivatable-ribonucleoside–enhanced cross-linking and immunoprecipitation (PAR-CLIP) technique (Hafner et al. 2010) was adapted to yeast using 4-thiouracil (4SU) as the cross-linking nucleoside (Creamer et al. 2011). By using this approach, we show that Nrd1, Nab3, Sen1, and Rpb2 cross-link to a variety of yeast transcripts (Table 1). In addition to the expected Pol II transcripts, we also observe cross-linking to the ribosomal RNA (rRNA) locus and to tRNA transcripts. A similar distribution was recently observed for Nrd1 and Nab3 cross-linked to RNA using 254 nm light (Wlotzka et al. 2011). The most frequently Nrd1 cross-linked Pol II transcribed RNAs are snoRNA 3′ ends (35 of 100 most frequently cross-linked sites), CUTs (17 of the 100 most frequently cross-linked sites), and the 5′ ends of mRNAs like NRD1 and IMD3 that are regulated by attenuation (17 of the 100 most frequently cross-linked sites). Nab3 binds to a similar distribution of targets, while Sen1 displays a different pattern, binding preferentially to box H/ACA snoRNAs and to the 3′ ends of highly transcribed mRNAs. Our Nrd1 and Nab3 binding sites significantly overlap as shown in Figure 1. Sen1 and Rpb2 also show significant overlap, but surprisingly, Sen1 does not overlap with Nrd1 and only slightly with Nab3.
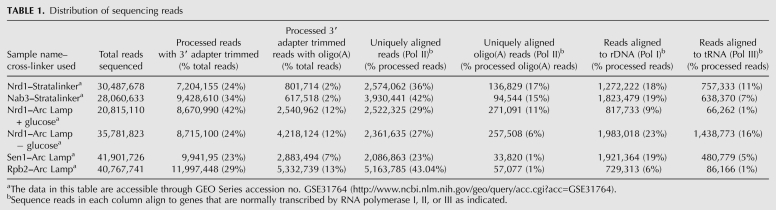
TABLE 1.
Distribution of sequencing reads
FIGURE 1.
Euler plots of cross-linked regions. (Left) Overlap between Nrd1, Nab3, and Sen1 cross-linked regions. (Right) Overlap between Rpb2, Sen1, and Nab3 cross-linked regions. The 100 most frequent T-to-C cross-linked sites in each data set were selected for this analysis. A region extending 20 nt in each direction from the cross-linked site was mapped to the genome, and values in the plot represent the number of overlapping or nonoverlapping nucleotides.
Nrd1, Nab3, and Sen1 play roles in processing snoRNA transcripts
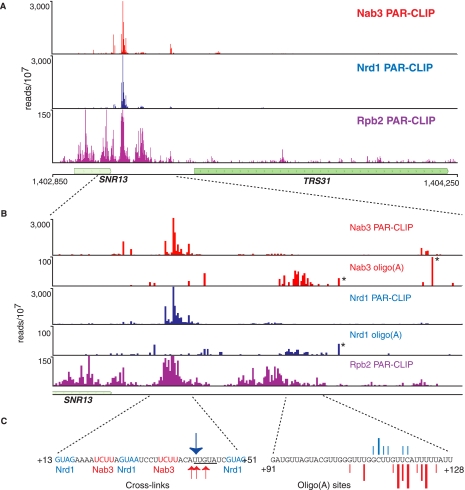
For every Pol II transcribed snoRNA, we observed peaks of Nrd1, Nab3, and Rpb2 cross-linking downstream from the mature 3′ end of the RNA, consistent with the previously demonstrated role in termination. For SNR13 (Fig. 2A), we show that Nrd1 and Nab3 cross-link within a tight cluster between 10 and 50 nucleotides (nt) downstream from the mature 3′ end. This region contains sequences previously shown to be required for termination (Steinmetz et al. 2001, 2006; Steinmetz and Brow 2003; Carroll et al. 2004, 2007). The actual cross-linking sites are not exactly in the genetically identified sequences but rather center on the sequence UGUAU, which is related to the Nrd1-binding consensus sequence UGUAG determined from analyzing the most frequently cross-linked sites in the yeast transcriptome (the underlined U residues indicate the site of cross-linking) (Creamer et al. 2011). The Pol II subunit Rpb2 cross-links to transcripts across the SNR13 gene extending 150 nt downstream. There is a very small amount of Pol II further downstream, which likely represents read-through transcripts that are terminated through the cleavage/polyadenylation pathway (Steinmetz and Brow 2003; Grzechnik and Kufel 2008).
FIGURE 2.
In vivo Nrd1, Nab3, and Rpb2 cross-linking to snR13 transcripts. (A) Cross-linking to transcripts from a 1400-nt region of chromosome 4 encoding the SNR13 and TRS31 genes. (B) Higher-resolution mapping of cross-linked reads to the SNR13 downstream region. The number of reads per 107 reads is indicated on the y-axis. The position on the x-axis is the center of the read for the PAR-CLIP experiment but is the 3′ end of the read for oligo(A)-containing reads. Asterisks designate the positions of naturally occurring genomic oligo(A) sequences that were not filtered in our bioinformatic analysis. (C) Sequences of the major cross-linked regions. The arrows in the left panel correspond to the most frequent T-to-C transitions in the Nrd1 (blue) or Nab3 (red) data sets. In the right panel, lines represent the exact position of oligo(A) addition. The size of the line corresponds to the frequency of the reads at that position.
As previously reported (Wlotzka et al. 2011), the Nrd1 and Nab3 cross-linked RNAs include sequences with short (2- to 4-nt) runs of A residues at the 3′ end. In our Nrd1, Nab3, and Sen1 data sets, we observe that between 6% and 17% of processed reads contain an oligo(A) tail. Oligo(A) runs that do not map to the genome likely originate from the activity of the poly(A) Pols Trf4 and/or Trf5, which are part of the TRAMP complex (Anderson and Wang 2009). In Figure 2B we show that oligo(A) sequences in the Nrd1 and Nab3 data sets derived from the snR13 transcript are most abundant just downstream from the Pol II peak located ∼80 nt downstream from the mature 3′ end. These oligo(A) sequences are not located on RNA fragments corresponding to the major Nrd1 and Nab3 cross-linking sites, but rather are found on a minor set of cross-linked fragments centered ∼80 nt further downstream. The position of our observed oligo(A) additions just downstream from the final Pol II peak suggests that these may represent the initial Pol II termination products.
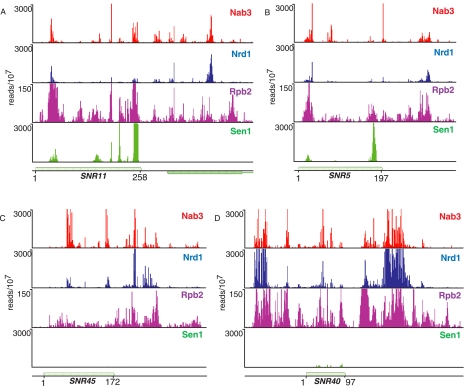
In Figure 3 we extend our analysis of snoRNAs to include cross-linking of the putative RNA helicase Sen1 (DeMarini et al. 1992). Sen1 is required for termination and processing of a number of snoRNAs (Ursic et al. 1997; Rasmussen and Culbertson 1998; Steinmetz et al. 2001; Finkel et al. 2010). Surprisingly, we observe efficient cross-linking of Sen1 only to box H/ACA snoRNAs. The 24 snoRNAs with the most Sen1 reads are all box H/ACA such as snR5 and snR11, shown in Figure 3. Figure 3 also shows several box C/D snoRNAs that cross-link poorly to Sen1, although they have prominent Nrd1 and Nab3 binding peaks. This is unexpected because we have previously shown that Pol II reads through several box C/D snoRNA terminators (snR39b, snR45, snR47, snR50, snR71, and snR128) in a sen1-E1597K mutant at the nonpermissive temperature (Steinmetz et al. 2001). The low level of Sen1 cross-linking to these RNAs suggests that Sen1 may not act as a helicase in Box C/D snoRNA termination or may not interact with this form of snoRNA in a manner that allows efficient cross-linking.
FIGURE 3.
In vivo Nrd1, Nab3, Sen1, and Rpb2 cross-linking to box H/ACA and box C/D snoRNA transcripts. (A,B) Cross-linking to snR11 and snR5 box H/ACA transcripts. (C,D) Cross-linking to snR45 and snR40 box C/D snoRNA transcripts. The y-axis is the number of sequence reads per 107 reads. The x-axis scale is given as the start and end of the snoRNA.
Unlike the case of Nrd1, Sen1 cross-links to sequences corresponding to the 3′ end of mature H/ACA snoRNA transcripts. This may indicate a role for Sen1 in 3′-end processing or in degradation of incorrectly processed transcripts. We also observe a subset of sequences cross-linked to Sen1 that contain oligo(A) residues not encoded at the snoRNA locus (data not shown). These oligo(A)-containing intermediates are most often shorter than the mature RNA, indicating that Sen1 plays a role in removing aberrantly processed box H/ACA snoRNAs.
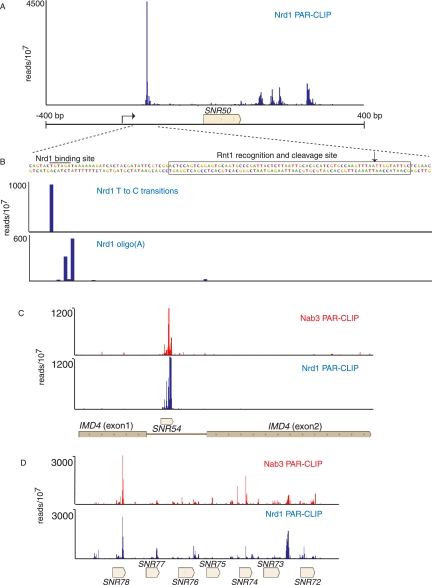
In addition to binding downstream from snoRNA terminator sequences, Nrd1 also cross-links to the upstream region of some snoRNA transcripts as shown in Figure 4A for SNR50. Binding to upstream sites may lead to cooperative binding to downstream sites (Carroll et al. 2007); however in most cases, the upstream binding sites are located in box C/D snoRNA transcripts upstream of previously characterized Rnt1 binding sites (Chanfreau et al. 1998; Lee et al. 2003; Ghazal et al. 2005). This positioning suggests Nrd1 may play a role in directing the exosome to the 3′ end of the Rnt1-cleaved 5′ leader RNA. Indeed, in Figure 4B we observe oligo(A) sequences on these upstream fragments, suggesting that these are TRAMP-modified degradation intermediates. This upstream processing pathway is consistent with the presence of Rnt1 in the Pol II-Nrd1-Nab3-Sen1 complex (Vasiljeva and Buratowski 2006). We also note that Nrd1 and Nab3 cross-link to snoRNA transcripts processed from introns and from longer precursor RNAs encoding multiple snoRNAs (Fig. 4C,D). In these cases as well, the role of Nrd1 and Nab3 is likely not termination but rather the coupling of transcription to cleavage by Rnt1 and subsequent exosome processing.
FIGURE 4.
In vivo Nrd1 and Nab3 cross-linking to snoRNA transcripts associated with Rnt1. (A) Nrd1 cross-linking to transcripts from the SNR50 gene. (B) Higher-resolution map showing the positions of T-to-C transitions and oligo(A) sites relative to the Rnt1 cleavage site. (C) Nrd1 and Nab3 cross-linking to snR54 transcripts in the intron of IMD4. (D) Nrd1 cross-linking to sequences in the polycistronic transcript that is processed to yield seven snoRNAs.
Glucose depletion alters Nrd1-Nab3 binding to snoRNAs
RNA sequencing has revealed widespread antisense transcription in yeast (David et al. 2006; Neil et al. 2009; Xu et al. 2009), and many of these antisense transcripts bind Nrd1 (Creamer et al. 2011; Wlotzka et al. 2011). Recent studies have shown that these unstable antisense transcripts are regulated by nutrient limitation and stress (Yassour et al. 2010), and our preliminary genetic studies have linked Nrd1 to the yeast glucose signaling pathway (M Darby, X Pan, L Serebreni, J Boeke, and J Corden, in prep.). Given these connections, we mapped the in vivo binding sites for Nrd1 in the immediate aftermath of glucose depletion.
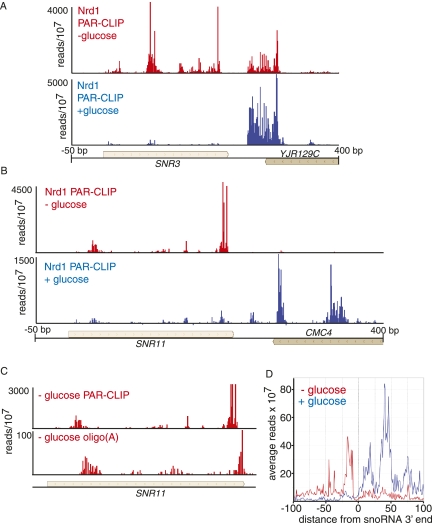
In cells adapting to the absence of glucose, there is a marked upstream shift in the binding of Nrd1 to snoRNAs. In Figure 5, A and B, we show that for SNR3 and SNR11 minor upstream peaks in the presence of glucose become the prominent peaks in the absence of glucose. Notably, all of these upstream binding sites are within the mature snoRNA. We obtained abundant sequence reads with oligo(A) tails associated with these upstream peaks (Fig. 5C). Figure 5D shows the relative distribution of all reads on 77 snoRNAs. The peak of binding is, on average, shifted ∼100 nt upstream. This shift could be due to earlier termination directed by an alteration in Nrd1-Nab3 binding specificity. Alternatively, this binding may be due to Nrd1 and Nab3 binding to and directing the degradation of mature snoRNAs in the absence of glucose. In either case this observation is consistent with a shut-down of ribosome biosynthesis in the absence of glucose.
FIGURE 5.
In vivo Nrd1 cross-linking to snoRNA transcripts in cells growing in the presence or absence of glucose. (A,B) Cross-linking of Nrd1 in vivo to snR3 and snR11 transcripts in the presence (blue) or absence (red) of glucose. (C) Frequency of Nrd1 cross-linked reads from the SNR11 gene, showing the presence of oligo(A) reads within the mature snR11 sequence. (D) Plot of all reads on the 77 snoRNAs on the genomic plus strand. The y-axis is median frequency over a 25-nt window. The x-axis is plotted relative to the 3′ end of the snoRNA.
Small nuclear RNAs (snRNAs)
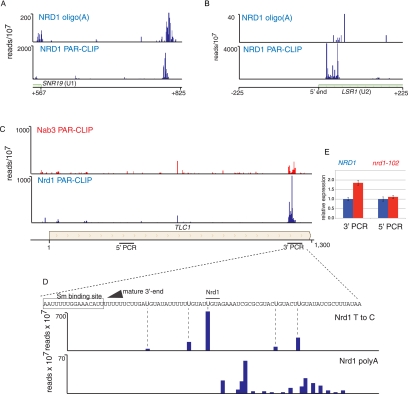
We have previously shown that the Nrd1-Nab3-Sen1 pathway is required for U4 3′-end formation (Steinmetz et al. 2001). The current data sets support this earlier work by pinpointing terminator sequences to a region 600 nt downstream from the start of the mature U4 RNA. We observe a similar peak of Nrd1 and Nab3 downstream from the SNR19 gene encoding U1 RNA (Fig. 6A). U2 (LSR1) and U5 (SNR7) 3′-end formation requires Rnt1 (Chanfreau et al. 1997; Abou Elela and Ares 1998), but we see very little cross-linking of Nrd1 and Nab3 downstream from these genes. However, we do see major peaks of Nrd1 and Nab3 at the 5′ end of the U2 transcript, suggesting that this RNA is regulated at the 5′ end by premature termination (Fig. 6B). Consistent with the model, we observe a peak of oligo(A) transcripts cross-linked to Nrd1 and originating close to the 5′ end of the U1 transcript.
FIGURE 6.
In vivo cross-linking to snRNA and telomerase RNA. (A) Nrd1 cross-linking to the 3′ end of U1 RNA. (B) Nrd1 cross-linking to transcripts from the 5′ end of U2 RNA. (C) Nrd1 and Nab3 cross-linking to transcripts from the 3′ end of the TLC1 gene encoding telomerase RNA. Lines below the gene map indicate the positions of PCR amplicons used to quantify TLC1 transcripts. (D) High-resolution cross-linking map showing the position of the mature 3′ end (arrow), the Sm binding site (box), the precise positions of cross-linked U residues (Nrd1 T to C), and the positions where oligo(A) is added (Nrd1 polyA). (E) Quantitative RT-PCR of TLC1 RNA derived from wild-type cells or nrd1-102 cells grown at the nonpermissive temperature. The relative abundance of each target transcript is normalized to the abundance of ACT1 in the same sample.
Nrd1 is required for efficient processing of telomerase RNA
The yeast telomerase RNA TLC1 is like snRNAs in that it binds SM proteins and contains a hypermethylated cap (Seto et al. 1999). TLC1 RNA is transcribed by Pol II and is polyadenylated at its 3′ end (Chapon et al. 1997). This precursor transcript is then processed at the 3′ end by the removal of 94 nt (Bosoy et al. 2003). We observe a peak of Nrd1 binding at the 3′ end of the transcript derived from the TLC1 gene that encodes the telomerase RNA. Figure 6, C and D, shows that Nrd1 binding sites are located downstream from the mature 3′ end but within the precursor transcript. The presence of Nrd1 cross-linked sequences containing oligo(A) tracts (Fig. 6D) indicates that Nrd1 directs 3′-processing of telomerase RNA by the exosome. To confirm that Nrd1 plays a role in TLC1 RNA processing, we carried out real-time quantitative PCR using two primer pairs: one in the mature RNA and one in the processed 3′ region. Figure 6E shows that in a nrd1-102 mutant at the nonpermissive temperature, there is a nearly twofold increase in the amount of unprocessed TLC1 RNA, indicating that Nrd1 is required for efficient 3′ processing of TLC1 precursor RNA.
Nrd1 cross-links to the 3′ UTR of some mRNAs
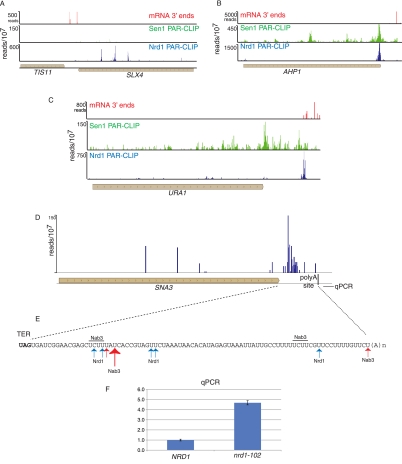
Our data also provide evidence that Nrd1 plays a role in processing the 3′ end of several mRNAs. Previous studies showed that TIS11/CTH2 mRNA is processed from a 3′-extended transcript by a mechanism that involves Nrd1 and Nab3 (Ciais et al. 2008; Wlotzka et al. 2011). We observe Nrd1 cross-linking sites 730, 991, 1249, and 1462 nt downstream from the TIS11/CTH2 ORF (Fig. 7A). The first two of these sites correspond to Nrd1 binding sites predicted by Ciais et al. (2008). In addition to TIS11/CTH2, we also observe major sites of Nrd1 binding in the 3′ UTRs of several other mRNAs, including URA1 (encoding a pyrimidine biosynthetic enzyme), AHP1 (encoding an antioxidant protein), and SNA3 (encoding a vacuolar membrane protein). The major Nrd1 cross-linking site on the AHP1 transcript is located right at the 3′ end of the ORF (Fig. 7B) in the sequence UGUAG, where the cross-linked U is underlined and the termination codon is in bold. Nrd1 cross-linked sequences containing oligo(A) tracts are located just downstream from this site, indicating that the exosome degrades this RNA close to the end of the ORF, likely precluding transport and translation. In the URA1 case (Fig. 7C), the binding site is located ∼250 nt downstream from the ORF near a cluster of minor polyadenylation sites and ∼40 nt before the major polyadenylation site determined from direct RNA sequencing (Ozsolak et al. 2010). In both the AHP1 and URA2 cases, the peak of 3′ Nrd1 cross-linking corresponds to a peak of Sen1 cross-linking. This is one of the few sites where these data sets overlap. In the SNA3 case (Fig. 7D,E), the major Nrd1 binding sites are >50 nt upstream of the major poly(A) site. Quantitative RT-PCR of total RNA derived from a wild-type and nrd1-102 strain indicates that Pol II reads through this poly(A) site in the mutant strain at the nonpermissive temperature (Fig. 7F), suggesting a role for Nrd1 in termination of this transcript.
FIGURE 7.
In vivo Nrd1 and Sen1 cross-linking downstream from protein-coding genes. The distribution of Nrd1 cross-linked RNA is shown in blue and Sen1 in green above the gene map. The position of poly(A) sites derived from RNA sequencing (Ozsolak et al. 2010) is shown in red. (A) Nrd1 and Sen1 cross-linking downstream from the TIS11 (CTH2) mRNA 3′ end. (B,C) Binding of Nrd1 and Sen1 in the 3′ UTR of the AHP1 and URA1 genes. (D) Binding of Nrd1 in the SNA3 3′ UTR. (E) Map of binding sites in the 3′ UTR. (F) Quantitative RT-PCR of RNA derived from wild-type or nrd1-102 cells grown at the nonpermissive temperature. The amplicon is located downstream from the SNA3 polyA site as indicated in D, and its relative abundance was normalized to the abundance of ACT1 in the same sample.
Nrd1 binding to Pol III transcripts?
As has been recently reported by Wlotzka et al. (2011), we also observe numerous tRNA reads among the cross-linked cDNA libraries, suggesting that Nrd1 and Nab3 may play a role in quality control of these RNAs. This is unexpected since neither Nrd1 nor Nab3 has been shown to associate with Pol III. In addition, deletion of the Nrd1 CTD-interacting domain (CID) does not interfere with tRNA processing, arguing against a role for Pol II (Wlotzka et al. 2011). However, we observe Pol II cross-linking to transcripts corresponding to some Pol III genes (Fig. 8), suggesting that some of the Nrd1 and Nab3 cross-linking could be due to surveillance of transcripts transcribed by the wrong Pol.
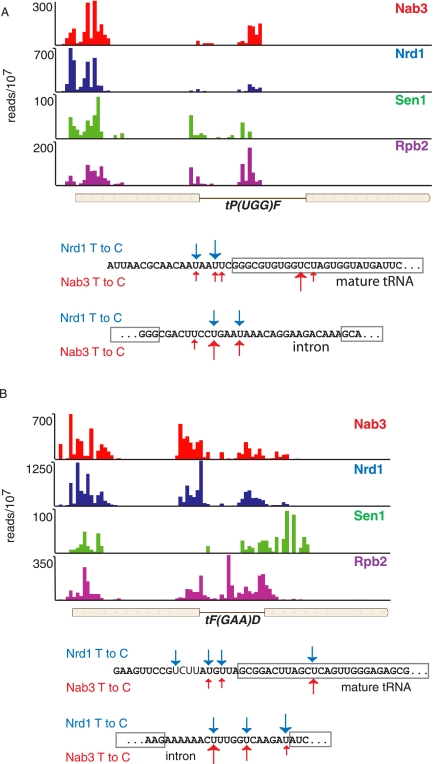
FIGURE 8.
In vivo Nrd1, Nab3, Sen1, and Rpb2 cross-linking to tRNA transcripts. (A,B) Cross-linking to tRNA Pro and Phe, respectively. Sequences below each map show the positions of Nrd1 (blue) and Nab3 (red) cross-links. Boxed sequences represent the mature tRNA. Bold sequence indicates a conserved Nab3 binding site upstream of tRNA Phe.
In Figure 8 we show that cross-linking to tRNA sequences occurs in the tRNA 5′ leader and intron sequences. Both the Pro and Phe tRNAs shown in Figure 8 are inefficiently processed when Nrd1 expression is repressed (Wlotzka et al. 2011). We propose that the Nrd1-Nab3-Sen1 complex binds to tRNA precursor transcripts and directs destruction of improperly processed transcripts. This would be consistent with observations that TRAMP components are involved in degrading improperly folded or modified tRNA, snRNA, and 5S RNA precursors in vitro (Kadaba et al. 2004, 2006; Vanacova et al. 2005). Interestingly, we note that the Nrd1 cross-linked U residues in tRNA often do not match the consensus sequence UGUAG derived from snoRNA and mRNA sense and antisense transcripts (see, e.g., the tP(UGG)F 5′ leader and the tF(GAAD) intron). The binding of Nrd1 and Nab3 to some tRNAs and the RNA component of RNase P (RPR1) is greatly enhanced in the absence of glucose (Fig. 9), indicating a possible role for the Nrd1 pathway in removing nascent tRNAs and other Pol III transcripts under conditions in which they are not needed for biogenesis of the translation machinery. This may also be the case for snR52, a snoRNA transcribed by Pol III. We see a peak of Nab3 cross-linking to the mature snR52 RNA, although in this case the contribution of an upstream CUT cannot be ruled out.
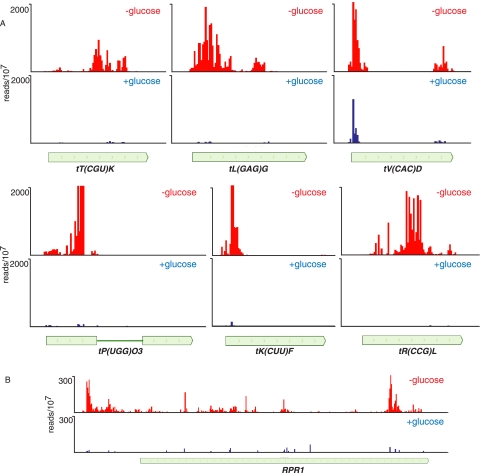
FIGURE 9.
In vivo cross-linking of Nrd1 to transcripts from Pol III genes in the presence and absence of glucose. (A) Cross-linking to tRNA genes. (B) Cross-linking to the transcript of the RPR1 gene encoding the RNase P RNA.
We also observe two major cross-linking sites on 5S RNA at nucleotides 83 and 86 (data not shown). Both of these sites are in a Nrd1 consensus binding site UGUAGU, where the cross-linked sites are underlined. Although this is a consensus binding sequence (Creamer et al. 2011), the most frequently cross-linked residue at other sites (the 5′ U) is not cross-linked in the 5S, again supporting the idea that binding is slightly different on transcripts derived from Pol III genes. We believe that the interaction with 5S RNA is post-transcriptional because previous studies have shown that antibody to Nrd1 fails to ChIP to the 5S gene (Houseley et al. 2007; Vasiljeva et al. 2008b). As has been previously reported (Wlotzka et al. 2011), we also observe oligo(A) tracts on Nrd1 and Nab3 cross-linked 5S RNA sequences, indicating that the Nrd1-Nab3 complex is involved in removing aberrantly processed 5S transcripts.
DISCUSSION
In this article, we present and analyze RNA-binding maps for components of the yeast non-poly(A) termination machinery. These maps provide a higher-resolution look at the sequences that direct 3′-end processing of a number of different classes of yeast ncRNAs. In addition, we discuss a number of novel binding targets for Nrd1 and Nab3 and show that the putative RNA helicase Sen1 preferentially cross-links to box H/ACA snoRNAs. The picture that emerges from these maps is that the Nrd1-Nab3-Sen1 pathway plays a wide range of roles in yeast RNA processing. Changes in the pattern of Nrd1 binding suggest that these roles change under conditions of nutrient depletion, arguing that the non-poly(A) termination pathway is under metabolic control.
snoRNA processing
The most efficiently bound targets of Nrd1 and Nab3 are ncRNAs, predominantly snoRNAs that are terminated downstream and trimmed back to the mature 3′ end (Allmang et al. 1999; van Hoof et al. 2000). Nrd1 and Nab3 play dual roles in this process both by facilitating termination and by recruiting the 3′-end processing machinery (Steinmetz et al. 2001; Arigo et al. 2006b; Thiebaut et al. 2006; Vasiljeva and Buratowski 2006; Wlotzka et al. 2011). We show here that the well-studied snR13 snoRNA precursor transcript binds Nrd1 and Nab3 in a short region located from ∼10–50 nt downstream from the mature 3′ end. This binding region contains previously identified SNR13 terminator elements (Steinmetz et al. 2001, 2006; Steinmetz and Brow 2003; Carroll et al. 2004, 2007). Pol II is cross-linked to snR13 transcripts covering the coding region and extending downstream ∼100–130 nt. Just beyond this point, we observe oligo(A)-containing sequences cross-linked to Nrd1 and Nab3. These modified RNAs are likely intermediates in the 3′ processing pathway. Because they are further downstream from previously observed intermediates (Grzechnik and Kufel 2008; Wlotzka et al. 2011) and because they are just downstream from the most distal peak of Pol II, we propose that these oligo(A)-containing fragments are primary termination products.
Nrd1 and Nab3 also cross-link upstream of some independently transcribed snoRNAs, and this binding seems well positioned to degrade the upstream leader RNAs produced by Rnt1 cleavage. We also observe Nrd1 cross-linking to intronic snoRNAs and to snoRNAs expressed as long poly-cistronic precursors. In all of these cases, we envision the role of Nrd1 as localizing the Rnt1 endonuclease and exosome components on the transcript to facilitate cotranscriptional processing. If this binding occurs cotranscriptionally, then there must be some signal that prevents Nrd1 pathway-mediated termination.
Sen1 and snoRNA 3′-end formation
Sen1 is a 252-kDa superfamily I helicase that has been implicated in synthesis and processing of both C/D and H/ACA snoRNAs (DeMarini et al. 1992; Ursic et al. 1997; Rasmussen and Culbertson 1998; Steinmetz et al. 2001). While helicase activity has not yet been demonstrated for Saccharomyces cerevisiae Sen1, the Schizosaccharomyces pombe homolog has been shown to possess 5′–3′ RNA helicase activity (Kim et al. 1999). Because the only predicted Sen1 RNA-interaction motifs are in the helicase domain, we infer that cross-linking is due to engagement of the helicase on RNA substrates. This is supported by the observation that immunoprecipitation of snoRNAs is reduced 10-fold in a temperature-sensitive sen1-1 mutant containing an amino acid substitution (G1747D) in the helicase domain (DeMarini et al. 1992; Ursic et al. 1997).
One of the major questions raised by our data is why Sen1 shows a preference for cross-linking to box H/ACA over box C/D snoRNAs. These two classes of snoRNA have distinct structures and functions (Kiss 2002; Henras et al. 2004). While both classes of snoRNA are assembled cotranscriptionally, they employ different assembly factors. Box C/D snoRNA assembly requires Nop1, the methyltransferase component that associates with box C/D snoRNA genes and is required for 3′-end formation (Lafontaine and Tollervey 2000; Galardi et al. 2002; Omer et al. 2002; Morlando et al. 2004). Nop1 also interacts with Ref2, a component of the APT complex that forms part of the cleavage polyadenylation factor (CPF) (Morlando et al. 2004). H/ACA snoRNA genes have been shown to interact with Cbf5 and the assembly factor Naf1 by ChIP analysis, suggesting that these proteins may bind snoRNAs cotranscriptionally (Ballarino et al. 2005; Yang et al. 2005). As yet, no interactions between these proteins and 3′-end formation factors have been presented. Interestingly, Cbf5 interacts with the ACA sequence at the 3′ end of the mature snoRNA, close to where we observe Sen1 cross-linking. One possible model is that Sen1 preferentially interacts with box H/ACA assembly factors and is recruited to these snoRNAs to assist in processing. Binding of Sen1 to the 3′ end may help recruit the TRAMP complex and facilitate 3′-end trimming by the exosome.
The presence of CPF at box C/D snoRNAs implies that Sen1 is recruited to these genes through interaction with Glc7 (Nedea et al. 2008). If so, why does Sen1 not efficiently cross-link to these RNAs? Sen1 is a large protein and certainly could have functions other than helicase activity. Perhaps the read-through phenotype seen on C/D snoRNAs in the sen1-E1597K mutant is due to an alteration in the structure of the Nrd1-Nab3-Sen1 complex that disrupts its ability to direct Pol II termination.
Processing the 3′ ends of longer transcripts
The prevailing model of Nrd1-Nab3-Sen1 function is that interaction of Nrd1 with Ser5 phosphorylated CTD repeats restricts function to the 5′ end of genes. In our data we observe numerous cases where Nrd1 seems to function more than a kilobase downstream, including the snRNAs U4 and U1, the telomerase RNA TLC1, and several mRNAs (Fig. 6). One question that needs to be asked is whether the Pol II transcribing the 3′ end of these genes has a Ser5 phosphorylated CTD typical of Pol II near the 5′ end of genes.
Pol III transcripts
As previous work from the Tollervey laboratory has shown, Nrd1 and Nab3 bind to a variety of Pol III transcripts (Wlotzka et al. 2011). Consistent with a role in processing of tRNAs, binding to precursor sites was observed, including the 5′ leader and intron sequences. Whether these cross-links represent intermediates in processing to produce mature tRNA or are the products of a surveillance mechanism designed to remove aberrant transcripts is not clear. We show here that the pol II subunit Rpb2 also cross-links to many Pol III–transcribed genes. Thus, the Nrd1-Nab3-Sen1 cross-linking may be taking place in Pol II transcription complexes aberrantly transcribing Pol III genes. How Pol II might initiate on Pol III genes is not clear. We do not see cross-linked Pol II through intergenic regions flanking tRNA genes, arguing that initiation must take place near the gene. Perhaps there is some affinity of Pol II for Pol III transcription factors.
ncRNA degradation in response to glucose deprivation
Data presented here suggest that the Nrd1-Nab3-Sen1 termination pathway may play a role in lowering ncRNA levels in response to nutrient limitation. For both snoRNAs and tRNAs, we observe large increases in the relative cross-linking to regions of these transcripts that are present in the mature RNA. This binding is accompanied by cross-linking of oligo(A) sequences, indicating ongoing degradation. These results can be obtained in several ways. First, the specificity of the Nrd1-Nab3 sensors may change such that sequences occurring earlier in the transcript are recognized cotranscriptionally and are thus prematurely terminated and degraded before assembly is completed. An alternative explanation is that Nrd1 and Nab3 bind to mature snoRNAs after transcription is shut off in response to glucose deprivation. This may be due to an increased pool of unbound Nrd1 and Nab3 caused by the shut-off of snoRNA transcription and the resulting lack of high-affinity Nrd1-Nab3 binding sites located in snoRNA terminators. In either case, the binding specificity of Nrd1 and Nab3 is proposed to change. We note subtle changes in the motif search among the top 100 Nrd1 binding peaks derived in the presence and absence of glucose. In the presence of glucose, the sequence UGUAG is the most prevalent with the U in position one (underlined) the major cross-linking site (Creamer et al. 2011). In the absence of glucose, the same consensus is observed, but the expectancy value is markedly increased (data not shown), indicating that binding of Nrd1 to RNA in the absence of glucose is less constrained by sequence. Taken together, our results indicate that the Nrd1-Nab3-Sen1 pathway plays a variety of roles in regulating the formation and stability of yeast noncoding RNAs.
MATERIALS AND METHODS
Yeast strains
Genomic NRD1, NAB3, SEN1, and RPB2 genes in BY4733 were tagged with both hexahistidine and biotin signal peptide tags separated by a TEV protease cleavage site (Tagwerker et al. 2006). The resulting HTB strains displayed no abnormal growth phenotypes and expressed fusion proteins with the expected increase in molecular weight (Creamer et al. 2011).
Cell growth and 365-nm UV cross-linking
Yeast cells expressing HTB tagged protein were grown at 30° from OD600 ∼ 0.1 to mid-exponential phase (OD600 ∼ 0.5) in 2 L of cross-linking medium made up of synthetic complete medium lacking uracil (SC-URA) supplemented with 2% dextrose, 120 μM uracil, and 0.01 μM biotin. 4SU was added to a final concentration of 300 μM and growth continued at 30° until OD600 ∼ 1.5. Two different irradiation schemes were employed.
In the first approach, cells were harvested before irradiation, washed in ice-cold water, and irradiated for 30 min in a Stratalinker 2400 at 365 nm (0.15 J/cm2) with shaking after 10 and 20 min. Irradiated cells were centrifuged, resuspended in 5 mL of buffer 1 (8 M urea, 300 mM NaCl, 0.5% Nonidet P-40, 50 mM sodium phosphate, 50 mM Tris-HCl at pH 8.0, and EDTA-free protease inhibitor mix for His-Tag sequences [RPI]), and then frozen in liquid nitrogen and stored at −80° until protein–RNA purification detailed below. Analysis of RNAs cross-linked to Nrd1 and Nab3 in these first data sets revealed expression of heat shock genes, suggesting that washing cells in cold water before irradiation may trigger a stress response. We therefore developed a second irradiation protocol.
In the second irradiation protocol, 2 L of growing culture was irradiated at 365 nm for 12.5 min with an arc lamp (UV Power–Shot 1100 Lamp [365 nm, 1 W/cm2]) while being continuously mixed. To eliminate shorter wavelength light, a Pyrex baking dish was placed between the beaker and the lamp. Irradiated cells in culture media were collected by centrifugation, washed in water, resuspended in 5 mL of buffer 1, and frozen as described above.
Glucose deprivation
Cells were grown in cross-linking medium (described above) at 30° to an OD600 of about 1. Cells were collected by centrifugation at 30°C and resuspended in cross-linking medium lacking glucose and supplemented with 4SU. The resuspended culture was incubated for a further 20 min at 30°C and irradiated according to the arc lamp protocol described above.
Protein–RNA purification
Cross-linked protein was purified by a modification of a previously published protocol (Tagwerker et al. 2006). Frozen cells were lysed in liquid nitrogen using a Spex SamplePrep 6870 freezer mill with 10 cycles of 1 min of breakage at a frequency setting of 15 cycles per second with 2 min of cooling between cycles. Lysates were thawed at room temperature, diluted with 5 mL of buffer 1, and then sonicated using a 1/8″ microprobe tip of Branson sonifer cell disruptor Model 250/450. Sonication was performed three times at 50% power for 5 sec with 30-sec intervals at room temperature. Cell lysates were cleared by centrifugation at 40,000 rpm in a Beckman L-80 ultracentrifuge at room temperature for 30 min using Ti 70.1 rotor. Cleared lysates were pooled and incubated with Ni-NTA agarose (Qiagen, 500 μL slurry pre-equilibrated in buffer 1) for 3 h at room temperature. Ni-NTA agarose was then washed in 5 mL of buffer 1; 5 mL of buffer 1 (pH 6.3); and 5 mL of buffer 1 (pH 6.3), + 10 mM imidazole. Proteins were eluted in 8 mL of buffer 2 (8 M urea, 200 mM NaCl, 2% SDS, 50 mM sodium phosphate, 10 mM EDTA, 100 mM Tris-HCl at pH 4.3, and EDTA-free protease inhibitor mix for His-Tag sequences [RPI]). The eluate was neutralized with 1 M Tris base and loaded onto streptavidin magnetic beads (New England Biolabs S1420S) from a 200 μL slurry that was pre-equilibrated in buffer 3 (8 M urea, 200 mM NaCl, 0.2% SDS, 100 mM Tris-HCl at pH 8.0, and EDTA-free protease inhibitor mix for His-Tag sequences). After incubation overnight at room temperature, the streptavidin magnetic beads were washed in 3 × 500 μL of buffer 3, 3 × 500 μL of buffer 3 with 2% SDS, 3 × 500 μL of buffer 3 without SDS, and then 3 × 500 μL of T1 ribonuclease buffer (150 mM KCl, 2 mM EDTA, 0.5 mM DTT, 50 mM Tris-HCl at pH 7.4, and EDTA-free protease inhibitor mix for His-Tag sequences [RPI]). The washed beads were resuspended in 1 mL of T1 buffer. RNase T1 (Fermentas) was added to obtain a final concentration of 40 U/mL, and the bead suspension was incubated for 15 min at room temperature. Beads were washed three times with 500 μL of T1 wash buffer (500 mM KCl, 0.05% NP40, 0.5 mM DTT, 50 mM Tris-HCl at pH 7.8, and EDTA-free protease inhibitor mix for His-Tag sequences [RPI]) and three times with 500 μL of polynucleotide kinase (PNK) buffer (50 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM Tris-HCl at pH 7.4, and EDTA-free protease inhibitor mix for His-Tag sequences). Beads were resuspended with 160 μL of PNK buffer and thermosensitive Alkaline Phosphate (TSAP; Promega) was added to obtain a final concentration of 0.15 U/μL, and SuperRase inhibitor (Ambion) was added to a final concentration of 1 U/μL. The bead suspension was incubated for 30 min at 37°C then was washed once with 500 μL of Buffer 3 without SDS, and three times with 500 μL of PNK buffer.
cDNA library construction from cross-linked RNA
RNA bound to protein on streptavidin magnetic beads was treated with T4 PNK (NEB) to create 5′ phosphate and 3′ hydroxyl ends. The RNA-containing beads were resuspended with 100 μL PNK buffer and incubated with 1 U/μL of T4 PNK and 1 mM ATP for 30 min at 37°C. The beads were then washed three times in 1× T4 Rnl2, truncated buffer (NEB). The washed beads were resuspended with 19 μL of T4 Rnl2, truncated buffer, and 19 μL of 50% polyethylene glycol 8000 (PEG 8000) (Promega). Two microliters of 100 μM adenylated 3′ adapter oligodeoxynucleotide (AppATCTCGTATGCCGTCTTCTGCTTGTC; IDT) was added with 0.4 μL of 1 M MgCl2, 2.5 μL of 200 U/μL T4 Rnl2, truncated (NEB), and 1.25 μL of RNaseOUT (Invitrogen). The bead suspension was incubated at room temperature for 4 h then washed once with 500 μL of buffer 3 without SDS, and three times with 500 μL of PNK buffer. The washed beads were resuspended in 38 μL of PNK buffer with 2 μL 100 μM 5′ adapter oligonucleotide (GUUCAGAGUUCUACAGUCCGACGAUC; IDT), 1.25 μL of RNaseOUT, 2.5 μL of 10 U/μL T4 RNA ligase (Fermentas), and 3 μL of 10 mM ATP (NEB). The bead suspension was incubated overnight at 16°C. The beads were then washed once with 500 μL of buffer 3 without SDS or protease inhibitors, and three times with 500 μL of Proteinase K buffer (75 mM NaCl, 6.25 mM EDTA, 1% SDS, 50 mM Tris-HCl at pH 7.5). The beads were resuspended with 200 μL of proteinase K buffer with 1.2 mg/mL of Proteinase K (Ambion). After incubation for 30 min at 55°, the supernatant was transferred to a new tube and another 200 μL of proteinase K buffer was added to the beads and incubated for a further 5 min at 55°C. The supernatants were combined, and the RNA was recovered by acidic phenol/chloroform extraction followed by an ethanol precipitation. The pellet was dried and dissolved with 12 μL of DPEC water. The recovered RNA was aliquoted into 3 × 4 μL tubes and kept in −80° until further preparation.
The recovered RNA was used to synthesize a cDNA library. One aliquot of recovered RNA was thawed on ice and added to 1 μL of 33 μM reverse transcription primer (CAAGCAGAAGACGGCATACGA; IDT). The mixture was briefly centrifuged and heated at 70°C on a preset thermal cycler for 2 min, and the tube was then placed on ice. Four microliters of a premixed reverse transcription reaction mixture containing 2 μL of 5× first strand buffer (Invitrogen), 0.5 μL of 12.5 mM dNTP mix (Fermentas), 1 μL of 0.1 M DTT (Invitrogen), and 0.5 μL of RNaseOUT was added to the chilled tube containing the primer-annealed template RNA. The tube was heated for 3 min to 48°C, then Superscript II reverse transcriptase (Invitrogen) was added to the final concentration of 20 U/μL, and the incubation was continued for 1 h at 44°C.
A time course PCR amplification was performed in order to determine the optimum number of cycles for amplifying the cDNA library. Using 10 μL of the cDNA from the previous step, PCR was carried out on a 50 μL scale with 1 U Phusion DNA Pol (Finnzymes), 0.5 μL of 25 mM dNTP mix, 10 μL l of 5× Phusion HF buffer, and 1 μmol of each primer (primer 1: CAAGCAGAAGACGGCATACGA; primer 2: AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA; IDT). PCR cycle conditions of 10 sec at 98°C, 30 sec at 60°C, 15 sec at 72°C were used. Aliquots of 6 μL were removed every other cycle, starting with cycle number 14 by temporarily pausing the PCR cycle at the end of the 72°C step. The maximum cycle of the PCR was set at 30 cycles. PCR aliquots were analyzed on a 6% Novex TBE PAGE gel (Invitrogen), and the optimal cycle number for cDNA amplification was chosen as five cycles prior to reaching the saturation level of PCR amplification. This level appeared to be within the exponential amplification.
To create the DNA library, PCR was performed for the optimal number of cycles on a 50 μL scale using cDNA prepared from another aliquot of recovered RNA (4 μL). The amplified product was separated on a 6% Novex TBE PAGE gel (Invitrogen), and a band corresponding to 100–150 nt was excised and eluted using 1× gel elution buffer (Illumina). RNA fragments with both adapters and an insert between 28 and 78 nt long produce PCR fragments that span a range of 100–150 nt in length. The 5′-adapter–3′-adapter products without inserts may be detected after amplification of the cDNA as an additional PCR band at ∼75 nt in length. The eluted DNA from gel extraction was ethanol precipitated followed by DNA analysis using the Agilent 2100 Bioanalyzer. DNA was sequenced using an Illumina GAII sequencer.
Bioinformatic analysis
Sequences were trimmed using the function trimLRPatterns from the ShortRead package in Bioconductor (Gentleman et al. 2004). Reads were separated into three files: trimmed, notrim, and oligo(A). The trimmed file contained reads that had a minimum length of 3 nt of adapter sequence at the 3′ end. The oligo(A) file contained reads from the trim file that contained a string of As at the 3′ end (minimum of 3 consecutive As). These As were removed for subsequent alignment. The notrim file contained the reads with no adapter on the 3′ end of the sequence. These notrim sequences were not used in our analysis.
The trimmed and oligo(A) reads were aligned to the S. cerevisiae genome using the short-read aligner Bowtie (Langmead et al. 2009). The bowtie alignment allowed the read to have up to one mismatch and align once to the genome. The reads that aligned more than once were not used in the genome browser but were aligned to a custom genome containing individual copies of repeated elements in the genome for statistical analysis. The non-oligo(A) trimmed reads were mapped from the midpoint of the sequence, and the oligo(A) reads were mapped at the 3′ terminus of the sequences. All figures were made using the MochiView genome browser (Homann and Johnson 2010). An overview of the data sets is provided in Table 1. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession no. GSE31764 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31764).
Yeast RNA analysis
Total RNA was extracted from yeast with hot acid phenol and run on a 1% denaturing formaldehyde MOPS agarose gel to test quality. Samples with clear rRNA bands and no visible degradation were analyzed by quantitative real-time RT-PCR. RNA was treated with turbo-DNA-free (Ambion) according to the manufacturer's instructions for the most stringent treatment. Reverse transcription was performed using the iScript cDNA Synthesis Kit (BioRad). Real-time PCR was performed in triplicate 20 μL reactions on a CFX96 real-time PCR detection system (BioRad) using iQ SYBR Green supermix (BioRad) according to the manufacturer's instructions. Data from at least two replicate experiments were pooled using the Gene Study feature of the CFX96 real-time software, which normalizes for fluorescence intensity differences between plates. Expression was normalized to ACT1 unless otherwise indicated, and the ratios were graphed relative to the wild-type control sample, which was set to 1 for each gene. Error bars represent the positive and negative range of the standard error of the mean.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM R01GM066108 and by an ARRA administrative supplement to this grant. We thank Leo Serebreni for technical assistance and Jon Lorsch for helpful comments on the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2840711.
REFERENCES
- Abou Elela S, Ares M Jr 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J 17: 3738–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Wang X 2009. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol 44: 16–24 [DOI] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL 2006a. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL 2006b. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I 2005. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol 25: 5396–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosoy D, Peng Y, Mian IS, Lue NF 2003. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J Biol Chem 278: 3882–3890 [DOI] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL 2004. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol 24: 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL 2007. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Elela SA, Ares M Jr, Guthrie C 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev 11: 2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Legrain P, Jacquier A 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol 284: 975–988 [DOI] [PubMed] [Google Scholar]
- Chapon C, Cech TR, Zaug AJ 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3: 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- Ciais D, Bohnsack MT, Tollervey D 2008. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res 36: 3075–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, Corden JL 2011. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3 and Sen1. PLoS Genet (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM 2006. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Winey M, Ursic D, Webb F, Culbertson MR 1992. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol 12: 2154–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Chinchilla K, Ursic D, Culbertson MR 2010. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics 184: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi S, Fatica A, Bachi A, Scaloni A, Presutti C, Bozzoni I 2002. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol Cell Biol 22: 6663–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazal G, Ge D, Gervais-Bird J, Gagnon J, Abou Elela S 2005. Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals. Mol Cell Biol 25: 2981–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 32: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D 2008. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol 15: 786–794 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Dez C, Henry Y 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol 14: 335–343 [DOI] [PubMed] [Google Scholar]
- Homann OR, Johnson AD 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8: 49 doi: 10.1186/1741-7007-8-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MH, O'Rourke TW, Reines D 2008. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol Cell Biol 28: 3883–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Choe J, Seo YS 1999. The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38: 14697–14710 [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S 2006. Distinct pathways for snoRNA and mRNA termination. Mol Cell 24: 723–734 [DOI] [PubMed] [Google Scholar]
- Kiss T 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopcewicz KA, O'Rourke TW, Reines D 2007. Metabolic regulation of IMD2 transcription and an unusual DNA element that generates short transcripts. Mol Cell Biol 27: 2821–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA 2008. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell 31: 201–211 [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol 20: 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Lee A, Chanfreau G 2003. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. RNA 9: 1362–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Hazelbaker DZ, Buratowski S 2011. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription 2: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I 2004. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J 23: 2392–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL 2008. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell 29: 577–587 [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci 99: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TP, Culbertson MR 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol 18: 6885–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 23: 6339–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Ng SB, Cloute JP, Brow DA 2006. cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol 26: 2688–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P 2006. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol Cell Proteomics 5: 737–748 [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the Nrd1-Nab3 pathway in genome surveillance. Mol Cell 23: 853–864 [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D 2008. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 31: 671–682 [DOI] [PubMed] [Google Scholar]
- Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR 1997. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res 25: 4778–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A 2008a. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S 2008b. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell 29: 313–323 [DOI] [PubMed] [Google Scholar]
- Wlotzka W, Kudla G, Granneman S, Tollervey D 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30: 1790–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G 2005. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol 25: 3295–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A 2010. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11: R87 doi: 10.1186/gb-2010-11-8-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]