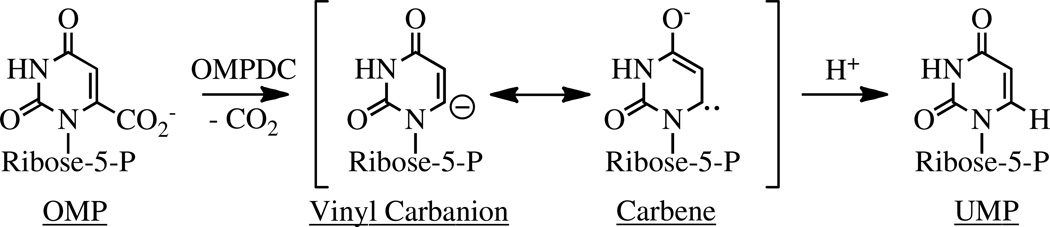
Abstract
The reaction catalyzed by orotidine 5’-monophosphate decarboxylase (OMPDC) is accompanied by exceptional values for the rate enhancement [kcat/knon = 7.1 × 1016] and catalytic proficiency [(kcat/KM)/knon = 4.8 × 1022 M−1]. Although a stabilized vinyl carbanion/carbene intermediate is located on the reaction coordinate, the structural strategies by which the reduction in the activation energy barrier is realized remain incompletely understood. This laboratory recently reported that “substrate destabilization” by Asp 70 in the OMPDC from Methanothermobacter thermoautotrophicus (MtOMPDC) lowers the activation energy barrier by ~5 kcal/mol (contributing ~2.7 × 103 to the rate enhancement) [K. K. Chan, B. M. Wood, A. A. Fedorov, E. V. Fedorov, H. J. Imker, T. L. Amyes, J. P. Richard, S. C. Almo, and J. A. Gerlt (2009) Biochemistry 48, 5518–31]. We now report that substitutions of hydrophobic residues in a pocket proximal to the carboxylate group of the substrate (Ile 96, Leu 123, and Val 155) with neutral hydrophilic residues decrease the value of kcat by as much as 400-fold but have minimal effect on the value of kex for exchange of H6 of the FUMP product analog with solvent deuterium; we hypothesize that this pocket destabilizes the substrate by preventing hydration of the substrate carboxylate group. We also report that substitutions for Ser 127 that is proximal to O4 of the orotate ring decrease the value of kcat/KM, with the S127P substitution that eliminates hydrogen-bonding interactions with O4 producing a 2.5 × 106-fold reduction in the value of kcat/KM; this effect is consistent with delocalization of the negative charge of the carbanionic intermediate on O4 to produce an anionic carbene intermediate and thereby provide a structural strategy for stabilization of the intermediate. These observations provide additional information on the identities of the active site residues that contribute to the rate enhancement and, therefore, insights into the structural strategies for catalysis.
The reaction catalyzed by orotidine 5’-monophosphate decarboxylase (OMPDC) continues to attract attention because the structural bases for its impressive rate enhancement [kcat/knon = 7.1 × 1016] and catalytic proficiency [(kcat/KM)/knon = 4.8 × 1022 M−1] (1) are not well understood. A “vinyl carbanion” intermediate is on the reaction coordinate (Scheme 1): 1) the reactions catalyzed by the OMPDCs from Saccharomyces cerevisiae (ScOMPDC) and Methanothermobacter thermoautotrophicus (MtOMPDC) proceed with a product isotope effect of 1.0 (measured when the reaction is conducted in a 1:1 mixture of H2O and D2O) (2). Both enzymes also catalyze the exchange of H6 of the UMP product and the activated 5-fluoroUMP (FUMP) product analog with solvent hydrogen (3, 4), although at rates that are less than those observed for decarboxylation of either OMP or 5-fluoroOMP. However, the structural strategies by which the activation energy barrier (ΔG‡) for decarboxylation of OMP is reduced by 23 kcal/mol have not yet been completely described.
Scheme 1.
Although “ground state destabilization” of the substrate’s carboxylate group by its presumed proximity to Asp 70 in a conserved hydrogen-bonding network (residue numbering for MtOMPDC) was proposed when structures for four OMPDCs were reported in 2000 [MtOMPDC (5), ScOMPDC (6), and the OMPDCs from Bacillus subtilis (7) and Escherichia coli (8)], that proposal has been criticized (9–11). Recently, we provided experimental evidence in support of “substrate destabilization” in which destabilizing interactions develop as the reaction coordinate is traversed (the value of the KM for the OMP is less than the Ki for the UMP product in which the carboxylate group is absent, thereby discounting “ground state destabilization”) (4). Substitution of Asp 70 with an isosteric uncharged residue (D70N) or a less sterically demanding, uncharged residue (D70G) reduces the value of kcat for decarboxylation by a factor of ~2,700 (D70G) without significantly affecting the value of kex for exchange of H6 of FUMP with solvent hydrogen.
Wolfenden estimated the value of the pKa of H6 of the UMP product in aqueous solution (~32) by measuring the temperature dependence of the rate constant for nonenzymatic exchange in a model compound (12). Richard and Amyes used the value of kex for the OMPDC-catalyzed exchange of H6 to calculate an upper limit on the value of the pKa of H6 of UMP in the active site (~22) (3). The difference (~10 pKa units) provides a lower limit on the stabilization of the intermediate by its interactions with the active site (≥14 kcal/mol).
On the basis of the structure of ScOMPDC complexed with 1-(5’-phospho-β-d-ribofuranosyl)barbiturate (BMP; alternatively, 6-hydroxyUMP), Wolfenden and Short and their coworkers proposed that Lys 93 (Lys 72 in MtOMPDC) is positioned to both provide electrostatic stabilization of the (localized) negative charge at C6 and then protonate the intermediate to form the UMP product (6). This proposal was reiterated by Cleland and coworkers, who concluded that “the mechanism of this enzyme is fairly well understood” (13).
However, in 1997, before the availability of a structure for any OMPDC, Houk and Lee proposed that the negative charge of the “vinyl carbanion” intermediate is not localized to C6 as implied in the proposed role of Lys 72 (MtOMPDC) as an electrostatic catalyst but can be resonance delocalized to O4 to generate a “carbene” at C6 (C6 is neutral with an electron pair in the sp2 orbital) (14). Furthermore, protonation of O4 by an active site acid was predicted to provide substantial stabilization of the “carbene” intermediate. However, the structures for divergent OMPDCs reveal the absence of an acidic functional group proximal to O4; instead, based on the structures of liganded complexes with anionic intermediate analogs (BMP and azaUMP), O4 of the OMP substrate is assumed to participate in hydrogen-bonding interactions with the backbone amide of Ser 127 as well as a structurally conserved water molecule. Delocalization of the negative charge on O4 as the anionic transition state/intermediate is formed should increase the strengths of these hydrogen bonds as the proton affinity of O4 increases (15–19). Houk and coworkers noted this environment could stabilize the charge-delocalized anionic intermediate (20). However, the importance of these hydrogen bonding interactions in the OMPDC-catalyzed reactions has not been investigated.
Also, Wolfenden and Lewis recently reported that the rate of decarboxylation of 1-methylorotate, a model of OMP, is increased in solvents less polar than water (21). At the extreme, the rate enhancement was reported to be 6,100-fold in tetrahydrofuran, dioxane, or acetone. The slower reaction in water is attributed to more effective solvation of the anionic carboxylate group in the reactant than in the anionic transition state/vinyl carbanion intermediate. These effects are similar to a rate enhancement of >104 reported by Lienhard and coworkers for the decarboxylation of 2-(1-carboxy-1-hydroxyethyl)thiamin pyrophosphate to yield 2-(1-hydroxyethyl)thiamin pyrophosphate performed in dimethylsulfoxide instead of water (22, 23). Again, these effects were attributed to diminished solvation of the reactant in the nonpolar solvents relative to the solvation of the transition state.
Although not noted by Wolfenden and Lewis, the active sites of all OMPDCs contain a hydrophobic cavity proximal to the presumed location for the carboxylate group of the bound OMP substrate. A reasonable hypothesis is that this hydrophobic cavity prevents solvation of the carboxylate group; because the negative charge of the anionic intermediate is delocalized to O4, the hydrophobic cavity is not expected to influence solvation of the transition state/intermediate. However, the importance of this hydrophobic cluster has not been previously noted, so its potential importance in “substrate destabilization” has not been tested.
In this manuscript we report the effects of site-directed substitutions for residues in the MtOMPDC that form the hydrophobic pocket proximal to the C6 carboxylate group as well as Ser 127 that hydrogen bonds to O4 of the substrate/vinyl carbanion intermediate via its backbone amide. Our results provide evidence that contradict the proposal that “the mechanism of this enzyme is fairly well understood” (13).
MATERIALS AND METHODS
Materials
azaUMP was purchased from R.I. Chemicals Inc. Barbiturate was purchased from Sigma. All solutions were prepared with Millipore Ultrapure filtered water. OMP, FOMP, FUMP, and BMP were prepared by published procedures (4, 13, 24, 25).
Cloning, Site-Directed Mutagenesis, and Protein Expression and Purification
The gene encoding MtOMPDC was previously cloned from Methanothermobacter thermoautotrophicus strain Delta H genomic DNA into pET-15b vector (Novagen) (4, 24). This plasmid served as the template for site-directed mutagenesis by the overlap extension procedure. Primers with the desired mutations (Eurofins MWG Operon) were combined with external primers to generate two gene fragments. The PCR product was digested by Nde I and Bam HI (Promega) followed by ligation into pET-15b. DNA sequencing at the University of Illinois Core Sequencing Facility confirmed the desired sequences. The mutants were purified from a kanamycin-resistant pyrF− strain of E. coli generated using Wanner’s method for chromosomal disruptions (26); a chloramphenicol-resistant auxiliary plasmid, pTara, a gift from Professor John Cronan at the University of Illinois, supplied the T7 RNA polymerase for transcription from pET-15b. The mutant proteins were isolated by published procedures (4, 24).
Enzymatic Assays
All assays were performed at 25°C and in 10 mM MOPS, pH 7.1, containing 100 mM NaCl following published procedures (4, 24). Because the values of KM for wild type and most mutants are < 5 µM, they cannot be quantitated by measuring the initial velocity as a function of initial substrate concentration. Instead, as described elsewhere (24), the values of kcat/KM were determined from the first-order decay of the absorbance when the concentration of remaining substrate was ≤ 20% of the estimated KM value; the values of kcat were determined in separate assays with substrate concentrations exceeding the estimated values of KM by a factor ≥10. The kinetic constants for mutants with values of KM values > 20 µM were measured by initial velocity measurements.
Exchange of H6 of FUMP
Exchange of the H6 proton of FUMP (5 mM) was measured in 100 mM glycylglycine (pD 9.35, I = 0.1 M with NaCl) using 1H and 19F NMR spectroscopies as described elsewhere (3, 4).
Crystallization and Data Collection
Eight different crystal forms (Table 1) of mutants of MtOMPDC liganded with azaUMP were grown by the hanging drop method at room temperature: 1) S127G•azaUMP, 2) S127A•azaUMP, 3) S127P•azaUMP, 4) I96S•azaUMP, 5) I96T•azaUMP, 6) L123N•azaUMP, 7) L123S•azaUMP, and 8) V155S•azaUMP. The protein solutions contained the mutant protein (30 mg/mL) in 20 mM Hepes, pH 7.5, 150 mM NaCl, 3 mM DTT, and 40 mM azaUMP. The precipitants were the following:
For S127G•azaUMP, the precipitant contained 20% PEG 3350 and 0.15 M DL-malate, pH 7.0.
For S127A•azaUMP, the precipitant contained 30% PEG 400, 0.1 M Hepes (pH 7.5), and 0.2 M sodium chloride.
For S127P•azaUMP, the precipitant contained 60% tacsimate, pH 7.0.
For I96S•azaUMP, the precipitant contained 30% PEG 4000, 0.1 M sodium citrate, pH 5.6, and 0.2 M ammonium acetate.
For I96T•azaUMP, the precipitant contained 30% PEG 4000, 0.1 M sodium citrate, pH 5.6, and 0.2 M ammonium acetate.
For L123N•azaUMP, the precipitant contained 25% PEG 3350, 0.1 M Bis-Tris, pH 5.5, and 0.2 M lithium sulfate.
For L123S•azaUMP, the precipitant contained 20% PEG 8000, 0.1 M sodium cacodylate, pH 6.5, and 0.2 M magnesium acetate.
For V155S•azaUMP, the precipitant contained 30% PEG ME 5000, 0.1 M MES, pH 6.5, and 0.2 M ammonium sulfate.
Table 1.
Data collection and refinement statistics for azaUMP complexes of MtOMPDC mutants
| S127G | S127A | S127P | I96S | I96T | L123N | L123S | V155S | |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | P21 | P21 | P41 | P21 | P21 | P21 | P21 | P21 |
| No. of molecules | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| in asym. unit | ||||||||
| Cell dimensions | ||||||||
| a (Å) | 59.78 | 59.71 | 56.67 | 59.79 | 59.78 | 59.78 | 59.77 | 59.71 |
| b (Å) | 63.64 | 63.60 | 56.67 | 64.09 | 63.95 | 64.14 | 64.04 | 63.83 |
| c (Å) | 61.12 | 61.20 | 125.95 | 61.60 | 61.64 | 61.61 | 61.61 | 61.70 |
| β (°) | 115.20 | 115.19 | 115.60 | 115.47 | 115.55 | 115.50 | 115.22 | |
| Resolution (Å) | 1.45 | 1.32 | 1.30 | 1.30 | 1.3 | 1.3 | 1.3 | 1.3 |
| No. of unique | 68702 | 96918 | 91113 | 99507 | 97354 | 101238 | 97097 | 92236 |
| reflections | ||||||||
| Rmerge | 0.064 | 0.059 | 0.071 | 0.092 | 0.083 | 0.083 | 0.074 | 0.069 |
| Completeness (%) | 93.4 | 99.8 | 93.6 | 96.3 | 94.6 | 98.2 | 94.3 | 89.4 |
| Refinement | ||||||||
| Resolution (Å) | 25.0-1.45 | 25.0-1.32 | 25.0-1.30 | 25.0-1.30 | 25.0-1.3 | 25.0-1.30 | 25-1.3 | 25-1.3 |
| Rcryst | 0.184 | 0.196 | 0.234 | 0.223 | 0.161 | 0.161 | 0.160 | 0.175 |
| Rfree | 0.216 | 0.218 | 0.258 | 0.247 | 0.183 | 0.175 | 0.169 | 0.193 |
| No. atoms | ||||||||
| Protein | 3296 | 3396 | 3294 | 3378 | 3404 | 3464 | 3410 | 3402 |
| Waters | 522 | 405 | 308 | 348 | 463 | 482 | 419 | 398 |
| Bound ligand | 6azaUMP | 6azaUMP | 6azaUMP | 6azaUMP | 6azaUMP | 6azaUMP | 6azaUMP | 6azaUMP |
| Ligand atoms | 42 | 42 | 42 | 42 | 42 | 42 | 42 | 42 |
| R.m.s deviations | ||||||||
| Bond lengths (Å) | 0.004 | 0.006 | 0.005 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.2 | 1.0 | 1.2 | 1.1 | 1.1 | 1.1 | 1.1 | 1.1 |
| PDB entry | 3LLD | 3SY5 | 3LLF | 3PBU | 3RPV | 3PBW | 3PBY | 3PC0 |
Seven different crystal forms (Table 2) of mutants of MtOMPDC liganded with BMP were grown by the hanging drop method at room temperature: 1) S127A•BMP, 2) I96S•BMP, 3) I96T•BMP, 4) L123N•BMP, 5) L123S•BMP, 6) V155D•BMP, and 7) V155S•BMP. The protein solutions contained the mutant protein (30 mg/mL) in 20 mM Hepes, pH 7.5, 150 mM NaCl, 3mM DTT, and 40mM BMP. The precipitants were the following:
For S127A•BMP, the precipitant contained 30% PEG 4000, 0.1 M sodium citrate (pH 5.6), and 0.2 M ammonium acetate.
For I96S•BMP, the precipitant contained 60% tacsimate, pH 7.0.
For I96T•BMP, the precipitant contained 25% PEG 3350, 0.1 M Bis-Tris, pH 6.5, and 0.2 M MgCl2.
For L123N•BMP, the precipitant contained 30% PEG ME 2000, and 0.1 M potassium thiocyanate.
For L123S•BMP, the precipitant contained 25% PEG 3350, 0.1M Bis-Tris, pH 6.5, and 0.2 M MgCl2.
For V155D•BMP, the precipitant contained 1.4M sodium citrate, and 0.1 M Hepes, pH 7.5.
For V155S•BMP, the precipitant contained 2.4 M sodium malonate, pH 7.0.
Table 2.
Data collection and refinement statistics for BMP complexes MtOMPDC mutants
| S127A | I96S | I96T | L123N | L123S | V155D | V155S | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P21 | P21 | P21 | P21 | P21 | P21 | P21 |
| No. of molecules | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| in asym. unit | |||||||
| Cell dimensions | |||||||
| a (Å) | 59.89 | 59.93 | 59.97 | 59.92 | 59.85 | 59.74 | 59.71 |
| b (Å) | 64.16 | 64.34 | 63.84 | 63.98 | 63.93 | 64.33 | 64.28 |
| c (Å) | 61.57 | 61.58 | 61.58 | 61.75 | 61.74 | 61.55 | 61.33 |
| β (°) | 115.60 | 115.86 | 115.54 | 115.56 | 115.57 | 115.53 | 115.54 |
| Resolution (Å) | 1.32 | 1.53 | 1.42 | 1.42 | 1.31 | 1.42 | 1.32 |
| No. of unique | 98290 | 63155 | 76659 | 78628 | 95704 | 77594 | 97409 |
| reflections | |||||||
| Rmerge | 0.071 | 0.087 | 0.063 | 0.079 | 0.089 | 0.058 | 0.091 |
| Completeness (%) | 99.8 | 99.4 | 97.7 | 99.3 | 95.5 | 97.9 | 99.26 |
| Refinement | |||||||
| Resolution (Å) | 25.0-1.32 | 25.0-1.53 | 25.0-1.42 | 25.0-1.42 | 25.0-1.31 | 25.0-1.42 | 25-1.32 |
| Rcryst | 0.172 | 0.161 | 0.164 | 0.180 | 0.165 | 0.174 | 0.164 |
| Rfree | 0.193 | 0.186 | 0.185 | 0.205 | 0.177 | 0.202 | 0.177 |
| No. atoms | |||||||
| Protein | 3422 | 3388 | 3364 | 3388 | 3414 | 3404 | 3386 |
| Waters | 424 | 358 | 393 | 432 | 468 | 387 | 465 |
| Bound ligand | BMP | BMP | BMP | BMP | BMP | BMP | BMP |
| Ligand atoms | 44 | 44 | 44 | 44 | 44 | 44 | 44 |
| R.m.s deviations | |||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.05 | 1.08 | 1.09 | 1.10 | 1.11 | 1.07 | 1.11 |
| PDB entry | 3SIZ | 3NQC | 3NQD | 3NQE | 3NQF | 3NQG | 3NQM |
Prior to data collection, the crystals were transferred to cryoprotectant solutions composed of their mother liquor and 20% glycerol and flash-cooled in a nitrogen stream. All X-ray diffraction data sets (Tables 1 and 2) were collected at the NSLS X4A beamline (Brookhaven National Laboratory) on an ADSC CCD detector. Diffraction intensities were integrated and scaled with programs DENZO and SCALEPACK (27). The data collection statistics are given in Tables 1 and 2.
Structure Determination and Model Refinement
All 15 structures (Tables 1 and 2) were solved by molecular replacement with the fully automated molecular replacement pipeline BALBES (28), using only input diffraction and sequence data. Partially refined structures of all 15 crystal forms (Tables 1, 2) were the outputs from BALBES without any manual intervention. Subsequent several iterative cycles of refinement were performed for each crystal form including model rebuilding with COOT (29), refinement with PHENIX (30), and automatic model rebuilding with ARP (31).
In each structure, the polypeptides are arranged as dimers with the monomers connected by a noncrystallographic two-fold axis. The active site loops, chain segment 181–189, are well-defined in each structure. Each structure also has well-ordered density for the inhibitor in each active site. No residues (other than Gly) lie in disallowed regions of the Ramachandran plots in the various structures.
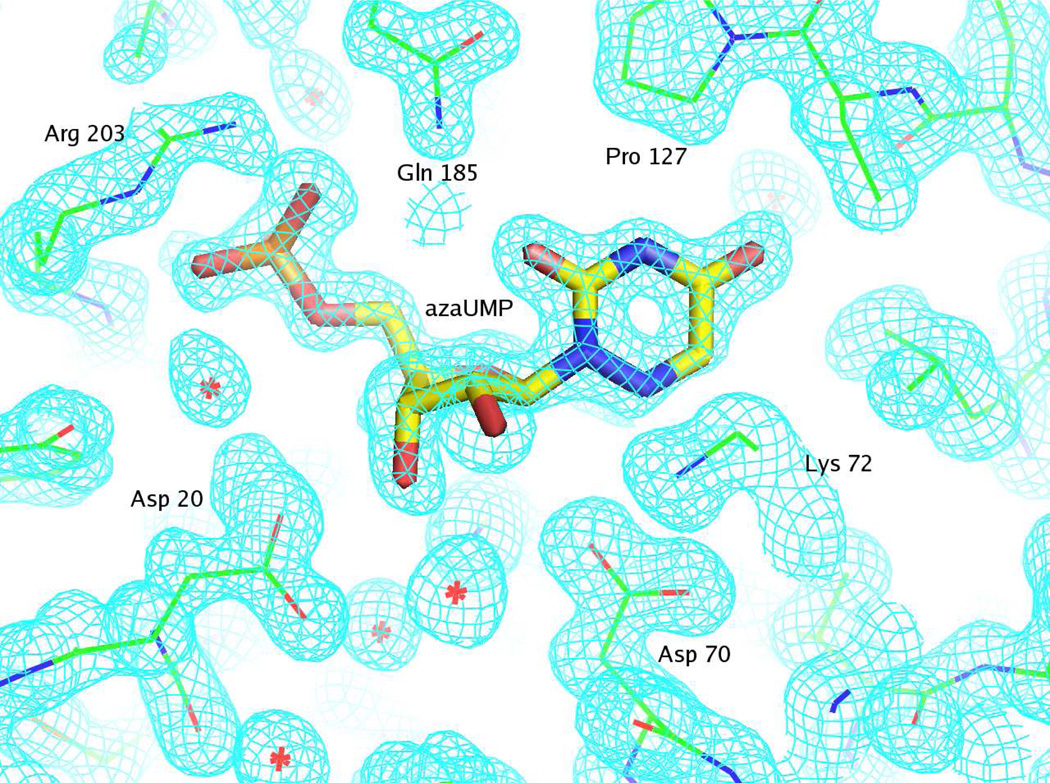
Representative electron density for the S127P mutant liganded with azaUMP is shown in Figure 1.
Figure 1.
Representative electron density map for the active site of S127P mutant complexed with azaUMP and countered at 1.5 σ. The figure was produced with PyMOL (34). The details of the interactions between 6-azaUMP and the active site are described in the text.
Final crystallographic refinement statistics for all determined OMPDC structures are provided in the Tables 1 and 2.
RESULTS AND DISCUSSION
As summarized in the Introduction, the value of ΔG‡ for decarboxylation of OMP is reduced by 23 kcal/mol in the active sites of homologous, but divergent, OMPDCs, although the structural strategies that accomplish this reduction remain uncertain. In this manuscript we describe studies of two features of the active site of MtOMPDC that have not been previously investigated: 1) a hydrophobic pocket proximal to C6, and 2) Ser 127 that interacts with N3 and O4 of the orotate moiety. On the basis of the experiments described in this manuscript, we conclude that both features contribute to the reduction in ΔG‡.
Importance of the Hydrophobic Pocket
We previously reported structures of wild type MtOMPDC liganded with BMP and azaUMP, analogs of the transition state/carbanion intermediate, as well as with 5,6-dihydroOMP (H2OMP), a stable analog of OMP that may be a mimic of “substrate destabilization” (4, 32). The structures of the orotate binding pockets are superimposable, although the active site of the complex with BMP contains an “extra” water molecule hydrogen-bonded to O6 of the 6-OH uracil (barbiturate) ring.
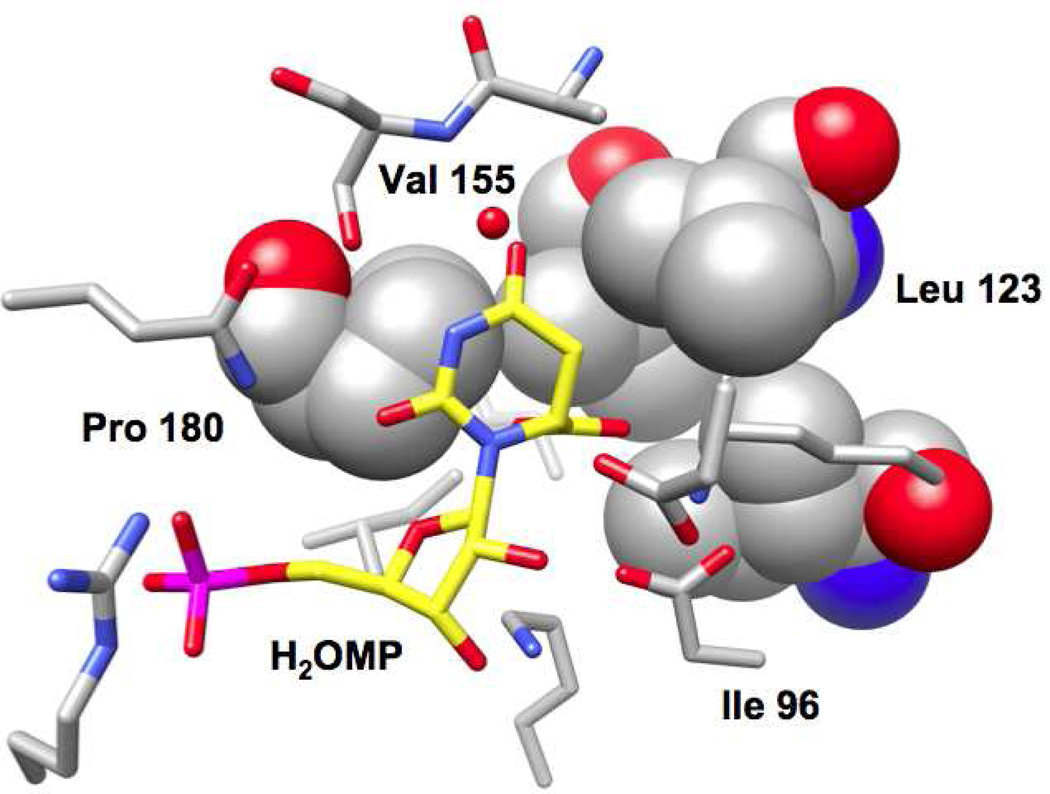
The active site of the previously reported complex of wild type MtOMPDC with H2OMP is shown in Figure 2. The C6-carboxylate group “points” into the hydrophobic cavity that is formed by the side chains of Ile 96, Leu 123, Val 155, and Pro 180. No water molecules are observed in the hydrophobic cavity. Although these could be brought into the active site by hydrogen bonding interactions with the carboxylate group, the hydrophobic side cavity would prevent completion of their preferred tetrahedral hydrogen bonding interactions, thereby making their presence in the active site unfavorable.
Figure 2.
Active site of MtOMPDC liganded with H2OMP (4) with the residues that form the hydrophobic pocket represented with space-filling atoms.
We constructed polar substitutions for Ile 96, Leu 123, and Val 155, i.e., I96S, I96T, L123N, L123S, V155D, and V155S, to increase the polarity of the active site cavity; these “targets” for study were based on the report from Wolfenden and Lewis that the nonenzymatic decarboxylation of OMP analogs is increased as the solvent polarity is decreased because the negative charge of the departing carboxylate group is not stabilized (21). Therefore, we hypothesized that introduction of polar residues in this cavity (and possible “new” water molecules that can interact with hydrogen bond donors/acceptors) would decrease the value of kcat. We did not construct substitutions for Pro 180 because it is located at the N-terminus of the active site loop: we recently reported that the interaction of the sequence-proximal Val 182 in the loop with the (β/α)8-barrel scaffold favors formation of the closed, active conformation of the enzyme (32); therefore, we were concerned that polar substitutions for Pro 180 could influence the kinetic constants by perturbing the distribution between the open and closed conformations of the enzyme.
Structures of the Mutants with Polar Substitutions for Hydrophobic Pocket Residues
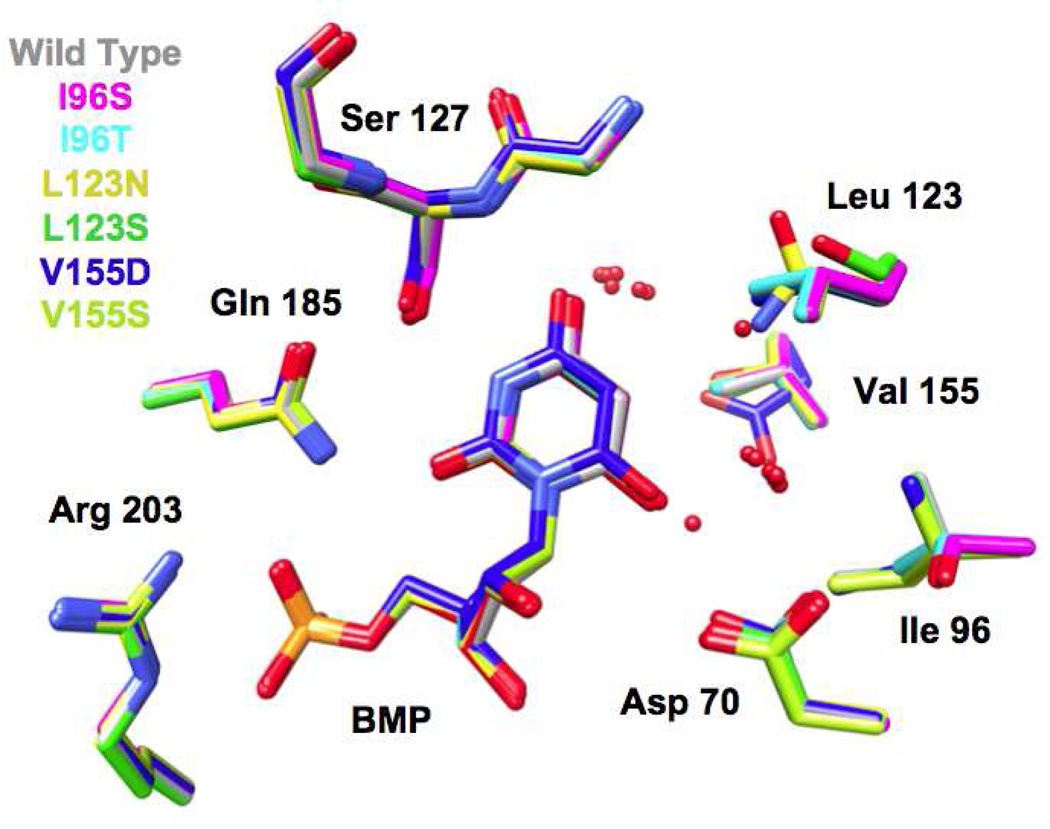
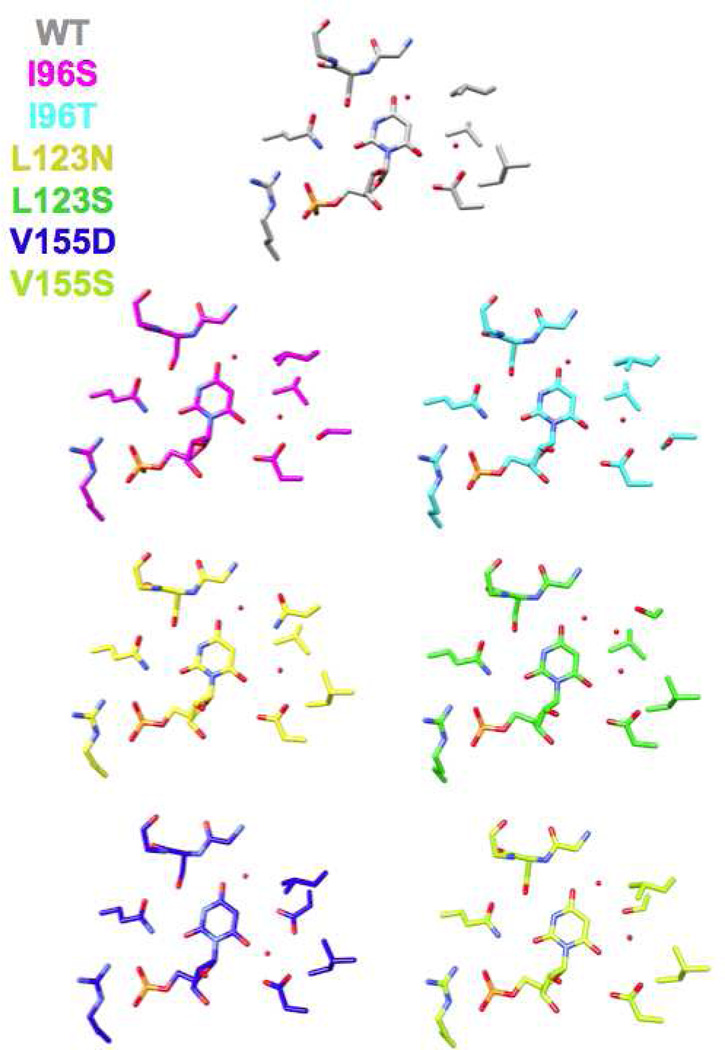
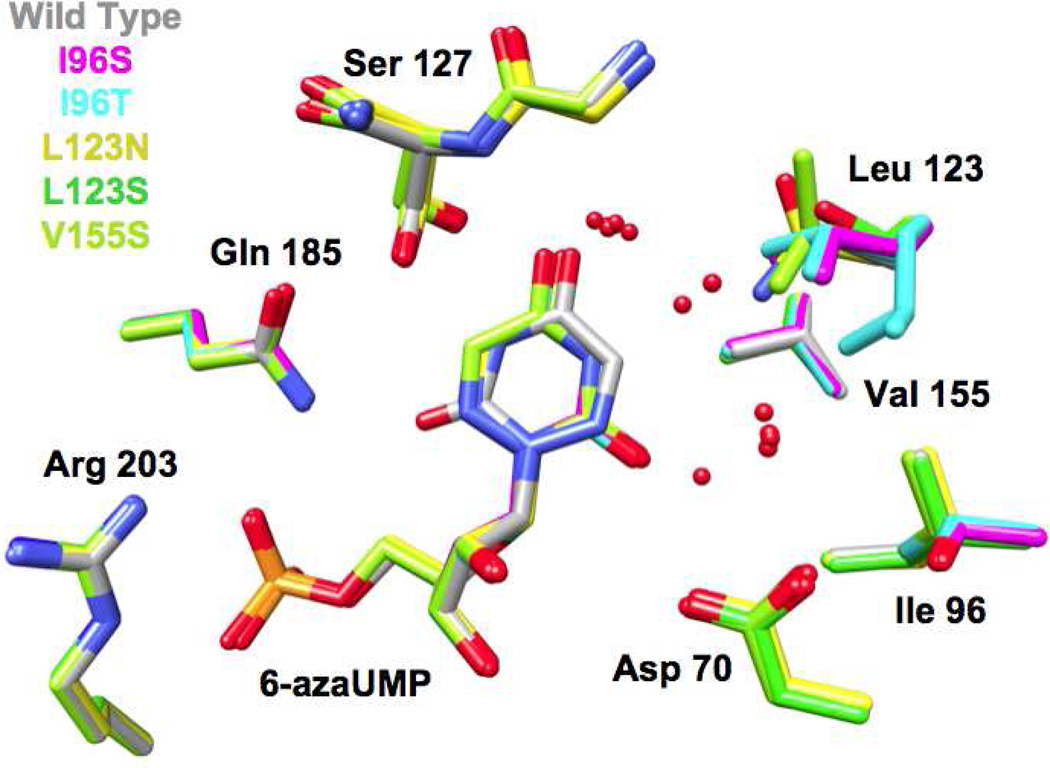
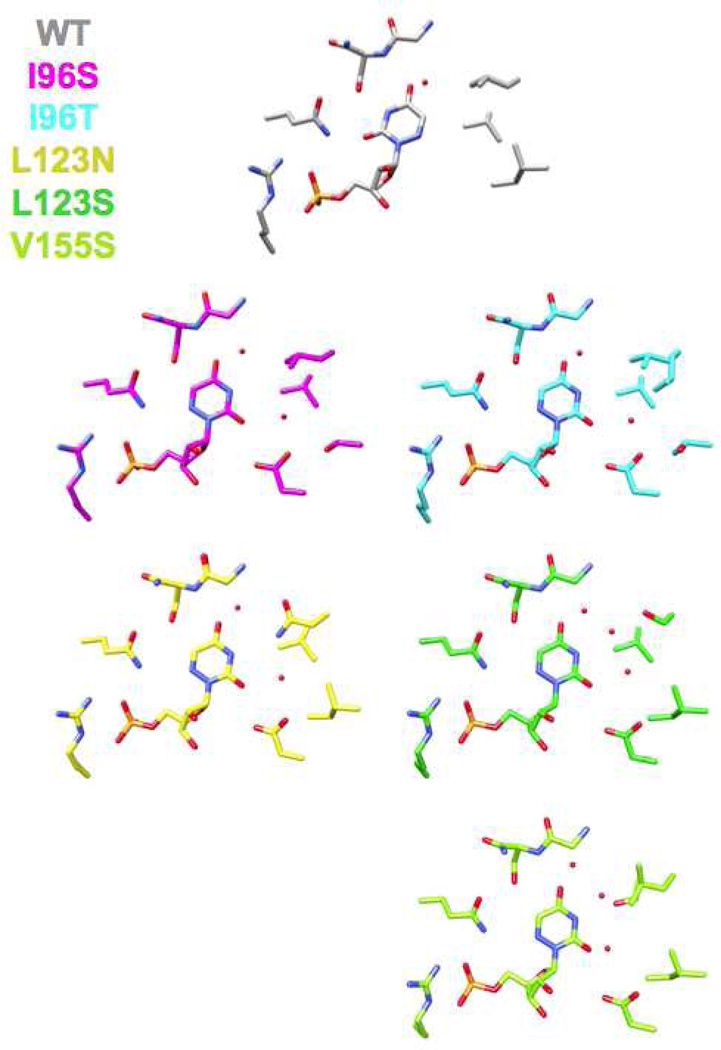
We determined structures for all six hydrophobic pocket mutants in the presence of BMP (Figure 3 and 4); we also determined structures for five of the mutants (except V155D) in the presence of azaUMP (Figures 5 and 6). In all of the structures, the side chains of the active site residues, including Asp 70, Lys 72, and Asp 75 (from the adjacent polypeptide in the dimer) superimpose well with those of the BMP/azaUMP-liganded structures of wild type MtOMPDC (Figures 3 and 5), providing evidence that the interactions of the transition state/vinyl carbanion intermediate with the active site are preserved in each mutant. Therefore, in our interpretation of their kinetic constants, we assume that changes in kcat arise from the altered environment provided by the now polar cavity adjacent to the carboxylate group of OMP.
Figure 3.
Superposition of the active sites of the hydrophobic pocket mutants liganded with BMP. The red spheres represent water molecules.
Figure 4.
Active sites of the hydrophobic pocket mutants liganded with BMP. The red spheres represent water molecules. All of the active sites contain two spatially conserved water molecules; the active of L123S contains an additional water molecule hydrogen bonded to the OH group of Ser 123.
Figure 5.
Superposition of the active sites of the hydrophobic pocket mutants liganded with azaUMP. The red spheres represent water molecules.
Figure 6.
Active sites of the hydrophobic pocket mutants liganded with azaUMP. The red spheres represent water molecules. All of the active sites contain two spatially conserved water molecules; the active sites of L123S and V155S contain an additional water molecule hydrogen bonded to the OH group of Ser 123.
In five of the six BMP-liganded mutant structures (I96S, I96T, L123N, L123S, and V155S), two “conserved” water molecules which hydrogen bond to O4 and O6 of BMP undergo small changes in position (Figures 3 and 4). The water proximal to O4 is conserved in all structurally characterized OMPDCs and may participate in stabilization of the negative charge that is localized on O4 of the vinyl carbanion/carbene intermediate; the water proximal to O6 of BMP likely is not present when the OMP substrate binds (recall that the complex of wild type MtOMPDC with H2OMP does not contain any water molecules). The active site of the BMP-liganded L123S contains a third (“new”) water that is stabilized by its proximity to the waters adjacent to O4 and O6, the O6 oxyanion of BMP, and the hydroxyl group of the L123S side chain.
Wild type MtOMPDC binds azaUMP in the syn-conformation, in which O2 is hydrogen bonded to the carboxamide NH2 group of Gln 185 and N3 is hydrogen bonded to the OH group of Ser 127. However, in the mutants, azaUMP is bound in the anti-conformation in which the pyrimidine ring is rotated by 180° around the glycosidic bond (Figure 6). With this geometry and in each structure, O2 is hydrogen bonded to a “new” water molecule that is not observed in the wild type azaUMP complex (analogous to the “conserved” water molecules proximal to O6 in the BMP-liganded structures). The substitutions for Val 155 and Ile 96 perturb the position of the Leu 123 side chain, thereby altering the shape of the pocket. Also, in the L123N and V155S mutants, a second “new” water is hydrogen bonded to N3 of the 6-azauracil moiety.
Kinetic Constants for the Mutants with Polar Substitutions for Hydrophobic Pocket Residues
For wild type MtOMPDC, decarboxylation of OMP is fully rate-determining for kcat, although kcat/KM has a small inverse dependence on viscosity, indicating that both chemical and physical steps determine its value (24). Therefore, changes in kcat report on changes in ΔG‡ for decarboxylation. We also previously demonstrated that substitutions for Asp 70 decrease the value of kcat but do not alter the value of kex, (exchange of H6 of the UMP product or the FUMP product analog with solvent), thereby providing support for the occurrence of the vinyl carbanion intermediate (obtained either from decarboxylation of OMP or exchange of H6 of the UMP product) on the reaction coordinate (4). We interpreted the effects of the substitutions for Asp 70 as evidence for “substrate destabilization” during the transition from the open, inactive conformation to the closed, active conformation, presumably by unfavorable electrostatic (and steric) interactions with the substrate carboxylate group.
We measured the effects of the various polar substitutions for the hydrophobic pocket residues on the values of 1) kcat and KM for decarboxylation of OMP, and 2) kex for exchange of H6 of the more reactive product analog 5-fluoroUMP; these are presented in Table 3.
Table 3.
Kinetics of Hydrophobic Pocket Mutants
| OMPDC | kcat (s−1) | KM (µM) | kcat/KM (M−1s−1) | kex (s−1) |
|---|---|---|---|---|
| WT | 4.0 ± 0.2 | 1.6 ± 0.1 | 2.0 × 106 | 0.015 ± 0.004 |
| I96T | 0.69 ± 0.05 | 2.3 ± 0.06 | 3.0 × 105 | 0.0047 ± 0.0004 |
| I96S | 0.011 ± 0.002 | 1.0 ± 0.04 | 1.0 × 104 | 0.006 ± 0.001 |
| L123N | 0.64 ± 0.01 | 3.5 ± 0.2 | 1.9 × 105 | 0.0007 ± 0.0001 |
| L123S | 0.36 ± 0.04 | 3.0 ± 0.5 | 1.2 × 105 | 0.00039 ± 0.00001 |
| V155S | 0.011 ± 0.003 | 4.7 ± 0.1 | 2.2 × 104 | 0.006 ± 0.003 |
| V155D | 5 ± 2 × 10−4 | 4.6 ± 0.5 | 4.6 × 102 | 0.057 ± 0.002 |
| D70N | 0.024 ± 0.05 | 6.0 ± 0.3 | 4.0 × 103 | 0.005 ± 0.002 |
| D70N/I96S | 6.0 (± 1.0) × 10−5 (5)a |
- | - | 0.002 ± 0.0003 (0.13)d (22)a |
| D70N/L123S | 9.3 (± 0.1) × 10−4 (0.1)b |
- | - | 0.00001 ± 0.000007 (1500)e (1.7)b |
| D70N/V155S | 1.8 ± (0.4) × 10−4 (1.5)c |
- | - | 0.0019 ± 0.0006 (0.13)f (44)c |
(D70N/I96S) / (D70N)(I96S)
(D70N/L123S) / (D70N)(L123S)
(D70N/V155S) / (D70N)(V155S)
(D70N/I96S) / (D70N)
(D70N/L123S) / (D70N)
(D70N/V155S) / (D70N)
The values of kcat are decreased from 6- to 400-fold for the various neutral polar substitutions; for the V155D substitution the value of kcat is decreased by ~10,000-fold. However, the values of KM for the mutants are increased < 3-fold, including for the V155D mutant. Thus, each of the polar substitutions decreases the rate at which the vinyl carbanion intermediate is formed by decarboxylation but do not alter the conformational equilibrium between the open, inactive and closed, active conformations (32).
However, for Ile 96 and Val 155 (including V155D for which the value of kcat is substantially decreased), the values of kex are not significantly altered by the substitutions. The simplest interpretation is that the stability of the anionic intermediate is not affected by the substitutions. That these substitutions increase ΔG‡ for decarboxylation but not exchange suggests that the hydrophobic pocket contributes to the reduction in ΔG‡ by placing the substrate carboxylate in a destabilizing, hydrophobic environment (“hydrophobic stress”).
For L123N and L123S the values of kex are decreased more than the values of kcat. A possible explanation is provided by the structures of azaUMP complexes of these mutants in which an “extra” water is located proximal to the pyrimidine moiety in each active site, providing the potential to influence either the rate of formation of the anion intermediate by proton abstraction or the stability of the intermediate.
Thus, for the neutral polar substitutions, the decreases in kcat support the hypothesis that enhanced polarity proximal to C6 of the orotate substrate decreases the rate of decarboxylation by stabilizing the anionic carboxylate group relative to the anionic intermediate, as suggested by Wolfenden and Lewis to explain the enhanced rate of decarboxylation in solvents less polar than water. The very substantial effect of the V155D substitution likely can be best explained by unfavorable electrostatic interactions with the substrate carboxylate group as the reaction coordinate for decarboxylation is traversed.
We note that a hydrophobic pocket proximal to the substrate carboxylate group is conserved in all OMPDCs, despite considerable divergence in sequence, so all OMPDCs presumably adopt this structural strategy for reducing the value of ΔG‡.
Independent Mechanisms for “Ground State Destabilization”?
As summarized in the Introduction, we recently reported that the D70N and D70G substitutions also decrease the value of kcat without influencing the value of kex; these effects were interpreted as evidence for “ground state destabilization” of the substrate’s carboxylate by its proximity to the anionic carboxylate group of Asp 70 (4). We now have concluded that the hydrophobic pocket prevents stabilization of the substrate carboxylate group relative to the transition state/vinyl carbanion intermediate. Therefore, we constructed the D70N/I96S, D70N/L123S, and D70N/V155S double mutants in an attempt to obtain evidence that these structurally distinct effects are independent contributors to the reduction in ΔG‡.
The values of kcat, KM, and kex for the double mutants also are reported in Table 3. The values of kcat for the single D70N, V155S and I96S mutants are reduced by factors between 200- and 400-fold. For the double mutants with these residues, the values of kcat are reduced by factors that approximate the reductions observed for the single mutants, i.e., the increases in ΔG‡ are additive, supporting the conclusion that electrostatic repulsion by Asp 70 and “hydrophobic stress” by the hydrophobic pocket independently contribute to “substrate destabilization” as the transition state for decarboxylation is achieved (we could not measure the values of KM by analysis of the first order decay in absorbance at low substrate concentrations because of the very slow rates of reaction).
The values for kex for the D70N/I96S and D70N/V155S double mutants are each less than that for the D70N mutant (by a factor of ~8) but are significant greater than those expected if the effects of the individual mutants were additive (by 22- and 44-fold, respectively). Although not “perfect”, these data support the suggestion that the increases in ΔG‡ for exchange of H6 are not additive, i.e., the effects of the hydrophobic pocket and D70N are independent contributors to “substrate destabilization” with neither contributing significantly to intermediate/transition state destabilization.
Like the L123S single mutant, the D70N/L123S double mutant shows “anomalous” behavior: the value of kcat is 10-fold lower than the value predicted if the effects on ΔG‡ were additive; however, the value of kex is reduced 1500-fold from that observed for wild type MtOMPDC and 300-fold from those observed for D70N/I96S and D70N/V155S double mutants. Again, these lower than expected values for kcat and kex may result from the “extra” water molecule proximal to the pyrimidine moiety.
Thus, we conclude that the reduction in ΔG‡ by MtOMPDC involves at least two structurally independent mechanisms to achieve “substrate destabilization”.
Importance of Ser 127 in Stabilization of an Anionic Transition State/Intermediate
As proposed by Houk and coworkers (14, 20), a hydrogen bond donor adjacent to O4 of the OMP substrate could contribute significantly to the reduction in ΔG‡ by stabilization of the transition state/anionic intermediate as developing negative charge in the pyrimidine moiety is delocalized to O4. Structural studies of divergent OMPDCs reveal that O4 of the substrate always is hydrogen bonded to the backbone NH group of a conserved Ser (or, occasionally, Thr) in the loop following the fifth β-strand of the (β/α)8-barrel domain (Ser 127 in MtOMPDC) as well as the spatially conserved water molecule. In addition, the side chain OH group of the Ser residue is hydrogen bonded to the proton on N3. The quantitative importance of these interactions in the reduction of ΔG‡ has not been reported.
We constructed and kinetically characterized the S127A, S127G, and S127P mutants of MtOMPDC to obtain information about the importance of these interactions; we also obtained structures of the S127A, S127G, and S127P mutants liganded with azaUMP. The S127A and S127G substitutions allow the importance of the interactions between the side chain OH group and N3 of the orotate moiety to be investigated. The S127P substitution replaces the backbone NH group with Cγ of the side chain; although we recognized that this likely would introduce a steric clash with O4 of the OMP substrate, we considered this mutant to be worthwhile to characterize.
Structures of the S127A, S127G, and S127P Mutants
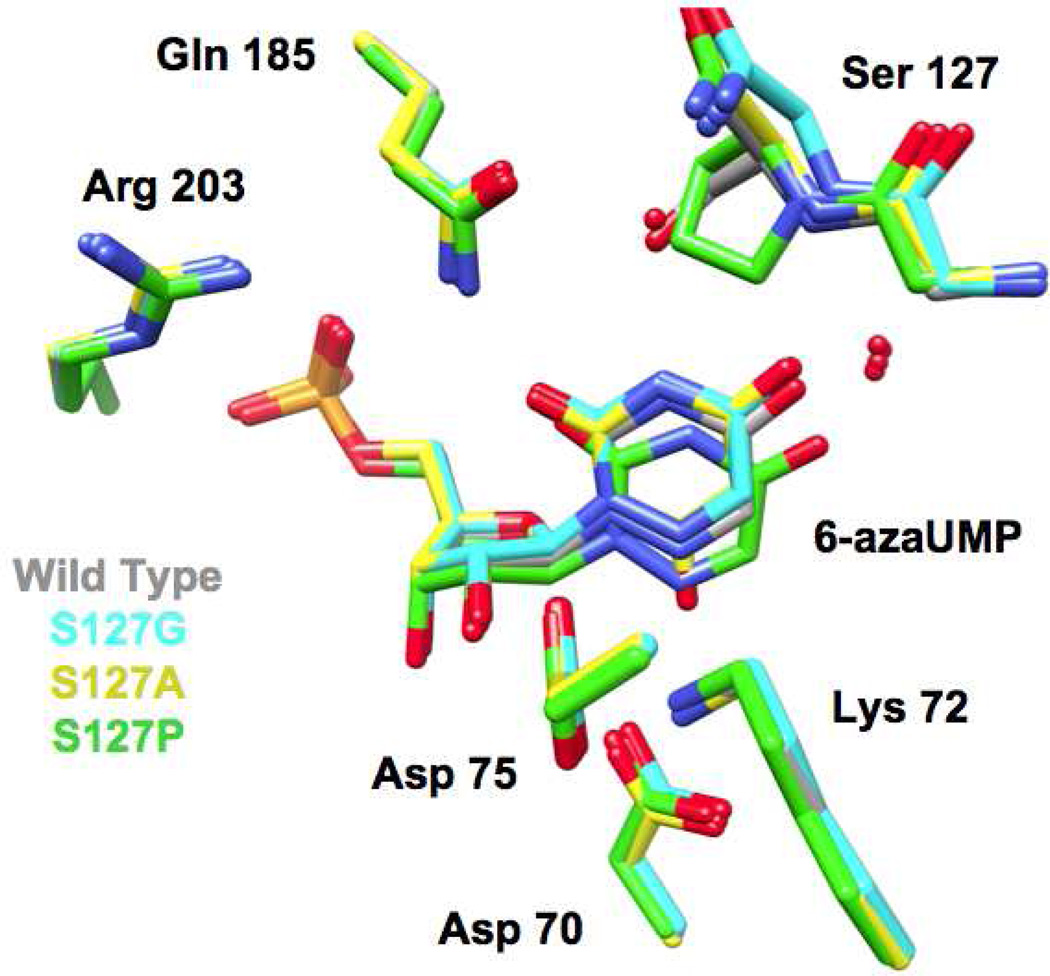
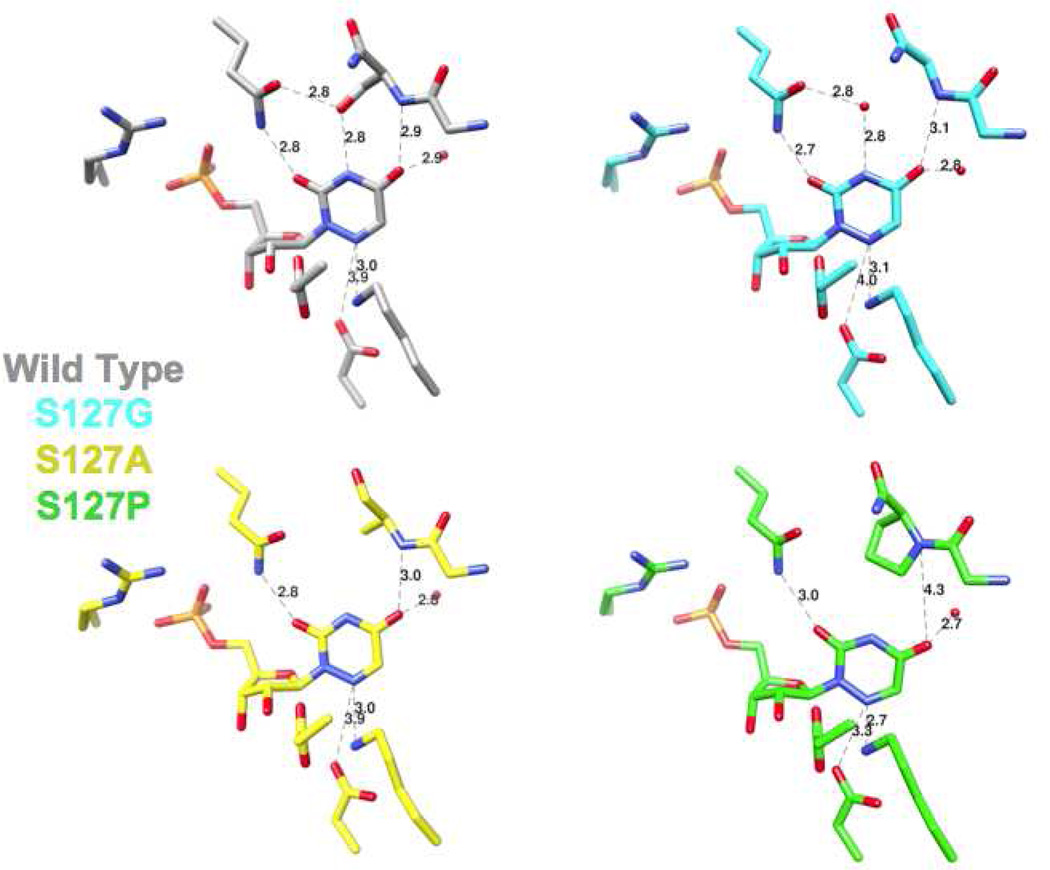
We determined structures for the Ala, Gly, and Pro substitutions for Ser 127 in the presence of azaUMP. The structures of the active sites of the mutants superimpose well with that of wild type MtOMPDC complexed with azaUMP (Figure 7), with small deviations in backbone geometry observed for residue 127 and the two flanking residues (Met 126 and His 128). In the structure of the S127G mutant, a water molecule occupies the position of the OH group of the Ser residue in wild type MtOMPDC and is hydrogen bonded to both N3 of the pyrimidine ring and the carboxamide group of Gln 185; this water molecule is not present in the S127A, presumably because of steric exclusion. The positions of the side chains of the active site residues, including Asp 70, Lys 72, and Asp 75 (from the adjacent polypeptide), are not affected by the substitutions for Ser 127.
Figure 7.
Superposition of the active sites of the mutants of Ser 127 liganded with azaUMP. The red spheres represent water molecules.
The positions of the ligands in the various azaUMP-liganded complexes are shifted from the position in wild type MtOMPDC, with the effects most pronounced for the 6-azauracil moiety (Figures 6 and 7). For example, in the S127P mutant, O4 is displaced by 1.1 Å from the positions of O4 in the wild type and by 1.5 Å from the positions of O4 in the S127A and S127G mutants because of the steric clash with the aliphatic Pro ring; also, N6 is positioned 0.6 Å closer to the carboxylate group of Asp 70 and 0.3 Å closer to the ε-ammonium group of Lys 72 than the wild type. Although these changes complicate precise structure-based interpretation of the kinetic constants (vide infra), we decided that characterization of this substitution would provide relevant information about the role of Ser 127.
Kinetic Constants for the S127A, S127G, and S127P Mutants
The S127A and S127G substitutions retain the interaction between O4 of the pyrimidine ring and the backbone NH of residue 127, although these alter the interactions between the side chain and N3 of the pyrimidine ring as well as the carboxamide group of Gln 185.
For both mutants, the values of KM are increased (Table 4), consistent with the proposal that the hydrogen bond between the OH group of Ser 127 and the carboxamide group of Gln 185 provides a “clamp” for stabilizing the closed, active conformation of the enzyme (33). The value of kcat is reduced ~130-fold for the S127A mutant and ~18-fold for the S127G mutant. The reduction for the Ala substitution suggests that a hydrogen bond between the OH group of Ser 127 and the proton of N3 provides stabilization of the enhanced, delocalized anionic charge in the pyrimidine ring that occurs as the vinyl anion/carbene intermediate is formed. The partial “rescue” observed for the Gly substitution is explained by the presence of a water molecule that occupies the position of the Ser OH group, thereby mimicking the OH group.
Table4.
Kinetics of Ser 127 Mutants
| OMPDC | kcat (s−1) | KM (µM) | kcat/KM (M−1s−1) |
|---|---|---|---|
| WT | 4.6 ± 0.4 | 2.1 ± 0.1 | 2.2 (± 0.2) × 106 |
| S127A | 0.035 ± 0.002 | 96 ± 5 | 3.6 (± 0.3) × 102 |
| S127G | 0.26 ± 0.007 | 59 ± 5 | 4.3 (± 0.3) × 103 |
| S127P | - | - | 0.88 ± 0.16a |
Velocity is directly proportional to substrate concentration.
The values of kcat and KM cannot be measured for the S127P mutant; the value of kcat/KM is determined from the linear dependence of velocity on substrate concentration. The value so determined is reduced by an impressive factor of 2.5 × 106. Quantitative interpretation of this effect in the context of the interaction between O4 and the backbone NH group of Ser 127 is equivocal. The structure reveals, as expected, that the Pro substitution alters the position of the pyrimidine ring in the active site: O4 of the ligand is displaced 1.1 Å from the position of O4 in the wild type and 1.5 Å from those in the S127A and S127G mutants; it obviously cannot form a hydrogen bond with the backbone. However, the position of N6 (homologue of C6 in the vinyl anion/carbene) relative to Nε of Lys 72 is decreased by 0.3 Å in the mutant relative to wild type (2.7 Å vs. 3.0 Å, respectively). If electrostatic effects between Lys 72 and the anionic intermediate are of primary importance in reducing the value of ΔG‡, the decrease in the distance in the S127P mutant would be expected to have only a “minor” effect on kcat (23% assuming Coulomb’s law applies to the interaction between the ε-ammonium group of Lys 72 and the anionic C6, i.e., no charge delocalization to form a carbene in the anionic intermediate). Thus, the very large reduction in the value of kcat observed for the S127P mutant is an indication that the interaction of O4 with the backbone NH of Ser 127 contributes significantly to stabilization of the vinyl anion/carbene intermediate.
Houk and Lee proposed an anionic carbene resonance structure for the anionic intermediate in which negative charge is delocalized to O4 (14, 20). Making the reasonable assumption that the strength of the hydrogen bond to the backbone NH of Ser 127 increases as negative charge is delocalized to O4, this interaction could provide significant stabilization of the intermediate and contribute to the reduction in ΔG‡ (17). However, quantitation of this stabilization is difficult because 1) removal of the O4 hydrogen bond acceptor, i.e., 4-deoxyuracil, will also result in loss of a proton from N3, thereby eliminating the hydrogen bond to Oβ of Ser 127, and 2) alteration of the hydrogen bond donor (the NH of Ser 127) cannot be readily accomplished, even with unnatural mutagenesis (in principle, replacement of the peptide bond linkage with an ester linkage may be possible, but, in practice, ribosome-mediated synthesis of depsipeptide linkages in protein is difficult).
Conclusions
The structural strategies by which the remarkable rate enhancement achieved by OMPDCs [kcat/knon = 7.1 × 1016] are not well-defined. Although electrostatic stabilization of the anionic intermediate by Lys 72 is frequently used to explain the rate acceleration, we are investigating the role of other active site residues. We previously demonstrated that Asp 70 contributes modestly to the rate enhancement by destabilization of the substrate. We now conclude that a hydrophobic cavity proximal to the carboxylate group of the substrate also destabilizes the substrate, presumably by preventing the carboxylate group from being stabilized by waters of hydration when it is bound in the active site. This effect is also modest, but likely is employed by all OMPDCs given that residues in the hydrophobic cavity are conserved. The importance of hydrogen bonding interactions with O4 of the pyrimidine moiety of the substrate is more difficult to investigate experimentally because the hydrogen bond donor is a peptide backbone NH group of Ser 127. Replacement of Ser 127 with proline (S127P) has a large effect on kcat/KM (a reduction of 2.5 × 106), supporting the proposal by Lee and Houk that the hydrogen bond between O4 of the substrate and the backbone NH is important in reducing ΔG‡.
Figure 8.
Active sites of the mutants of Ser 127 liganded with azaUMP. The red spheres represent water molecules. Distances for important interactions of the ligand with active site residues are shown.
Abbreviations
- FOMP
5-fluoroorotidine 5’-monophosphate
- FUMP
5-fluoroUMP
- azaUMP
6-azauridine 5’-monophosphate
- BMP
1-(5’-phospho-β-D-ribofuranosyl)barbituric acid
- MtOMPDC
OMPDC from Methanothermobacter thermoautotrophicus
- OMP
orotidine 5’-monophosphate
- OMPDC
orotidine 5’-monophosphate decarboxylase
- ScOMPDC
OMPDC from Saccharomyces cerevisiae
- WT
wild type
Footnotes
This research was supported by an NIH Chemistry Biology Interface Training Grant under Ruth L. Kirschstein National Research Service Award T32GM070241 (to V.I.) and NIH GM065155 (to J.A.G.). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). The X-ray coordinates and structure factors for the following structures have been deposited in the Protein Data Bank with the indicated accession codes: the S127G mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3LLD), the S127A mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3SY5), the S127A mutant of MtOMPDC in the presence of BMP (PDB accession code 3SIZ), the S127P mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3LLF), the I96S mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3PBU), the I96S mutant of MtOMPDC in the presence of BMP (PDB accession code 3NQC), the I96T mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3RPV), the I96T mutant of MtOMPDC in the presence of BMP (PDB accession code 3NQD), the L123N mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3PBW), the L123N mutant of MtOMPDC in the presence of BMP (PDB accession code 3NQE), the L123S mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3PBY), the L123S mutant of MtOMPDC in the presence of BMP (PDB accession code 3NQF), the V155D mutant of MtOMPDC in the presence of BMP (PDB accession code 3NQG), the V155S mutant of MtOMPDC in the presence of azaUMP (PDB accession code 3PC0), and the 155SS mutant of MtOMPDC in the presence of BMP (PDB accession code 3NQM).
REFERENCES
- 1.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Toth K, Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. Product deuterium isotope effect for orotidine 5'-monophosphate decarboxylase: evidence for the existence of a short-lived carbanion intermediate. J Am Chem Soc. 2007;129:12946–12947. doi: 10.1021/ja076222f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. Formation and stability of a vinyl carbanion at the active site of orotidine 5'-monophosphate decarboxylase: pKa of the C-6 proton of enzyme-bound UMP. J Am Chem Soc. 2008;130:1574–1575. doi: 10.1021/ja710384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan KK, Wood BM, Fedorov AA, Fedorov EV, Imker HJ, Amyes TL, Richard JP, Almo SC, Gerlt JA. Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization. Biochemistry. 2009;48:5518–5531. doi: 10.1021/bi900623r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu N, Mo Y, Gao J, Pai EF. Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc Natl Acad Sci U S A. 2000;97:2017–2022. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Anatomy of a proficient enzyme: the structure of orotidine 5'-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc Natl Acad Sci U S A. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appleby TC, Kinsland C, Begley TP, Ealick SE. The crystal structure and mechanism of orotidine 5'-monophosphate decarboxylase. Proc Natl Acad Sci U S A. 2000;97:2005–2010. doi: 10.1073/pnas.259441296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris P, Navarro Poulsen JC, Jensen KF, Larsen S. Structural basis for the catalytic mechanism of a proficient enzyme: orotidine 5'-monophosphate decarboxylase. Biochemistry. 2000;39:4217–4224. doi: 10.1021/bi992952r. [DOI] [PubMed] [Google Scholar]
- 9.Warshel A, Strajbl M, Villa J, Florian J. Remarkable rate enhancement of orotidine 5'-monophosphate decarboxylase is due to transition-state stabilization rather than to ground-state destabilization. Biochemistry. 2000;39:14728–14738. doi: 10.1021/bi000987h. [DOI] [PubMed] [Google Scholar]
- 10.Warshel A, Florian J, Strajbl M, Villa J. Circe effect versus enzyme preorganization: what can be learned from the structure of the most proficient enzyme? Chembiochem. 2001;2:109–111. doi: 10.1002/1439-7633(20010202)2:2<109::AID-CBIC109>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Kamerlin SC, Chu ZT, Warshel A. On catalytic preorganization in oxyanion holes: highlighting the problems with the gas-phase modeling of oxyanion holes and illustrating the need for complete enzyme models. J Org Chem. 2010;75:6391–6401. doi: 10.1021/jo100651s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sievers A, Wolfenden R. Equilibrium of formation of the 6-carbanion of UMP, a potential intermediate in the action of OMP decarboxylase. J Am Chem Soc. 2002;124:13986–13987. doi: 10.1021/ja021073g. [DOI] [PubMed] [Google Scholar]
- 13.Van Vleet JL, Reinhardt LA, Miller BG, Sievers A, Cleland WW. Carbon isotope effect study on orotidine 5'-monophosphate decarboxylase: support for an anionic intermediate. Biochemistry. 2008;47:798–803. doi: 10.1021/bi701664n. [DOI] [PubMed] [Google Scholar]
- 14.Lee JK, Houk KN. A proficient enzyme revisited: the predicted mechanism for orotidine monophosphate decarboxylase. Science. 1997;276:942–945. doi: 10.1126/science.276.5314.942. [DOI] [PubMed] [Google Scholar]
- 15.Gerlt JA, Gassman PG. An Explanation for Rapid Enzyme-Catalyzed Proton Abstraction from Carbon Acids: The Importance of Late Transition States in Concerted Mechanisms. J. Am. Chem. Soc. 1993;115:11552–11569. [Google Scholar]
- 16.Cleland WW, Kreevoy MM. Low-barrier hydrogen bonds and enzymic catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [see comments] [DOI] [PubMed] [Google Scholar]
- 17.Shan SO, Herschlag D. The change in hydrogen bond strength accompanying charge rearrangement: implications for enzymatic catalysis. Proc Natl Acad Sci U S A. 1996;93:14474–14479. doi: 10.1073/pnas.93.25.14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlt JA, Kreevoy MM, Cleland W, Frey PA. Understanding enzymic catalysis: the importance of short, strong hydrogen bonds. Chem Biol. 1997;4:259–267. doi: 10.1016/s1074-5521(97)90069-7. [DOI] [PubMed] [Google Scholar]
- 19.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 20.Houk KN, Lee JK, Tantillo DJ, Bahmanyar S, Hietbrink BN. Crystal structures of orotidine monophosphate decarboxylase: does the structure reveal the mechanism of nature's most proficient enzyme? Chembiochem. 2001;2:113–118. doi: 10.1002/1439-7633(20010202)2:2<113::AID-CBIC113>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Lewis CA, Jr, Wolfenden R. Orotic acid decarboxylation in water and nonpolar solvents: a potential role for desolvation in the action of OMP decarboxylase. Biochemistry. 2009;48:8738–8745. doi: 10.1021/bi901085m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crosby J, Lienhard GE. Mechanisms of thiamine-catalyzed reactions. A kinetic analysis of the decarboxylation of pyruvate by 3,4-dimethylthiazolium ion in water and ethanol. J Am Chem Soc. 1970;92:5707–5716. doi: 10.1021/ja00722a027. [DOI] [PubMed] [Google Scholar]
- 23.Crosby J, Stone R, Lienhard GE. Mechanisms of thiamine-catalyzed reactions. Decarboxylation of 2-(1-carboxy-1-hydroxyethyl)-3,4-dimethylthiazolium chloride. J Am Chem Soc. 1970;92:2891–2900. doi: 10.1021/ja00712a048. [DOI] [PubMed] [Google Scholar]
- 24.Wood BM, Chan KK, Amyes TL, Richard JP, Gerlt JA. Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: effect of solvent viscosity on kinetic constants. Biochemistry. 2009;48:5510–5517. doi: 10.1021/bi9006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine HL, Brody RS, Westheimer FH. Inhibition of orotidine-5'-phosphate decarboxylase by 1-(5'-phospho-beta-d-ribofuranosyl)barbituric acid, 6-azauridine 5'-phosphate, and uridine 5'-phosphate. Biochemistry. 1980;19:4993–4999. doi: 10.1021/bi00563a010. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM, Abelson JN, Simon MI, editors. Methods in Enzymology. New York: Academic Pres; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 28.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamzin VS, Wilson KS. Automated refinement for protein crystallography. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 32.Wood BM, Amyes TL, Fedorov AA, Fedorov EV, Shabila A, Almo SC, Richard JP, Gerlt JA. Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation. Biochemistry. 2010;49:3514–3516. doi: 10.1021/bi100443a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnett SA, Amyes TL, Wood BM, Gerlt JA, Richard JP. Dissecting the total transition state stabilization provided by amino acid side chains at orotidine 5'-monophosphate decarboxylase: a two-part substrate approach. Biochemistry. 2008;47:7785–7787. doi: 10.1021/bi800939k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific LLC; 2002. [Google Scholar]