Abstract
Why isn’t random variation always deleterious? Are there factors that sometimes make adaptation easier? Biological systems are extraordinarily robust to perturbation by mutations, recombination, and the environment. It has been proposed that this robustness might make them more evolvable. Robustness to mutation allows genetic variation to accumulate in a cryptic state. Switching mechanisms known as evolutionary capacitors mean that the amount of heritable phenotypic variation available can be correlated to the degree of stress and hence to the novelty of the environment and remaining potential for adaptation. There have been two somewhat separate literatures relating robustness to evolvability. One has focused on molecular phenotypes and new mutations, the other on morphology and cryptic genetic variation. Here we review both literatures, and show that the true distinction is whether recombination rates are high or low. In both cases, the evidence supports the claim that robustness promotes evolvability.
But nature has not been kind enough to endow the organism with the ability to react purposefully to the needs of the changing environment by producing only beneficial mutations where and when needed. Mutations are random changes. Hence the necessity for the species to possess at all times a store of concealed, potential, variability. —Dobzhansky, 1937 [1]
Robustness affects evolvability in different ways
Biological organisms and computer programs are both encoded by a string of characters: DNA in organisms, binary code in programs. In biological systems, random mutations to DNA sequences are the fundamental source of long term evolutionary adaptation. However, if one reduces a typical computer program to its binary sequence, making a random change to a single digit might literally have zero probability of improving the performance of the program [2]. But why? What makes biological systems evolvable when so many other systems are not?
Much has been said about the proper definition of evolvability (see [3] for an excellent review). On the longest geological timescales, one can discuss the evolvability of major morphological features or other radical innovations [4], whereas at the short time scale of a single generation evolvability is captured entirely by quantitative genetics [5, 6]. In this review, we focus on evolvability at the intermediate timescales best described by population genetics. At this level, we define evolvability as the capacity of a population to produce heritable phenotypic variation of a kind that is not unconditionally deleterious [7, 8]. This definition includes both evolution from standing variation and the ability of the population to produce new variants.
Three quite different mechanisms have been proposed to explain the high evolvability of biological systems: modularity [2, 9], robustness [10] and recombination [11–14]. Robustness can be defined and measured as the average effect of a specified perturbation on a specified phenotype. Here we focus on genetic robustness, i.e. robustness to perturbations both in the form of new mutations and in the form of the creation of new combinations of existing alleles by recombination (Box 1). We isolate the effect of robustness on evolvability from that of recombination on evolvability by considering high and low recombination rates as distinct cases.
Box 1. Robustness.
There is no single, universal measure of robustness. Robustness can only be defined unambiguously as the average effect of a particular perturbation on a particular phenotype, relative to some control. Here we consider genetic robustness, i.e. robustness to mutation and to recombination. High levels of genetic robustness might be adaptive (i.e. have evolved due to selection against the deleterious effects of mutation and recombination), they might be congruent (e.g. have evolved as a byproduct of selection for robustness to environmental perturbations) or they might be an intrinsic consequence of genetic and physiological constraints [80,81].
At first, robustness and evolvability appear to be opposites – if most mutations have no effect, then there will be less variation for selection to act on. Indeed, “robustness” mechanisms that prevent genetic change from occurring in the first place, such as proofreading and DNA repair, necessarily impede evolvability [15]. When many mutations occur, but phenotypes are robust to them, populations tend to spread out over a larger region of genotype space. The population (although not a single individual within it) can then access a greater range of genotypic possibilities, increasing evolvability [16]. Even when mutation rates are very low and populations typically monomorphic, robustness can increase evolvability by allowing the population to visit a greater number of genotypes over a given period of time [16].
When genetic variation is present, potential evolutionary innovations might already be present, but cryptic, in the population. For example, an environmental change or other major perturbation such as a gene knockout might cause existing genotypes to switch from being neutral to being adaptive. When recombination is common, hybridization or simply shifting allele frequencies might change the genetic background such that an allele switches from being neutral to being adaptive. We call all of these examples the “post-mutation” case because they involve adaptation that takes advantage of mutations that have already occurred. Alternatively, consider the adaptive potential of genotypes not yet present or accessible through recombination, but which can be produced through a single round of new mutations. We call this the “pre-mutation” case because it considers evolvability via future mutations that have yet to occur. As we will see, most asexual models have focused on pre-mutation evolvability and most sexual models on post-mutation evolvability. This has created two somewhat disjoint literatures, whose true distinction is low versus high recombination rates rather than, for example, molecular versus morphological phenotypes. Here we systematically examine all four permutations of low versus high recombination rates and pre- versus post-mutation.
As we go, keep in mind one last distinction. Robustness might modulate both the quantity and quality of heritable phenotypic variation, and these two arguments will be treated separately. In a typical quantity argument, the amount of heritable phenotypic variation might vary, and be correlated with the potential for adaptation, for example as signaled by stress. The existence of this correlation between quantity of variation and opportunity for adaptation promotes evolvability.
In addition, robustness might also have effects on the quality of variation. For example, lethal mutations will clearly never be adaptive in any environment, whereas mutations of small effect might be adaptive in at least some environments. Although quality arguments have been described by some authors in terms of “preadaptation” [8, 17], others deny that significant variation in quality exists [18]. However, the distribution of fitness effects of new mutations is strongly bimodal, with one mode near lethality and the other near neutrality [19]. Any process that reduces the proportion of variation that is lethal will increase the relative quality of variation that remains [8]. We will discuss both quantity- and quality-based arguments about the relationship between robustness and evolvability in the context of pre- and post-mutation effects on evolvability in both high and low recombination rate systems.
Post-mutation evolvability under high recombination rates
When a system is robust to mutations, selection does not prevent their accumulation. Later genetic or environmental changes might, however, cause changes to robustness which trigger the revelation of previously cryptic genetic variation. Cryptic genetic variation is abundant and ubiquitous [20, 21]. The classic literature on cryptic genetic variation refers to high recombination: genetic assimilation, i.e. loss of crypticity, is envisaged to happen when many alleles, each cryptic on their own, are brought together by recombination, until collectively they exceed a quantitative threshold [22]. In this section we focus on cryptic genetic variation that can be assimilated through recombination, although note that genetic assimilation might alternatively be driven by mutation, especially in low recombination systems.
Cryptic genetic variation might be revealed in response to stress [23]. Because stress is likely correlated to opportunities for adaptation, this ability to modulate the quantity of heritable phenotypic variation can increase evolvability [24]. Cryptic genetic variation is currently experiencing a resurgence of research interest, although there is not yet firm evidence regarding the extent to which it contributes to adaptation in natural populations [25].
Evolutionary capacitance is a relatively new term that is used to describe the hide and release of cryptic genetic variation. Stress might act as a signal that the current phenotype is not well adapted, and that opportunities for adaptation are therefore likely to exist. A capacitor is a switch, whose change in state in response to stress might act to adjust the quantity of variation in response to the environment, thereby promoting evolvability [17, 26–29]. To promote evolvability, switching must not happen at too rapid a rate, both to allow sufficient time for cryptic variation to accumulate, and for assimilation to occur before variation is returned to the cryptic state [26]. Capacitors can also promote evolvability even when their switching is random rather than correlated to stress, by hedging bets as to whether opportunities for adaptation via cryptic genetic variation exist at a given point in time [27,30–31].
Previously cryptic genetic variation might also be of higher quality than variation generated by new mutations. Cryptic genetic variation might be biased towards phenotypes adaptive in environments at the spatial or temporal margins of the current population, which might correlate with the direction of environmental change [17]. In addition, when robustness has not been 100% complete, cryptic variation might have gone through a process of preadaptation by experiencing weak purifying selection to purge unconditionally deleterious alleles while they were in a partially hidden state [8]. Preadaptation in the form of incomplete robustness is particularly effective in facilitating adaptations that occur through combinations of mutations [8, 26].
The best studied putative evolutionary capacitor in a high recombination system is the heat shock protein HSP90 [28, 32–34], a molecular chaperone that might provide genetic robustness. When HSP90 action is inhibited (by stress, or by manipulation in the lab), diverse new phenotypes appear, although the quality of revealed variation was low, with most phenotypes obviously deleterious. Because the identity of the phenotypes depends on the genetic background, HSP90 inhibition was inferred to tap into stocks of cryptic genetic variation. However, when HSP90 is inhibited, transposons are reactivated: this genetic-background-dependent mutagenesis rather than capacitance might also explain these results [35].
Gene knockout mutations might in general be potent evolutionary capacitors. In simulations, many regulatory genes act as capacitors when deleted, promoting evolvability in the process [36]. A recent exhaustive study of knockout mutations in Saccharomyces cerevisiae [37] identified more than 300 gene products, with exceptionally high capacities to stabilize morphological variation, which released previously cryptic phenotypic variation when silenced. Similar experimental protocols could be used to assess whether this result extends, as theory predicts [38, 39], to the provision of robustness to genetic variation.
Hybridization between two dissimilar lineages can also be thought of as a major one-off perturbation in genetic background that triggers the revelation of cryptic genetic variation. This often manifests as “transgressive segregation” in which hybrid populations show phenotypes more extreme than either parent [40,41]. For example, cryptic genetic variation for phenotypically invariant traits in teosinte (a wild relative of domestic maize) was discovered [42] by producing hybrid crosses with maize. This cryptic variation was proposed to have been important for the divergence of maize from teosinte. Hybridization in wild sunflowers seems to have promoted ecological divergence into extreme habitats, where the advantages of hybridization can also be demonstrated using synthetic hybrids [43].
Post-mutation evolvability under low recombination rates
One can envision the neutral region of a genotype space (Boxes 2 and 3) as a “rug” placed on a dusty floor [44], and each genotype present in a population as dust on that floor. Beyond the rug, genotypes are deleterious and swept away by selection, whereas under the rug, variation is hidden from selection (and is also free to accumulate further). Genetic robustness is determined in part by the size of the rug. In more extreme environments, or under the influence of an evolutionary capacitor, the rug itself might either shrink in a process of decanalization, or shift position without necessarily shrinking [44]. In either case, this exposes previously hidden variation to selection, and evolvability might be facilitated by a sudden increase in the quantity of selectable variation in a novel environment.
Box 2. Genotype Spaces and Adaptive Landscapes.
Wright [82] introduced the adaptive landscape metaphor as a tool to help non-mathematicians visualize the connection between high dimensional genotype space and fitness. Wright’s original allele space (Figure I) had two points per genotypic dimension – wild type and mutant. A landscape in DNA sequence space would have four points per dimension (CGAT) and in protein sequence space [83] 20 points per dimension. Despite Wright’s original drawing, the word “landscape” has been criticized for conjuring an image of a low-dimensional continuous space rather than a high-dimensional discrete space [84–86].
“Adaptive landscape” also has a second meaning, relating allele frequencies to mean fitness of an entire population [82]. This “landscape” is continuous, but has limited utility. Here we refer instead to adaptive landscapes that relate individual genotype to individual fitness.
Moving on from the merely heuristic metaphorical sketch of Wright, it is very difficult to model arbitrary genotype-fitness spaces, and so several simplified models have been influential. For example, the NK model [87] describes genotypes as sequences of N binary sites whose contributions to fitness depend on the state of K other sites. The features of the landscape are determined entirely by N and K, producing a single peak when K=0 and an uncorrelated space when K=N.
Quasispecies theory [88] describes the evolution of an infinitely large population of asexual replicators. The key assumption of infinite population size has been criticized as being unrealistic, even for RNA viruses [89]. Although the equations for quasispecies are otherwise very general [90], most quasispecies research has focused on one particular formulation where fitness has a single global optimum that decays monotonically as a function of the number of mutations.
More recently, an alternative formulation known as a ‘holey adaptive landscape’ or neutral network model (Box 3) has begun to acknowledge the geometrical complexity of the genotype-phenotype map. These models focus on phenotypically-equivalent ridges or plateaux in genotype space, simplifying the fitness ‘dimension’ to a binary choice between zero and one. Neutral network models have been particularly influential on the study of robustness and evolvability.
Box 3. Neutral Networks.
The neutral network model [52] is based on the general quasispecies equations, but has no ‘peak’, focusing instead on plateaux of the fitness landscape (Box 2). A neutral network is a set of mutationally connected, phenotypically equivalent genotypes. All or most off-network genotypes are lethal. Neutral networks are equivalent to Gavrilets’ ‘holey adaptive landscape’ [91,92] where connected genotypes of equivalent phenotype are interspersed with ‘hole’genotypes of low fitness. The same genotype space might contain multiple neutral networks, each of which might correspond to a different phenotype and hence fitness. Evolvability can then be defined as the ability to find a new neutral network of higher fitness.
Evolutionary behavior on a neutral network depends on the product of the mutation rate μ and the population size N [52]. When µN≫1, natural selection drives the population towards more mutationally robust parts of the genotype space. When µN≪1 the population is usually monomorphic for a single random genotype. These two limiting cases are mathematically tractable, and they bracket the solution for intermediate values of µN [52]. Simulations show that realistically large, but not infinite, populations have summary statistics that match the infinite-population predictions of quasispecies theory [52]. This has reinvigorated interest in quasispecies approaches to study questions of robustness and evolvability.
Mathematically, neutral network models apply only to asexual populations [52]. As a metaphor, a “neutral space” of equivalent phenotypes can also be defined within the set of sexual genotypes [10], although its connectivity patterns are not mathematically tractable, because each offspring genotype depends on two rather than a single parental genotype.
The neutral network metaphor is sometimes extended so that “neutrality” refers to equivalence at one phenotype level, e.g. RNA or protein folding, but not necessarily at all others [10, 53]. Although useful as a metaphor, once neutrality no longer refers to fitness, the advantages of mathematical tractability no longer apply.
This rug metaphor was originally used in conjunction with a quantitative genetic model [44], which assumes high recombination and also a very simple, additive relationship between genotype and phenotype. Robustness up to a point (i.e. the edge of the rug) is then the optimal solution to a fluctuating environment [17]. The metaphor applies perhaps even better in the post-mutation low-recombination case [45] because the “boundaries” of a neutral genotype space remain well defined even in non-additive models if they are breached only by mutation and not also by recombination.
The rug metaphor shows how robustness can promote evolvability post mutation with low recombination, according to a quantity-based argument. The post mutation arguments for both low and high recombination depend entirely on the existence of some form of evolutionary capacitance, i.e. the hiding and release of cryptic genetic variation. And indeed, capacitance promotes evolvability in simulations of asexual populations [36]. However, the details of both the theoretical models and the empirical examples are obviously quite different with and without recombination.
An empirical low recombination model capacitor system for which post-mutation evolvability might be important is the yeast prion [PSI+] [29, 46–48]. When cells switch to the [PSI+] state, stop codon readthrough errors increase from ~0.3% to ~1% [49]. This shrinks the rug that previously obscured the impact of sequences beyond stop codons. Genetic assimilation for any adaptive phenotypes might then occur, for example through loss of appropriate stop codons. Saccharomyces, unlike other taxa that do not have the ability to form [PSI+], shows increased rates of in-frame loss of stop codons, consistent with [PSI+] mediating evolvability in wild populations [50]. These properties position [PSI+] as the leading candidate for a capacitor whose evolvability properties might have been favored by natural selection (Box 4).
Box 4. Evolution of Evolvability.
Some authors have argued that it is impossible for natural selection to favor evolvability [93,94]. These general arguments have been comprehensively rebutted elsewhere [95], leaving the possible extent and nature of the evolution of evolvability an open question.
The most significant criticism comes from studies of mutators. When a mutator generates an adaptive mutation, recombination may rapidly separate the adaptive allele from the mutator allele, and so the mutator derives little benefit via hitchhiking. Indeed, although theoretical models have predicted the evolution of evolvability via capacitance switches, they have done so only in the absence of sex [27,31,94,96] or with realistically low rates of outcrossing such as can be found in many non-metazoan species [30]. The best candidate capacitor to date is the yeast prion [PSI+], which most likely evolved due to selection favoring its evolvability properties [7]. Evolvability can evolve even with obligate sex, however, when it is a distributed property across the genotype or across a population in the genotype space, rather than residing in a single modifier locus [55, 95].
Evolvability is sometimes studied in the context of fluctuation between two previously seen environments, rather than the ability to innovate in response to an entirely novel environment [54, 63]. Evolvability in a simple model of this sort might consist of a genotype poised at the interface between two neutral networks in genotype space, and this precarious position might come at the expense of robustness [54]. This tradeoff does not always apply, however, for more complex genotype-phenotype maps with more interfacial options [63].
In general, the most parsimonious explanation for robustness is that it reduces the effect of deleterious perturbations, rather than occasionally promoting evolvability. Similarly, incomplete robustness might be a result of the imperfect nature of molecular and developmental processes, and the diminishing returns on expensive error-correction mechanisms. Biological evolvability might often be a fortuitous side effect both of robustness and of its limits.
Pre-mutation evolvability under low recombination rates
The dominant model for this category is the neutral network (Box 3) of phenotypically equivalent genotypes connected by single mutations. A population is typically spread out over some portion of this genotype space. Genetic robustness is defined as the probability that a mutation in an individual causes no change in phenotype, i.e. that the individual remains on the neutral network after a single mutational step in the genotype space. Different phenotypes are specified by different neutral networks within the same overall genotype space. Evolvability is the ability of a population on one neutral network to discover another network of higher fitness.
Evolvability obviously depends on the details of the relationship between all genotypes and all phenotypes. A common simplifying assumption is that genotypes of higher fitness are uniformly distributed with respect to the topology of a single ancestral neutral network. In this case, the evolvability of a population is directly related to the number of distinct genotypes that can be accessed through single new mutations. Very small neutral networks inevitably have low evolvability. When the product of the mutation rate μ and the effective population size N is small, the population is genetically homogeneous, and evolvability is generally low. However, increased robustness can still facilitate evolvability in the low mutation case when robust populations explore their neutral networks more quickly than low-robustness populations [16].
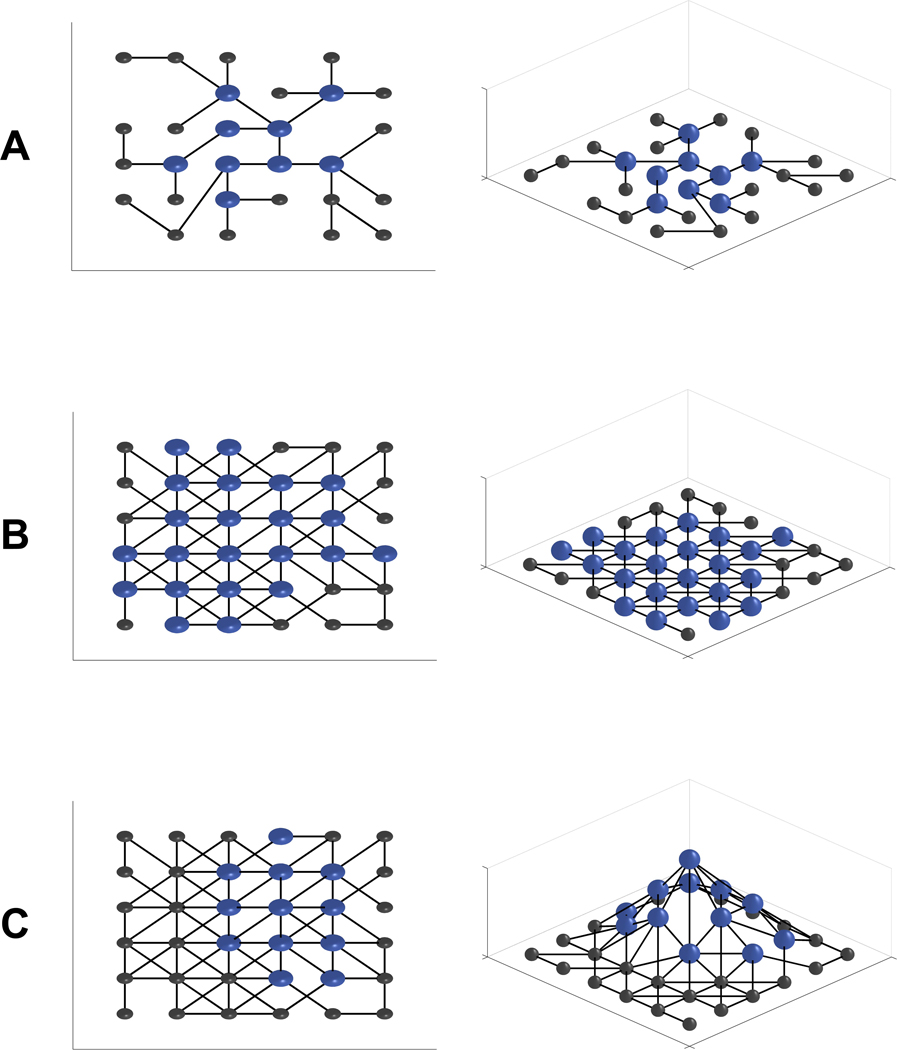
When µN>1, genetic robustness allows a population to spread out over more genotypes in a neutral network, and hence increases evolvability so long as the total number of possible alternative phenotypes is not so tiny as to be soon exhausted [51]. Populations inhabiting more densely connected and perhaps larger neutral networks will have higher average robustness. This robustness is intrinsic (Box 1) to the genotype-phenotype map, although over long time scales it is possible that natural selection has acted to favor more robust neutral networks among all possible networks of the same phenotype. Therefore, intrinsic robustness (i.e. a larger neutral network) increases evolvability [16] (Figure 1) so long as robustness is not complete. In the limit, extremely robust populations will have lowered evolvability as they almost never produce novel phenotypes; although a theoretical possibility, robustness this high is not normally encountered in biology.
Figure 1.
Interconnected nodes represent genotypes on a neutral network. Blue spheres represent genotypes present in the population and black spheres represent unoccupied nodes. (a) A neutral network with low intrinsic robustness has low spread and low evolvability. (b) A neutral network with high intrinsic robustness has its occupancy spread out across a larger portion of the network, gaining access to more potential variability. (c) In the presence of environmental perturbations some genotypes might generate the target phenotype more reliably than others, creating slight fitness differences. Selection for environmental robustness might hinder the spread of a population through a “nearly neutral network”. In our schematic, more environmentally robust genotypes (shown as higher) are more fit and so have higher occupancy than neighboring nodes.
Additional genetic robustness might be a consequence of natural selection. Large populations are subject to selection favoring the regions of a given neutral network that are less likely to be harmed by mutation. When all on-network genotypes have completely identical phenotypes, this increased robustness reaches a plateau for values of µN modestly greater than 1 [52]. However, some on-network genotypes might have higher fitness than others, e.g. due to differences in environmental robustness. When, as is commonly the case, there is congruence between genetic and environmental robustness, still higher levels of genetic robustness might evolve [53]. This selection for only the most environmentally robust genotypes might purge genetic variation from the population (Figure 1), restricting its ability to explore genotype space [53]. Genetic robustness only promotes evolvability when it is associated with increased spread of a population across genotype space. Anything that increases the spread of a population across genotype space, such as fluctuating selection, also increases evolvability, even if it also reduces genetic robustness.
The geometry of the relationship between genotype and phenotype is important for the relationship between robustness and evolvability. Intuitively, genotypes and hence populations near the “center” of a neutral network would seem to have fewer off-network options, whereas those on the “edge” would have more [54]. However, alternative non-lethal phenotypes are not necessarily distributed uniformly throughout genotype space, and different regions of genotype space might access different degrees of neighboring phenotypic richness. The argument that robustness promotes evolvability requires the existence of neighboring phenotypic richness. There has therefore been a substantial effort to determine the genotype-phenotype geometry for well-characterized model systems [10], and to find simple correlates of neighborhood richness and evolvability [55]. Spread across a neutral network is a quantity argument for evolvability, whereas characterizations of the genotype-phenotype maps might also provide quality-based arguments.
The leading model system for neutral networks takes an RNA sequence as the genotype, and assumes that the phenotype can be reduced entirely to folding at the level of secondary structure [56]. However, for proteins, a basic fold is clearly only a prerequisite for a protein’s phenotype, as function can vary dramatically across proteins with the same basic fold. The “neutral network” of a protein’s fold is not necessarily neutral with respect to function and fitness. The number of possible amino acid sequences that have a given fold is known as the “designability” of a protein. Over long evolutionary times, more designable proteins show greater diversity of molecular function, in agreement with the predicted relationship between intrinsic robustness and evolvability [57].
Proteins possess only marginal thermostability, such that a single mutation might destabilize them. Excess thermostability is therefore a biophysical mechanism of both genetic and environmental robustness. Marginal protein thermostability seems to be a consequence of mutation-selection balance [58]. Mutants derived from highly stable variants of a given protein are more likely to retain their fold, and are consequently more likely to develop novel or improved protein functions [59,60]. Ancestral and/or consensus protein sequences tend to be highly stable, and such stability can also evolve under strong pressure from mutations [59] or transcriptional errors [61]. This quality-based relationship whereby genetic robustness (excess thermostability) promotes evolvability applies to single genotypes as well as to populations.
Each genotype-phenotype map needs to be assessed separately in this way to estimate whether more “central” genotypes are less evolvable, due to the fact that many mutations do nothing (quantity), or more evolvable, due to the fact that more mutations have small effects of the kind that are most likely to be favored by selection (quality). As in the high-recombination case [8], the observed bimodal distribution of fitness effects of new mutations [19] creates the possibility of quality-based arguments. In the case of protein sequences, this bimodality might often correspond to the destruction of a protein fold versus tinkering with function within the same overall fold.
Simple gene regulatory networks can also be analyzed as neutral networks [62–64]. Gene regulatory networks show immense phenotypic richness in the neighborhood of a typical neutral network [62]. Similar results were found for prokaryotic metabolic networks [65]. The genotypic network of transcription factors recognizing alternative binding sites, however, is small, with low phenotypic richness [66]. Differential phenotypic richness of high robustness versus low robustness regions of the same genotype space has not yet been established for these and many other cases. However, a finding of substantial richness in general for a particular class of genotype-phenotype map creates the potential for any robustness-related differences in quality to have evolutionary significance.
Toy genotype-phenotype models can yield interesting results even when they treat incredibly simple examples, such as codon evolution at a single amino acid site [54]. One toy model suggests that robustness via simple redundancy might yield a poorer phenotypic neighborhood than when the proximate cause of robustness is distributed across the system [67].
Accessible phenotypic richness might be still higher if alternative phenotypes that are two mutations away can be reached. This is the classic problem of crossing an adaptive valley, which could potentially be facilitated by flattening the landscape [68]. Molecular processes subject to natural error rates can “look-ahead” across adaptive valleys, and so this partial robustness to molecular errors can further increase evolvability [30, 69,70]. The two mutations in question can be either in the same gene [70], or in different genes in an organism with low recombination [7, 30].
Two empirical studies on RNA viruses have tried to assess the relationship between robustness and evolvability, but unfortunately neither is conclusive. One found that robustness promoted the evolvability of thermotolerance in an RNA virus [71]. However, thermotolerance itself is often directly correlated to robustness, and limited replicates at a single temperature make it hard to rule out this relationship conclusively. This study is therefore hard to interpret in the context of evolvability in general.
The second study found that an artificially assembled low-robustness virus adapted faster than the wild type to a novel host cell type [72]. However, a population spread across a neutral network was excluded in this experiment by the use of clones. Low environmental robustness might have led to a larger proportion of strongly deleterious mutations, inflating the relative advantage of higher fitness genotypes and hence accelerating the rate of adaptation even within a comparable set of genotypic options [72,73].
Although evolutionary capacitance is primarily thought of as a post-mutation mechanism of tapping into “stored” cryptic genetic variation (sometimes known as “genetic charge”, in an extension of the capacitance analogy [74]), pre-mutation evolvability can also increase through switches such as [PSI+] that increase the rate of molecular errors [7, 27, 30]. Indeed, it is not clear to what extent [PSI+] mediates evolvability through pre-mutation versus post-mutation effects. For systems such as [PSI+], “robustness” refers to the normally low error rates of molecular processes such as transcription and translation. Evolvability comes about both because residual error rates steer the population away from proximity to the most damaging genotypes (quality) and because the presence of switches allows higher error rates specifically during stressful episodes when opportunities for adaptation are most present (quantity).
Pre-mutation evolvability under high recombination rates
Consider evolution in the same genotypic space, but now with significant recombination. Selection for robustness to recombination is now a strong force, with greater robustness to single mutations evolving as a correlated byproduct [75–78]. Selection for increased robustness within a given primary phenotype might constrain the range of genotypes and hence decrease evolvability in an asexual population [53]. But although genetic diversity at single sites decreases with sex due to the purging effects of strong selection for increased robustness, diversity at the level of the entire genotype nevertheless increases through recombination [76,78]. Pre-mutation evolvability (i.e. spread of genotypes within a neutral space) might therefore increase with recombination, especially as recombination might also promote modularity [78, 79]. This increased evolvability might, however, be in spite of rather than because of increased adaptive robustness. Greater intrinsic robustness (size of the neutral genotype space itself) should increase pre-mutation evolvability in both low recombination and high recombination cases.
In any case, recombination itself promotes evolvability through new combinations of existing mutations [11–14]. Given the role of existing mutations, it seems likely that post-mutation rather than pre-mutation mechanisms are the driving force in any link between robustness and evolvability with high recombination.
Concluding remarks
There have been two somewhat separate literatures, both claiming that robustness promotes evolvability. One focuses largely on single genes and molecular phenotypes, uses the metaphor of neutral networks, and considers the effects of new mutations [10,16,51,54, 56–61]. The other focuses on gene networks and morphological traits in multicellular organisms, and uses the Waddingtonian metaphors of canalization, cryptic genetic variation, and genetic assimilation [8,17,18,20,21,23–25, 28, 32–34, 41–44, 74].
Here we reconcile these views by showing that the primary distinction is not molecular versus morphological, but simply low versus high recombination rate. Scenarios with different recombination rates not only invoke different verbal metaphors, but are underpinned by fundamentally distinct theoretical models, which have different implications. Perhaps coincidently, robustness seems to promote evolvability in both cases.
Other distinctions are secondary to recombination. For example, both gene networks in asexual organisms and single genes in sexual organisms show low recombination. Cryptic genetic variation (post-mutation effects), while often considered only in the context of high recombination, might also be important with low recombination [36,37], and can be studied using the metaphors of the capacitor or “rug”. Similarly, new mutations could, in principle if not in practice, contribute to differential evolvability even with high recombination (Table 1).
Table 1.
Metaphors and model systems for each of our four conceptual subcategories.
| Post-Mutation | Pre-Mutation | |||
|---|---|---|---|---|
| Metaphors | Examples | Metaphors | Examples | |
| Low Recombination |
The Rug | Single genes | Neutral networks | Single genes |
| Capacitance | Asexual organisms yeast prion [PSI+] |
Look-ahead effect | Asexual organisms yeast prion [PSI+] |
|
| High Recombination |
Canalization, cryptic genetic variation and genetic assimilation |
Gene networks in sexual organisms |
None | None (because recombination makes post- mutation effects more important) |
| Capacitance | ||||
|
If the assumptions of quantitative genetics hold: The Rug |
||||
In all cases, the ability to modulate the quantity of variation via a capacitor can increase variation [7,17, 26–31, 36, 44, 46–48, 80]. Intrinsic robustness, meaning the size of the relevant genotypic space, also promotes evolvability via a quantity-based argument [16]. Whether additional adaptive robustness further promotes evolvability is a quality argument that depends strongly on the nature of the genotype-phenotype map. Indeed, the nature of the genotype-phenotype map can be thought of as the central question of biology. Model systems for this map have been drivers of progress in this field, and the development of both existing and new model systems is expected to advance the field. Obviously, the nature of the genotype-phenotype map cannot be predicted a priori from theoretical considerations alone. But nor are experiments alone sufficient: the combinatorics of genotypes that could vary at any site are prohibitive. The most promising approach from here involves exhaustive computational models of the genotype-phenotype map that are informed by rich but not comprehensive empirical data, and from which experimental predictions of higher-order phenomena can be derived.
Box 2 Figure I.
Wright’s original adaptive landscape from 1932. Wright’s diagram illustrates the rapid increase in complexity of genotype space as number of loci increases. Conceptualizing a high-dimensional genotype space is very difficult. (Figure taken from [82]).
Acknowledgements
We thank Ben Wilson for technical help with Figure 1 and Jeremy Draghi, Etienne Rajon, Mark Siegal, and three anonymous reviewers for helpful comments on the manuscript. Work was supported by the National Institutes of Health (R01GM076041). J.M. is a Pew Scholar in the Biomedical Sciences.
Glossary
- Adaptive valley
When any mutational path between two high fitness genotypes or ‘adaptive peaks’ must pass through lower fitness mutational intermediates, the low fitness genotypes represent an adaptive valley
- Canalization and Decanalization
Canalization is biological robustness that evolves in the context of developmental processes [24]. Robustness or canalization is the extent to which phenotypes remain constant in the face of specified environmental and/or genetic perturbations. Decanalization is the breakdown of this robustness by a large perturbation, leading to a sudden increase in phenotypic variation
- Capacitor
Analogous to electrical capacitors that store and release charge, phenotypic capacitors are biological switches that hide and reveal heritable phenotypic variation. Evolutionary capacitors have been defined as that subset of phenotypic capacitors that can promote adaptation [80]
- Cryptic genetic variation
Standing genetic variation that does not ordinarily contribute to the phenotype. Following a major perturbation, a system might lose its robustness so that the phenotypic effects of cryptic genetic variation are revealed
- Evolvability
The ability of a population to generate heritable phenotypic variation that could be adaptive in some contexts [7,8]. For a systematic discussion of alternative definitions, see Ref [3]
- Genetic assimilation
A process by which a phenotype initially produced in response to an environmental factor loses its dependence on that factor. Genetic assimilation occurs when the induced phenotype is selected for several or many generations [24]
- Genotype space
The set of all possible genotypes. Two genotypes which differ by a single mutation are referred to as ‘neighbors’ in genotype space
- Genotype-phenotype map
Which genotypes encode which phenotypes
- Neutral network
A set of genotypes, all encoding the same phenotype, which are all connected by single mutational steps [10]. When this phenotype is fitness, neutral networks are mathematically tractable [52]
- Phenotypic neighborhood
The range of alternative phenotypes that can be accessed by single mutations from a population residing in a given location in genotype space
- Post-mutation evolvability
Refers to adaptation involving only alleles that are already present in a population
- Pre-mutation evolvability
Refers to the adaptive potential of alleles that are not yet present but which are one or maybe two mutational steps away from an existing genotype
- Redundancy
Robustness arising from two interchangeable copies of a functional unit such as a gene, such that one compensates for deletion of the other
- Preadaptation
Any process that systematically increases the probability that a given variant will be adaptive
- Quantitative genetics
A model in which the phenotype is a function of a continuous phenotypic value, which is the sum of many discrete effects. These include both genetic and environmental effects. Genetic effects are further decomposed into those which are heritable from parent to sexual offspring and those which are only passed along through clones. The simplest models assume independent, additive effects for each allele
- Toy model
A model that is not intended to capture realistic biology. Sometimes only one core aspect of a scenario, e.g. the nature of natural selection, is realistic, and the aim is to discover very general, model-independent insights
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Works Cited
- 1.Dobzhansky T. Genetics and the Origin of Species. NY: Columbia University Press; 1937. p. 126. [Google Scholar]
- 2.Wagner GP, Altenberg L. Perspective: Complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 3.Pigliucci M. Is evolvability evolvable? Nat. Rev. Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- 4.Kirschner M, Gerhart J. Evolvability. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen TF, Houle D. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 2008;21:1201–1219. doi: 10.1111/j.1420-9101.2008.01573.x. [DOI] [PubMed] [Google Scholar]
- 6.Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masel J, Bergman A. The evolution of the evolvability properties of the yeast prion [PSI+] Evolution. 2003;57:1498–1512. doi: 10.1111/j.0014-3820.2003.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 8.Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TF. Is modularity necessary for evolvability? Remarks on the relationship between pleiotropy and evolvability. BioSystems. 2003;69 doi: 10.1016/s0303-2647(02)00132-6. 83-04. [DOI] [PubMed] [Google Scholar]
- 10.Wagner A. Robustness and evolvability in living systems. Princeton University Press; 2005. [Google Scholar]
- 11.Fisher RA. The Genetical Theory of Natural Selection. Oxford University Press; 1930. [Google Scholar]
- 12.Muller HJ. Some genetic aspects of sex. Am. Nat. 1932;66:118–138. [Google Scholar]
- 13.Neher RA, Shraiman BI, Fisher DS. Rate of adaptation in large sexual populations. Genetics. 2010;184:467–481. doi: 10.1534/genetics.109.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto SP. The evolutionary enigma of sex. Am. Nat. 2009;174:s1–s14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- 15.Lenski RE, Barrick JE, Ofria C. Balancing robustness and evolvability. PLoS Biol. 2006;4:2190–2192. doi: 10.1371/journal.pbio.0040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner A. Robustness and evolvability: a paradox resolved. Proc. R. Soc. Biol. Sci. Ser. B. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eshel I, Matessi C. Canalization, genetic assimilation and preadaptation. A quantitative genetic model. Genetics. 1998;149:2119–2133. doi: 10.1093/genetics/149.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moczek AP. Developmental capacitance, genetic accommodation, and adaptive evolution. Evol. and Devel. 2007;9:299–305. doi: 10.1111/j.1525-142X.2007.00162.x. [DOI] [PubMed] [Google Scholar]
- 19.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 2007;8:610–619. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 20.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat. Rev. Genet. 2004;5:681–691. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 21.Schlichting CD. Hidden reaction norms, cryptic genetic variation, and evolvability. Ann. N.Y. Acad. Sci. 2008;1133:187–203. doi: 10.1196/annals.1438.010. [DOI] [PubMed] [Google Scholar]
- 22.Falconer D, Mackay T. Introduction to Quantitative Genetics. 4th Edition. Prentice Hall; 1996. [Google Scholar]
- 23.Badyaev A. Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proc. R. Soc. Biol. Sci. Ser. B. 2005;272:877–886. doi: 10.1098/rspb.2004.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waddington CH. The Strategy of the Genes. George Allen & Unwin Ltd; 1957. [Google Scholar]
- 25.McGuigan K, Sgro CM. Evolutionary consequences of cryptic genetic variation. Trends Ecol. Evol. 2009;24:305–311. doi: 10.1016/j.tree.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y. Rate of adaptive peak shifts with partial genetic robustness. Evolution. 2007;61:1847–1856. doi: 10.1111/j.1558-5646.2007.00166.x. [DOI] [PubMed] [Google Scholar]
- 27.Masel J. Evolutionary capacitance may be favoured by natural selection. Genetics. 2005;170:1359–1371. doi: 10.1534/genetics.105.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 29.Tyedmers J, Madariaga M, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6(11):e394. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI+], even with realistic rates of yeast sex. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000517. e1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King OD, Masel J. The evolution of bet-hedging adaptations to rare scenarios. Theoretical Population Biology. 2007;72:560–575. doi: 10.1016/j.tpb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debat V, et al. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evolution. 2007;60:2529–2538. [PubMed] [Google Scholar]
- 33.Kellermann VM, Hoffmann AA, Sgro CM. Hsp90 inhibition and the expression of phenotypic variability in the rainforest species Drosophila birchii. Biol. J. Linn Soc. 2007;92:457–465. [Google Scholar]
- 34.Rutherford SL. Between genotype and phenotype: Protein chaperones and evolvability. Nat. Rev. Genet. 2003;4:263–274. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- 35.Specchia V, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 36.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 37.Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6:2588–2604. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehner B. Genes confer similar robustness to environmental, stochastic and genetic perturbations in Yeast. PLoS ONE. 2010;5(2):e9035. doi: 10.1371/journal.pone.0009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol. Evol. 2002;17:468–473. [Google Scholar]
- 40.Brem R, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Nat. Acad. Sci. U.S.A. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieseberg L, Archer M, Wayne R. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- 42.Lauter N, Doebley J. Genetic variation for phenotypically invariant traits detected in Teosinte: Implications for the evolution of novel forms. Genetics. 2002;160:333–342. doi: 10.1093/genetics/160.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieseberg L, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 44.Hermisson J, Wagner G. The population genetic theory of hidden variation and genetic robustness. Genetics. 2004;168:2271–2284. doi: 10.1534/genetics.104.029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masel J, Maughan H. Mutations leading to loss of sporulation ability in Bacillus subtilis are sufficiently frequent to favor genetic canalization. Genetics. 2007;175:453–457. doi: 10.1534/genetics.106.065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joseph SB, Kirkpatrick M. Effects of the [PSI+] prion on rates of adaptation in yeast. J. Evol. Biol. 2008;21:773–780. doi: 10.1111/j.1420-9101.2008.01515.x. [DOI] [PubMed] [Google Scholar]
- 47.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 48.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–484. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 49.Firoozan M, et al. Quantitation of readthrough of terminations codons in yeast using a novel gene fusion assay. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- 50.Giacomelli M, Hancock A, Masel J. The conversion of 3' UTRs into coding regions. Mol. Biol. Evol. 2007;24:457–464. doi: 10.1093/molbev/msl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Draghi J, et al. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–355. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Nimwegen E, Crutchfield JP, Huynen M. Neutral evolution of mutational robustness. Proc. Nat. Acad. Sci. U.S.A. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ancel LW, Fontana W. Plasticity, evolvability and modularity in RNA. J. Exp. Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 54.Meyers LA, Ancel FD, Lachmann M. Evolution of genetic potential. PLoS Comp. Biol. 2005;1(3):e32. doi: 10.1371/journal.pcbi.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Draghi J, Wagner GP. The evolutionary dynamics of evolvability in a gene network model. J. Evol. Biol. 2009;22:599–611. doi: 10.1111/j.1420-9101.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- 56.Fontana W. Modelling 'evo-devo' with RNA. BioEssays. 2002;24:1164–1177. doi: 10.1002/bies.10190. [DOI] [PubMed] [Google Scholar]
- 57.Ferrada E, Wagner A. Protein robustness promotes evolutionary innovations on large evolutionary time-scales. Proc. R. Soc. Biol. Sci. Ser. B. 2008;275:1595–1602. doi: 10.1098/rspb.2007.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bloom JD, Raval A, Wilke CO. Thermodynamics of neutral protein evolution. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J. Mol. Biol. 2008;379:1029–1104. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 60.Bloom JD, et al. Protein stability promotes evolvability. Proc. Nat. Acad. Sci. U.S.A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Nat. Acad. Sci. U.S.A. 2009;105:6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciliberti S, Martin OC, Wagner A. Innovation and robustness in complex regulatory gene networks. Proc. Nat. Acad. Sci. U.S.A. 2007;104:13591–13596. doi: 10.1073/pnas.0705396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crombach A, Hogeweg P. Evolution of evolvability in gene regulatory networks. PLoS Comp. Biol. 2008;4(7):e1000112. doi: 10.1371/journal.pcbi.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huerta-Sanchez E, Durrett R. Wagner's canalization model. Theor. Popul. Biol. 2007;71:121–130. doi: 10.1016/j.tpb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 65.Rodrigues JM, Wagner A. Evolutionary plasticity and innovations in complex metabolic reaction networks. PLoS Comp. Biol. 2009;5(12):e1000613. doi: 10.1371/journal.pcbi.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maerkl SJ, Quake SR. Experimental determination of the evolvability of a transcription factor. Proc. Nat. Acad. Sci.U.S.A. 2009;106:18650–18655. doi: 10.1073/pnas.0907688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitacre J, Bender A. Degeneracy: a design principle for achieving robustness and evolvability. J. Theor. Biol. 2010;63(1):143–145. doi: 10.1016/j.jtbi.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 68.Borenstein E, Meilijson I, Ruppin E. The effect of phenotypic plasticity on evolution in multipeaked fitness landscapes. J. Evol. Biol. 2006;19:1555–1570. doi: 10.1111/j.1420-9101.2006.01125.x. [DOI] [PubMed] [Google Scholar]
- 69.Drummond DA, Wilke C. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitehead DJ, et al. The look-ahead effect of phenotypic mutations. Biology Direct. 2008;3(1):18. doi: 10.1186/1745-6150-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McBride RC, Ogbunugafor CB, Turner PE. Robustness promotes evolvability of thermotolerance in an RNA virus. BMC Evol. Biol. 2008;8(1):231. doi: 10.1186/1471-2148-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuevas JM, Moya A, Sanjuan R. A genetic background with low mutational robustness is associated with increased adaptability to a novel host in an RNA virus. J. Evol. Biol. 2009;22:2041–2048. doi: 10.1111/j.1420-9101.2009.01817.x. [DOI] [PubMed] [Google Scholar]
- 73.Desai MM, Fisher DS. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics. 2007;176(3):1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le Rouzic A, Carlborg O. Evolutionary potential of hidden genetic variation. Trends Ecol. Evol. 2008;23:33–37. doi: 10.1016/j.tree.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 75.Azevedo RBR, et al. Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature. 2006;440:87–90. doi: 10.1038/nature04488. [DOI] [PubMed] [Google Scholar]
- 76.Lohaus R, Burch CL, Azevedo RBR. Genetic architecture and the evolution of sex. Heredity. 2010;101:S142–S157. doi: 10.1093/jhered/esq013. [DOI] [PubMed] [Google Scholar]
- 77.Lynch M. The evolution of genetic networks by non-adaptive processes. Nat. Rev. Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- 78.Martin OC, Wagner A. Effects of recombination on complex regulatory circuits. Genetics. 2009;183(2):673–684. doi: 10.1534/genetics.109.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Misevic D, Ofria C, Lenski RE. Sexual reproduction reshapes the genetic architecture of digital organisms. Proc. Roy. Soc. Biol. Sci. Ser. B. 2006;273:457–464. doi: 10.1098/rspb.2005.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25(9):395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Visser JAGM, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 82.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proceedings of the 6th International Congress of Genetics. 1932;1:356–366. [Google Scholar]
- 83.Maynard Smith J. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 84.Kaplan J. The end of the adaptive landscape metaphor? Biology & Philosophy. 2008;23:625–638. [Google Scholar]
- 85.Pigliucci M. Sewall Wright's adaptive landscapes: 1932 vs. 1988. Biology & Philosophy. 2008;23:591–603. [Google Scholar]
- 86.Provine WB. Sewall Wright and evolutionary biology. University of Chicago Press; 1986. [Google Scholar]
- 87.Kauffman S. The Origins of Order: Self-Organization and Selection in Evolution. Oxford University Press; 1993. [Google Scholar]
- 88.Eigen M, Schuster P. A principle of natural self-organization. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 89.Jenkins GM, et al. Evidence for the non-quasispecies evolution of RNA viruses. Mol. Biol. Evol. 2001;18:987–994. doi: 10.1093/oxfordjournals.molbev.a003900. [DOI] [PubMed] [Google Scholar]
- 90.Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol. Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gavrilets S. A dynamical theory of speciation on holey adaptive landscapes. Am. Nat. 1999;154:1–22. doi: 10.1086/303217. [DOI] [PubMed] [Google Scholar]
- 92.Gavrilets S. Evolution and speciation on holey adaptive landscapes. Trends Ecol. Evol. 1997;12:307–312. doi: 10.1016/S0169-5347(97)01098-7. [DOI] [PubMed] [Google Scholar]
- 93.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Nat. Acad. Sci.U.S.A. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sniegowski PD, Murphy HA. Evolvability. Curr. Biol. 2006;16:R831–R834. doi: 10.1016/j.cub.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 95.Draghi J, Wagner G. Evolution of evolvability in a developmental model. Evolution. 2008;62:301–315. doi: 10.1111/j.1558-5646.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 96.Lancaster AK, et al. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]