Abstract
The modification of proteins by lipid peroxidation products has been linked to numerous diseases and age-related disorders. Here we report on the identification of endogenous protein targets of electrophilic 2-alkenals in cardiac mitochondria. An aldehyde/keto-specific chemical labeling and affinity strategy in combination with LC-MS/MS resulted in 39 unique lipoxidation sites on 27 proteins. Several of the target sites were modified by a variety of 2-alkenal products including acrolein, β-hydroxyacrolein, crotonaldehyde, 4-hydroxy-2-hexenal, 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Many of the adduction sites are implicated in the catalytic function of key mitochondrial enzymes suggesting potential impact on pathways and overall mitochondrial function.
Keywords: Carbonyls, aldehyde-reactive probe, mitochondria, proteomics, lipid peroxidation product, acrolein, hydroxynonenal
Introduction
Since the 1950’s when Harman’s “free radical theory of aging” was introduced there have been a considerable number of studies focused on understanding the molecular mechanisms of oxidative stress and its impact on living systems [1, 2]. It is now widely accepted that oxidative stress plays a role in an increasing number of human health disorders and the aging process [3-9]. Within the rapidly advancing field of proteomics, several efforts have been devoted to identifying chemical changes to the proteome in relation to oxidative stress [10-15].
Mitochondria are central to studies of oxidative stress, as they are known to be generators of reactive oxygen species (ROS) within the cell and play a pivotal role in cell death [7, 8, 16, 17]. There are a number of ROS producing sites within the mitochondria including Complex I and Complex III of the mitochondrial electron transport chain [18, 19]. Because the ROS are generated in close proximity to the mitochondrial membrane, which is rich in polyunsaturated fatty acids, a whole host of reactive lipid peroxidation products are formed. These lipid peroxidation products are relatively long lived compared to the highly reactive radical oxygen species which led to their formation, allowing them to diffuse further from their site of production to potentially react with proteins and other biomolecules located in the inner membrane space and mitochondrial matrix. Among the most abundant and studied of these lipid peroxidation products are the highly electrophilic α,β-unsaturated aldehydes, such as 4-hydroxy-2-nonenal (HNE), 4-oxo-2-nonenal (ONE), 4-hydroxy-2-hexenal (HHE), and acrolein [20, 21].
A number of techniques exist for measuring oxidative modifications to proteins and other biomolecules. Among the most widely used of these techniques are assays based on the use of either 2,4–dinitrophenylhydrazine (DNPH) or thiobarbituric acid (TBARS) derivatization for protein carbonyl determinations. While these methods are able to provide researchers with a general overview of levels of protein carbonyls present, they fail to provide specific information such as the identity of the modified proteins, and the site and chemical nature of the modification. In addition to the traditional colorimetric and immunochemical-based methods of analysis, mass spectrometry has emerged as a powerful technique for studying protein carbonylations [22-24]. Towards the goal of identifying the sites of protein carbonyls in proteins, multiple methods involving chemical derivatization followed by mass spectrometric detection have been developed. Chemical tools that have been employed to label and detect carbonyl sites in proteins include biotin-based hydrazide-functionalized reagents, Girard’s P reagent, and Click probes [25-27]. We recently demonstrated the use of a hydroxylamine-functionalized biotin derivative, i.e. N’-amino-oxymethylcarbonylhydrazino-D-biotin, for selective labeling of aldehydic proteins and subsequent mass spectrometry-based profiling studies of complex biological matrices [13].
Numerous in vitro studies have demonstrated that α,β-unsaturated aldehydes, due to their electrophilic nature, covalently modify proteins at nucleophilic sites, in particular at cysteine, histidine and lysine residues [28-30]. Several recent studies have been devoted to identifying the protein targets of lipid-derived electrophiles, in particular targets of HNE [31-33]. While there has been much success in identifying protein targets from in vitro exposure studies, there has been only a few studies reported attempting the site-specific identification of endogenous oxidative modifications in complex proteome samples [24, 34]. Here we report the use of a chemical aldehyde/keto specific labeling and affinity enrichment strategy in combination with LC-MS/MS that enabled the site-specific identification of major sites and the concomitant detection of multiple endogenous 2-alkenal modifications to mitochondrial proteins.
Materials and Methods
Chemicals
N’-aminooxymethylcarbonyl hydrazino-D-biotin (Aldehyde Reactive Probe, ARP) was purchased from Dojindo Molecular Technologies Inc. (Rockville, MD). Ultralink Monomeric Avidin, Zeba Spin Desalting columns, SuperSignal West Pico Chemiluminescent substrate were purchased from Thermo Scientific (Rockford, IL). Neutravidin-HRP was purchased from Pierce. Sequencing grade modified trypsin was purchased from Promega (Madison, WI). Ready Gels (12% Tris-HCl) were purchased from Bio-Rad. Immobilon-P PVDF membranes were purchased from Millipore.
Preparation of rat cardiac mitochondria
The Institutional Animal Care and Use Committee (IACUC) at Oregon State University approved the experimental protocol for the use of animals in this study. Male Fischer 344 rats, obtained from the National Institute of Aging (Bethesda, MD), were housed in individual plastic cages covered with Hepa filters and allowed free access to food and water. Rats were anesthetized with diethyl ether and a midlateral incision was made in the chest to remove the heart. Subsarcolemmal mitochondria were isolated from the rat hearts by differential centrifugation according to the procedures of Palmer with minor modifications by Suh et al and stored at -80°C until needed [35, 36]. Mitochondrial samples containing approximately 1mg total protein were washed twice with 0°C 10mM NaH2PO4, pH 7.4. The mitochondria were then re-suspended in 200μl of 10mM ice-cold NaH2PO4, pH 7.4. The mitochondria were ruptured by four rounds of freeze/thaw cycles which involved rapid freezing with liquid N2 followed by thawing in a cold water bath with 10 minutes of sonication. Soluble and membrane proteins were separated by centrifugation at 14,000g for 15 min. Pierce Coomassie Plus protein assay was used to determine protein concentrations.
ARP-labeling of mitochondrial proteins
Labeling of mitochondrial proteins with the ARP was accomplished by simultaneously adjusting the protein concentration to ~1 μg/μl and the ARP concentration to 5 mM in 10 mM NaH2PO4, pH 7.4. The mixture was then allowed to react for up to one hour at room temperature. Excess ARP was removed using Zeba desalting spin columns with a molecular weight cut-off of 7 kDa. Protein samples were digested with a 1:50 w/w ratio of modified trypsin in 100 mM NH4HCO3, pH 8.3, for 6-18 hours at 37 °C.
SDS-PAGE and Western blotting
For 1D SDS-PAGE analyses Bio-Rad 12 % Tris-HCl ‘readymade’ gels were used. Mitochondrial protein samples (20 μg per well) was used and gels were run at a constant voltage of 130 V for 60 min. For visualization of the protein bands the gels were incubated with Biosafe Coomassie dye for 1 hr. Western blotting was accomplished by transferring the proteins from the gel to a PVDF membrane by applying a constant current of 150 mA for 2 hours. The blot was then blocked with 5 % milk in TBS buffer with 0.1% Tween-20 and then washed extensively before being incubated with 40 ng/mL of HRP-NeutrAvidin for one hour. After additional wash steps the gel was exposed to the PicoWest chemiluminescent substrate for 10 minutes and developed using Kodak BioMax X-Ray film.
Affinity enrichment of ARP-labeled peptides
Ultralink monomeric avidin (100-300 μl) was packed into Handee Mini Spin columns which accommodate solvent addition with a Luer Lok syringe. The column was washed with 1.5 ml of 10 mM NaH2PO4, pH 7.4. Irreversible binding sites were blocked by washing with three column volumes of 2 mM D-biotin. To remove excess D-biotin the column was washed with five column volumes of 2 M glycine-HCl, pH 2.8. The column was then re-equilibrated by washing with 2 mL of 2x PBS (20 mM NaH2PO4, 300 mM NaCl). The mitochondrial peptide sample was then slowly added to the affinity column and the flow-through collected. To remove non-labeled and non-specifically bound peptides the column was washed with 1 mL 2 × PBS followed by 1 mL of 10 mM NaH2PO4 and finally 1.5 mL of 50 mM NH4HCO3 20 % CH3OH. The column was then rinsed with 1 mL of MilliQ H 2O before eluting the ARP-labeled peptides with 30 % aqueous acetonitrile acidified with 0.4 % formic acid. Collected fractions were concentrated using vacuum centrifugation and stored at -20 °C before mass spectrometric analysis. For each mitochondrial protein preparation a new affinity column was used.
LC-MS/MS analysis
The affinity-enriched ARP-labeled peptides were analyzed by LC-MS/MS using a quadrupole orthogonal time-of-flight mass spectrometer (Q-TOF Ultima Global, Micromass/ Waters, Manchester, UK) mass spectrometer equipped with a NanoAcquity UPLC system. Peptides were fractionated by reversed-phase chromatography on a 100 μm i.d. × 200 mm long BEH column (Waters, Milford, MA) using a linear gradient of a binary solvent system consisting of solvent A (2 % aqueous acetonitrile with 0.1% formic acid) and solvent B (acetonitrile containing 0.1% formic acid). The electrospray source was operated in the positive mode with a spray voltage set to 3.5 kV. The mass spectrometer was operated in the data-dependant MS/MS mode, performing 0.6 second MS scans followed by 2.4 second MS/MS scans, on the three most abundant precursor ions in the MS scan with a 60 second dynamic exclusion of previously selected ions. The MS/MS collision energy (25-65 eV) was dynamically selected based on the charge state of the precursor ion selected by the quadrupole analyzer. Mass spectra were calibrated using fragment ions of Glu1-fibrinopeptide B (MH+ 1570.6774 Th, monoisotopic mass). Additionally, lock spray mass correction was performed on the doubly charged ion of Glu1-fibrinopeptide ([M+2H]2+ 785.8426 Th) every 30 seconds during the MS/MS run.
Peptide samples were also analyzed by LC-MALDI-MS/MS by performing off-line reversed-phase fractionation on a Dionex/LC Packings Ultimate nanoLC system coupled to a Probot target spotter. Peptides were fractionated with a 75 μm i.d. × 150 mm C18 PepMap 100 column (LC-Packings) using a binary gradient consisting of solvent A (5% aqueous acetonitrile with 0.1% trifluoroacetic acid (TFA)) and solvent B (80% aqueous acetonitrile with 0.1% TFA). Eluting peptides were mixed with α-cyano-4-hydroxycinnamic acid (2 mg/mL in 50 % acetonitrile containing 0.1 % TFA) and spotted to a 144-spot stainless steel MALDI target plate. MALDI-MS/MS analysis was performed on an ABI 4700 proteomics analyzer with TOF/TOF optics equipped with a 200-Hz frequency-tripled Nd:YAG laser operating at a wavelength of 355 nm (Applied Biosystems, Inc., Framingham, MA). Operating in a data-dependent reflectron mode, full MS scans were obtained over a range of m/z 700-4000 followed by MS/MS scans on the 10 most abundant ion signals detected in the MS survey scan. Collision-induced dissociation was performed using a collision energy of 1 keV in a collision cell with a gas pressure of 6 × 10-7 Torr. External mass calibration was applied using the ABI 4700 calibration mixture consisting of the following peptides des-Arg1-bradykinin ([M+H]+ 904.4675 Th), angiotensin I ([M+H]+ 1296.6847 Th), Glu1-fibrinopeptide B ([M+H]+ 1570.6768 Th), and ACTH 18-39 ([M+H]+ 2465.1983 Th).
MS/MS data analysis and identification of ARP-labeled peptides
Tandem mass spectrometric data generated on the Q-TOF were processed into peak list files using ProteinLynx Global Server v2.3 (PLGS Waters, Manchester, UK). Both the MS survey and MS/MS scans were calibrated using the lock mass of m/z 785.8426 with a lock spray tolerance of 0.1 Da. MS/MS spectra were smoothed using the Savitzky-Golay algorithm performing 2 iterations over a smoothing window of 3 channels. The survey scan was deisotoped and centroided using a fast algorithm performing 30 iterations. MS and MS/MS spectra were centroided with a minimum peak width of 4 channels centroiding the top 80 % with a TOF resolution set at 10,000 and a number of pulses (NP) multiplier of 0.7.
MALDI-MS/MS data were processed into peak list files using the Peaks-to-Mascot tool in 4000 Series Explorer V3.0 software (Applied Biosystems). Default settings were used except that the peak density for MS/MS spectra was set to a max of 10 peaks per 200 Da, the minimum S/N was set to 10 with a minimum peak area of 500 and a max peak/precursor set to 40.
Peak list files of the tandem mass spectrometric data were analyzed with Mascot v2.1 (Matrix Science, London, UK) and Peaks Studio v4.5 (Bioinformatic Solutions Inc., Waterloo, ON Canada). The Swiss Prot database v50 (270778 sequences, 99412397 residues) was searched using the following parameters: taxonomy rodentia (20991 sequences), ± 0.2 Da mass tolerances for the precursor and fragment ions, possibility of 3 missed proteolytic cleavage sites, with trypsin/P or semitrypsin selected as the digesting enzyme, and ARP-HHE (CHK), ARP-HNE (CHK), ARP-Acrolein (CHK), ARP-ONE (CHK), ARP-MDA (KR), ARP-β-hydroxyacrolein, ARP-crotonaldehyde and methionine oxidation (M) selected as variable modifications at the residues specified in parenthesis.
Calculation of B-factors and solvent accessible areas (SAA)
The 3D coordinates of porcine heart mitochondrial type II malate dehydrogenase (MDH) was retrieved from the Protein Data Bank (PDB ID: 1MLD) [37]. The B-factor and SAA values for all cysteine residues of MDH protein in the tetramer conformation were calculated using the program Molsoft ICM v3.5-1p. The mean B-factor values were derived from the atomic crystallographic data for each cysteine residue in each of the four monomers of the tetramer complex of MDH. The reported B-Factor values for each cysteine residue are the average of the four monomers. The solvent accessible areas were calculated using a faster modification of the Shrake and Rupley alogorithm accounting for the presence of hydrogen atoms in the protein structure and using a water probe radius of 1.4 Å [38]. SAA values were calculated as the average of a span of three residues, including ±1 residue from the cysteine for each of the monomers of the tetrameric complex of MDH. The reported SAA values for each cysteine are the average of the four monomers.
Functional pathway network analysis
The carbonyl-modified proteins identified by LC-MS/MS analysis were evaluated for their functional biological pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Cytoscape (v. 2.6.2) was used to construct a visual representation of the network connections between the modified proteins and their functional pathways.
Results
Global analysis of carbonyl-modified proteins
Mitochondria are well known sources of ROS and oxidative modifications of mitochondrial proteins are associated with many human health disorders as well as the aging process [39, 40]. The relatively low abundance of carbonylated proteins under physiological conditions requires the use of an affinity enrichment approach. To identify mitochondrial protein targets of electrophilic 2-alkenal lipid peroxidation products we employed a chemical labeling and affinity enrichment technique using an aldehyde/keto reactive probe. The hydroxylamine functionality of the probe reacts with the carbonyl group on the oxidized protein to form an oxime product which is sufficiently stable during trypsinolysis and directly amenable to mass spectral analysis. This is a practical advantage of the ARP-based approach over similar affinity labeling methods, such as those employing hydrazide-functionalized probes, e.g. biotin hydrazide, which require reduction of the hydrazone bond to form a stable adduct [12, 32]. The derivatization reaction with ARP adds a mass of 313.1 Da to the carbonylated peptide [41]. Cardiac mitochondria from 24-month old rats were used for this study based on related studies indicating that oxidative modifications to proteins may play a role in mitochondrial decay observed in aging and age-related diseases [42].
First, we applied a previously reported method to produce an “ARP-blot” to confirm the presence of oxidatively modified proteins amenable to labeling with ARP in the mitochondrial preparations [43]. Mitochondrial protein samples were reacted with 5 mM ARP, then separated by 1D-SDS-PAGE and detected using Western blotting in combination with HRP-conjugated NeutrAvidin and PicoWest chemiluminescent reagents (Pierce) to visualize the ARP-reactive bands (Figure 1). Several ARP-reactive protein bands are visible in the Western blot from the mitochondrial sample that was reacted with ARP. A mitochondrial protein sample, to which no ARP was added, served as control and showed no signal except for a single HRP-NeutrAvidin-reactive protein band visible in the control lane which is likely due to a naturally biotinylated protein, such as one of the mitochondrial carboxylase enzymes.
Figure 1.
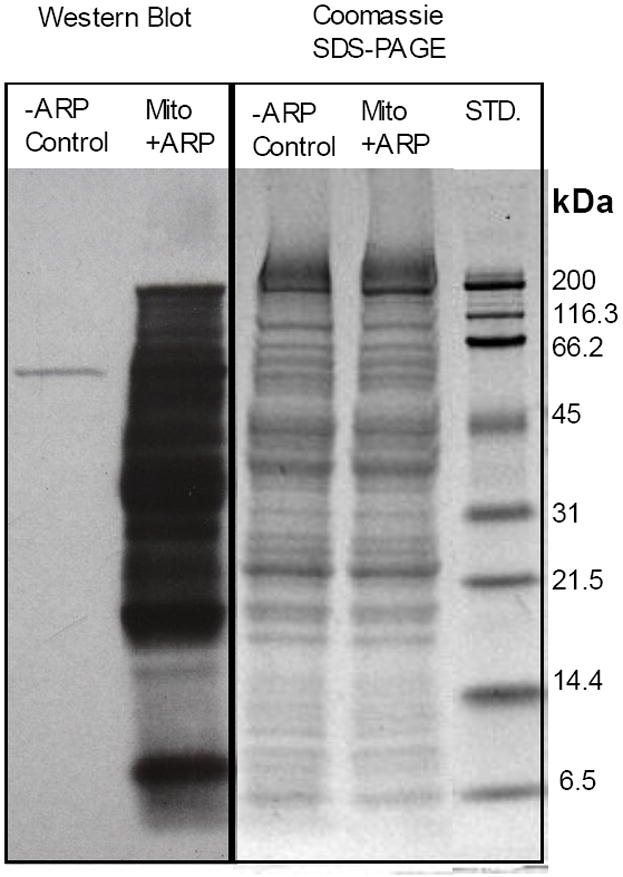
SDS-PAGE and Western blot of mitochondrial proteins from rat hearts. The difference in signal between −ARP control and the +ARP mitochondrial samples demonstrates the presence of a large number or ARP reactive proteins, as well as the selectivity of NeutrAvidin-HRP for the ARP-labeled proteins.
Subsequently, mitochondrial protein preparations were labeled with ARP according to the previously described procedure [31, 41]. ARP-labeled mitochondrial proteins were tryptically digested, enriched with monomeric avidin and analyzed by LC-MS/MS using ESI-Q-TOF and MALDI-TOF/TOF instruments. The resulting MS/MS data were searched against the Swiss Prot database using Mascot and Peaks software. Database search parameters allowed for the possibility of the previously specified ARP-labeled oxidative modifications listed in the methods section. It should be noted that in targeted proteomics experiments, such as the ones described in this study, the datasets are usually too small to allow an accurate determination of the false discovery rate.
Site-specific identification of ARP-labeled peptide 2-alkenal adducts by affinity enrichment and tandem mass spectrometry
Modified peptides often display atypical and complicated fragmentation patterns that are difficult to annotate for automated search algorithms, such as Mascot and Peaks. In our previous studies we identified several unique fragment ion signatures for ARP-labeled peptides, which aid in their identification by manual inspection [31, 41]. Identification of ARP-labeled aldehydic peptides was accomplished by first putatively identifying MS/MS spectra of ARP-labeled 2-alkenal-modified peptides in the Mascot and Peaks search results. These spectra were then subjected to manual inspection before being considered as positive identifications. Manual analysis included assigning a continuity of b- and y- type ions, comparison to homologous peptides with high scores, as well as inspection for the presence of fragment ions characteristic for ARP-labeled peptides. Using this approach we identified a variety of oxidative modifications to peptides originating from several mitochondrial proteins (summarized in table 1). A complete set of annotated tandem mass spectra from the ARP-labeled 2-alkenal modified peptides with fragment ion annotation are provided as supplemental materials (Figures S1-S77). In total, we identified 39 unique sites of modification in 27 mitochondrial proteins. The masses of the modifications were consistent with Michael-type adduction involving acrolein, β-hydroxyacrolein, crotonaldehyde, 4-hydroxy-2-hexenal, 4-hydroxy-2-nonenal, and 4-oxo-2-nonenal. Three of the modified peptides - NALANPLYCPDYR from ubiquinol-cytochrome-c reductase core protein 2, ETECTYFSTPLLLGK from malate dehydrogenase, and CIGAIAMTEPGAGDLQGVR from long-chain specific acyl-CoA dehydrogenase - were identified with more than one type of modification to the same residue (table 1).
Table 1. ARP-labeled 2-alkenal modified peptides identified from rat cardiac mitochondrial proteins.
Summary of the mitochondrial proteins modified by lipid peroxidation derived 2-alkenals and subsequently labeled with ARP and identified by tandem mass spectrometry. The MS/MS identified peptide sequences containing the ARP label and 2-alkenal modification from each of the corresponding carbonylated proteins is shown. The amino acid residue and chemical nature of the modification are indicated for each case. Biological and functional assignments were made on the basis of information from the NCBI (www.ncbi.nlm.nih.gov/Pubmed) and the UniProt Knowledgebase (http: http://www.uniprot.org) websites.
| Biological Function | SwissProt ID1 | UniProt Accession | Protein Name | Peptide Sequence2 | Modified Residue | Modification |
|---|---|---|---|---|---|---|
| Respiratory chain | AT5F1_RAT | P19511 | ATP synthase B chain | C*IGDLKa,b | C239 | ARP-acrolein |
| ATP5H_RAT | P31399 | ATP synthase D chain | NC*AQFVTGSQARa,b,c | C100 | ARP-acrolein | |
| ATPA_RAT | P15999 | ATP synthase subunit alpha | GCSMGEYFRa,b,c,d | C294 | ARP-acrolein | |
| SGC*SMGEYFRa,b,c,d | C294 | ARP-acrolein | ||||
| YSGC*SMGEYFRa,b,d | C294 | ARP-acrolein | ||||
| LAPYSGC*SMGEYFRa,c | C294 | ARP-acrolein | ||||
| ATPB_RAT | P10719 | ATP synthase subunit beta | H*GGYSVFAGVGERa,b,c,d | H227 | ARP-crotonaldehyde | |
| ATPG_RAT | P35435 | ATP synthase gamma chain | GLC*GAIHSSVAKa,b,c | C78 | ARP-acrolein | |
| H*SDQFLVSFKa,b | H120 | ARP-acrolein | ||||
| ATPO_RAT | Q06647 | ATP synthase O subunit | GEVPC*TVTTAFPLDEAVLSELKa,b,c | C141 | ARP-acrolein | |
| CX6B1_MOUSE (EDM07730) | Q91YT0 | Cytochrome c oxidase subunit VIb isoform 1 | GGDVSVC*EWYRa,c | C54 | ARP-acrolein | |
| NDUV1_MOUSE (AAH83709) | Q91YT0 | NADH dehydrogenase [ubiquinone] flavoprotein 1 | QIEGHTIC*ALGDGAAWPVQGLIRa | C425 | ARP-acrolein | |
| QCR1_RAT | Q68FY0 | Ubiquinol-cytochrome-c reductase complex core protein 1 | NALISHLDGTTPVC*EDIGRa,b,c | C410 | ARP-acrolein | |
| YFYDQC*PAVAGYGPIEQLSDYNRb | C453 | ARP-acrolein | ||||
| QCR2_RAT | P32551 | Ubiquinol-cytochrome-c reductase complex core protein 2 | NALANPLYC*PDYRa,b,c | C191 | ARP-acrolein, ARP-HNE | |
| NPLYC*PDYRa,d | C191 | ARP-acrolein | ||||
| DHSA_RAT | Q920L2 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit | GHSLLHTLYGRa,b | H190 | ARP-acrolein | |
| Transport | Q6IRH6_RAT | Q6IRH6 | Slc25a3 protein | AVEEQYSC*EYGSGRb,d | C52 | ARP-acrolein |
| ADT1_RAT | Q05962 | ADP/ATP translocase 1 | GADIMYTGTVDC*WRa,b,c | C256 | M(ox), ARP-acrolein | |
| TGTVDC*WRa,c,d | C256 | ARP-acrolein | ||||
| EFNGLGDC*LTKa,c | C159 | ARP-acrolein | ||||
| YFAGNLASGGAAGATSLC* a,d | C128 | ARP-acrolein | ||||
| C*FVYPLDFARa,b,c | C128 | ARP-acrolein | ||||
| DIMYTGTVDC*WRb | C256 | ARP-acrolein | ||||
| KGADIMYTGTVDC*WRb | C256 | ARP-acrolein | ||||
| ADT2_RAT | Q09073 | ADP/ATP translocase 2 | VSFAK*DFLAGGVAAAISKa,c | K10 | ARP-HNE | |
| TGTLDC*WRa,d | C257 | ARP-acrolein | ||||
| VDAC1_RAT | Q9Z2L0 | Voltage-dependent anion-selective channel protein 1 | YQVDPC*ACFSAKa,b,c | C232 | ARP-acrolein | |
| VDAC3_RAT | Q9R1Z0 | Voltage-dependent anion-selective channel protein 3 | VC*NYGLIFTQKa | C65 | ARP-acrolein | |
| TCA cycle | ACON_RAT | Q9ER34 | Aconitate hydratase | VGLIGSC*TNSSYEDMGRa,b | C385 | ARP-acrolein |
| VAVPSTIHC*DHLIEAQLGGEKa | C126 | ARP-acrolein | ||||
| IDHP_RAT | P56574 | Isocitrate dehydrogenase [NADP] | C*ATITPDEARb | C184 | ARP-acrolein | |
| DLAGC*IHGLSNVKa | C416 | ARP-acrolein | ||||
| MDHM_RAT | P04636 | Malate dehydrogenase | ETEC*TYFSTPLLLGKa,b,c | C285 | ARP-acrolein, ARP-HHE | |
| TEC*TYFSTPLLLGKa,b,d | C285 | ARP-acrolein | ||||
| EGVIEC*SFVQSKa | C275 | ARP-acrolein | ||||
| GYLGPEQLPDC*LKa | C89 | ARP-acrolein | ||||
| TIIPLISQC*TPKa | C212 | ARP-acrolein | ||||
| GC*DVVVIPAGVPRa | C93 | ARP-acrolein | ||||
| QDPA_RAT | P26284 | Pyruvate dehydrogenase E1 component alpha subunit | VDGMDILC*VRb | C261 | ARP-acrolein | |
| H*GFTFTRa | H121 | ARP-crotonaldehyde | ||||
| QDPB_RAT | P49432 | Pyruvate dehydrogenase E1 component subunit beta | EGIEC*EVINLRa | C263 | ARP-acrolein | |
| Oxidoreductase | ETFD_RAT | Q6UPE1 | Electron transfer flavoprotein-ubiquinone oxidoreductase | AAQIGHTLSGAC*LDPAAFKa,b | C263 | ARP-acrolein |
| MMSA_RAT | Q02253 | Methylmalonate-semialdehyde dehydrogenase | C*MALSTAVLVGEAKa,b | C317 | ARP-acrolein | |
| NNTM_MOUSE (AAH91271) | Q61941 | NAD(P) transhydrogenase | EANSIVITPGYGLC*AAKa,b,c | C936 | ARP-acrolein | |
| Chaperone | CH60_RAT | P63039 | 60 kDa heat shock protein | C*EFQDAYVLLSEKb | C237 | ARP-acrolein |
| Lipid Metabolism | ACADL_RAT | P15650 | Long-chain specific acyl-CoA dehydrogenase | C*IGAIAMTEPGAGSDLQGVRa,b,c | C166 | ARP-acrolein,ARP-βHA, ARP-HHE, ARP-HNE, ARP-ONE |
| Kinase | KCRB_RAT | P07335 | Creatine Kinase B Type | LGYILTCPSNLGTGLRa,c | C283 | ARP-acrolein |
| ILTCPSNLGTGLRa,d | C283 | ARP-acrolein |
Rat protein was not present in SwissProt database; thus gene bank accession number for the rat protein containing the equivalent peptide sequence is given.
An * marks the ARP-labeled 2-alkenal modified residue. All MS/MS spectra were visually inspected and fragment ions annotated manually; MS/MS spectra are available in the supplemental material.
Peptide identified by ESI-Q-TOF;
peptide identified by MALDI-TOF/TOF;
peptide identified in repeated biological sample preparations;
identified peptide sequence is semi-tryptic.
Several of the proteins identified are components of the mitochondrial electron transport chain (ETC). Many of these protein subunits have been previously identified as being carbonylated or modified by HNE in gel-based studies, which employed Western blot detection using anti-DNP or anti-HNE antibodies. For example, succinate dehydrogenase [ubiquinone] flavoprotein subunit, ATP synthase α-chain, ATP synthase β-chain, ATP synthase γ-chain, cytochrome c oxidase subunit VIb isoform 1, and NADH dehydrogenase [ubiquinone] flavoprotein 1 were identified with carbonylation and/or HNE modifications. However, the sites and types of modifications were not conclusively characterized [44, 45].
By comparing the results in table 1 with other reports aimed at identifying sites of oxidative modification to proteins by the exogenous addition of electrophiles, there is some overlap of the sites and proteins identified. This is particularly interesting in that it suggests that the modified residues represent major target sites susceptible to adduction by lipid peroxidation products. For example, in comparison with previous results from our group in which we utilized a novel synthetic probe (HICAT) to label and quantify mitochondrial protein targets of HNE, we identified eight of the same proteins and sites of modification: His-227 of ATP synthase β-subunit, Cys-100 of ATP synthase D chain, Cys-141 of ATP synthase O subunit, Cys-256 of ADP/ATP translocase 1, Cys-166 of long-chain specific acyl-CoA dehydrogenase, Cys-93 and Cys-285 of malate dehydrogenase, and Cys-191 of ubiquinol-cytochrome-c reductase complex core protein 2 [27]. In a recent study by Wong and Liebler, in which they added two thiol-reactive probes, N-iodoacetyl- N-biotinylhexylene-diamine (IAB) and 1-biotinamido-4-(4’-[maleimidoethylcyclohexane]-carboxamido)butane (BMCC), to a mitochondrial protein fraction obtained from HEK293 cells, 1693 sites of adduction belonging two 809 proteins were identified [28]. A comparison of their results with the modified protein sites identified in the present study reveals seven cases of overlap. These sites are Cys-385 of aconitase, Cys-256 of ADP/ATP translocase, Cys-100 of ATP synthase D chain, Cys-317 of methylmalonate-semialdehyde dehydrogenase, Cys-936 of NAD(P) transhydrogenase, Cys-191 of ubiquinol-cytochrome-c-reductase complex core protein 2, and Cys-231 of voltage-dependant anion selective channel protein 1. The fact that only seven common sites exist out of the 1693 residues identified by Wong and Liebler and the 39 residues identified in this study raises some interesting questions as to what actually represent biologically significant targets of oxidative stress, and what is the best approach to identify them. Clearly in vitro studies in which relatively high concentrations of electrophilic probes are added are capable of identifying a large number of potential target sites. However, such artificial situations may not accurately reflect the most biologically relevant sites. There is also the question of the site specificity of various electrophiles based on their differing chemical structures and physicochemical properties [46]. Our results include a few examples that indicate that despite the variation in size and chemical structure of these 2-alkenals, their common electrophilic properties allow them to modify the same nucleophilic sites on proteins. Perhaps the most striking of these examples is the case of the long chain-specific acyl-CoA dehydrogenase, which was identified with modifications from an unprecedented, five different types of 2-alkenal lipid peroxidation products to Cys-166. The masses of these modifications indicated adduction by acrolein, β-hydroxyacrolein, 4-hydroxy-2-hexenal, 4-hydroxy-2-nonenal, and 4-oxo-2-nonenal. Figure 2 displays the MALDI-TOF/TOF generated tandem mass spectra of the tryptic peptide CIGAIAMTEPGAGSDLQGVR spanning residues 166 to 185 illustrating the diversity of modifications identified on the cysteine residue.
Figure 2.
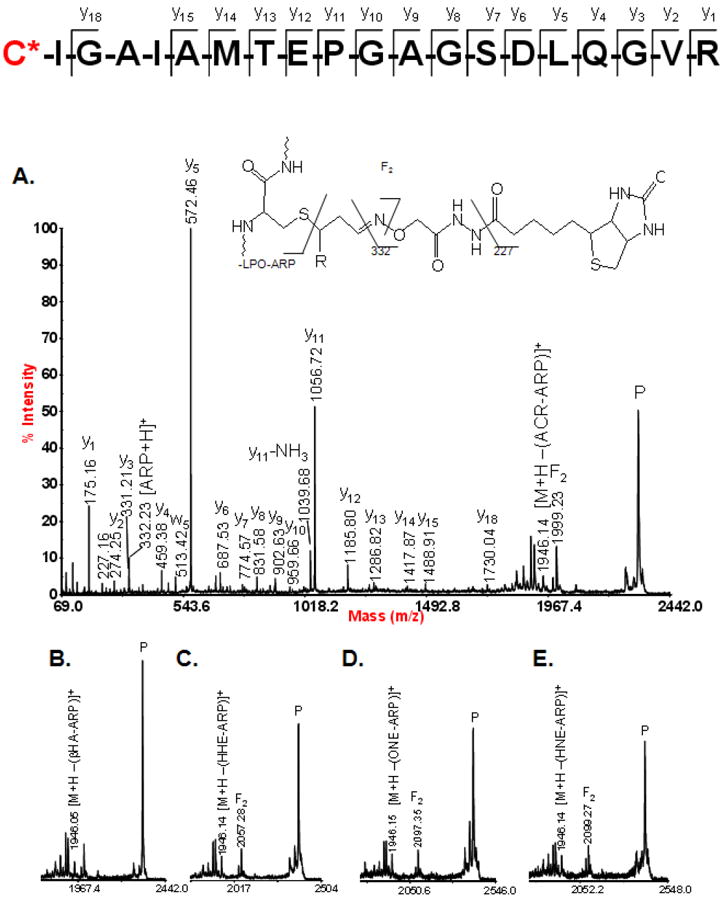
MALDI tandem mass spectra from the peptide spanning residues 166 to 185 from the long chain-specific acyl-CoA dehydrogenase which was found to be modified on Cys-166 by a variety of lipid peroxidation products. A. Full tandem mass spectrum of the ARP-acrolein modified version of the peptide. B, C, D, and E display the high m/z region of the tandem mass spectra for the ARP-β-hydroxyacrolein modified, ARP-HHE modified, ARP-ONE modified, and ARP-HNE modified versions of the peptide, respectively.
Several of the sites identified in this study are known to be crucial for the function of the specific proteins. For example, the tryptic peptide encompassing residue Cys-166 from the long chain-specific acyl-CoA dehydrogenase contains several residues which make up part of a FAD binding site, and modification of this site by lipid peroxidation products would likely interfere with the protein’s ability to bind FAD and affect the catalysis of the first step in the β-oxidation of fatty acids.
Another example is Cys-385 of aconitase, which was identified with an ARP-acrolein modification. Aconitase has been shown to be inhibited by acrolein in a concentration dependent manner [47]. Aconitase resides in the mitochondrial matrix and is involved in the second step of the citric acid cycle, catalyzing the reversible isomerization of citrate and isocitrate with cis-aconitate as a reaction intermediate. We identified the tryptic peptide VGLIGSCTNSSYEDMGR to be modified on Cys-385 by ARP-acrolein. In a separate study performed by our group, this cysteine residue was also found to be susceptible to modification by the thiol reactive probe (4-iodobutyl)triphenyl-phosphonium (IBTP) [13]. The susceptibility of Cys-385 to modification by diverse electrophilic species is in line with functional studies that describe the loss of activity of aconitase under conditions of oxidative stress [48]. Cys-385 is part of the 4Fe-4S iron sulfur cluster which is required for the catalytic function of aconitase [49, 50].
The peptide spanning residues 317 to 330 from methylmalonate-semialdehyde dehydrogenase was identified by both ESI-Q-TOF and MALDI-TOF/TOF analysis as containing an acrolein modification to Cys-317. This residue is the nucleophilic active site of this enzyme, which catalyzes the irreversible oxidative decarboxylation of malonsemialdehyde and methylmalonate semialdehyde to acetyl-CoA and propionyl-CoA respectively. The ESI-Q-TOF generated MS/MS is shown in Figure 3. Note the presence of fragment ions at m/z 227.09 and m/z 332.14, which indicate fragmentation of the ARP tag and are useful as diagnostic ion peaks to assist in the identification of ARP-labeled peptides [31, 41].
Figure 3.
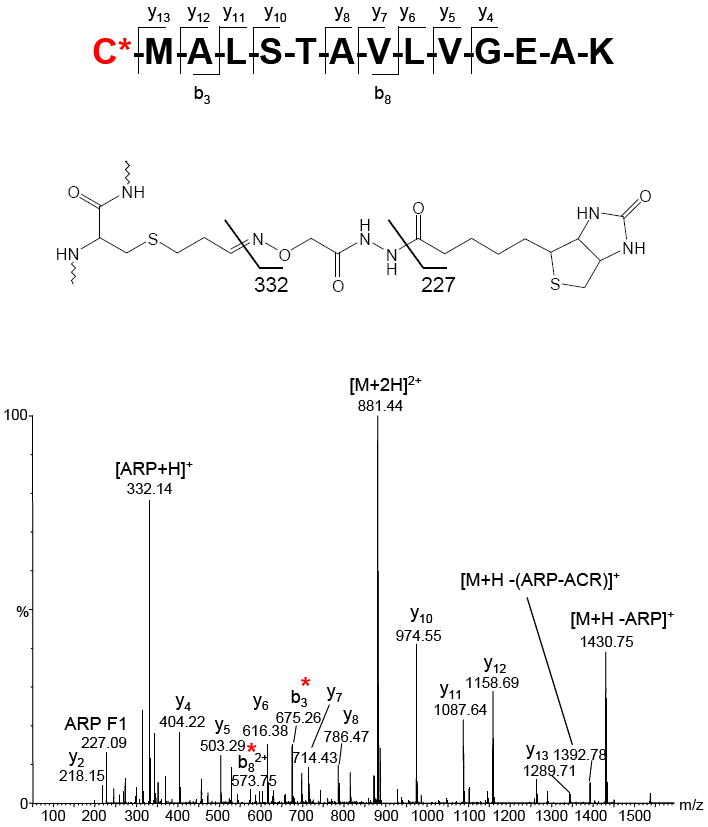
ESI-Q-TOF generated MS/MS of the peptide CMALSTAVLVGEAK, spanning residues 317-330 of methylmalonate-semialdehyde dehydrogenase containing an ARP-acrolein modification at Cys-317. Cys-317 is the nucleophilic active site for this enzyme. This spectrum also contains the fragment ions at 332.22 m/z and 227.15 m/z, which are generated from the ARP tag and are diagnostic of ARP labeling.
The fact that proteins contain sites with differential susceptibility to modification by lipid peroxidation products raises the question to what extent the structure of the protein impacts the reactivity towards electrophiles. Why are certain residues targeted over others in a certain protein? A possible factor contributing to the susceptibility of a functional site would likely be solvent accessibility and at least transient exposure to the surrounding environment. A residue’s susceptibility to modification would also depend on the side chain’s pKa value and the chemical microenvironment in which the functional group is located. In our limited dataset, the modification pattern observed for malate dehydrogenase allowed us at least partly to address our hypothesis that accessibility of a nucleophilic site may contribute to the susceptibility towards electrophilic attacks.
Mitochondrial malate dehydrogenase (MDH) was found to be modified by lipid peroxidation products on five cysteine residues, Cys 89, 93, 212, 275, and 285 (Figures 4). Structural analysis was performed on the crystal structure of porcine heart (MDH) using the protein modeling software Molsoft ICM V3.5-1p. Normalized B-factor values, which serve as a measure of local flexibility of the protein structure over a given region, were obtained for all eight cysteine residues in a monomer of MDH in the tetramer conformation. The solvent accessible areas (SAA) for each of the eight Cys residues, which provide a measure of how exposed each residue is to the surrounding solvent, were also calculated using a water probe radius of 1.4 Å. Four out of the five Cys residues we identified as modified correspond with the Cys residues with the largest B-factor values. Cys-285, which has the largest B-factor and second largest SAA of all of the Cys residues in MDH, was identified as being modified by acrolein and HHE. The MALDI tandem mass spectra of the ARP-acrolein and ARP-HHE modified peptides spanning residues 282-296 are shown in Figures 4A and 4B along with the ribbon structure of MDH illustrating the position of Cys-285 and a table with the corresponding average B-factor and SAA values for all Cys residues in the MDH tetramer (Figures 4C and 4D). Thus, for the case of malate dehydrogenase it seems that B-factor and SAA values relate the susceptibility of specific residues towards electrophilic modification.
Figure 4.
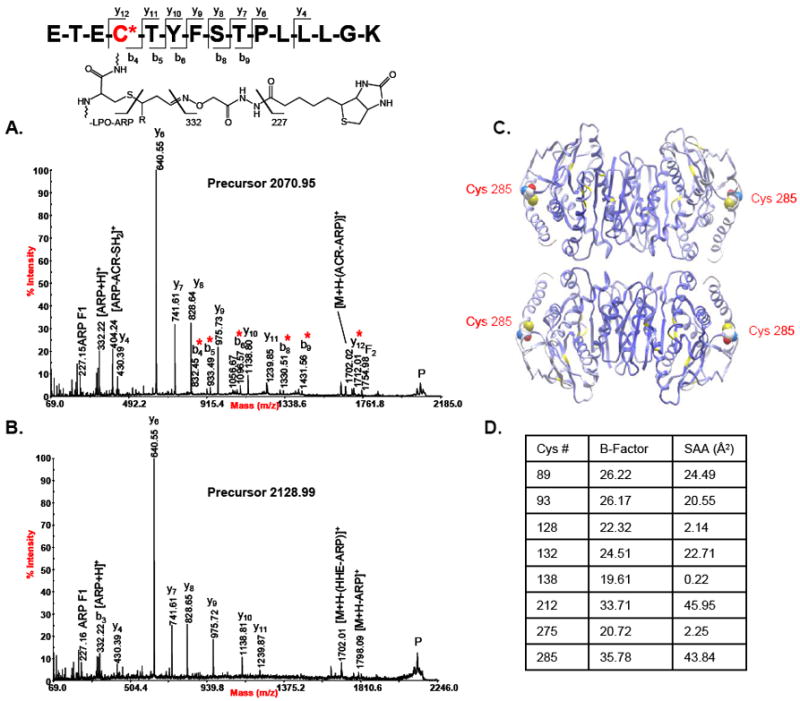
A. Tandem mass spectrum of the peptide spanning residues 282-296 from malate dehydrogenase which was found with the ARP-acrolein and ARP-HHE modifications on Cys-285. Cys-285 is the most solvent accessible Cys residue according to solvent accessibility analysis. B. Tandem mass spectrum of the ARP-HHE modified version of the same peptide. C. Porcine heart MDH tetramer (PDB:1MLD) with the protein backbone displayed as ribbon (cysteine residues colored in yellow) and colored by B-Factor (ICM v3.6-1d, Molsoft). The residue Cys-285 is displayed as CPK and colored by atom type with the carbon atoms in white. D. Table listing the average B-Factor and solvent accessible area values of all Cys residues of tetrameric MDH.
Functional network analysis of 2-alkenal-modified proteins
In order to get an understanding of the potential impact of 2-alkenal modification to mitochondrial proteins on a systems level we performed a network analysis of the functional pathways of our identified proteins. The KEGG database was queried to obtain the functional pathway information and Cytoscape was used to visualize the connections between the proteins and their pathways (Figure S79). The network analysis shows that many of the proteins identified in this study are involved in the functional pathways of degenerative diseases including Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease, all in which oxidative stress induced modification to proteins has been implicated to play an important role. Many of the identified proteins are also involved in calcium signaling, which is involved in mitochondrial regulation of cell death.
Discussion
In this study, protein adducts of diverse lipid peroxidation products were simultaneously labeled, enriched, characterized by LC-MS/MS analysis and assigned to specific protein sites. In total, we detected 49 distinct Michael-type protein adducts of lipoxidation products, identified 27 proteins containing 39 unique nucleophilic sites that were modified by six varieties of 2-alkenals. The proteins and modified sites identified in this study represent targets within the mitochondrial proteome that are susceptible to modification by electrophilic lipid peroxidation products. The majority of modified proteins identified (40 %) were members of the respiratory chain. The electron transport enzymes are both a major source of ROS within the mitochondria and in close proximity to a high concentration of polyunsaturated fatty acids present in the inner mitochondrial membrane. TCA cycle proteins comprised the second most abundant category of identified modified proteins at 19 %. A pie chart displaying the percentages of all identified proteins and modified sites is shown in Figure 5A. Eighty five percent of the 2-alkenal Michael type additions characterized were present on cysteine residues, while 12 % of the modification sites were found on histidine residue. Adduction to lysine residue was only once observed (Figure 5B). Our findings are in agreement with previous reactivity studies that indicate that the cysteine thiol is usually the most nucleophilic site in proteins compared to the imidazole and amino group. In the current study six different lipid peroxidation-derived 2-alkenals were identified with the following distribution: acrolein ≫ HHE, HNE >ONE, crotonaldehyde, hydroxyacrolein (Figure 5C).
Figure 5.
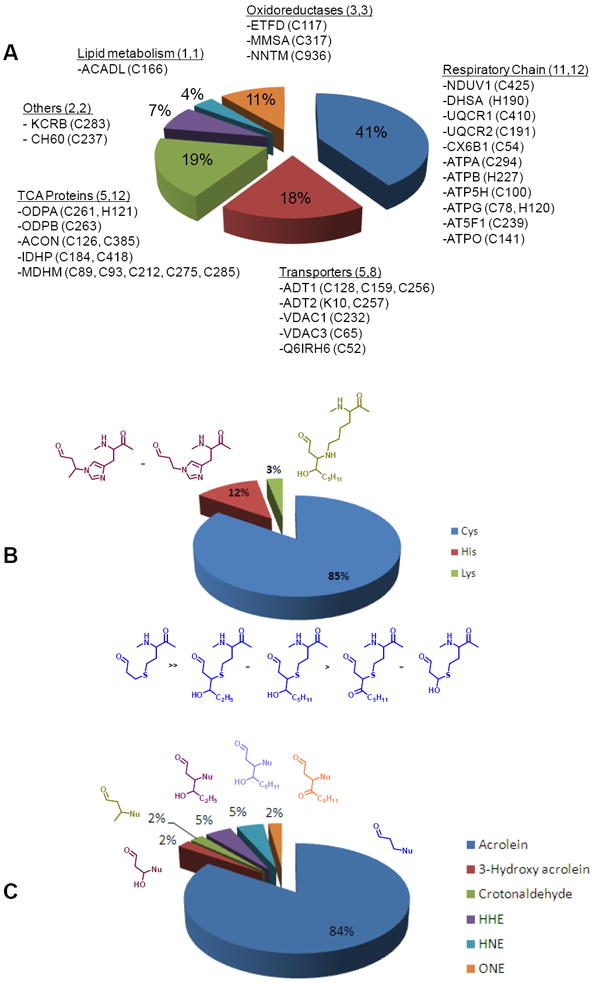
Summary of the identified lipoxidation modifications on mitochondrial proteins. A. Pie chart displaying the mitochondrial proteins and amino acid residue sites identified with modifications by lipid peroxidation-derived 2-alkenals. B. Pie chart illustrating the percentage breakdown of the types of residues identified with Michael-type additions of lipid peroxidation-derived 2-alkenals. C. Pie chart indicating the percentages of the different types of lipid peroxidation derived 2-alkenals identified in this study.
Acrolein modification of mitochondrial proteins
The Michael-type addition of acrolein to nucleophilic sites in proteins was the most common modification we identified in this profiling analysis. We observed acrolein modification on 26 out of the 27 proteins that were observed. Modification by acrolein comprised a total of 84% of the 2-alkenal modifications observed (Figure 5C). Acrolein is formed via multiple routes, e.g. during metabolism of allyl compounds, lipid peroxidation of polyunsaturated fatty acids, and the amine-oxidase mediated degradation of polyamines [20, 51, 52]. Acrolein is known to be the most reactive of the α,β–unsaturated aldehydes [21]. In the current study, ARP-labeled acrolein adducts made up the majority of Michael-type adducts identified. A possible reason that contributed likely to the screwed dataset is that protein adducts of acrolein cannot form cyclic hemiacetals. Whereas, Michael-type adducts of 4-hydroxy 2-enal are partly present as cyclic hemiacetals that are in equilibrium with the open-form aldehyde [21]. The ARP labeling strategy will only label the open-form aldehydic adduct which may lead to a possible underrepresentation of 4-hydroxy 2-enal adducts in the current study. Uchida et al. reported the formation of Nε-(3-formyl-3,4-dehydropiperidino) lysine (FDP-Lys) as a reaction product between acrolein and Lys residues [51]; however, in the present study we were not able to detect FDP-Lys adducts.
The toxicity of acrolein has been shown to be approximately one to three orders of magnitude greater than formaldehyde, acetaldehyde, and HNE [53]. Exposure to acrolein induces conditions of oxidative stress by depleting protective nucleophiles such as glutathione (GSH) and ascorbic acid [52, 54]. Recent studies have shown concentrations of acrolein as low as 1 μM can induce oxidative stress damage resulting in functional impairment of mitochondria [47]. Recent studies have shown that acrolein is a mitochondrial toxin inhibiting electron transport and increasing overall oxidative stress damage in brain and liver mitochondria [47, 53]. Several important mitochondrial enzymes including aconitase, ADP/ATP translocase, Complex I, Complex II, pyruvate dehydrogenase and alpha ketogluterate dehydrogenase have been shown to be inhibited by acrolein [47, 53]. In this LC-MS/MS-based proteome analysis we were able to identify distinct sites of acrolein modifications in Complex I, Complex II, aconitase, and ADP/ATP translocase (Table 1). Site-specific information of acrolein adductions may at least partly advance our understanding of the mechanisms that contribute to enzyme inactivation and mitochondrial dysfunction in aging and under condition of oxidative stress.
Protein modification by other lipid peroxidation products
In addition to acrolein, we also identified other lipid peroxidation products adducted to mitochondrial proteins. These other lipoxidation-derived products were HNE, ONE, HHE, crotonaldehyde and β-hydroxyacrolein. It should be noted that HNE (C9H16O2, monoisotopic mass 156.115) is isobaric with the reduction product of ONE, namely 1-hydroxy-non-2-en-4-one (ONO; C9H16O2, monoisotopic mass 156.115) (also known as 1-hydroxynon-2-en-4-one, 1HNO [55]), and, thus, the current mass spectrometry-based identification of the modifying lipoxidation product does not truly allow for the ability to distinguish these two modifications. Assignments as HNE adducts were based on the assumption that HNE is the lipid peroxidation product which is predominantly studied and reported in literature. The 4-hydroxy-2-nonenal and 4-oxo-2-nonenal arise primarily from the lipid peroxidation of the ω–6 fatty acids, linoleic and arachidonic acids, while 4-hydroxy-2-hexenal is an end product of the peroxidation of ω-3 fatty acids, including α-linolenic, eicosapentaenoic, and docosahexanoic acids. Combined, these other 2-alkenal products comprised 16% of the modifications identified (Figure 5C). A remarkable outcome of the current study is that distinct protein sites were found to be targeted by variety of LPOs, emphasizing the beauty of a mass spectrometry-based proteome analyses strategy. For example, Cys-166 of the long-chain specific acyl CoA dehydrogenase was found to be adducted by five different lipoxidation products (table 1, figure 2).
To assess the structural implications of why certain residues are susceptible to modification we performed structural analysis of malate dehydrogenase by integrating our mass spectrometry data identifying sites of modification with the x-ray crystal dataset of MDH. This proved useful in that the sites of acrolein and HHE adduction correspond to the cysteine residues with high B-factors and solvent accessible areas. Further studies will be necessary to see if this correlation extends to other protein systems and if B-factor and SAA values may be useful in predicting sites susceptible to modification by lipid-derived and other electrophiles.
Some comments towards the applicability of the ARP approach for profiling aldehydic protein modifications
With the rapidly increasing number of new analytical proteomics techniques it is of critical importance to perform validation of the analytical method to ensure the results are scientifically sound. Due to the sample complexity, biological variability and absence of reference standards we made an attempt to assess the robustness of the ARP labeling and enrichment method. We tested the reproducibility of our method by repeating our analysis several times using new mitochondrial protein preparations from different animals and analyzing the samples on multiple instruments. We found that 20 of the ARP-labeled oxylipid modified peptides belonging to 15 proteins were reproducibly identifiable in repeated sample preparations. The ARP label strategy, as do other hydrazide-based labeling strategies, necessitates the presence of the aldehydic form of the Michel-type adduct of reactive lipid peroxidation products, which may at least partly explain the possible underrepresentation of protein adducts of 4-hydroxy 2-enals, such as HHE and HNE adducts, because these adducts are present in equilibrium with their cyclic hemiacetals [21]. Hemiacetals cannot be derivatized with aldehyde-reactive probes and, thus, aldehydic labeling strategies are depended on the presence of the open-form aldehydic adduct.
Oxidative modifications can be introduced during the processing of the sample. However, even if such modifications do occur as artifacts of sample preparation the proteins and amino acid residue sites that were modified still represent susceptible targets to oxidative damage and are therefore likely still relevant. The majority of modified proteins identified in this study are located on the matrix side of the mitochondrial inner membrane, supporting the notion that the observed protein adducts are truly susceptible in vivo targets.
In summary, we provide a unique compilation of endogenous mitochondrial protein targets modified by α,β-unsaturated lipoxidation products. Our data set reveals the susceptibility of distinct sites to modification by a variety of lipid peroxidation-derived 2-alkenals and, as such, provides insight on the chemical consequences of an aerobic lifestyle and oxidative stress on the proteome of subsarcolemmal mitochondria from heart. The described affinity-based approach in combination with LC-MS/MS is broadly applicable to the study of protein carbonylation and has been also successfully used for the site-specific assignment of semialdehydic protein modifications linked to metal-catalyzed oxidations [22].
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Dr. Cristobal Miranda with animal husbandry and mitochondrial isolation. The authors also thank Brian Arbogast (1945-2010) for assistance with the Q-TOF mass spectrometer. This work was supported by grants from the NIH/NIA (AG025372). The OSU EHSC mass spectrometry core facility is supported in part by the Environmental Health Science Center (ES00210).
Footnotes
Supporting information available
Annotated tandem mass spectra for all ARP-labeled peptides are provided in figures S1 through S77. Figure S78 displays the network of biological pathways in which the identified carbonylated proteins are involved.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harman D. Aging: a theory based on free radical and radiation chemistry. Journal of Gerontology. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. The aging process. Proc Natl Acad Sci U S A. 1981;78:7124–7128. doi: 10.1073/pnas.78.11.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 4.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 5.Gibson BW. The human mitochondrial proteome: oxidative stress, protein modifications and oxidative phosphorylation. Int J Biochem Cell Biol. 2005;37:927–934. doi: 10.1016/j.biocel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Liebler DC. The poisons within: application of toxicity mechanisms to fundamental disease processes. Chem Res Toxicol. 2006;19:610–613. doi: 10.1021/tx0600292. [DOI] [PubMed] [Google Scholar]
- 7.Sastre J, Pallardo FV, Vina J. Mitochondrial oxidative stress plays a key role in aging and apoptosis. IUBMB Life. 2000;49:427–435. doi: 10.1080/152165400410281. [DOI] [PubMed] [Google Scholar]
- 8.Linford NJ, Schriner SE, Rabinovitch PS. Oxidative damage and aging: spotlight on mitochondria. Cancer Res. 2006;66:2497–2499. doi: 10.1158/0008-5472.CAN-05-3163. [DOI] [PubMed] [Google Scholar]
- 9.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 10.Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, et al. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 11.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. J Biol Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier CS, Chavez J, Wang J, Wu J. Protein adducts of aldehydic lipid peroxidation products identification and characterization of protein adducts using an aldehyde/keto-reactive probe in combination with mass spectrometry. Methods Enzymol. 2010;473:305–330. doi: 10.1016/S0076-6879(10)73016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoneich C. Mass spectrometry in aging research. Mass Spectrom Rev. 2005;24:701–718. doi: 10.1002/mas.20035. [DOI] [PubMed] [Google Scholar]
- 15.Ying J, Clavreul N, Sethuraman M, Adachi T, Cohen RA. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic Biol Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orrenius S, Gogvadze V, Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 17.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 18.Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 19.O’Malley Y, Fink BD, Ross NC, Prisinzano TE, Sivitz WI. Reactive oxygen and targeted antioxidant administration in endothelial cell mitochondria. J Biol Chem. 2006;281:39766–39775. doi: 10.1074/jbc.M608268200. [DOI] [PubMed] [Google Scholar]
- 20.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 21.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 22.Chavez JD, Bisson WH, Maier CS. A targeted mass spectrometry-based approach for the identification and characterization of proteins containing alpha-aminoadipic and gamma-glutamic semialdehyde residues. Anal Bioanal Chem. 2010;398:2905–2914. doi: 10.1007/s00216-010-4289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal Chem. 2009;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madian AG, Regnier FE. Profiling carbonylated proteins in human plasma. J Proteome Res. 2010;9:1330–1343. doi: 10.1021/pr900890k. [DOI] [PubMed] [Google Scholar]
- 25.Vila A, Tallman KA, Jacobs AT, Liebler DC, et al. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzaei H, Regnier F. Identification and quantification of protein carbonylation using light and heavy isotope labeled Girard’s P reagent. J Chromatogr A. 2006;1134:122–133. doi: 10.1016/j.chroma.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 27.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal Chem. 2007;79:3342–3354. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 28.Wong HL, Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landar A, Oh JY, Giles NM, Isom A, et al. A sensitive method for the quantitative measurement of protein thiol modification in response to oxidative stress. Free Radic Biol Med. 2006;40:459–468. doi: 10.1016/j.freeradbiomed.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 30.Marley K, Mooney DT, Clark-Scannell G, Tong TT, et al. Mass tagging approach for mitochondrial thiol proteins. J Proteome Res. 2005;4:1403–1412. doi: 10.1021/pr050078k. [DOI] [PubMed] [Google Scholar]
- 31.Chavez J, Chung WG, Miranda CL, Singhal M, et al. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP 1 monocytic cells: Protein carbonylation is diminished by ascorbic acid. Chem Res Toxicol. 2009 doi: 10.1021/tx9002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Prot. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 34.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Prot. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- 36.Suh JH, Heath SH, Hagen TM. Two subpopulations of mitochondria in the aging rat heart display heterogenous levels of oxidative stress. Free Radic Biol Med. 2003;35:1064–1072. doi: 10.1016/s0891-5849(03)00468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gleason WB, Fu Z, Birktoft J, Banaszak L. Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases. Biochemistry. 1994;33:2078–2088. doi: 10.1021/bi00174a014. [DOI] [PubMed] [Google Scholar]
- 38.Shrake A, Rupley JA. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J Mol Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- 39.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 40.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 41.Chavez J, Wu J, Han B, Chung WG, Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N’-aminooxymethylcarbonylhydrazino d-biotin. Analytical Chemistry. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 42.Hagen TM, Moreau R, Suh JH, Visioli F. Mitochondrial decay in the aging rat heart: evidence for improvement by dietary supplementation with acetyl-L-carnitine and/or lipoic acid. Ann N Y Acad Sci. 2002;959:491–507. doi: 10.1111/j.1749-6632.2002.tb02119.x. [DOI] [PubMed] [Google Scholar]
- 43.Chung WG, Miranda CL, Maier CS. Detection of carbonyl-modified proteins in interfibrillar rat mitochondria using N’-aminooxymethylcarbonylhydrazino-D-biotin as an aldehyde/keto-reactive probe in combination with Western blot analysis and tandem mass spectrometry. Electrophoresis. 2008;29:1317–1324. doi: 10.1002/elps.200700606. [DOI] [PubMed] [Google Scholar]
- 44.Choksi KB, Boylston WH, Rabek JP, Widger WR, Papaconstantinou J. Oxidatively damaged proteins of heart mitochondrial electron transport complexes. Biochim Biophys Acta. 2004;1688:95–101. doi: 10.1016/j.bbadis.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Wen JJ, Garg N. Oxidative modification of mitochondrial respiratory complexes in response to the stress of Trypanosoma cruzi infection. Free Radic Biol Med. 2004;37:2072–2081. doi: 10.1016/j.freeradbiomed.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 46.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J, Shi R. Acrolein induces oxidative stress in brain mitochondria. Neurochem Int. 2005;46:243–252. doi: 10.1016/j.neuint.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- 49.Bulteau AL, Lundberg KC, Ikeda-Saito M, Isaya G, Szweda LI. Reversible redox-dependent modulation of mitochondrial aconitase and proteolytic activity during in vivo cardiac ischemia/reperfusion. Proc Natl Acad Sci U S A. 2005;102:5987–5991. doi: 10.1073/pnas.0501519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matasova LV, Popova TN. Aconitate hydratase of mammals under oxidative stress. Biochemistry (Mosc) 2008;73:957–964. doi: 10.1134/S0006297908090010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida K, Kanematsu M, Morimitsu Y, Osawa T, et al. Acrolein is a product of lipid peroxidation reaction. Formation of free acrolein and its conjugate with lysine residues in oxidized low density lipoproteins. J Biol Chem. 1998;273:16058–16066. doi: 10.1074/jbc.273.26.16058. [DOI] [PubMed] [Google Scholar]
- 52.Stevens JF, Maier CS. Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol Nutr Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun L, Luo C, Long J, Wei D, Liu J. Acrolein is a mitochondrial toxin: effects on respiratory function and enzyme activities in isolated rat liver mitochondria. Mitochondrion. 2006;6:136–142. doi: 10.1016/j.mito.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Horton ND, Mamiya BM, Kehrer JP. Relationships between cell density, glutathione and proliferation of A549 human lung adenocarcinoma cells treated with acrolein. Toxicology. 1997;122:111–122. doi: 10.1016/s0300-483x(97)00086-3. [DOI] [PubMed] [Google Scholar]
- 55.Doorn JA, Maser E, Blum A, Claffey DJ, Petersen DR. Human carbonyl reductase catalyzes reduction of 4-oxonon-2-enal. Biochemistry. 2004;43:13106–13114. doi: 10.1021/bi049136q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


