Abstract
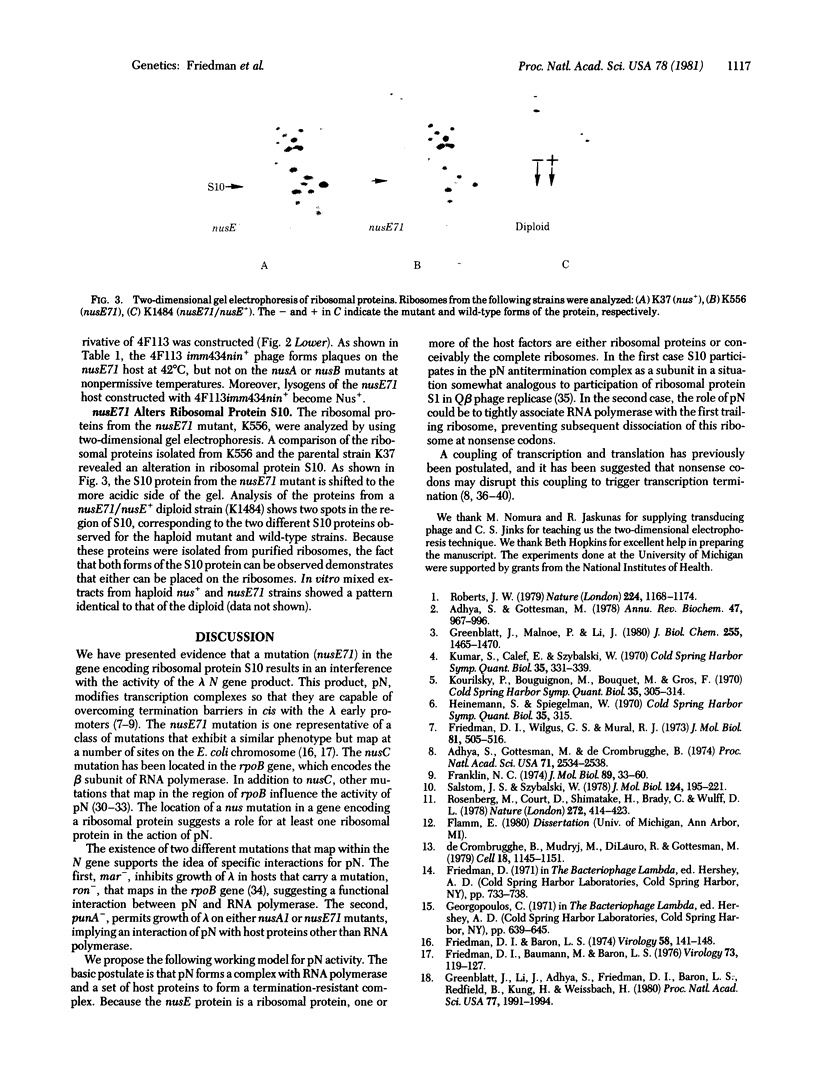
We report the isolation of an Escherichia coli K-12 strain with a mutation, nusE71, that results in a change in ribosomal protein S10. Phage lambda fails to grow in hosts carrying the nusE71 mutation because the lambda N gene product is not active. The N product regulates phage gene expression by altering transcription complexes so that they can overcome termination barriers. This suggests that a ribosomal protein is involved in antitermination of transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Adhya S., Gottesman M., De Crombrugghe B. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2534–2538. doi: 10.1073/pnas.71.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artz S. W., Broach J. R. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3453–3457. doi: 10.1073/pnas.72.9.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barritault D., Expert-Bezancon A., Guérin M. F., Hayes D. The use of acetone precipitation in the isolation of ribosomal proteins. Eur J Biochem. 1976 Mar 16;63(1):131–135. doi: 10.1111/j.1432-1033.1976.tb10215.x. [DOI] [PubMed] [Google Scholar]
- Baumann M. F., Friedman D. I. Cooperative effects of bacterial mutations affecting lambda N gene expression. II. Isolation and characterization of mutations in the rif region. Virology. 1976 Aug;73(1):128–138. doi: 10.1016/0042-6822(76)90067-2. [DOI] [PubMed] [Google Scholar]
- Franklin N. C. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J Mol Biol. 1974 Oct 15;89(1):33–48. doi: 10.1016/0022-2836(74)90161-2. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baron L. S. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974 Mar;58(1):141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baumann M., Baron L. S. Cooperative effects of bacterial mutations affecting lambda N gene expression. I. Isolation and characterization of a nusB mutant. Virology. 1976 Aug;73(1):119–127. doi: 10.1016/0042-6822(76)90066-0. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Jolly C. A., Mural R. J., Ponce-Campos R., Baumann M. F. Growth of lambda variants with added or altered promoters in N-limiting bacterial mutants: evidence that an N recognition site lies in the PR promoter. Virology. 1976 May;71(1):61–73. doi: 10.1016/0042-6822(76)90094-5. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Ponce-Campos R. Differential effect of phage regulator functions on transcription from various promoters: evidence that the P22 gene 24 and the lambda gene N products distinguish three classes of promoters. J Mol Biol. 1975 Nov 5;98(3):537–549. doi: 10.1016/s0022-2836(75)80085-4. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Wilgus G. S., Mural R. J. Gene N regulator function of phage lambda immun21: evidence that a site of N action differs from a site of N recognition. J Mol Biol. 1973 Dec 25;81(4):505–516. doi: 10.1016/0022-2836(73)90519-6. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A., Pironio M. Relationship between the N function of bacteriophage lambda and host RNA polymerase. J Mol Biol. 1972 Mar 28;65(2):259–272. doi: 10.1016/0022-2836(72)90281-1. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J., Adhya S., Friedman D. I., Baron L. S., Redfield B., Kung H. F., Weissbach H. L factor that is required for beta-galactosidase synthesis is the nusA gene product involved in transcription termination. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1991–1994. doi: 10.1073/pnas.77.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Malnoe P., Li J. Purification of the gene N transcription anti-termination protein of bacteriophage lambda. J Biol Chem. 1980 Feb 25;255(4):1465–1470. [PubMed] [Google Scholar]
- Howard G. A., Traut R. R. Separation and radioautography of microgram quantities of ribosomal proteins by two-dimensional polyacrylamide gel electrophoresis. FEBS Lett. 1973 Jan 15;29(2):177–180. doi: 10.1016/0014-5793(73)80555-1. [DOI] [PubMed] [Google Scholar]
- Isono K., Krauss J., Hirota Y. Isolation and characterization of temperature-sensitive mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1976 Dec 22;149(3):297–302. doi: 10.1007/BF00268531. [DOI] [PubMed] [Google Scholar]
- Jaskunas S. R., Fallon A. M., Nomura M. Identification and organization of ribosomal protein genes of Escherichia coli carried by lambdafus2 transducing phage. J Biol Chem. 1977 Oct 25;252(20):7323–7336. [PubMed] [Google Scholar]
- Jaskunas S. R., Fallon A. M., Nomura M., Williams B. G., Blattner F. R. Expression of ribosomal protein genes cloned in Charon vector phages and identification of their promoters. J Biol Chem. 1977 Oct 25;252(20):7355–7364. [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D., JACOB F. Recombination between related temperate bacteriophages and the genetic control of immunity and prophage localization. Virology. 1957 Dec;4(3):509–521. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt E., Stöffler G., Dzionara M., Wittmann H. G. Ribosomal proteins. XVII. Comparative studies on ribosomal proteins of four strains of Escherichia coli. Mol Gen Genet. 1970;109(4):303–308. doi: 10.1007/BF00267700. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pironio M., Ghysen A. A bacterial mutation which affects recognition of the N gene product of bacteriophage lambda. Mol Gen Genet. 1970;108(4):374–375. doi: 10.1007/BF00267775. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Grimley C., Lowery C. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci U S A. 1975 May;72(5):1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Szybalski W. Coliphage lambdanutL-: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978 Sep 5;124(1):195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- Sternberg N. A class of rifR RNA polymerase mutations that interferes with the expression of coliphage lambda late gene. Virology. 1976 Aug;73(1):139–154. doi: 10.1016/0042-6822(76)90068-4. [DOI] [PubMed] [Google Scholar]
- Williams B. G., Blattner F. R., Jaskunas S. R., Nomura M. Insertion of DNA carrying ribosomal protein genes of Escherichia coli into Charon vector phages. J Biol Chem. 1977 Oct 25;252(20):7344–7354. [PubMed] [Google Scholar]
- de Crombrugghe B., Mudryj M., DiLauro R., Gottesman M. Specificity of the bacteriophage lambda N gene product (pN): nut sequences are necessary and sufficient for antitermination by pN. Cell. 1979 Dec;18(4):1145–1151. doi: 10.1016/0092-8674(79)90227-7. [DOI] [PubMed] [Google Scholar]



