Abstract
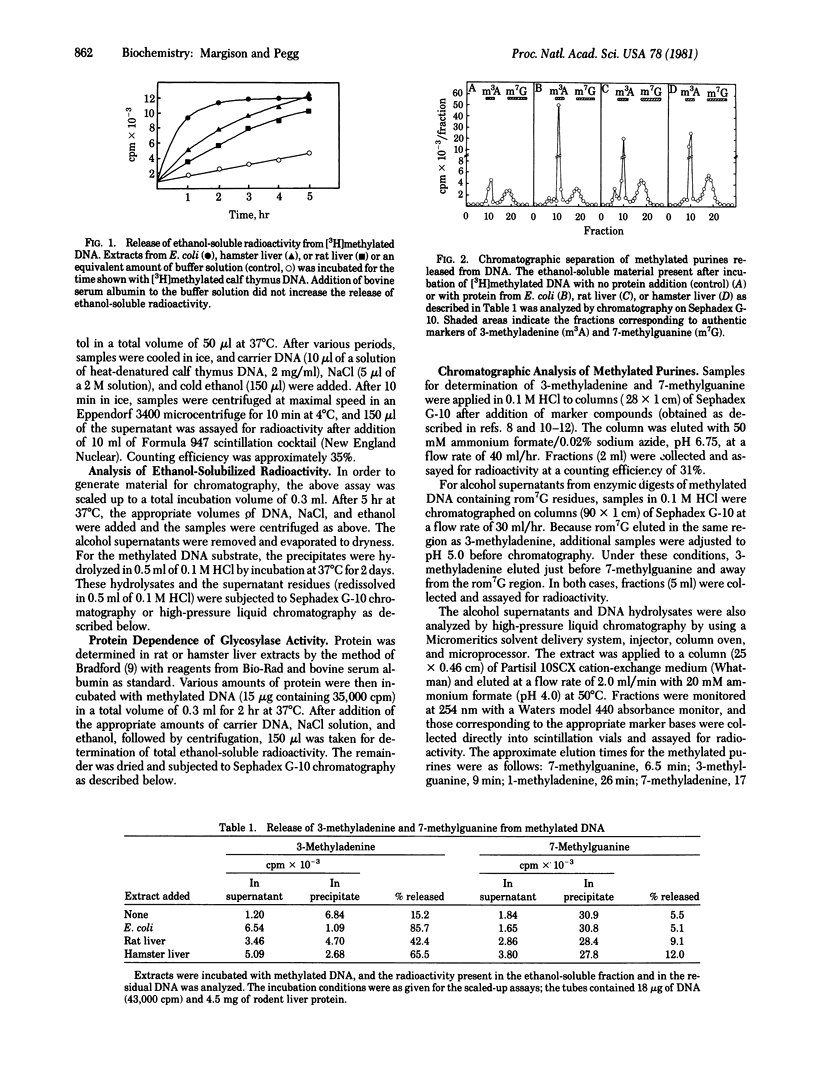
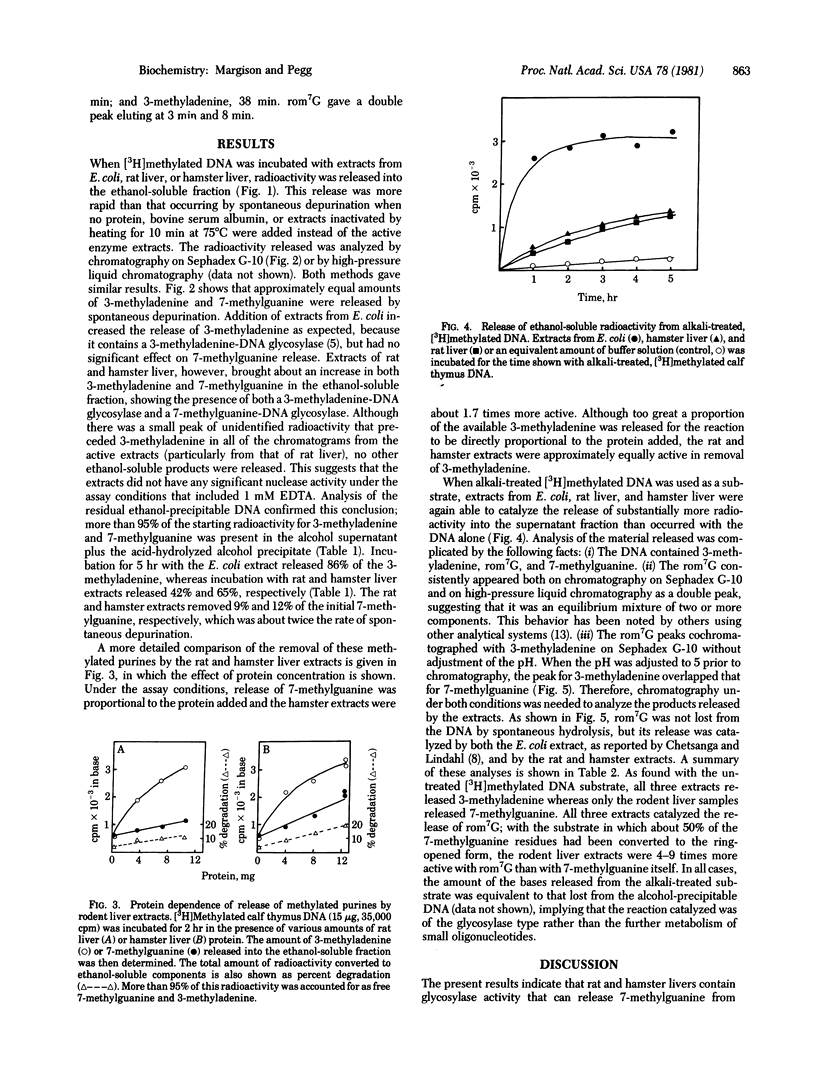
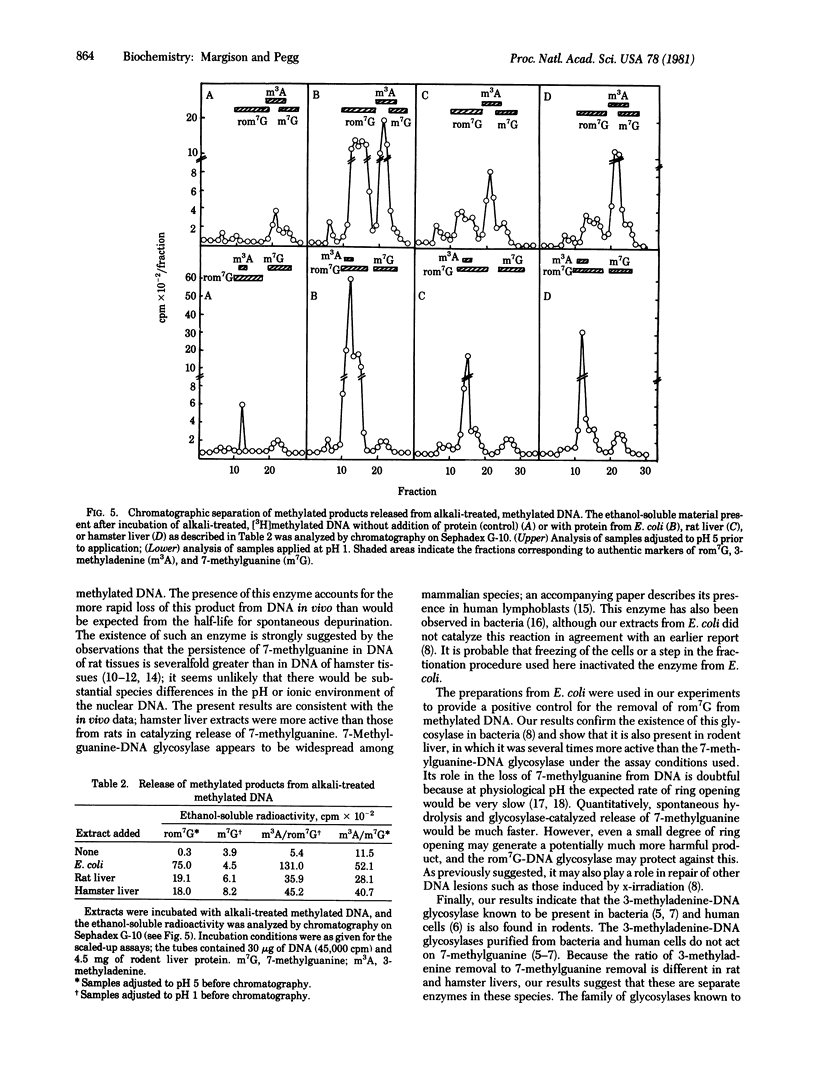
Rat and hamster liver extracts were found to contain DNA glycosylases capable of removing 3-methyladenine and 7-methylguanine from methylated DNA. The activity of 7-methylguanine-DNA glycosylase was greater tin hamster than in rat liver extracts. This finding is consistent with previous reports that the half-life of 7-methylguanine in DNA after treatment with the carcinogen dimethylnitrosamine is longer in rats than in hamsters. These enzymes may, therefore, play an important role in the removal of abnormal alkylation products from mammalian cell DNA. Rodent liver extracts also contained a DNA glycosylase able to remove from alkylated DNA the imidazole-ring-opened form of 7-methylguanine which is produced by treatment with alkali. Although this product may occur in vivo after treatment with alkylating agents to only a very small extent, the enzyme may be needed to minimize its potentially harmful biological effects.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brent T. P. Partial purification and characterization of a human 3-methyladenine-DNA glycosylase. Biochemistry. 1979 Mar 6;18(5):911–916. doi: 10.1021/bi00572a028. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock V. M. Stability of deoxyribonucleic acid methylated in the intact animal by administration of dimethylnitrosamine. Rate of breakdown in vivo and in vitro at different dosages. Biochem J. 1969 Feb;111(4):497–502. doi: 10.1042/bj1110497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. FURTHER STUDIES ON THE ALKYLATION OF NUCLEIC ACIDS AND THEIR CONSTITUENT NUCLEOTIDES. Biochem J. 1963 Oct;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval J., Pierre J., Laval F. Release of 7-methylguanine residues from alkylated DNA by extracts of Micrococcus luteus and Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):852–855. doi: 10.1073/pnas.78.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval J. Two enzymes are required from strand incision in repair of alkylated DNA. Nature. 1977 Oct 27;269(5631):829–832. doi: 10.1038/269829a0. [DOI] [PubMed] [Google Scholar]
- Lawley P. D., Shah S. A. Methylation of ribonucleic acid by the carcinogens dimethyl sulphate, N-methyl-N-nitrosourea and N-methyl-N'-nitro-N-nitrosoguanidine. Comparisons of chemical analyses at the nucleoside and base levels. Biochem J. 1972 Jun;128(1):117–132. doi: 10.1042/bj1280117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley P. D. Some chemical aspects of dose-response relationships in alkylation mutagenesis. Mutat Res. 1974 Jun;23(3):283–295. doi: 10.1016/0027-5107(74)90102-x. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Margison J. M., Montesano R. Methylated purines in the deoxyribonucleic acid of various Syrian-golden-hamster tissues after administration of a hepatocarcinogenic dose of dimethylnitrosamine. Biochem J. 1976 Sep 1;157(3):627–634. doi: 10.1042/bj1570627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E. Alkylation of rat liver DNA by dimethylnitrosamine: effect of dosage on O6-methylguanine levels. J Natl Cancer Inst. 1977 Mar;58(3):681–687. doi: 10.1093/jnci/58.3.681. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. Formation and metabolism of alkylated nucleosides: possible role in carcinogenesis by nitroso compounds and alkylating agents. Adv Cancer Res. 1977;25:195–269. doi: 10.1016/s0065-230x(08)60635-1. [DOI] [PubMed] [Google Scholar]
- Singer B., Brent T. P. Human lymphoblasts contain DNA glycosylase activity excising N-3 and N-7 methyl and ethyl purines but not O6-alkylguanines or 1-alkyladenines. Proc Natl Acad Sci U S A. 1981 Feb;78(2):856–860. doi: 10.1073/pnas.78.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. N-nitroso alkylating agents: formation and persistence of alkyl derivatives in mammalian nucleic acids as contributing factors in carcinogenesis. J Natl Cancer Inst. 1979 Jun;62(6):1329–1339. [PubMed] [Google Scholar]
- Stumpf R., Margison G. P., Montesano R., Pegg A. E. Formation and loss of alkylated purines from DNA of hamster liver after administration of dimethylnitrosamine. Cancer Res. 1979 Jan;39(1):50–54. [PubMed] [Google Scholar]


