Abstract
The RNA-binding proteins TAR DNA-binding protein (TDP-43) and fused in sarcoma (FUS) play central roles in neurodegeneration associated with familial amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U). Normally localized in the nucleus, in sites affected by ALS and FTLD-U they are mislocalized to the cytoplasm and form cytoplasmic inclusions. TDP-43 and FUS are transported to the nucleus in a Ran-GTPase-dependent manner via nuclear import receptors, but they also contribute to the formation of stress granules (SGs), which are intracytoplasmic structures incorporating RNA. C-terminal truncations of TDP-43 eliminate the nuclear transport signal and cause mislocalization of the protein to the cytoplasm, where it accumulates and forms SGs. ALS-associated FUS mutations impair nuclear transport and cause mislocalization of FUS to the cytoplasm, where it also contributes to assembly of SGs. Furthermore, the ALS susceptibility factor ataxin-2, recently identified as a potent modifier of TDP-43 toxicity, is also a predicted cytoplasmic RNA-binding protein and a constituent protein of SGs, suggesting that it is a part of the common pathologic cascade formed by TDP-43 and FUS. Thus, we propose that excessive mislocalization of the RNA-binding proteins TDP-43, FUS, and ataxin-2 into the cytoplasm leads to impairment of the RNA quality control system, forming the core of the ALS/FTLD-U degenerative cascade. In this review, we discuss the molecular basis of the novel disease spectrum of ALS/FTLD-U, including the neurodegenerative mechanism of the cytoplasmic RNA-binding proteins TDP-43 and FUS and the possibility of a novel therapeutic strategy.
Through advanced genetic approaches, the causative genes of major hereditary neurodegeneration such as Alzheimer disease, spinocerebellar ataxia, and Huntington disease have been successively identified, leading to a greater understanding of their pathologic mechanisms. Recently, critical molecules that are directly associated with the pathogenesis of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U) have also been identified, including TAR DNA-binding protein (TDP-43),1,2 fused in sarcoma/translated in liposarcoma (FUS/TLS) protein,3,4 and a new entrant, ataxin-2.5 In this review, we outline advances in the understanding of the molecular basis of the ALS/FTLD-U disease spectrum, and discuss the neurodegenerative effects of the RNA-binding proteins TDP-43, FUS, and ataxin-2.
ESTABLISHMENT OF THE ALS AND FTLD-U NEURODEGENERATIVE DISEASE SPECTRUM
Since Rosen et al. associated mutations in the Cu/Zn superoxide dismutase (SOD1) gene with familial ALS (ALS1) in 1993, other proteins involved in the pathogenesis of this disease have been successively identified, including angiogenin and optineurin. Point mutations in SOD1 occur most frequently in familial ALS, and many studies are focusing on the pathogenesis of SOD1. However, there are clear differences in the pathologic findings between ALS1 and sporadic ALS, suggesting the possibility that different mechanisms underlie these diseases.6,7 The knowledge obtained to date from analyses of SOD1 mutations has not contributed significantly to the development of therapeutic strategies to combat sporadic ALS.
In 2006, 2 groups identified TDP-43 as a component of inclusions in FTLD-U.1,2 TDP-43 was also identified as the main constituent protein of ubiquitin-positive skein-like inclusions and round inclusions, which are pathologic hallmarks in the spinal anterior horn cells characteristic of ALS. Moreover, TDP-43-positive inclusions have been observed outside the motor neuron system in the cerebral cortex and basal ganglia in patients with ALS with dementia (ALS-D). Evidence suggests that FTLD-U, ALS-D, and ALS may constitute a disease spectrum that shares a common molecular basis with TDP-43.1,2 The relationship between these diseases has been verified by genetic approaches as well, and in 2008, several groups detected TDP-43 mutations in familial ALS.8–12 TDP-43 mutations were also detected in 2 families that exhibited the FTLD phenotype,13 suggesting that mutations in TDP-43 are the primary cause of the neurodegeneration associated with ALS and possibly FTLD-U. These findings led to the establishment of a novel disease concept, TDP-43 proteinopathy.
Recent studies have resulted in an even greater broadening of our understanding of the ALS/FTLD-U spectrum. In 2009, mutations in the gene encoding the RNA-binding protein FUS, which is similar to TDP-43, were identified as the cause of familial ALS (ALS6), which was linked to chromosome 16.3,4 Subsequently, Neumann et al.14 performed extensive FUS immunostaining in cases of FTLD-U that had ubiquitin-positive tau and TDP-43-negative inclusions and detected cytoplasmic accumulation of FUS protein in the frontotemporal lobe. In addition, several investigators reported observing numerous FUS-positive inclusions in FTLD that was classified as basophilic inclusion body disease (BIBD) and neuronal intermediate filament inclusion disease (NIFID).15,16 Thus, it has been proposed that mutant-FUS linked ALS, atypical FTLD-U, BIBD, and NIFID compose a novel disease spectrum, FUS proteinopathy.
Because no abnormal localization or accumulation of TDP-43 is observed in ALS6,3,4 and because even in FUS-accumulated FTLD-U the FUS-positive inclusions are TDP-43-negative, FUS proteinopathy was initially considered to be independent of TDP-43 proteinopathy.14,16 However, the observation of inclusions containing both FUS and TDP-43 in sporadic ALS was recently reported,17 and it is conceivable that there is crosstalk between the 2 RNA-binding proteins and that they may constitute a common pathologic cascade of the ALS/FTLD-U disease spectrum.
THE MOLECULAR BASIS OF TDP-43 PROTEINOPATHY
The TDP-43 protein has been identified as a factor that binds the transactivation-response region, which is a transcription activation region within the long terminal repeat in the HIV-1 gene. TDP-43 possesses 2 RNA-recognition motifs and was originally linked to splicing of the cystic fibrosis transmembrane conductance regulator.18 A recent proteomic analysis revealed that TDP-43 complexes with components of Drosha microprocessor complexes, consistent with roles for TDP-43 in both mRNA processing and microRNA biogenesis.19 Furthermore, comprehensive analyses aimed at identifying TDP-43 RNA binding targets demonstrated that TDP-43 is crucial for maintaining normal levels and splicing patterns of various mRNAs, including premRNAs with very long introns and noncoding RNAs.20,21 Importantly, TDP-43 also regulates its own expression level in a negative feedback loop by binding to the 3′UTR of its own mRNA.21 However, it remains unclear whether ALS mutations directly affect the normal RNA quality control function of TDP-43 and consequently lead to the pathogenesis of TDP-43 proteinopathy.
TDP-43-positive inclusions found in FTLD-U are formed in the neuronal cytoplasm, dystrophic neuritis, neuronal nucleus, and glial cells.22 ALS is characterized by prominent TDP-43-positive round Lewy body–like and skein-like inclusions in the cytoplasm of motor neurons, and also by depletion of intact TDP-43 protein in the nuclei of inclusion-bearing neurons.1,22 Therefore, one hypothesis concerning the pathogenesis of TDP-43 proteinopathy asserts that inclusions in affected neurons pull out intact TDP-43 from the nucleus, resulting in the depletion of nuclear TDP-43 and subsequent motor neuron degeneration.1,22,23
To date, in vivo TDP-43 knockout (KO) or transgenic (Tg) models have been developed in mice, worms, and flies.24–31 Flies lacking Drosophila TDP-43 are born normal, but develop deficient locomotive behaviors with defects at the neuromuscular junctions, suggesting TDP-43 is necessary for regulating synaptic terminals.32 Null or conditional KO mice exhibit embryonic lethal or postnatal death,24,25 indicating that TDP-43 is essential for viability in mice. Two groups reported that TDP-43 ± heterozygous mice express a similar level of TDP-43 as wild-type mice, suggesting that the level of endogenous TDP-43 is tightly controlled and compensated.24,33 Interestingly, Kraemer et al.33 showed that while TDP-43 ± mice exhibit no evidence of neurodegeneration, they do have forelimb weakness, though additional studies are needed to uncover the underlying mechanism. Tg mice that overexpress wild-type TDP-43, as well as mice that express mutant TDP-43, have been reported to develop motor neuron degeneration reminiscent of TDP-43 pathologies.27–29 Xu et al. demonstrated that overexpression of human TDP-43 in mice downregulates TDP-43 protein and RNA,29 and a recent conditional Tg mouse model study revealed that exogenous human TDP-43 lacking the nuclear localization signal (NLS) leads to a reduction of endogenous mouse TDP-43 in the nucleus.27 These studies suggested that the disruption of mouse TDP-43 expression in the nucleus may be a major contributor to neurodegeneration. However, because the level of TDP-43 expression is tightly controlled, it is possible that a toxic gain of function due to the presence of excessive TDP-43 protein beyond autoregulation artificially leads to motor neuron degeneration. The evidence from studies in Tg mice does not explain how ALS mutations contribute to the molecular mechanism of neurodegeneration in familial ALS, nor have these studies identified the trigger for cytoplasmic localization of endogenous wild-type TDP-43 in age-related sporadic ALS. Studies involving knock-in mice harboring a mutant TDP-43 gene and studies involving transgenic primate models are needed in order to determine whether neurodegeneration results from a loss of TDP-43 function, toxicity associated with overexpression, or both.
CELLULAR AND BIOCHEMICAL ASPECTS OF TDP-43 PROTEIN
Pathologic examination utilizing an antibody specific to phosphorylated TDP-43 revealed that aggregated TDP-43 in inclusions is abnormally phosphorylated.34 Furthermore, SDS-PAGE analysis of TDP-43 from normal brain tissue revealed multiple C-terminal fragments (CTF) between 35 and 25 kDa in addition to the full-length 43 kDa molecule. These CTFs were highly phosphorylated and accumulated in the brains of ALS/FTLD-U patients, suggesting that abnormal TDP-43 localization, phosphorylation, or fragmentation play a prominent role in the pathogenesis of ALS/FTLD-U.
Zhang et al.35 identified 2 caspase-3 cleavage consensus sites within TDP-43, and demonstrated that caspase-3 mediates cleavage of TDP-43 to generate 25 and 35 kDa fragments. Nishimoto et al.36 observed a marked decrease in caspase-3-dependent production of the 35 kDa (p35f) and 25 kDa (p25f) fragments in caspase-3-deficient mouse embryonic fibroblasts (MEFs), but also found a caspase-3-independent 35 kDa CTF in caspase-3(−/−) MEFs. Alanine scanning of this CTF indicated it is a novel isoform (p35iso) translated from an alternate in-frame translation start site (Met85) downstream of the original initiation codon. Importantly, while both p35f (aa 90–414) and p35iso (aa 85–414) lack the NLS, the 2 RNA recognition motifs (RRMs) have been completely preserved. Conversely, p25f, which lacks a NLS and also the first RRM that is essential for RNA binding,18 seems to have lost its RNA-binding function. Recent studies in yeast,37 worms,26 and flies30 demonstrated that the toxicity associated with TDP-43 overexpression depends on the protein retaining its ability to bind RNA, suggesting that p35 is more critical for development of neurodegeneration than p25.
When the p35 CTF is expressed, it mislocalizes in the cytoplasm, where it assembles into cytoplasmic inclusions. Nishimoto et al.36 investigated the biochemical properties of these inclusions and reported that they exhibit properties of SGs (figure e-1, A–F, on the Neurology® Web site at www.neurology.org),36,38 cellular structures that package mRNA and RNA-binding proteins during cell stress. SGs are thought to protect mRNA during cellular stress, heat shock, and oxidation, and to arrest translation, thereby preventing further damage by limiting the accumulation of misfolded proteins.39 Although a knock-down study showed that endogenous TDP-43 is unnecessary for SG formation,38 Nishimoto et al.36 proposed that truncated p35 facilitates SG assembly and promotes SG formation, thus suggesting a previously undescribed function for TDP-43. A recent pathologic study also demonstrated colocalization of TDP-43 inclusions with SG markers (eIF3 and TIA-1) in ALS and FTLD-U-affected brains,40 indicating a possible association between accumulation of intracytoplasmic RNA-binding proteins, SG formation, and the pathogenesis of ALS/FTLD-U. The validity of this assumption is strongly supported by identification of ALS mutations in FUS and subsequent studies described below.
THE MOLECULAR BASIS OF FUS PROTEINOPATHY
The FUS/TLS gene was originally identified as part of a fusion protein with the transcription factor CHOP, arising from chromosomal translocation in liposarcoma. The FUS/TLS gene also encodes an RNA-binding protein equipped with an RRM, similar to TDP-43. Together with the highly homologous Ewing sarcoma protein (EWS) and TAF15, FUS/TLS forms the TET (TLS/FUS, EWS, TAF15) family, and may be involved in RNA processing, splicing, and RNA metabolism. The mRNA targets of FUS and its role in RNA regulation are largely unknown. FUS knockout mice show perinatal lethality and defects in B lymphocyte development,41 while Kuroda et al.42 reported that survival KO mice exhibit male sterility, but the neurodegenerative phenotype has thus far not been reported.
Pathologic analyses have shown that FUS cytoplasmic inclusions are immunopositive for ubiquitin in immunohistologic evaluation.4,14 However, in contrast to TDP-43, there is no immunoblotting evidence for posttranslational modification of FUS through ubiquitination, hyperphosphorylation, or truncation.14,43 These results suggest that FUS may not be preferentially modified, or that other components in the FUS inclusions are the actual ubiquitination targets.43
In a study involving an EWS-deletion mutant, Zakaryan et al.44 discovered that the C-terminus of EWS is a NLS, and, in particular, that basic arginine residues in the C-terminus are essential for nuclear localization. Both EWS and FUS have high C-terminal homology, and ALS mutations have accumulated at these arginines in the C-terminus.45 Several groups43,46,47 have confirmed that nuclear transport is severely impaired in cell lines lacking the C-terminus of FUS, and that, as is the case with the p35f and p35iso of TDP-43, ALS mutations result in the mislocalization of FUS from the nucleus to the cytoplasm and enhance SG formation in an additive manner. The C-terminus of FUS thus represents a NLS, and ALS mutations directly impede nuclear localization mediated by the C-terminus. Furthermore, subcellular analyses have indicated that compared to other mutations, the P525L and R522G mutations result in stronger cytoplasmic localization of FUS and cause juvenile ALS, with a mean disease onset in the second and third decades of life.3 Thus, the increased mislocalization of mutant protein into the cytoplasm seems to decrease the age at disease onset.43,46
Recent pathologic findings revealed that inclusion bodies found in BIBD, a subtype of FUS proteinopathy, contain RNA and SG component proteins,46,48 indicating that SGs induced by expression of mutant FUS share unique features with pathologic FUS inclusions. Therefore, these findings provide direct evidence of an association between the accumulation of cytoplasmic RNA-binding proteins and SG formation in ALS/FTLD-U brain.
TDP-43 and FUS have several characteristics in common. Because both are RNA-binding proteins, are components of SGs, and show cytoplasmic mislocalization in affected tissues, they are conceivably part of a cooperative pathologic process in the development of ALS/FTLD-U. Importantly, it remains to be determined whether the formation of SGs is pathogenic or protective in neurodegeneration. SGs are basically thought to function in the protection of mRNA against harmful stress events and to arrest translation, preventing the accumulation of misfolded proteins.39 Additionally, SGs likely protect neurons by assembling cytoplasmic RNA-binding proteins and reducing toxic levels of RNA-binding proteins. It has also been speculated that inappropriate/excessive SG assembly results in chronic disturbance of RNA metabolism or entrapment and depletion of nuclear FUS and TDP-43, thereby leading to the development of neurodegeneration. Further study is needed to clarify the relationship between SG formation and cell fate in TDP-43/FUS proteinopathy in order to better understand the pathogenesis of ALS/FTLD-U.
THE NUCLEAR-CYTOPLASMIC PROTEIN TRANSPORT MECHANISM OF TDP-43 AND FUS
In intranuclear transport mechanisms, nuclear proteins that contain a NLS form a nuclear pore-targeting complex together with the nuclear import receptors importin α, β, or transportin. Proteins are then transported into the nucleus via mediation of the small Ras-related GTPase, Ran. A GTP-bound Ran gradient exists between the nucleus and the cytoplasm, created by active transport resulting from hydrolysis of the GTP bound to Ran. Subcellular localization studies have demonstrated that nuclear localization of both TDP-43 and FUS is impaired in the Ran dominant-negative strain Q69L, indicating that nuclear import is mediated by Ran-dependent machinery (figure e-2).43
Using RNA interference screening, Nishimura et al.49 discovered that the importin β pathway is involved in the nuclear import of TDP-43. Based on the NLS consensus sequence [R/H/KX(2–5)PY] of the nuclear import receptor transportin, Lee et al.50 predicted that transportin targets FUS. In a study involving small-interfering RNA, Dormann et al.46 showed that transportins are essential for nuclear localization of FUS.
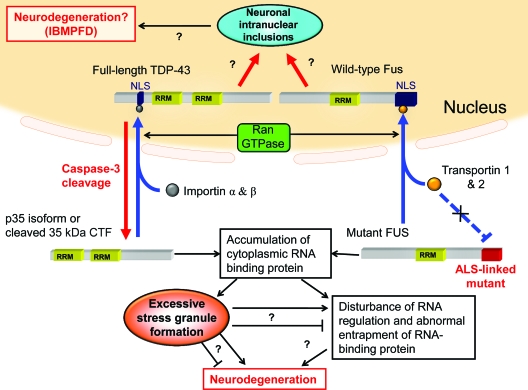
Figure 1 summarizes current information regarding TDP-43 and FUS nuclear-cytoplasmic protein transport and formation of SGs, as well as our hypothetical model of TDP-43 and FUS proteinopathy. Full-length TDP-43 is transported to the nucleus by importin and Ran-dependent transport machinery, where it normally functions in promoting RNA stability and regulating splicing. Truncated p35iso (translated from an alternate initiation codon), p35f (generated by caspase-3 during apoptosis), and mislocalized full-length TDP-43 (due to an unknown mechanism) accumulate in the cytoplasm. Wild-type FUS localizes in the nucleus in a transportin- and Ran-dependent manner. ALS mutations in FUS disturb the interactions with transportin, resulting in interference with nuclear traffic. TDP-43 and FUS subsequently accumulate in the cytoplasm, linking to assembly of SGs. The presence of excessive RNA-binding protein chronically disturbs RNA quality control or leads to the entrapment and depletion of intact nuclear TDP-43 and FUS, which in turn initiates the neurodegeneration that leads to ALS/FTLD-U.
Figure 1.
Hypothetical common pathology of TAR DNA-binding protein (TDP-43) proteinopathy and fused in sarcoma (FUS) proteinopathy
Nuclear import of TDP-43 is mediated by importin-α, importin-β, and Ran-GTPase. p35iso (which is derived from an alternate initiation codon) and p35f (proteolytically cleaved by caspase-3) do not have a nuclear localization signal (NLS) and are mislocalized to the cytoplasm. Wild-type FUS protein is transferred to the nucleus via transportin-1, transportin-2, and Ran-dependent transport machinery. Amyotrophic lateral sclerosis (ALS)-linked mutants of FUS interfere with the interaction with transportin and lead to localization in the cytoplasm. Under certain pathologic stresses or aging-related nuclear leakiness, cytoplasmic accumulation of TDP-43 and FUS lead to a common pathologic process, the formation of stress granules (SGs), or dysregulation of the RNA quality control system that may then be associated with motor neuron degeneration. Similar to inclusions in other neurodegenerative diseases, whether SGs are cytotoxic or cytoprotective remains unclear. An alternative pathway suggests that neuronal intranuclear inclusions (NIIs) may be associated with pathogenesis in some types of frontotemporal lobar degeneration (FTLD)-TDP and -FUS, especially Paget disease of bone and frontotemporal dementia (IBMPFD).
The findings described above suggest that mislocalization of TDP-43 and FUS from the nucleus and their accumulation in the cytoplasm is the initial step in the development of ALS/FTLD-U. The mechanism in sporadic ALS/FTLD-U cases whereby TDP-43 and FUS proteins that lack mutations in the NLS induce cytoplasmic mislocalization is unknown. However, an interesting recent report regarding the function of nuclear pores in relation to this point was published by D'Angelo et al.,51 who found that some nuclear pore proteins, such as those of the Nup107/160 complex, are extremely long-lived and do not turn over during the life of nondividing cells, such as neurons. A subset of nucleoporins is oxidatively damaged in aging cells, leading to an increase in nuclear permeability and leakiness. Therefore, it can be hypothesized that aging-related nuclear leakiness is accelerated in patients with sporadic ALS/FTLD-U, which causes a mislocalization of TDP-43 and FUS in the cytoplasm and triggers the neurodegenerative cascade in ALS/FTLD-U. If this hypothesis is correct, it would help explain the mechanism of neurodegeneration in age-related sporadic ALS/FTLD-U. Particularly promising would be novel therapeutic approaches aimed at overcoming the impaired nuclear-cytoplasmic protein transport mechanism that leads to accumulation of TDP-43 and FUS in the cytoplasm, thereby preventing or delaying the neurodegeneration associated with ALS/FTLD-U.
GENETIC RISK OF ATAXIN-2 INTERMEDIATE-LENGTH POLYGLUTAMINE EXPANSIONS
Ataxin-2 normally contains a polyglutamine tract of 22 glutamine residues. Tracts of greater than 33 glutamine residues result in hereditary spinocerebellar ataxia type 2 (SCA2). Elden et al.5 recently demonstrated that ataxin-2 is also a potent modifier of TDP-43 toxicity. They showed that overexpression or reduced expression of ATXN2 enhances or attenuates TDP-43 toxicity, respectively, in the retina of flies, and that ataxin-2 and TDP-43 form a complex in an RNA-dependent manner. In a genomic analysis of patients with ALS, they also found that intermediate-length ataxin-2 polyglutamine expansions (27–33 glutamine residues) are associated with an increased risk for ALS, and that this is accompanied by an earlier age at onset. Interaction between ataxin-2 and TDP-43 as well as cytoplasmic mislocalization of TDP-43 are enhanced by longer polyglutamine expansions. It is noteworthy that ataxin-2 has an Lsm domain (Like Sm domain), which has an RNA binding motif, and that ataxin-2 is a constituent protein of SGs,52 similar to TDP-43 and FUS. Presumably, ataxin-2 contributes to the common pathologic cascade formed by TDP-43 and FUS.37 Notably, to date there have been no pathologic or clinical reports of motor neuron disease or FTLD phenotypes in SCA2 cases, illustrating the need for additional studies focusing on TDP-43 and FUS in SCA2 brain tissue as a means of disentangling the complex interactions between TDP-43, FUS, and ataxin-2.
TOWARD AN UNDERSTANDING OF THE ALS/FTLD-U MOLECULAR NETWORK
When interpreting the above-mentioned findings and supposition, several issues should be kept in mind. For instance, the characteristic pathologic findings in TDP-43 proteinopathy are TDP-43-positive neuronal cytoplasmic inclusions (NCIs) and neuritis, but neuronal intranuclear inclusions (NIIs) and glial inclusions are also observed in the FTLD-TDP-43 subtype.22 In cases involving mutations in the gene encoding valosin-containing protein (VCP), which has been linked to FTLD with inclusion body myopathy and Paget disease of bone, the characteristic neuropathologic feature is an abundance of TDP-43 positive NIIs with few NCIs.22 The mechanism of TDP-43 proteinopathy therefore appears to involve complicated degenerative processes, and further studies focusing on the pathologic consequences of nuclear aggregation and glial involvement are needed in addition to those focusing on cytoplasmic accumulation.
In addition, how ALS mutations in TDP-43 lead to neurodegeneration is currently unknown. Recent pulse-chase labeling studies showed that mutant TDP-43 is more stable than the wild-type and has enhanced binding to FUS/TLS.19 Dewey et al.53 demonstrated that mutant TDP-43 incorporates into SGs earlier and forms larger SGs than the wild-type. These results support our hypothetical model (figure 1) of the common pathology of TDP-43 and FUS proteinopathies, but are not conclusive enough to explain the pathologic mechanism induced by mutations in TDP-43. Clarifying the toxic properties associated with ALS-dominant mutations in the TDP-43 protein is critical for understanding the molecular basis of ALS.
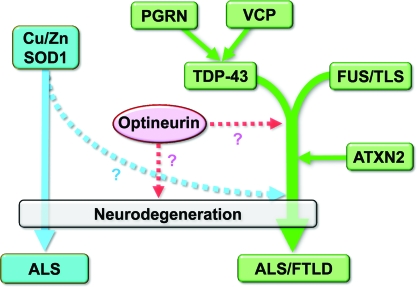
A diagram illustrating the relationships between the major molecules involved in ALS that have been identified to date is shown in figure 2. Mutations in the VCP and progranulin genes have also been identified as causes of familial FTLD-TDP.54–56 Pathologic accumulation of TDP-43 has been documented to occur with both mutations, and biochemical and pathologic studies suggested that the VCP and progranulin proteins are located upstream of TDP-43 in the degeneration cascade.22,35,57,58
Figure 2.
Possible pathologic cascade of amyotrophic lateral sclerosis (ALS)/frontotemporal lobar degeneration (FTLD)-linked molecules
TAR DNA-binding protein (TDP-43) and fused in sarcoma (FUS) induce a conjoint pathologic cascade of neurodegeneration that leads to ALS/FTLD with ubiquitin-positive inclusions. Ataxin-2 possibly promotes this cascade. Recent pathologic and biochemical evidence indicates that valosin containing protein (VCP) and progranulin (PGRN) may be located upstream of TDP-43 in the pathologic cascade. Cu/Zn SOD1 may contribute to an independent pathologic process or join the pathologic cascade downstream of TDP-43/FUS. The role of optineurin in the pathologic cascade remains to be elucidated.
A number of other genes are associated with familial ALS, including those encoding SOD1 and angiogenin, and a Japanese group recently identified novel mutations in optineurin as being associated with familial ALS.59 Familial ALS is most often associated with mutations in SOD1, but this seldom results in impairment of cognitive function in patients with ALS1 with this mutation,60 indicating that ALS1 differs clinically from the ALS/FTLD-U disease spectrum. Accumulating pathologic and biochemical evidence suggests that ALS associated with SOD1 mutations or TDP-43 proteinopathy result from distinct pathologic mechanisms.6,7 The current consensus is that ALS resulting from SOD1 mutations involves a pathologic process that is independent from the ALS/FTLD-U pathologic cascade or is joined downstream with that cascade.
Understanding the molecular network associated with ALS/FTLD-U and accurately elucidating the pathologic cascade are extremely important for establishing a treatment. Several questions must be addressed by future studies for such a strategy to become reality. For example, is mutant optineurin part of the same ALS/FTLD-U pathologic cascade as that involving mutations in TDP-43 and FUS, or do mutations in optineurin form an independent disease entity, similar to ALS1? Is FUS toxicity also modified by ataxin-2 with intermediate-length polyglutamine expansions? Despite these questions, recent studies indicate that steady progress is being made toward development of a therapeutic strategy to overcome ALS and FTLD-U.
Supplementary Material
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- ALS-D
amyotrophic lateral sclerosis with dementia
- BIBD
basophilic inclusion body disease
- CTF
C-terminal fragments
- EWS
Ewing sarcoma protein
- FTLD-U
frontotemporal lobar degeneration with ubiquitin-positive inclusions
- FUS/TLS
fused in sarcoma/translated in liposarcoma
- KO
knockout
- MEF
mouse embryonic fibroblast
- NCI
neuronal cytoplasmic inclusion
- NIFID
neuronal intermediate filament inclusion disease
- NII
neuronal intranuclear inclusion
- NLS
nuclear localization signal
- RRM
RNA recognition motifs
- SCA2
spinocerebellar ataxia type 2
- SG
stress granule
- TDP-43
TAR DNA-binding protein
- Tg
transgenic
- VCP
valosin-containing protein.
Footnotes
Disclosure: The authors report no disclosures.
Editorial, page 1588
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
D.I. and N.S. contributed to the preparation and writing of this article.
REFERENCES
- 1. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006;314:130–133 [DOI] [PubMed] [Google Scholar]
- 2. Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 2006;351:602–611 [DOI] [PubMed] [Google Scholar]
- 3. Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009;323:1205–1208 [DOI] [PubMed] [Google Scholar]
- 4. Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009;323:1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010;466:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tan CF, Eguchi H, Tagawa A, et al. TDP-43 immunoreactivity in neuronal inclusions in familial amyotrophic lateral sclerosis with or without SOD1 gene mutation. Acta Neuropathol 2007;113:535–542 [DOI] [PubMed] [Google Scholar]
- 7. Mackenzie IR, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol 2007;61:427–434 [DOI] [PubMed] [Google Scholar]
- 8. Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008;319:1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol 2008;7:409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yokoseki A, Shiga A, Tan CF, et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann Neurol 2008;63:538–542 [DOI] [PubMed] [Google Scholar]
- 11. Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 2008;40:572–574 [DOI] [PubMed] [Google Scholar]
- 12. Gitcho MA, Baloh RH, Chakraverty S, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol 2008;63:535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benajiba L, Le Ber I, Camuzat A, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol 2009;65:470–473 [DOI] [PubMed] [Google Scholar]
- 14. Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 2009;132:2922–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IR. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol 2009;118:605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Munoz DG, Neumann M, Kusaka H, et al. FUS pathology in basophilic inclusion body disease. Acta Neuropathol 2009;118:617–627 [DOI] [PubMed] [Google Scholar]
- 17. Deng HX, Zhai H, Bigio EH, et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol 2011;67:739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 2001;276:36337–36343 [DOI] [PubMed] [Google Scholar]
- 19. Ling SC, Albuquerque CP, Han JS, et al. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci USA 2010;107:13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 2011;14:452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Polymenidou M, Lagier-Tourenne C, Hutt KR, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 2011;14:459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neumann M, Kwong LK, Sampathu DM, Trojanowski JQ, Lee VM. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch Neurol 2007;64:1388–1394 [DOI] [PubMed] [Google Scholar]
- 23. Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 2010;9:995–1007 [DOI] [PubMed] [Google Scholar]
- 24. Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci USA 2010;107:16320–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sephton CF, Good SK, Atkin S, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem 2010;285:6826–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ash PE, Zhang YJ, Roberts CM, et al. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet 2010;19:3206–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Igaz LM, Kwong LK, Lee EB, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 2011;121:726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wils H, Kleinberger G, Janssens J, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 2010;107:3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu YF, Gendron TF, Zhang YJ, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci 2010;30:10851–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voigt A, Herholz D, Fiesel FC, et al. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One 2010;5:e12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 2009;106:18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett 2009;583:1586–1592 [DOI] [PubMed] [Google Scholar]
- 33. Kraemer BC, Schuck T, Wheeler JM, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol 2010;119:409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hasegawa M, Arai T, Nonaka T, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol 2008;64:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang YJ, Xu YF, Dickey CA, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci 2007;27:10530–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishimoto Y, Ito D, Yagi T, Nihei Y, Tsunoda Y, Suzuki N. Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J Biol Chem 2010;285:608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA 2008;105:6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colombrita C, Zennaro E, Fallini C, et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem 2009;111:1051–1061 [DOI] [PubMed] [Google Scholar]
- 39. Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol 2007;431:61–81 [DOI] [PubMed] [Google Scholar]
- 40. Liu-Yesucevitz L, Bilgutay A, Zhang YJ, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 2010;5:e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hicks GG, Singh N, Nashabi A, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet 2000;24:175–179 [DOI] [PubMed] [Google Scholar]
- 42. Kuroda M, Sok J, Webb L, et al. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J 2000;19:453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ito D, Seki M, Tsunoda Y, Uchiyama H, Suzuki N. Nuclear transport impairment of ALS-linked mutations in FUS/TLS. Ann Neurol 2010;69:152–162 [DOI] [PubMed] [Google Scholar]
- 44. Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J Mol Biol 2006;363:27–38 [DOI] [PubMed] [Google Scholar]
- 45. Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 2010;19:R46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dormann D, Rodde R, Edbauer D, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 2010;29:2841–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bosco DA, Lemay N, Ko HK, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 2010;19:4160–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fujita K, Ito H, Nakano S, Kinoshita Y, Wate R, Kusaka H. Immunohistochemical identification of messenger RNA-related proteins in basophilic inclusions of adult-onset atypical motor neuron disease. Acta Neuropathol 2008;116:439–445 [DOI] [PubMed] [Google Scholar]
- 49. Nishimura AL, Zupunski V, Troakes C, et al. Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 2010;133:1763–1771 [DOI] [PubMed] [Google Scholar]
- 50. Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006;126:543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009;136:284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nonhoff U, Ralser M, Welzel F, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 2007;18:1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dewey CM, Cenik B, Sephton CF, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol 2011;31:1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Watts GD, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 2004;36:377–381 [DOI] [PubMed] [Google Scholar]
- 55. Cruts M, Gijselinck I, van der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006;442:920–924 [DOI] [PubMed] [Google Scholar]
- 56. Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006;442:916–919 [DOI] [PubMed] [Google Scholar]
- 57. Ritson GP, Custer SK, Freibaum BD, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci 2010;30:7729–7739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gitcho MA, Strider J, Carter D, et al. VCP mutations causing frontotemporal lobar degeneration disrupt localization of TDP-43 and induce cell death. J Biol Chem 2009;284:12384–12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maruyama H, Morino H, Ito H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010;465:223–226 [DOI] [PubMed] [Google Scholar]
- 60. Wicks P, Abrahams S, Papps B, et al. SOD1 and cognitive dysfunction in familial amyotrophic lateral sclerosis. J Neurol 2009;256:234–241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.