Abstract
Severe injury activates many stress-related and inflammatory pathways that can lead to a systemic hyper-metabolic state. Prior studies using perfused hypermetabolic rat livers have identified intrinsic metabolic flux changes that were not dependent upon the continual presence of elevated stress hormones and substrate loads. We investigated the hypothesis that such changes may be due to persistent alterations in gene expression. A systemic hypermetabolic response was induced in rats by applying a moderate burn injury followed 2 days later by cecum ligation and puncture (CLP) to produce sepsis. Control animals received a sham-burn followed by CLP, or a sham-burn followed by sham-CLP. Two days after CLP, livers were analyzed for gene expression changes using DNA microarrays and for meta-bolism alterations by ex vivo perfusion coupled with Meta-bolic Flux Analysis. Burn injury prior to CLP increased fluxes while decreases in gene expression levels were observed. Conversely, CLP alone significantly increased metabolic gene expression, but decreased many of the corresponding meta-bolic fluxes. Burn injury combined with CLP led to the most dramatic changes, where concurrent changes in fluxes and gene expression levels occurred in about 1/3 of the reactions. The data are consistent with the notion that in this model, burn injury prior to CLP increased fluxes through post-translational mechanisms with little contribution of gene expression, while CLP treatment up-regulated the metabolic machinery by transcriptional mechanisms. Overall, these data show that mRNA changes measured at a single time point by DNA microarray analysis do not reliably predict metabolic flux changes in perfused livers.
Keywords: hypermetabolism, liver perfusion, metabolic flux analysis, DNA microarray analysis
Introduction
A common cause of hypermetabolism is severe trauma and burns, especially when patients are afflicted by complications, such as nosocomial infections. Although the primary insult is sufficient to trigger a systemic inflammatory response accompanied by hypermetabolism, it is believed that a second insult in the post-trauma period, due to infection or other insult, plays a major role in causing a more persistent inflammatory response with an ongoing hyper-metabolic and catabolic state leading to severe loss of lean body mass and increased risk of Multiple Organ Dysfunction Syndrome (Fitzwater et al., 2003; Sheridan et al., 1998). An important player in systemic hypermetabolism is the liver, which in large part controls circulating levels of metabolites, and it is the major site for gluconeogenesis as well as the disposal of amino acid nitrogen into urea. Thus, a better understanding of the regulation of the hypermetabolic response in the liver would provide clues for limiting its harmful consequences.
Prior studies using isolated perfused organs have identified differences in metabolic fluxes within liver (Banta et al., 2005; Lee et al., 2000; Yamaguchi et al., 1997) and muscle (Banta et al., 2004) after burn injury that were not dependent upon the continual presence of elevated stress hormones and substrate loads, which suggests that intrinsicchanges in the metabolism of these tissues had occurred. Such differences might be explained by changes in gene expression and enzyme protein levels; for example, sepsis-induced inhibition of gluconeogenesis has been attributed to decreased transcription of phosphoenolpyruvate carboxy-kinase and glucose 6-phosphatase (Deutschman et al., 1995; Maitra et al., 2000). However, no studies in the literature have reported the relationship between gene expression levels and metabolic flux alterations that occur during the response to systemic inflammation.
A holistic, systems-based approach has proven useful for the study of metabolic changes in complex biological systems (Cobb and O’Keefe, 2004; Lee et al., 1999; Nguyen and Yaffe, 2003; Yarmush and Banta, 2003; Yarmush and Berthiaume, 1997). This is especially true when characterizing hypermetabolic and catabolic states that involve many interorgan and intraorgan metabolic fluxes (Herndon and Tompkins, 2004; Tredget and Yu, 1992). Metabolomics-based studies, more specifically metabolic flux analysis (MFA) with or without isotopic tracers, have been used to characterize carbon and nitrogen metabolism in vivo (Hellerstein and Murphy, 2004; Yang and Brunengraber, 2000), in individual organs and tissues in isolated perfusion systems (Banta et al., 2004, 2005; Chatziioannou et al., 2003; Des Rosiers et al., 2004; Jin et al., 2004; Lee et al., 2000; Lee et al., 2003; Yokoyama et al., 2005), and isolated and cultured mammalian cells (Chan et al., 2003a,b; Marin et al., 2004; Zupke et al., 1995). Techniques to characterize system-wide changes in protein levels and gene expression include proteomic (Aulak et al., 2001; Duan et al., 2004) and DNA microarray analysis (Chinnaiyan et al., 2001; Dasu et al., 2004; Vemula et al., 2004). These techniques have been applied in combination to identify the contributions of translational and post-translational mechanisms to metabolic flux distributions primarily in prokaryotes (Hua et al., 2004; Kromer et al., 2004; Oh et al., 2002; Segre et al., 2003; Tummala et al., 2003), and more recently in liver cells (Wong et al., 2004).
In this study, we have used a dual insult model in the male rat, which consists of a ~20% total body surface area (TBSA) third degree scald burn, followed 2 days later by severe polymicrobial sepsis produced through the cecum ligation and puncture (CLP) technique (von Allmen et al., 1990), to induce a hypermetabolic and catabolic state. DNA micro-array analysis and in situ liver perfusions coupled with MFA were concomitantly carried out to determine whether the persistent changes in hepatic metabolism were due to changes in gene expression. The data show that burn injury prior to CLP increased metabolic fluxes while reductions in gene expression levels were observed. Conversely, CLP alone significantly increased the expression of key metabolic genes, but decreases were observed in the metabolic fluxes through these pathways. Combining a priming burn injury with subsequent CLP led to concurrent changes in fluxes and gene expression in about 1/3 of the fluxes. The data are consistent with the notion that in this animal model, burn injury prior to CLP increased fluxes through post-translational mechanisms with little contribution of gene expression, while CLP treatment up-regulated the metabolic machinery by transcriptional mechanisms. The greatest metabolic response in the liver was observed when both complementary changes occurred.
Materials and Methods
Animal Model
Male Sprague Dawley rats (Charles River Labs, Wilmington, MA) weighing between 150 and 200 g were housed, watered and fed in accordance with the National Research Council guidelines. The experimental protocol was approved by the Subcommittee on Research Animal Care, Committee on Research, Massachusetts General Hospital. The animals were randomly divided into three groups. Initially, one third of the rats received a ~20% TBSA dorsal scald burn through a 10 s immersion in boiling water under ketamine and xylazine anesthesia, as previously described (Yamaguchi et al., 1997). The remaining rats were treated identically, except that they were given a sham burn using 37°C water. All rats were resuscitated with saline (50 mL/kg) and placed in separate cages with food and water ad libitum. Two days later, the burned animals and half of the sham-treated animals were subjected to a cecal ligation and puncture (CLP) procedure under ketamine and xylazine anesthesia. Briefly, laparotomy was performed, and the cecum was ligated just distal to the iliocecal valve, and then punctured through and through 4 times with an 18-gauge needle. The cecum was gently squeezed to expel a small amount of fecal material through the holes, and then returned to the abdomen. The abdominal cavity was closed in layers. The remaining sham-treated animals were given a laparotomy only. All animals were resuscitated with saline (50 mL/kg) and allowed to recover. This created three groups of rats: Sham-Sham, Sham-CLP, and Burn-CLP.
Eighteen rats (six from each group) were used for weight gain studies only, and weighed daily (~12:00 p.m.) for 10 days after injury. Another 18 rats (six from each group) were used for liver perfusion experiments (described below). The remainder nine rats (three from each group) were used for liver DNA microarray analysis (described below).
Liver Perfusions and Metabolic Flux Analysis
Three days after induction of burn injury, the animals were fasted overnight to deplete hepatic glycogen stores (approximately 18 h), and on post-burn day 4, they were anesthetized and prepared for in situ liver perfusion as previously described (Banta et al., 2005; Yamaguchi et al., 1997; Yokoyama et al., 2005). Briefly, a laparotomy was performed and the liver was cannulated through the portal and hepatic veins, while the hepatic artery was ligated, thusisolating the liver from the rest of the animal. The liver was perfused for 1 h at 37°C in a recycling mode with Minimal Essential Medium supplemented with amino acids and 3% bovine serum albumin (BSA) as previously described (Yamaguchi et al., 1997).
For each perfused liver, perfusate samples were taken at regular intervals and the concentrations of 26 metabolites were measured off-line, as previously described (Banta et al., 2005; Yamaguchi et al., 1997). Metabolite concentrations were multiplied by the total perfusate volume at the time of sampling to obtain the total amount of each metabolite remaining at the time of sampling. This quantity was plotted as a function of time, and the slope was determined to obtain the rate of production or consumption of each metabolite. Partial pressure measurements for O2 and CO2 were made across the liver at 10, 30, and 50 min into the 1-h perfusion and averaged. The inlet-outlet differential in O2 and CO2 levels was converted to molar concentrations using Henry’s Law, and multiplied by the flow rate to obtain the total rates of O2 and CO2 consumption and uptake, respectively (Banta et al., 2005). All above rates were normalized to wet liver weight to obtain the final normalized value of the reported fluxes.
Since livers were metabolically stable during the perfusions, a pseudo steady-state could be assumed, and measured fluxes were then analyzed by MFA to estimate the unmeasured metabolic fluxes that involve strictly intracellular metabolites, as previously described (Arai et al., 2001; Banta et al., 2005; Lee et al., 2000). Briefly, a simplified metabolic network model of the liver that comprises 72 reactions involving 45 metabolites was constructed. A mass balance equation was written around each one of the 45 metabolites, and assuming steady-state, this yielded a system of 45 independent linear equations. Of the 72 fluxes in the model, 28 were measured (Table I), leaving 44 unknown reaction fluxes. Since the system was overdetermined by one degree of freedom (45 equations vs. 44 unknowns), a least-square method (Lee et al., 2000) was used to solve the linear system of equations, thus yielding the unknown fluxes.
Table I.
Metabolite fluxes measured across the perfused livers.
| Pathway number | Measured metabolite |
|---|---|
| 1 | Glucose |
| 14 | Lactate |
| 23 | Urea |
| 45 | β-hydroxybutyrate |
| 49 | Oxygen |
| 50 | Carbon dioxide |
| 51 | Acetoacetate |
| 52 | Ornithine |
| 53 | Ammonia |
| 54 | Alanine |
| 55 | Cystine |
| 56 | Aspartate |
| 57 | Glutamate |
| 58 | Phenylalanine |
| 59 | Glycine |
| 60 | Histidine |
| 61 | Isoleucine |
| 62 | Lysine |
| 63 | Leucine |
| 64 | Methionine |
| 65 | Asparagine |
| 66 | Proline |
| 67 | Glutamine |
| 68 | Arginine |
| 69 | Serine |
| 70 | Threonine |
| 71 | Valine |
| 72 | Tyrosine |
The metabolic network model was created that best reflected the major metabolic pathways that are known to be active in the liver as well as the measured metabolic fluxes (Table III). The observation of the net release of the branched chain amino acids (leucine, isoleucine, and valine) suggested that the perfused livers were actively degrading protein, and it was assumed that the protein was the BSA found in the perfusate. Therefore, a BSA degradation term was included in the model to account for this (Reaction 48) (Banta et al., 2005). A total carbon and nitrogen balance on the fluxes measured across the livers showed the net release of both carbon and nitrogen (Table II). The degradation of the BSA did not provide enough carbon to account for the large net release, and therefore a term for the oxidation of glycerol (Reaction 10), and a term for the β-oxidation of fatty acids, either from endogenous sources or from the lipids associated with the BSA, (Reaction 42) were added to the model.
Table III.
Metabolic flux analysis (MFA) results.
| Reaction number |
Enzyme (S) | Reaction | Sham-Sham (μmol/h/g) |
Sham-CLP (μmol/h/g) |
Burn-CLP (μmol/h/g) |
|---|---|---|---|---|---|
| 1 | Glucose 6-phosphatase | Glucose 6-phosphate→glucose | 58±4 | 41±3a | 58±8c |
| 2 | Glucose 6-P dehydrogenase, 6-P gluconolactonase, P-gluconate dehydrogenase |
Glucose 6-phosphate→2 NADPH+CO2+Ribulose 5-P | 180±30 | 150±20 | 200±30c |
| 3 | Ribose 5-P isomerase | Ribulose 5-P↔Ribose 5-P | 61±10 | 50±7 | 68±8c |
| 4 | Ribulose-P 3-epimerase | Ribulose 5-P↔xylulose 5-P | 120±20 | 99±13 | 130±20c |
| 5 | Transketolase, transaldolase | Ribose 5-P+xylulose 5-P↔fructose 6-P+erythrose 4-P | 61±10 | 50±7 | 67±8c |
| 6 | Transketolase | Erythrose 4-P+xylulose 5-P↔glyceraldehyde 3-P+fructose 6-P | 61±10 | 50±7 | 67±8c |
| 7 | Glucose phosphate isomerase | Fructose 6-phosphate↔Glucose 6-phosphate | 230±30 | 180±20a | 230±20c |
| 8 | Fructose 1,6-bisphosphatase | Fructose 1,6-bisphosphate→Fructose 6-phosphate | 110±10 | 85±5a | 93±7 |
| 9 | Triose P-isomerase, fructose bisphosphate aldolase | 2 glyceraldehyde 3-P↔fructose 1,6-bisphosphate | 110±10 | 85±5a | 93±7 |
| 10 | Glycerol kinase, glycerol 3-P dehydrogenase, triose P-isomerase |
Glycerol+ATP→Glyceraldehyde 3-P+NADH | 13±4 | 5.8±5.7 | 32±13c |
| 11 | Glyceraldehyde-P dehydrogenase, 3-phosphoglycerate kinase, phosphoglyceromutase, enolase |
Phosphoenolpyruvate+ATP+NADH↔Glyceraldehyde 3-P | 140±10 | 120±10a | 110±10b |
| 12 | Phosphoenolpyruvate carboxykinase | Oxaloacetate+GTP↔CO2+phosphoenolpyruvate | 140±10 | 120±10a | 110±10b |
| 13 | Pyruvate carboxylase | Pyruvate+CO2+ATP↔oxaloacetate | 87±5 | 55±5a | 42±11b |
| 14 | Lactate dehydrogenase | Lactate↔pyruvate+NADH | 68±6 | 40±6a | 11±8b,c |
| 15 | Citrate synthase | Acetyl-CoA+oxaloacetate→citrate | 26±3 | 31±2 | 35±4b |
| 16 | Aconitase, isocitrate dehydrogenase | Citrate↔2-oxo-gluterate+NADH+CO2 | 26±3 | 31±2 | 35±4b |
| 17 | 2-oxo-gluterate dehydrogenase | 2-oxo-gluterate→succinyl-CoA+NADH+CO2 | 57±5 | 66±4 | 75±8b |
| 18 | Succinyl-CoA synthetase, succinate dehydrogenase | Succinyl-CoA↔GTP+FADH2+fumarate | 62±4 | 69±4 | 77±7b |
| 19 | Fumarase | Fumarate↔malate | 130±10 | 140±10 | 170±20b,c |
| 20 | Malate dehydrogenase | Malate↔oxaloacetate+NADH | 130±10 | 140±10 | 170±20b,c |
| 21 | Carbonate dehydratase, carbamoyl-P synthase, ornithine transcarbamylase |
Ornithine+CO2++2 ATP→citrulline | 68±4 | 67±3 | 90±9b,c |
| 22 | Argininosuccinate synthase, argininosuccinate lyase | Citrulline+aspartate+ATP→arginine+Fumarate | 65±4 | 64±3 | 84±9b,c |
| 23 | Arginase | Arginine→ornithine+urea | 80±4 | 79±4 | 110±10b,c |
| 24 | Alanine transaminase | Alanine+2-oxo-gluterate↔pyruvate+glutamate | 6.2±0.8 | 1.9±0.9a | 5.5±2.0c |
| 25 | Serine dehydratase | Serine→pyruvate+ | 10±1 | 12±2 | 21±3b,c |
| 26 | Cysteine dioxygenase, aspartate aminotransferase | Cysteine+2-oxo-gluterate+O2↔glutamate+pyruvate | 2.7±0.8 | 1.7±0.4 | 3.4±1.1c |
| 27 | Threonine 3-dehydrogenase, acetyl-CoA ligase | Threonine+ATP→NADH+glycine+acetyl-CoA | 0.89±0.46 | 1.2±0.6 | 3.0±0.8b,c |
| 28 | Glycine hydroxymethyltransferase, glycine cleavage system | 2 glycine↔serine+CO2++NADH | 2.7±0.3 | 2.9±0.4 | 5.3±0.7b,c |
| 29 | Valine metabolism | Valine+2-oxo-gluterate+ATP→glutamate +succinyl-CoA+2 NADH+FADH2+CO2 |
2.2±0.6 | 1.1±0.5a | 0.71±0.58 |
| 30 | Isoleucine metabolism | Isoleucine+2-oxo-gluterate+ATP→glutamate +succinyl-CoA+acetyl-CoA+2 NADH+FADH2 |
0.82±0.21 | 0.30±0.34 | −0.11±0.32b |
| 31 | Leucine metabolism | Leucine+2-oxo-gluterate→glutamate+NADH +FADH2+ATP+acetoacetate+acetyl-CoA |
3.1±0.9 | 0.96±0.77a | 1.1±1.4 |
| 32 | Lysine metabolism | Lysine+2 2-oxo-gluterate→2 glutamate+3 NADH+FADH2+2 CO2+acetoacetatyl-CoA |
6.6±0.9 | 3.4±0.7a | 5.6±1.0c |
| 33 | Phenylalanine 4-monooxygenase | Phenylalanine+O2→tyrosine | 5.6±0.4 | 4.1±0.3a | 5.5±0.8c |
| 34 | Tyrosine metabolism | Tyrosine+2-oxo-gluterate+2 O2→glutamate+ CO2+fumarate+acetoacetate |
6.2±0.5 | 4.8±0.5a | 5.3±1.3 |
| 35 | Glutamate dehydrogenase | Glutamate↔2-oxo-gluterate+NADH+ | 16±4 | 5.7±2.6a | 7.6±4.4 |
| 36 | Glutaminase | Glutamine→glutamate+ | 28±4 | 35±4 | 37±6 |
| 37 | Proline dehydrogenase, 1-pyrroline 5-carboxylate dehydrogenase |
Proline→glutamate+NADH | 4.4±0.4 | 3.3±0.5a | 5.8±0.7c |
| 38 | Histidine metabolism | Histidine→+Glutamate | 2.0±0.4 | 2.1±0.4 | 4.0±1.1b,c |
| 39 | Methionine metabolism | Methionine+3 ATP+serine→cysteine +NADH+succinyl-CoA+ |
1.5±0.1 | 1.3±0.1 | 2.0±0.2b,c |
| 40 | Aspartate aminotransferase | Oxaloacetate+glutamate↔aspartate+2-oxo-gluterate | 50±3 | 47±3 | 60±6c |
| 41 | Asparaginase | Asparagine↔aspartate+ | 12±1 | 16±1 | 23±3b,c |
| 42 | β Oxidation | Triacylglycerol+4 ATP→glyceraldehyde 3-P +24 acetyl-CoA+21 FADH2+22 NADH |
6.4±0.6 | 4.4±0.6a | 4.6±0.4b |
| 43 | Thiolase | 2 acetyl-CoA↔acetoacetyl-CoA | 67±7 | 39±8a | 40±4b |
| 44 | Acetoacetyl-CoA hydrolase | Acetoacetyl-CoA→acetoacetate | 73±7 | 42±7a | 46±3b |
| 45 | β Hydroxybutyrate dehydrogenase | Acetoacetate+NADH↔β hydroxybutyrate | 85±7 | 51±7a | 51±4b |
| 46 | Electron transport chain | NADH+0.5 O2→NAD | 250±10 | 230±10 | 270±20c |
| 47 | Electron transport chain | FADH2+0.5 O2→FAD | 210±10 | 170±10a | 180±10 |
| 48 | BSA degradation | →19 amino acids | 47±12 | 22±6a | 36±18 |
| 49 | O2 uptake | O2→ | 250±10 | 220±10a | 240±10c |
| 50 | CO2 output | →CO2 | 280±30 | 250±20 | 320±30c |
| 51 | Acetoacetate output | →Acetoacetate | −2.4±1.1 | −3.5±1.1 | 1.6±1.2b,c |
| 52 | Ornithine output | →Ornithine | 8.5±2.0 | 9.5±1.8 | 15±4c |
| 53 | Ammonia output | →Ammonia | 3.5±0.7 | 7.9±1.3 | 11±2b,c |
| 54 | Alanine output | →Alanine | −1.2±0.7 | 0.53±0.50 | −1.6±1.4 |
| 55 | Cysteine output | →Cysteine | 1.7±0.4 | 0.95±0.08a | 0.97±0.16 |
| 56 | Aspartate output | →Aspartate | 0.086±0.070 | 0.27±0.11 | 0.43±0.28 |
| 57 | Glutamate output | →Glutamate | 7.0±0.8 | 7.5±0.6 | 11±1b,c |
| 58 | Phenylalanine output | →Phenylalanine | −3.4±0.3 | −3.1±0.2 | −3.7±0.2c |
| 59 | Glycine output | →Glycine | −3.1±0.5 | −3.9±0.2a | −6.4±0.5b,c |
| 60 | Histidine output | →Histidine | 0.23±0.58 | −0.93±0.35a | −2.1±0.6b |
| 61 | Isoleucine output | →Isoleucine | 0.32±0.20 | 0.28±0.26 | 1.1±0.6 |
| 62 | Lysine output | →Lysine | −2.2±0.7 | −1.2±0.3 | −2.0±0.9 |
| 63 | Leucine output | →Leucine | 1.5±0.2 | 1.2±0.4 | 2.6±0.7c |
| 64 | Methionine output | →Methionine | −0.88±0.11 | −0.94±0.04 | −1.5±0.1b,c |
| 65 | Asparagine output | →Asparagine | −10±2 | −15±1a | −21±3b,c |
| 66 | Proline output | →Proline | −1.9±0.4 | −2.0±0.2 | −3.8±0.6b,c |
| 67 | Glutamine output | →Glutamine | −25±5 | −34±4a | −35±7 |
| 68 | Arginine output | →Arginine | −17±2 | −17±3 | −31±4b,c |
| 69 | Serine output | →Serine | −7.0±1.2 | −9.0±1.2 | −17±3b,c |
| 70 | Threonine output | →Threonine | 1.8±0.4 | 0.087±0.331a | −0.79±0.45b |
| 71 | Valine output | →Valine | 0.67±0.25 | 0.34±0.19 | 1.6±0.8c |
| 72 | Tyrosine output | →Tyrosine | 1.1±0.4 | 0.23±0.05a | 1.7±0.5c |
n=6 for all groups.
Values reported±standard error.
Sham-CLP values are significantly different from Sham-Sham values.
Burn-CLP values are significantly different from Sham-Sham values.
Burn-CLP values are significantly different from Sham-CLP values.
Table II.
Overall carbon and nitrogen balances across the perfused livers
| Sham-Sham (μmol/h/g) | Sham-CLP (μmol/h/g) | Burn-CLP (μmol/h/g) | |
|---|---|---|---|
| Carbon | −590±60 | −330±50a | −530±80 |
| Nitrogen | −35±16 | −5.7±17 | −1.7±36 |
n=6 for all groups.
Values reported±standard error.
Negative values indicate net release from the perfused livers.
Sham-CLP values are significantly different from Sham-Sham values by unpaired t-test (P< 0.01).
DNA Microarray Analysis
For each condition, whole livers were harvested from anesthetized rats. The livers were quickly rinsed in PBS buffer, wrapped in aluminum foil, and flash frozen in liquid nitrogen. The whole livers were pulverized, and total RNA was isolated from ~50 mg of liver tissue using the Nucleospin II RNA isolation kit from BD Biosciences (Palo Alto, CA). Briefly, the tissue was homogenized in buffer RA1 (supplied by the manufacturer) and β-mercaptoethanol. To clear the homogenate, 70% ethanol was added, and loaded onto a column to retain the RNA on the membrane. After DNase I treatment, the RNA was eluted with 30 μL RNase-free water and the integrity verified on a gel. Twenty micrograms of RNA were labeled and hybridized onto Affymetrix RAE230A chips using standard Affymetrix hybridization protocols as previously described (Vemula et al., 2004). The chips were scaled to a target intensity of 500 to account for the experimental variations between replicates using Affymetrix GENECHIP MAS V.0 software. The entire dataset is available at gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE1781.
The genes involved in the metabolic pathways used for the MFA analysis were searched in the Affymetrix website (www.affymetrix.com) and the corresponding 221 probe set IDs were identified. A search for amino acid transporters retrieved several probes out of which only those expressed in the liver were retained. ESTs were eliminated from the data set unless they represented genes in the metabolic pathways. This resulted in 156 probes that were used, and they are listed in Table IV.
Table IV.
Microarray analysis results.
| Reaction number |
Genbank number |
Gene IDS |
Gene name | Sham-Sham (SS) | Sham-CLP (SC) | Burn-CLP (BC) |
|---|---|---|---|---|---|---|
| 1 | U07993.1 | G6pc | Glucose-6-phosphatase | 11,000±2,000 | 6,500±4,000 | 14,000±4,000 |
| 1 | NM_013098.1 | G6pc | Glucose-6-phosphatase | 16,000±3,000 | 13,000±6,000 | 19,000±4,000 |
| 2 | NM_017006.1 | G6pdx | Glucose-6-P dehydrogenase | 440±170 | 290±80 | 300±120 |
| 3 | AW253884 | Ribose 5-P isomerase | 680±210 | 400±70 | 380±130 | |
| 5 | NM_031811.1 | Taldo1 | Transaldolase | 1,100±100 | 960±100 | 770±110b,c |
| 7 | BI283882 | Gpi | Glucose-6-phosphate isomerase | 1,200±200 | 1,300±200 | 1,600±200b |
| 8 | NM_012558.1 | Fbp1 | Fructose-1,6-biphosphatase | 11,000±1,000 | 12,000±1,000 | 12,000±1,000 |
| 9 | NM_012495.1 | Aldoa | Fructose bisphosphate aldolase a | 1,900±100 | 2,000±100a | 4,200±1,000b,c |
| 9 | M10149.1 | Aldob | Fructose bisphosphate aldolase b | 17,000±1,000 | 20,000±1,000a | 15,000±1,200 |
| 9 | NM_012497.1 | Aldoc | Fructose bisphosphate aldolase c | 50±41 | 23±5 | 29±2 |
| 10 | NM_022215.1 | Gpd3 | Glycerol 3-P dehydrogenase | 1,200±400 | 1,300±200 | 630±200c |
| 10 | NM_024381.1 | Gyk | Glycerol kinase | 1,500±500 | 1,000±500 | 470±60b |
| 11 | NM_017008.1 | Gapd | Glyceraldehyde-P dehydrogenase | 8,900±500 | 9,700±1,100 | 9,500±1,400 |
| 11 | NM_012554.1 | Eno1 | Enolase1 | 3,600±400 | 3,200±100 | 4,500±600b,c |
| 11 | NM_017328.1 | Pgam2 | Phosphoglycerate mutase 2 | 21±15 | 7±1 | 7±1 |
| 11 | NM_053291.1 | Pgk1 | Phosphoglycerate kinase | 50±41 | 29±3 | 24±7 |
| 11 | NM_023964.1 | Gapds | Glyceraldehyde-P dehydrogenase | 370±120 | 280±40 | 280±10 |
| 11 | NM_053290.1 | Pgam1 | Phosphoglycerate mutase 1 | 3,700±200 | 3,900±500 | 4,700±1,100 |
| 11 | NM_053291.1 | Pgk1 | Phosphoglycerate kinase | 2,100±200 | 1,800±200 | 1,900±300 |
| 12 | BI277460 | Pck | Phosphoenolpyruvate carboxykinase | 17,000±2,000 | 17,000±4,000 | 17,000±2,000 |
| 13 | NM_012744.1 | Pc | Pyruvate carboxylase | 1,800±200 | 2,000±100a | 1,700±200c |
| 14 | NM_01702 5.1 | Ldha | Lactate dehydrogenase A | 9,400±1,700 | 7,400±600 | 4,900±1,200b,c |
| 14 | AA848319 | Ldhb | Lactate dehydrogenase B | 300±90 | 260±20 | 360±70 |
| 14 | NM_017266.1 | Ldhc | Lactate dehydrogenase C | 150±50 | 96±13 | 94±27 |
| 15 | NM_130755.1 | Cs | Citrate synthase | 440±140 | 540±100 | 480±90 |
| 16 | NM_024398.1 | Aco2 | Aconitase | 1,800±200 | 1,900±300 | 2,600±500b |
| 16 | NM_053638.1 | Idh3a | Isocitrate dehydrogenase | 170±70 | 190±80 | 310±260 |
| 16 | NM_031510.1 | Idh1 | Isocitrate dehydrogenase | 8,200±1,400 | 8,200±1,200 | 4,100±2,200b,c |
| 16 | BI277627 | Idh3g | Isocitrate dehydrogenase | 740±210 | 650±60 | 740±80 |
| 16 | AI171793 | IDH3B | Isocitrate dehydrogenase | 1,700±100 | 1,700±100 | 1,800±200 |
| 17 | D90401.1 | Afadin | Dihydrolipamide succinyl transferase | 2,100±200 | 2,100±100 | 2,400±300 |
| 17 | AI410493 | Dld | Dihydrolipamide dehydrogenase | 1,400±200 | 1,600±200 | 1,500±100 |
| 18 | NM_053752.1 | Suclg1 | Succinyl-CoA ligase | 3,000±400 | 3,000±200 | 2,700±200 |
| 19 | NM_017005.1 | Fh1 | Fumarase | 4,700±300 | 5,100±300a | 7,500±1,700b |
| 20 | NM_033235.1 | Mdh1 | Malate dehydrogenase | 5,400±100 | 7,300±1,300a | 9,800±1,300b |
| 20 | NM_031151.1 | Mor1 | Malate dehydrogenase | 2,900±100 | 3,000±100 | 2,600±100b,c |
| 21 | NM_019291.1 | Ca2 | Carbonic anhydrase 2 | 230±150 | 290±100 | 220±160 |
| 21 | AB030829.1 | Ca3 | Carbonic anhydrase 3 | 5,800±1,700 | 3,100±2,300 | 590±720b |
| 21 | NM_019174.1 | Ca4 | Carbonic anhydrase 4 | 33±9 | 31±15 | 78±30 |
| 21 | K03040.1 | Otc | Ornithine transcarbamylase | 6,300±1,000 | 7,600±1,000a | 4,600±2,100c |
| 21 | NM_017072.1 | Cps1 | Carbamoyl-P Synthetase | 14,000±2,100 | 18,000±2,100a | 19,000±1,000 |
| 21 | AI408948 | Ca2 | Carbonic anhydrase 2 | 300±70 | 260±10 | 220±40 |
| 21 | NM_019292.1 | Ca3 | Carbonic anhydrase 3 | 1,300±300 | 900±300 | 210±120b,c |
| 21 | NM_019293.1 | Ca5 | Carbonic anhydrase 5 | 660±80 | 970±130 | 810±220 |
| 22 | NM_021577.1 | Asl | Argininosuccinate lyase | 4,300±1,100 | 5,500±700a | 8,600±2,200b |
| 22 | BF283456 | Ass | Argininosuccinate synthase | 22,000±1,000 | 28,000±3,000a | 26,000±2,000b |
| 23 | NM_017134.1 | Arg1 | Arginase 1 | 14,000±1,000 | 20,000±3,000a | 19,000±3,000b |
| 23 | NM_019168.1 | Arg2 | Arginase 2 | 270±90 | 160±10 | 230±50 |
| 24 | NM_031039.1 | Gpt | Alanine transaminase | 2,100±1,000 | 1,900±1,000 | 3,600±800b |
| 25 | NM_053962.1 | Sds | Serine dehydratase | 2,200±1,300 | 4,100±900a | 13,000±4,000b,c |
| 26 | NM_052809.1 | Cdo1 | Cysteine dioxygenase | 13,000±800 | 19,000±3,000a | 18,000±1,000b |
| 27 | AI013458 | Threonine dehydrogenase | 120±120 | 54±39 | 93±12 | |
| 27 | AA849497 | Acetyl-CoA ligase | 2,000±700 | 4,800±300a | 2,200±700c | |
| 28 | NM_133598.1 | Gcsh | Glycine cleavage system | 3,800±300 | 3,900±400 | 2,900±600b,c |
| 28 | AI412012 | Glycine hydroxymethyl transferase | 2,300±500 | 2,700±400 | 2,000±100c | |
| 28 | BF285150 | Glycine hydroxymethyl transferase | 2,100±100 | 1,900±200 | 1,500±400b | |
| 29 | AI176586 | Hibadh | 3-hydroxyisobutyrate dehydrog. (V) | 8,100±500 | 9,300±500a | 7,800±400c |
| 30 | AI502661 | Pccb | Propionyl-CoA carboxylase (I) | 2,000±200 | 2,200±200 | 1,700±400 |
| 31 | NM_024386.1 | Hmgcl | Hydroxymethylglutaryl-CoA lyase (L) | 3,200±500 | 2,700±100 | 2,200±500b |
| 31 | AI102838 | Ivd | Isovaleryl-CoA dehydrogenase (L) | 1,800±300 | 1,700±100 | 1,200±300b,c |
| 32 | AA944898 | Aminoadipate-semialdehyde synthase | 5,300±300 | 8,900±2,800a | 11,000±3,500b | |
| 33 | NM_012619.1 | Pah | Phenylalanine 4-monooxygenase | 10,000±1,000 | 12,000±1,000a | 13,000±2,000 |
| 34 | NM_017181.1 | Fah | Fumarylacetoacetate hydrolase | 7,700±100 | 8,500±400a | 7,500±400c |
| 34 | NM_017233.1 | Hpd | 4-hydroxyphenylpyruvate dioxygenase | 19,000±2,000 | 24,000±2,000a | 22,000±1,000 |
| 34 | M18340.1 | Tat | Tyrosine transaminase | 3,700±1,100 | 7,200±4,600 | 12,000±3,000 |
| 35 | BI284411 | Glud1 | Glutamate dehydrogenase | 5,300±1,100 | 7,000±1,600a | 5,900±100 |
| 35 | AW916644 | Glud1 | Glutamate dehydrogenase | 11,000±2,000 | 13,000±1,000a | 12,000±1,000 |
| 36 | NM_012569.1 | Gls | Glutaminase | 44±11 | 47±37 | 110±60 |
| 36 | M22586.1 | Gls | Glutaminase | 170±130 | 120±60 | 140±10 |
| 36 | J05499.1 | Ga | Glutaminase | 4,800±700 | 5,000±100 | 6,200±500b,c |
| 36 | AI577681 | Gls | Glutaminase | 87±45 | 62±18 | 84±22 |
| 37 | AI411345 | Proline dehydrogenase | 2,000±700 | 2,800±600a | 2,100±200 | |
| 37 | AI233266 | Proline dehydrogenase | 2,700±300 | 3,800±500a | 2,600±400c | |
| 37 | BE113304 | Prodh | Proline dehydrogenase | 22±10 | 19±3 | 18±5 |
| 38 | NM_017159.1 | Hal | Histidine ammonia lyase | 2,300±400 | 2,300±100 | 2,800±500 |
| 39 | AI454484 | Mat1a | Methionine adenosyltransferase 1 | 19,000±1,000 | 25,000±5,000a | 22,000±2,000b |
| 39 | NM_012522.1 | Cbs | Cystathionine beta synthase | 2,500±100 | 3,900±900a | 4,300±1,600 |
| 39 | NM_134351.1 | Mat2a | Methionine adenosyltransferase 2 | 350±80 | 280±20 | 290±20 |
| 41 | AB009372.1 | Asparaginase | 1,100±210 | 1,300±100a | 1,700±600 | |
| 42 | NM_133618.1 | Hadhb | Hydroxyacyl-CoA dehydrogenase | 4,300±900 | 3,400±100 | 2,800±400 |
| 42 | NM_016986.1 | Acadm | Acyl-CoA dehydrogenase medium | 10,000±1,000 | 7,600±900 | 5,100±1,700 ,c |
| 42 | NM_012819.1 | Acadl | Acyl-CoA dehydrogenase long | 6,400±700 | 5,600±800 | 4,100±700 ,c |
| 42 | NM_013200.1 | Cpt1b | Carnitine palmitoyltransferase 1 | 280±40 | 200±50 | 280±60 |
| 42 | D13921.1 | Acat1 | Acetyl-coenzyme A acetyltransferase | 4,000±500 | 3,200±300 | 2,700±800 |
| 42 | NM_022512.1 | Acads | Acyl-CoA dehydrogenase short | 2,200±200 | 2,000±100 | 1,700±200b,c |
| 42 | U88294.1 | Cpt1a | Carnitine palmitoyltransferase 1 | 4,400±2,100 | 2,900±500 | 5,100±1,400 |
| 42 | NM_012891.1 | Acadvl | Acyl-CoA dehydrogenase very long | 3,100±300 | 3,300±100a | 3,200±500 |
| 42 | NM_057107.1 | Facl3 | Fatty acid CoA ligase | 420±90 | 510±20a | 280±80c |
| 42 | NM_013084.1 | Acadsb | Acyl-CoA dehydrogenase short branched |
990±610 | 840±250 | 860±230 |
| 42 | NM_012597.1 | Lipc | Lipase | 4,700±400 | 3,500±1,300 | 1,600±300 ,c |
| 42 | AA800240 | Hadha | Hydroxyacyl-CoA dehydrogenase | 3,200±800 | 2,800±100 | 2,100±300c |
| 42 | D90109.1 | Facl2 | Fatty acid CoA ligase | 12,000±1,000 | 14,000±1,000a | 9,900±2,600c |
| 42 | NM_053607.1 | Facl5 | Fatty acid CoA ligase | 4,000±400 | 6,500±1,000a | 3,900±500c |
| 42 | NM_012930.1 | Cpt2 | Carnitine palmitoyltransferase 2 | 2,700±200 | 1,800±100a | 1,800±700 |
| 42 | NM_031559.1 | Cpt1a | Carnitine palmitoyltransferase 1 | 6,100±1,900 | 4,700±400 | 6,800±1,600 |
| 42 | NM_053623.1 | Facl4 | Fatty acid CoA ligase | 5,200±700 | 6,200±900a | 6,300±100 |
| 42 | BI277523 | Facl2 | Fatty acid CoA ligase | 11,000±1,000 | 13,000±1,000a | 7,300±3,000c |
| 42 | AA891362 | Hadhsc | Hydroxyacyl-CoA dehydrogenase | 140±20 | 69±23 | 96±61 |
| 43 | NM_130433.1 | Acaa2 | Acetyl-CoA C-acyltransferase 2 (I) | 8,200±800 | 8,200±500 | 6,100±1,800 |
| 43 | NM_012489.1 | Acaa1 | Acetyl-CoA C-acyltransferase 1 (I) | 7,900±1,200 | 7,300±700 | 4,800±600b,c |
| 45 | NM_053995.1 | Bdh | b Hydroxybutyrate dehydrogenase | 3,300±1,600 | 1,600±1,000 | 440±620b |
| 54 | NM_017206.1 | Slc6a6 | Taurinebeta-alanine transporter | 270±130 | 250±100 | 360±150 |
| 57 | NM_023972.1 | Eaac1 | Glutamate transport | 550±60 | 620±40a | 640±60 |
| 57 | NM_133554.1 | Slc17a1 | Glutamate transport | 1,200±200 | 1,200±300 | 700±300 |
| 60 | AB026665.1 | Pht2 | Peptide histidine transporter 1 homolog rPHT2 |
110±70 | 130±20 | 110±60 |
| 67 | AI177494 | Hnrpu | System N1; glutamine transport | 1,400±100 | 1,600±200a | 1,900±300 |
| 67 | AI058964 | Hnrpu | System N1; glutamine transport | 640±40 | 700±80 | 870±190 |
| 67 | AF075704.1 | Slc38a1 | Neuronal glutamine transporter | 150±40 | 130±50 | 140±40 |
| 68 | NM_022619.1 | Slc7a2 | Cationic amino acid transporter-2A | 180±60 | 200±40 | 370±100b |
| 18, 46, 47 | NM_130428.1 | Sdha | Succinate dehydrogenase | 3,300±300 | 2,900±400 | 3,200±600 |
| 26, 40 | NM_013177.1 | Got1 | Aspartate aminotransferase 1 | 6,000±400 | 6,800±200a | 5,400±300c |
| 26, 40 | D00252.1 | Got2 | Aspartate aminotransferase 2 | 3,100±400 | 9,200±2,500a | 16,000±2,000 ,c |
| 29, 30, 31 | NM_017253.1 | Bcat1 | Branched chain aminotransferase 1 (V,I,L) |
320±140 | 220±20 | 220±10 |
| 29, 30, 31 | AI102790 | Bcat1 | Branched chain aminotransferase 1 (V,I,L) |
43±8 | 28±10 | 35±8 |
| 29, 30, 31 | J02827.1 | Bckdha | Branched chain keto acid dehydrogenase (V,I,L) |
850±60 | 910±110 | 840±90 |
| 29, 30, 31 | BM385109 | Bckdhb | Branched chain keto acid dehydrogenase (V,I,L) |
2,400±400 | 2,200±100 | 2,100±500 |
| 29, 30, 31 | NM_022400.1 | Bcat2 | Branched chain aminotransferase 2 (V,I,L) |
20±12 | 15±9 | 23±16 |
| 29, 30, 42 | NM_078623.1 | Echs1 | Enoyl-CoA hydratase (V,I) | 2,800±300 | 4,200±300a | 1 3,300±500c |
| 29, 30, 42 | NM_022594.1 | Ech1 | Enoyl-CoA hydratase (V,I) | 7,600±1,800 | 4,800±900 | 4,600±1,500 |
| 30, 42 | AA799574 | Hadhsc | Hydroxyacyl-CoA dehydrogenase | 6,600±700 | 5,900±600 | 3,800±1,500b,c |
| 46, 47 | NM_017311.1 | Atp5g1 | ATP synthase | 3,800±600 | 4,900±200a | 5,000±200 |
| 46, 47 | NM_017202.1 | Cox4a | Cytochrome C oxidase | 9,400±700 | 8,600±900 | 9,000±800 |
| 46, 47 | NM_053756.1 | Atp5g3 | ATP synthase | 9,900±600 | 11,000±1,000a | 11,000±1,000 |
| 46, 47 | NM_019383.1 | Atp5jd | ATP synthase | 4,100±300 | 4,700±200a | 4,900±100 |
| 46, 47 | NM_022503.1 | Cox7a3 | Cytochrome C oxidase | 4,700±400 | 6,000±200a | 6,900±100 |
| 46, 47 | NM_012786.1 | Cox8h | Cytochrome C oxidase | 33±19 | 7±2 | 10±3 |
| 46, 47 | NM_019360.1 | Cox6c | Cytochrome C oxidase | 6,600±900 | 8,800±200a | 8,000±700 |
| 46, 47 | NM_012812.1 | Cox6a2 | Cytochrome C oxidase | 120±60 | 46±22 | 91±18 |
| 46, 47 | NM_053472.1 | Cox4b | Cytochrome C oxidase | 300±140 | 170±40 | 200±20 |
| 46, 47 | NM_019223.1 | Ndufs6 | NADH dehydrogenase | 2,900±500 | 3,100±300 | 2,700±300 |
| 46, 47 | AI170772 | Atp5g2 | ATP synthase | 3,500±900 | 3,300±600 | 3,300±500 |
| 46, 47 | BG666602 | Atp5j | ATP synthase | 2,000±600 | 2300±300 | 2500±100 |
| 46, 47 | M19044.1 | Atp5b | ATP synthase | 14,000±1,000 | 15,000±1,000a | 14,000±1,000 |
| 46, 47 | D13127.1 | Atp5o | ATP synthase | 3,000±200 | 3,800±400a | 3,400±100 |
| 46, 47 | U00926.1 | Atp5d | ATP synthase | 3,000±300 | 3,800±300a | 3,700±200 |
| 46, 47 | AF010323.1 | Atp5e | ATP synthase | 3,900±900 | 4,600±100a | 3,600±700 |
| 46, 47 | J05266.1 | Atp5a1 | ATP synthase | 7,200±600 | 8,500±300a | 7,800±500 |
| 46, 47 | BI282326 | Cox6a1 | Cytochrome C oxidase | 4,100±600 | 4,900±300a | 5,400±700 |
| 46, 47 | X15030.1 | Cox5a | Cytochrome C oxidase | 5,700±400 | 6,000±300 | 7,100±700 |
| 46, 47 | BI275939 | Atp5c1 | ATP synthase | 6,200±200 | 6,400±100a | 6,700±600 |
| 46, 47 | M22756.1 | Ndufv2 | NADH dehydrogenase | 3,100±700 | 3,500±100 | 3,300±100c |
| 46, 47 | BI395849 | Cox5a | Cytochrome C oxidase | 440±110 | 500±40 | 610±150 |
| 46, 47 | NM_053586.1 | Cox5b | Cytochrome C oxidase | 5,200±700 | 6,100±200a | 6,500±200 |
| 46, 47 | NM_012985.1 | Ndufa5 | NADH dehydrogenase | 240±120 | 440±60a | 390±80 |
| 46, 47 | NM_080481.1 | Atp5k | ATP synthase | 1,600±200 | 1,600±300 | 1,400±200 |
| 46, 47 | BG673321 | Cox8a | Cytochrome C oxidase | 7,600±1,700 | 9,300±1,000a | 9,300±700 |
| 46, 47 | AA893531 | Atp5f1 | ATP synthase | 6,800±1,000 | 7,300±400 | 7,700±500 |
| 5, 6 | BG667093 | Tkt | Transketolase | 22±14 | 10±1 | 13±1 |
| 5, 6 | NM_022592.1 | Tkt | Transketolase | 1,400±100 | 1,400±300 | 1,000±100 |
| 52, 54, 55, 59-71 | NM_130748.1 | Ata3 | Amino acid transport system A3 | 3,900±600 | 7,200±800 | 9,300±600 |
| 56, 57 | AF265360.1 | Slc1a3 | GLAST-1a | 190±50 | 130±50 | 170±40 |
| 56, 57 | NM_019225.1 | Slc1a3 | Solute carrier family 1, member 3 | 90±62 | 83±10 | 100±20 |
| 56, 57 | X63744.1 | Slc1a3 | Glutamate aspartate transporter | 280±120 | 170±40 | 180±60 |
| 57, 59 | AF276870.1 | System N2; glycine and glutamate | 250±40 | 210±70 | 250±30 | |
| 9, 10 | NM_022922.1 | Tpi1 | transport Triose P-Isomerase |
2,700±100 | 3,100±100 | 3,300±200 |
n = 3 for all groups.
Values reported±standard error.
Sham-CLP values are significantly different from Sham-Sham values.
Burn-CLP values are significantly different from Sham-Sham values.
Burn-CLP values are significantly different from Sham-CLP values.
RT-PCR Analysis of Selected Genes
The mRNA sequences for eight genes involved in metabolism (Ata3, Taldo1, Got2, Lipc, Sds, Arg1, Idh1, Pc) and 18S rRNA (housekeeping gene) were retrieved from the GenBank database and gene specific primers were designed for each transcript. RNA was extracted from ~50 mg of whole liver tissue, prepared as described above. RT-PCR was performed with ~100 ng of total RNA using the Superscript II one-step RT-PCR kit (Invitrogen, Carlsbad, CA) on a icycler real-time PCR machine (Bio-Rad, Hercules, CA). The cycle number at which the fluorescence in each amplification reaction increased beyond a threshold (in the exponential phase of amplification) was determined using the MyiQ software (Bio-Rad). Threshold cycle numbers for each gene was normalized to that of 18S rRNA as described earlier (Jayaraman et al., 2000). The fidelity of the amplified product was also verified on a 2% agarose gel.
Statistics
Weight loss data were compared pair-wise each day using 1-way ANOVA, and statistical significance was determined using post-hoc Tukey’s test with a 95% confidence level. The 72 fluxes quantified by the MFA and the 156 relevant probes from the microarray analysis were compared between control and experimental groups and the differences tested for statistical significance using significance analysis of microarrays (SAM) (Tusher et al., 2001), which uses a modified t-test and corrects for multiple testing using FDR (False discovery rate) approaches. By setting the FDR empirically at 10%, we identified 35 metabolic fluxes and 62 probe sets on the microarray chip to be significantly altered. To analyze the correlation between changes in gene expression and fluxes, each flux and gene expression change was labeled “ −1, 0, or 1” to indicate direction of change, that is, significant decrease, no significant change, or significant increase. Flux and gene expression changes were compared using the Mann-Whitney test, and P < 0.05 was considered a significant difference.
Results
Evaluation of the Burn and CLP Models
To assess the catabolic impact of the dual insult model, total body weight was monitored after administration of injury for 10 consecutive days (Fig. 1). Although blood samples were not taken for the measurement of bacteria types and levels, it has previously been shown that single needle punctures during CLP, using a range of different sized needles, always produced systemic bacteremia (Otero-Anton et al., 2001).
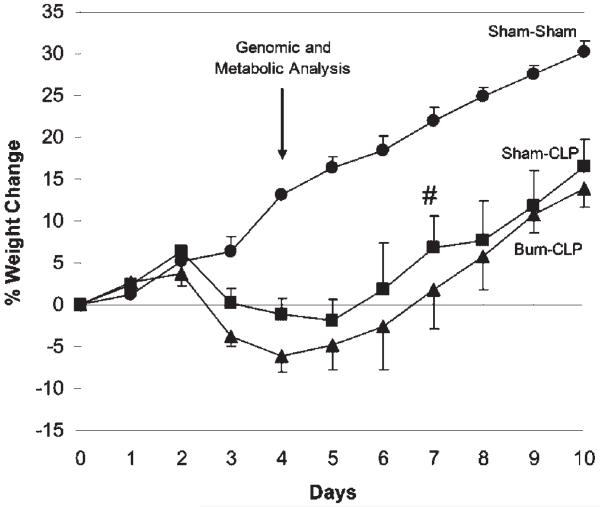
Figure 1.
Average percent weight changes following Sham-Sham (●), Sham-CLP (■), and Burn-CLP (▲) treatment. On post-burn day 7, two Sham-CLP rats died (#). n = 6 for all groups. Weights for the Sham-CLP and Burn-CLP groups were significantly different from the Sham-Sham controls starting on post-burn day 3 until the end of the experiment. The liver perfusions and microarray analyses were performed on post-burn day 4, as shown by the arrow.
The CLP procedure following sham (Sham-CLP) or burn (Burn-CLP) treatment resulted in a statistically significant weight loss compared to the sham-treated animals (Sham-Sham) from post-burn day 3 (i.e., 1 day after CLP treatment) onwards. The Burn-CLP group lost slightly more weight than the Sham-CLP group, although this difference was not statistically significant. The peak of the response occurred around post-burn day 4; consequently, livers were isolated at that time point for all subsequent metabolomic and genomic studies.
MFA and Microarray Analysis
The fluxes of 28 metabolites were measured across the perfused livers (Table I). As observed in prior studies, metabolites were found to change linearly with time, indicating that the perfused liver was metabolically stable for the duration of the 1 h perfusion period (Banta et al., 2005; Dahn et al., 1992; Lee et al., 2000, 2003; Mortimore and Surmacz, 1984; Yamaguchi et al., 1997). Examples of the oxygen uptake rates, urea production, glutamate production, and arginine consumption can be found in Figure 2. As described in the Materials and Methods section, an overall carbon and nitrogen balance using the measured metabolites across the perfused livers (Table II) suggests that protein degradation is occurring in the liver (Reaction #48) as well as the oxidation of stored fatty acids (Reaction #42) and glycerol (Reaction #10). The final results of the MFA can be found in Table III.
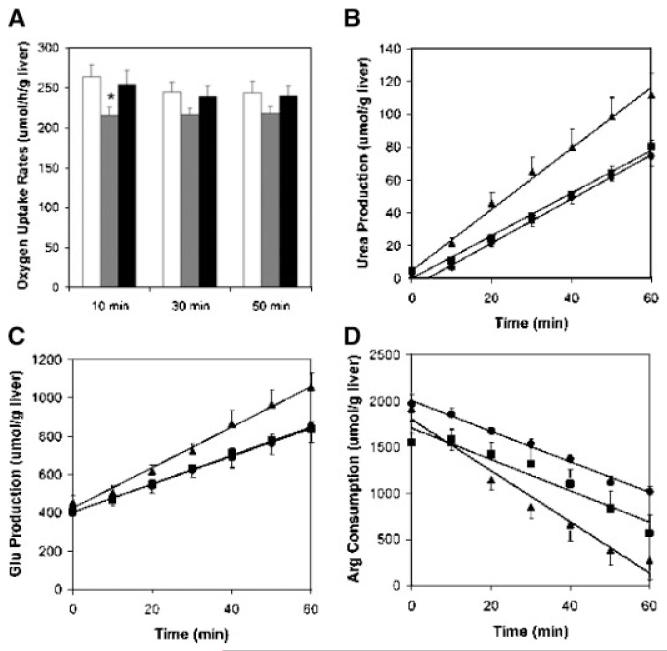
Figure 2.
Examples of averaged measured metabolic flux data across the perfused livers. Error bars represent ± SE, and n 6 for all measurements. A: Oxygen uptake rates measured at 10, 30, and 50 min into the liver perfusions for the Sham-Sham (white), Sham-CLP (gray), and Burn-CLP (black) groups.=There was not statistically significant difference between time points for any of the groups. At 10 min, the Sham-CLP oxygen uptake rate was statistically significantly lower than the Sham-Sham oxygen uptake rate (marked by *P < 0.05). B: Urea production by the perfused livers from the Sham-Sham (●), Sham-CLP (■), and Burn-CLP(▲) groups. C: Glutamate production by the perfused livers from the Sham-Sham (●), Sham-CLP (■), and Burn-CLP(▲) groups. D: Arginine uptake by the perfused livers from the Sham-Sham (●), Sham-CLP (■), and Burn-CLP(▲) groups. The concentrations were normalized to the weights of the perfused livers. The best-fit lines to the averaged data are shown, and the slope is used to obtain the metabolic flux.
The application of the MFA resulted in a protein degradation term that caused a release of the branched chain amino acids that was larger than the measured fluxesacross the livers. This could be a result of the fact that proteins other than BSA are being degraded, and thus the assumed ratio of the branched chain amino acids in the model protein is incorrect. Or, it could be that the liver is metabolizing the branched chain amino acids as they are released. It is well known that almost all branched chain amino acid oxidation occurs extrahepatically, but it has also been observed that isolated rat hepatocytes (Hutson et al., 1992) and whole rat liver tissue preparations (Shinnick and Harper, 1976) are able to oxidize the branched chain amino acids to some degree. Therefore the reactions for the metabolism of the branched chain amino acids were included in the metabolic network model (reactions 29-31). As expected, the fluxes through these reactions were found to be very small (Table III).
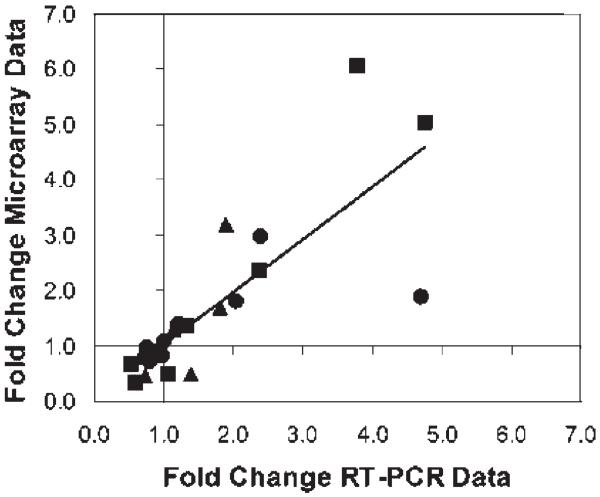
The microarray analysis was performed on liver tissue (Table IV) from animals that were treated identically to the animals used in perfusion studies, except the livers were never perfused. Microarray analysis using the Affymetrix RAE230A chips provides expression data on a large number of genes beyond what is used in the MFA analysis. In order to make a meaningful comparison, the available genes on thechip were searched for their relevance to metabolism, and only corresponding genes to the pathways in the MFA model were used. In the situation where multiple genes were identified for a given pathway, all of the genes were included in Table IV. RT-PCR was used to validate the microarray analysis results for several selected genes (Table V). The RT-PCR results were found to be very similar to the microarray results (Fig. 3)
Table V.
RT-PCR analysis of selected genes used in the microarray analysis.
| Gene name | Gene ID | Reaction number (Table III) |
Sham-CLP vs. Sham-Sham (fold- difference) |
Burn-CLP vs. Sham-Sham (fold- difference) |
Burn-CLP vs. Sham-CLP (fold- difference) |
|---|---|---|---|---|---|
| Transaldolase | Taldo1 | 5 | 0.97±0.11 | 0.52± 0.02 | 0.67±0.06 |
| Pyruvate carboxylase | Pc | 13 | 0.99± 0.08 | 0.89±0.08 | 0.85±0.10 |
| Isocitrate dehydrogenase | Idh1 | 16 | 0.75±0.05 | 1.1±0.1 | 1.4=1=0.1 |
| Arginase 1 | Arg1 | 23 | 1.2±0.1 | 1.3±0.1 | 0.96±0.07 |
| Serine dehydratase | Sds | 25 | 4.7±0.9 | 3.8±0.6 | 1.9±0.2 |
| Aspartate aminotransferase 2 | Got2 | 26, 40 | 2.4±0.1 | 4.8±0.1 | 1.8=1=0.1 |
| Lipase | Lipc | 42 | 0.78±0.07 | 0.58±0.10 | 0.72±0.07 |
| Amino acid transport system A3 | Ata3 | 52, 54, 55, 59-71 | 2.0±0.1 | 2.4±0.2 | 1.2=1=0.1 |
n=3 for all measurements.
Values reported±standard error.
Values less than 1.0 indicate down-regulation and values greater than 1.0 indicate up-regulation of the genes.
Figure 3.
Comparison of Microarray and RT-PCR data. The fold-differences between RT-PCR data from Table V are compared to the fold-difference values calculated from the DNA microarray data in Table IV. Sham-CLP versus Sham-Sham (●), Burn-CLP versus Sham-Sham (■), and Burn-CLP versus Sham-CLP (▲) comparisons are indicated. Values of fold-difference greater than 1.0 show up-regulation while values less than 1.0 indicate down-regulation. The best-fit line for all of the data combined has a slope of 0.95.
Effect of the Dual Injury Model on Hepatic Metabolism
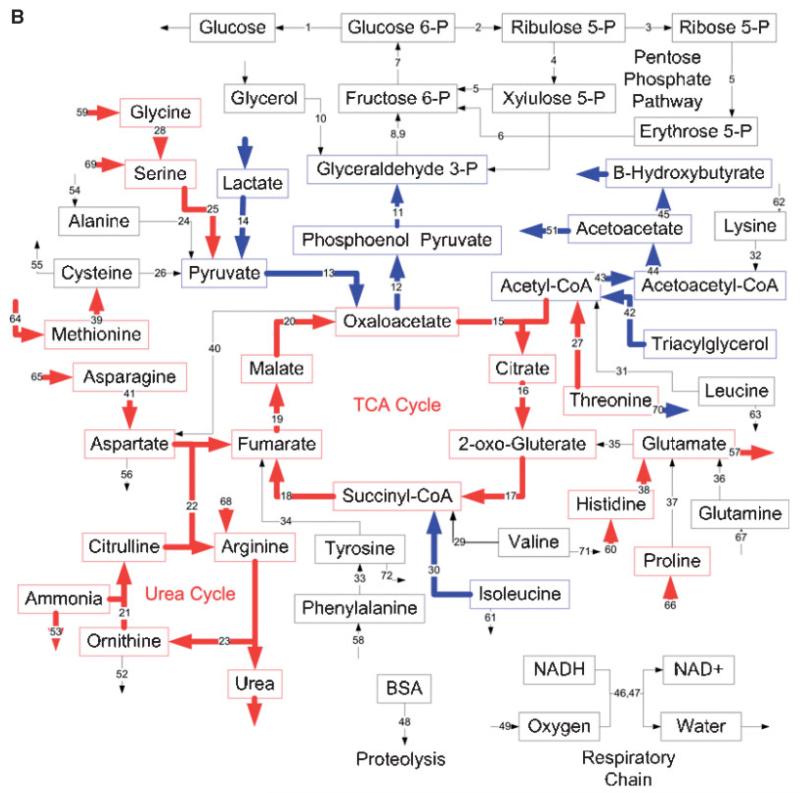
The overall impact of the dual insult was determined by comparing the results obtained from the Burn-CLP group to the Sham-Sham group. Figure 4A and B show this comparison for gene expression data and metabolic fluxes, respectively. Burn-CLP up-regulated mRNA levels for genes involved in the urea cycle, the respiratory chain, gluconeo-genesis, and the metabolism of several amino acids, such as alanine, serine, cysteine, methionine, glutamine, phenyla-lanine, tyrosine, and lysine. Furthermore, the specific transporters for glutamine and arginine, and the neutral and cationic amino acid Ata3 transporter (contributes to transport of all of the amino acids except glutamate, aspartate, phenylalanine, tyrosine, and tryptophan) had increased mRNA levels. A down-regulation was observed for the genes involved in ketone body production, the metabolism of lactate, glycine, isoleucine, and leucine, and some of the enzymes in the pentose phosphate pathway. Mixed results were observed for the genes involved in glycerol metabolism, the citric acid cycle, and β-oxidation.
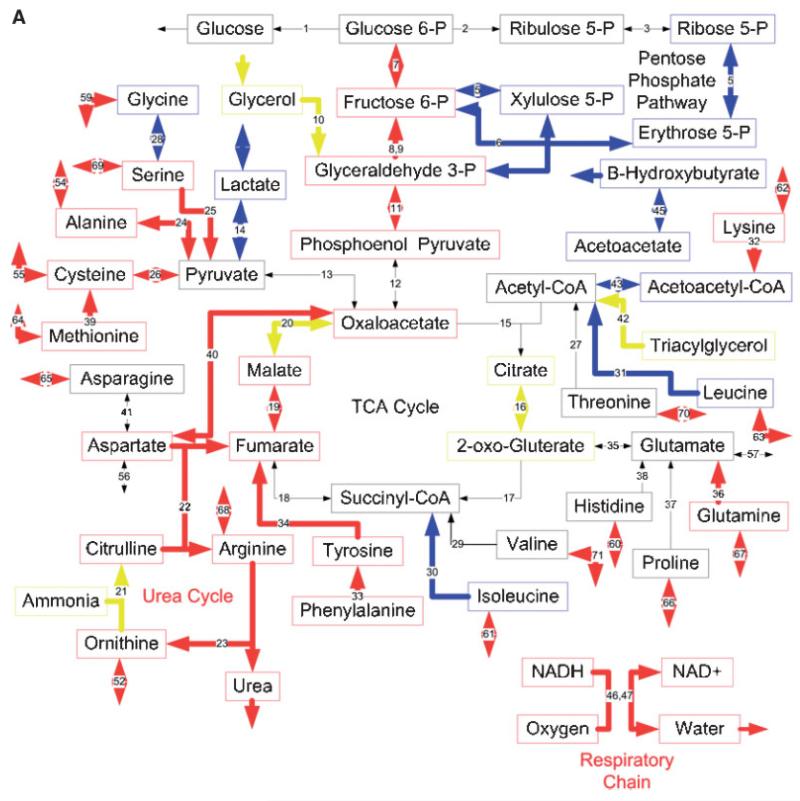
Figure 4.
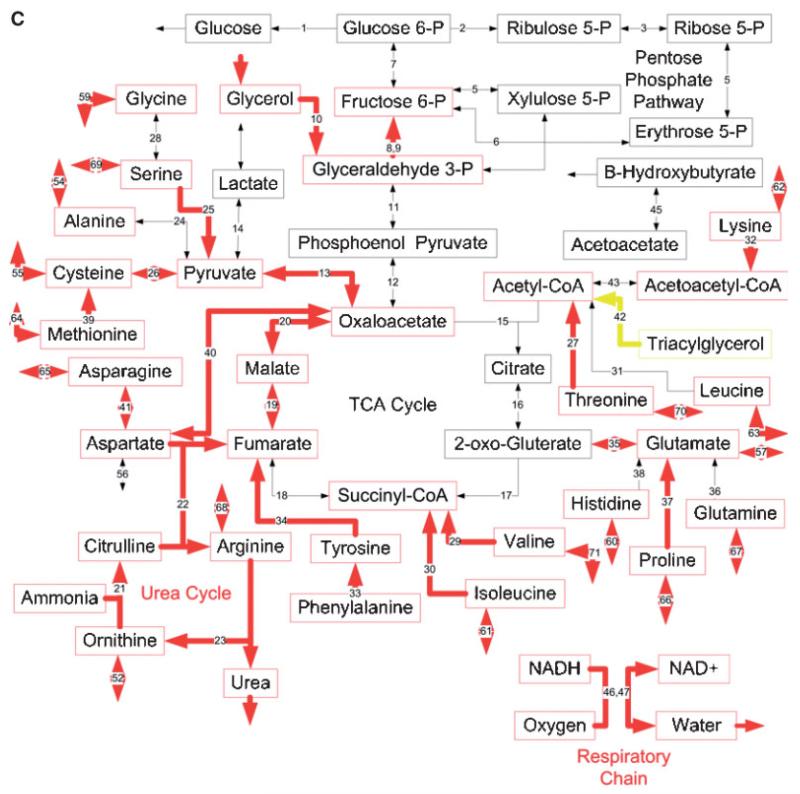
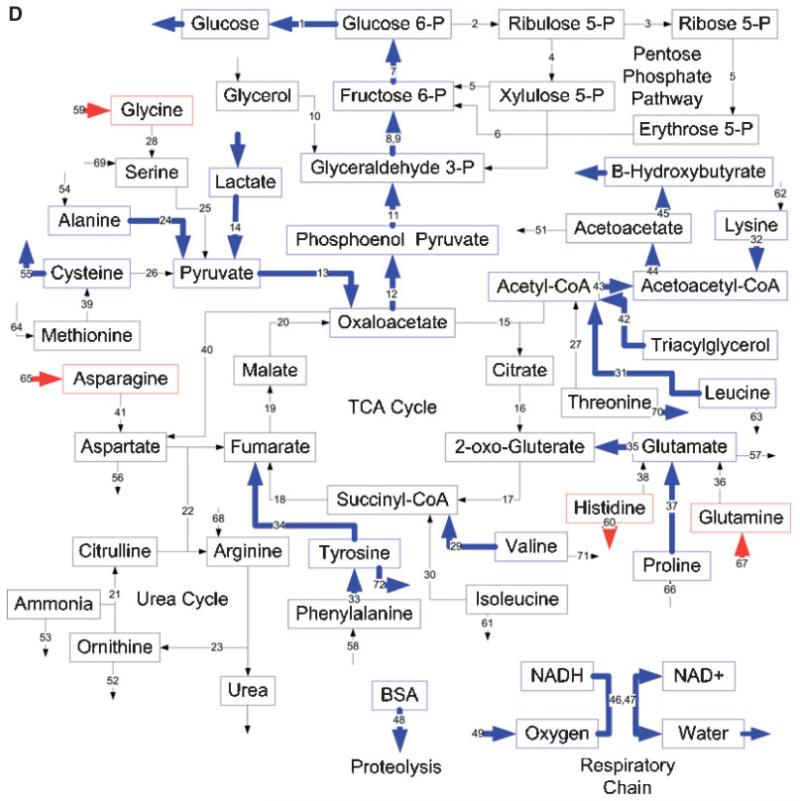
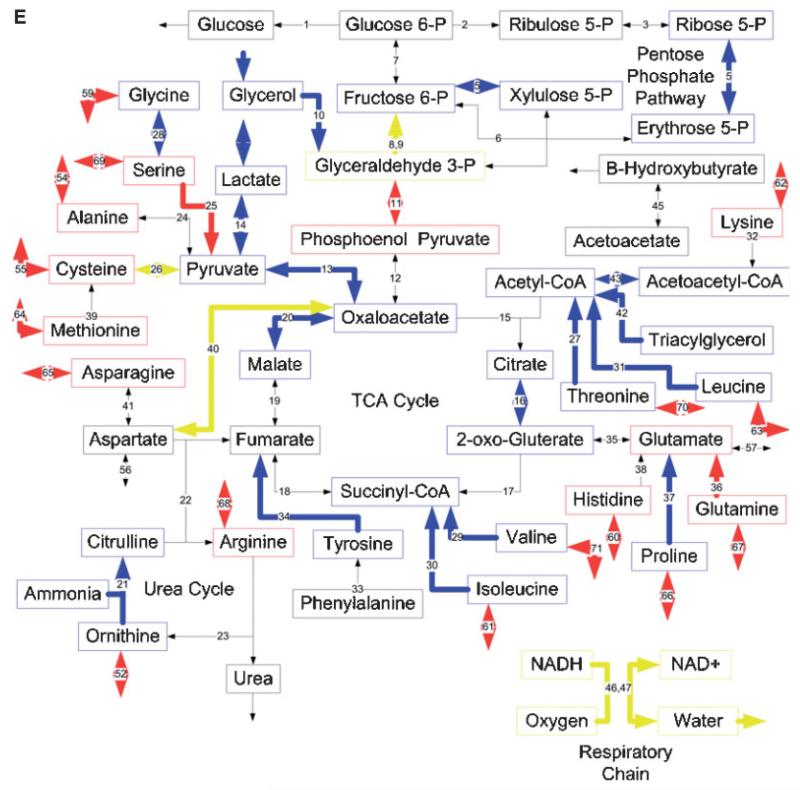
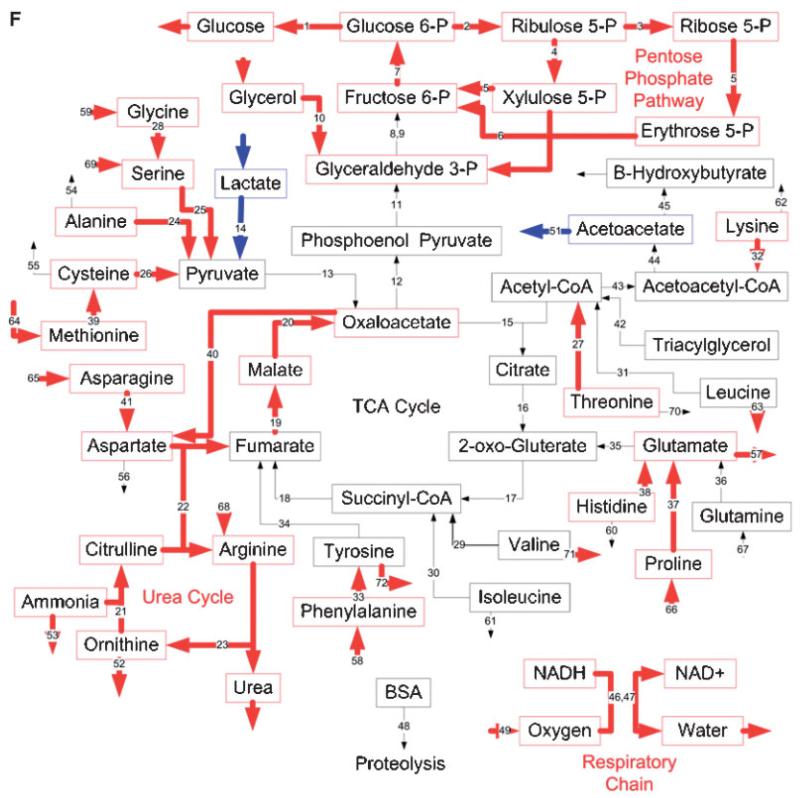
Effects of burn injury and CLP on the gene expression pattern and metabolic fluxes in the rat liver. A and B: Comparison between the Burn-CLP versus Sham-Sham groups using the DNA microarray and metabolic flux data, respectively. C and D: Comparison between the Sham-CLP versus Sham-Sham groups using the DNA microarray and metabolic flux data, respectively. E and F: Comparison between the Burn-CLP versus Sham-CLP groups using the DNA microarray and metabolic flux data, respectively. In panels A, C, and E, arrows in red indicate increased gene expression, blue indicates decreased gene expression, and yellow indicates mixed results. (“Mixed results” means that several genes on the array correspond to that particular pathway and changed in opposite directions.) In panels B, D, and F, arrows in red indicate increased metabolic flux, and blue indicates decreased metabolic flux.
MFA results (Fig. 4B) show several changes that are in qualitative agreement with gene expression data, such as increased urea cycle fluxes, increased metabolism of serine and methionine, decreased metabolism of lactate and isoleucine, and decreased production of ketone bodies. However, many changes in fluxes were not reflected in the DNA microarray data, such as increased metabolic fluxes through the citric acid cycle, and increased metabolism of glycine, histidine, asparagine, threonine, and the release of glutamate. In addition, decreases in β-oxidation fluxes, and the initial steps of gluconeogenesis were not seen at the gene expression level. The gene expression changes seen in the respiratory chain and the pentose phosphate pathway were not observed in the MFA results. A semi-quantitative Venn diagram shows that overall, about 1/3 of gene expression changes and flux changes were in agreement (Fig. 5A). Gene expression and flux changes were not significantly different (P = 0.2 per the Mann-Whitney test).
Figure 5.
Semi-quantitative Venn diagrams showing the overlap between the MFA and DNA microarray results described in Figure 4. The left hand column shows the number of pathways that were up-regulated based on gene expression (black) or metabolic fluxes (white). The right hand column shows the number of pathways that were down-regulated based on gene expression (black) or metabolic fluxes (white). The overlap areas show the number of pathways that had consistent changes in gene expression and metabolic flux (i.e. both up-regulated or both down-regulated). A: Burn-CLP versus Sham-Sham. B: Sham-CLP versus Sham-Sham. C: Burn-CLP versus Sham-CLP. In cases of mixed microarray results, (for example, respiratory chain enzymes in Fig. 4E), the pathway under consideration was assigned to the “up-regulated” or “down-regulated” category based on whether the largest number of genes on the microarray chip relevant to that pathway were up- or down-regulated.
Effect of CLP Treatment Alone
The Sham-CLP group was compared to the Sham-Sham treated group to determine the consequences of CLP alone. The DNA microarray results (Fig. 4C) show up-regulated mRNA levels for many genes, and no down-regulated genes. Some of the up-regulated genes were the same that were found up-regulated in the Burn-CLP group versus Sham-Sham comparison (Fig. 4A), including the urea cycle, the respiratory chain, the metabolism of several amino acids, including serine, cysteine, methionine, tyrosine, and lysine, the specific transporters for glutamine and arginine, and the neutral and cationic amino acid Ata3 transporter. In addition, other up-regulated genes included glycerol metabolism, the citric acid cycle, and the metabolism of a few more amino acids (aspartate, asparagine, valine, isoleucine, proline, glutamate, and threonine). As in the Burn-CLP group versus Sham-Sham comparison, the β-oxidation pathway showed mixed results.
MFA data (Fig. 4D) showed a dramatically different picture, with mostly decreased fluxes, including the respiratory chain, gluconeogenesis, lactate metabolism, β-oxidation, ketone body production, and proteolysis. The metabolism of most amino acids was affected, with a decrease in the metabolism of alanine, lysine, leucine, glutamate, proline, valine, tyrosine, and phenylalanine. In addition, there was an increase in the uptake of asparagine, glutamine, histidine and glycine, as well as a decrease in the release of cysteine and tyrosine. A few of these changes, namely lactate metabolism, the early steps in gluconeogenesis, β-oxidation and β-hydroxybutyrate production were previously observed in the Burn-CLP group versus Sham-Sham comparison (Fig. 4B).
Looking at the overall picture, very few gene expression and metabolic flux changes were in agreement (Fig. 5B), andthis difference was highly significant (P < 0.001 per Mann-Whitney test). Furthermore it is noteworthy that almost all up-regulated pathways changed at the gene expression level without a corresponding change in flux, while down-regulated pathways changed with respect to flux, but not gene expression.
Effect of a Priming Burn Injury Before CLP Treatment
The Burn-CLP group was compared to the Sham-CLP group to assess the effect of the burn injury before the CLP insult. The DNA microarray analysis (Fig. 4E) shows that the priming burn causes an increase in the expression of genesinvolved in the metabolism of serine as well as the Ata3 cationic amino acid transporter. A decrease was observed in the genes involved in the metabolism of glycine, lactate, isoleucine, proline, threonine, tyrosine, leucine, valine, glycerol, β-oxidation, and parts of the urea cycle, the citric acid cycle, and the pentose phosphate pathway. Mixed results were observed with the genes involved in gluconeo-genesis, the metabolism of aspartate, cysteine, valine, and the respiratory chain. Few of these changes were reflected in the Burn-CLP group versus Sham-Sham comparison (Fig. 4A), since the only common flux changes were leucine, isoleucine, glycine, serine, and lactate metabolism, as well as a few fluxes in the pentose phosphate pathway.
Flux data (Fig. 4F) showed very different results from the corresponding DNA microarray data. There was increased glucose production, pentose phosphate pathway fluxes, glycerol metabolism, urea cycle, and the respiratory chain, and decreased lactate metabolism. Amino acid metabolism was also affected, with an increased uptake of glycine, serine, methionine, asparagine, arginine, proline, and phenylalanine, increased release of valine, tyrosine, ornithine, and leucine, and increased metabolism of lysine, alanine, glycine, serine, cysteine, methionine, threonine, asparagine, histidine, proline, and phenylalanine. Interestingly, flux increases in the urea cycle, aspartate, and asparagine metabolism, as well as glycine, serine, methionine, and threonine metabolism, which were not seen in the Sham-CLP versus Sham-Sham comparison (Fig. 4D) were reflected in the Burn-CLP versus Sham-Sham comparison (Fig. 4B).
Figure 5C shows the overall correspondence between gene expression and metabolic flux change. More than half of the up-regulated genes were associated with an increased flux, although the majority of up-regulated fluxes did not exhibit up-regulated gene expression. The majority of the down-regulated pathways had reduced mRNA levels without decreases in their corresponding fluxes. Overall, flux and gene expression changes were significantly different (P < 0.001 per Mann-Whitney test).
Discussion
The purpose of this study was to characterize the effect of a dual insult consisting of a burn injury with subsequent bacterial infection through CLP on liver metabolism and to investigate the potential contribution of gene expression to the observed intrinsic metabolic flux changes. Liver metabolism was characterized using MFA, which uses a large set of metabolite measurements with a metabolic network model to provide an integrated view of the metabolic flux map within the tissue. Gene expression was analyzed using DNA microarrays, which provide a snap-shot of the gene expression pattern for the enzymes in the hepatic metabolic network. We found that burn plus CLP significantly altered fluxes through most pathways involved in central carbon and amino acid nitrogen metabolism in the liver. Only about 1/3 of these changes − 3/4 of which were increases—were qualitatively reflected at the gene expression level. CLP alone had a much different effect on hepatic metabolism and gene expression than burn plus CLP, causing increases in gene expression of metabolic enzymes while decreasing metabolic fluxes. The priming burn injury prior to CLP decreased the expression of many genes, but increased a large number of fluxes.
The use of metabolomics and genomics techniques provided a global picture of the consequences of the dual insult model on the major metabolic pathways in the rat liver. The two methods are complementary because gene expression data can be used to determine which enzymatic pathways are present in the tissue, and this information can be used to build a relevant hepatic metabolic network. For example, a previous study using isolated liver cells found that several genes were correlated or anti-correlated with fluxes in response to a glutamine withdrawal challenge (Wong et al., 2004). However, the data presented here indicate that single time point DNA microarray analysis cannot be used to predict metabolic fluxes through the metabolic network of whole perfused livers.
There are several possible explanations for the observed lack of correlation. mRNA transcripts are often subject to post-transcriptional regulation, and this can decrease the correlation between the mRNA abundance, protein expression (Anderson and Seilhamer, 1997) and in turn, enzymatic activity. In particular, the dynamics of gene expression may differ from enzyme protein levels due to the time required for gene transcription, and this should be addressed by timecourse studies in the future. Fluxes may also be altered in response to enzyme activity changes that are controlled through post-translational mechanisms, such as allosteric modulation. Furthermore, mass action effects are another potential factor that could affect fluxes through the metabolic reactions. For example, our Sham-CLP versus Sham-Sham comparison shows that fluxes are going down while gene expression is going up (Fig. 4C and D). A possible explanation would be lack of substrate availability, which would cause fluxes to go down, even when the metabolic machinery is upregulated. Interestingly, the priming burn (Burn-CLP versus Sham-CLP comparison) had a somewhatopposite effect, where fluxes were largely increased while gene expression data is evenly distributed between up-regulated and down-regulated genes (Fig. 4E and F).
Other explanations for the observed discrepancies may have a physiological basis. Conceivably, burn injury prior to CLP treatment could increase substrate availability to the liver, and this may explain the much higher fluxes observed in the Burn-CLP model compared to the Sham-CLP model. Consistent with this hypothesis, prior studies do indicate that burn injury significantly up-regulates amino acid transport mechanisms in the liver (Lohmann et al., 1998; Pawlick et al., 2000). However, CLP alone has also been shown to increase amino acid transport to the liver, at least at shorter time points (<24 h) post-CLP (Hasselgren et al., 1986). CLP has also been shown to reduce the hepatic energy charge due to decreased function of mitochondria (Chen et al., 2004; Hampton et al., 1987; Hsieh et al., 2004). Since many transport mechanisms indirectly derive their energyfrom ATP hydrolysis, it is plausible that the effectiveness of substrate transport processes is reduced after CLP. Further studies to clarify these potential mechanisms are warranted.
Prior studies have also suggested that in a hypermetabolic state, a large flux of amino acids is generated from the catabolism of the skeletal muscle (Banta et al., 2004; Demling and Seigne, 2000). Both Sham-CLP and Burn-CLP treatment induced delayed gain and even loss of weight (Fig. 1). The liver is the main site for urea synthesis, and increases in amino acid metabolism and urea cycle activity in the liver could negatively impact on the overall nitrogen balance. Interestingly, the Burn-CLP model showed elevated urea cycle activity and amino acid degradation both at the level of gene expression and metabolic fluxes (Fig. 4A and B), which suggests that in this model there is accelerated conversion of amino acid nitrogen into urea. It is plausible that these metabolic changes in liver contribute to the loss of body mass in this model. Sham-CLP treatment also induced significant weight loss, although slightly less than Burn-CLP. Compared to the Sham-Sham group, gene expressionchanges also indicate up-regulated urea cycle activity and amino acid metabolism (Fig. 4C); however, this was clearly not reflected at the level of fluxes (Fig. 4D).
Another major metabolic feature of systemic inflammation and hypermetabolism is elevated serum lactate levels, which increase in proportion with the severity of injury and are correlated with poor prognosis (Jeng et al., 2002). Hyperlactatemia is thought to be due to increased glycolysis at the wound site and in skeletal muscle. This is typically accompanied by increased lactate conversion into glucose by the liver to complete the Cori cycle (Dahn et al., 1995; Demling and Seigne, 2000). Our data show that the Sham-Sham control group was gluconeogenic (reactions 1, 7-12 in the network) with a net uptake of lactate and conversion to pyruvate (reaction 14). This was expected, because the livers were perfused after an overnight fast, and the fluxes therefore reflect fasting-induced gluconeogenesis. No change in glucose production, although a decrease in fluxes through the early steps of gluconeogenesis (lactate conversion into phosphoenolpyruvate via oxaloacetate), occurredas a result of Burn-CLP treatment. Sham-CLP treatment caused a more clear-cut decrease in fluxes through the gluconeogenic pathway. Prior studies have documented that CLP treatment can lead to a decrease in the activity of certain gluconeogenic enzymes, such as phosphoenolpyruvate carboxykinase (Deutschman et al., 1995). The priming burn may partially protect from the effect of CLP by inducing “endotoxin resistance” (Clancy et al., 1997). Thus, the lack of an increase in gluconeogenic fluxes in perfused livers from Burn-CLP and Sham-CLP animals is more likely due to the fact that lactate levels in the perfusate medium were not increased relative to the Sham-Sham perfusions. In addition, we cannot exclude the possibility that the fasting pre-conditioning could mask any stress-mediated induction of gluconeogenesis. Further liver perfusion studies that include non-fasted animals, and an injury-like substrate load and hormonal milieu in the perfusate, will be needed to help delineate the role of factors external to the liver.
In summary, we show that a moderate burn injury followed by CLP 2 days later induces transient weight loss inrats with eventual recovery on post-burn day 5. Evaluation of liver metabolism in this animal model of burn sepsis on post-burn day 4 (2 days after CLP) shows increased TCA and urea cycle activities, as well as up-regulated amino acid metabolism. Genomic analysis shows that only about 1/3 of these changes were consistent with gene expression changes. CLP alone had a much different effect on hepatic metabolism and gene expression than burn plus CLP, causing increases in gene expression of metabolic enzymes while decreasing metabolic fluxes. The priming burn injury prior to CLP decreased the expression of many genes, but increased a large number of fluxes.
Overall, these data show that mRNA changes measured at a single time point by DNA microarray analysis do not reliably predict metabolic flux changes through the metabolic network of whole perfused livers. This result may have large implications, as many authors use DNA microarray results to infer metabolic status under differentphysiological states in a variety of systems. Our work shows that drastic changes in metabolism can be measured with or without corresponding changes in the gene expression.
Acknowledgments
We thank Mr. Z Kelley for his excellent technical assistance. This work was partly carried out in the Boston Shriners Burns Hospital Special Shared Facility for Genomics and Proteomics.
Contract grant sponsor: National Institutes of Health
Contract grant number: RO1 DK 59766
Contract grant sponsor: Shriners Hospitals for Children
Footnotes
Scott Banta and Murali Vemula contributed equally to this work.
Scott Banta’s present address is Department of Chemical Engineering, Columbia University, 500 W. 120th St., New York, NY 10027.
References
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18(3-4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Arai K, Lee K, Berthiaume F, Tompkins RG, Yarmush ML. Intrahe-patic amino acid and glucose metabolism in a D-galactosamine-induced rat liver failure model. Hepatology. 2001;34(2):360–371. doi: 10.1053/jhep.2001.26515. [DOI] [PubMed] [Google Scholar]
- Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc Natl Acad Sci USA. 2001;98(21):12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta S, Yokoyama T, Berthiaume F, Yarmush ML. Quantitative effects of thermal injury and insulin on the metabolism of the skeletal muscle using the perfused rat hindquarter preparation. Biotechnol Bioeng. 2004;88(5):613–629. doi: 10.1002/bit.20258. [DOI] [PubMed] [Google Scholar]
- Banta S, Yokoyama T, Berthiaume F, Yarmush ML. Effects of dehydroepiandrosterone administration on rat hepatic metabolism following thermal injury. J Surg Res. 2005;127(2):93–105. doi: 10.1016/j.jss.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Chan C, Berthiaume F, Lee K, Yarmush ML. Metabolic flux analysis of cultured hepatocytes exposed to plasma. Biotechnol Bioeng. 2003a;81:33–49. doi: 10.1002/bit.10453. [DOI] [PubMed] [Google Scholar]
- Chan C, Berthiaume F, Lee K, Yarmush ML. Metabolic flux analysis of hepatocyte function in hormone- and amino acid-supplemented plasma. Metab Eng. 2003b;5:1–15. doi: 10.1016/s1096-7176(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Chatziioannou A, Palaiologos G, Kolisis FN. Metabolic flux analysis as a tool for the elucidation of the metabolism of neurotransmitter glutamate. Metab Eng. 2003;5:201–210. doi: 10.1016/s1096-7176(03)00029-6. [DOI] [PubMed] [Google Scholar]
- Chen HW, Kuo HT, Lu TS, Wang SJ, Yang RC. Cytochrome c oxidase as the target of the heat shock protective effect in septic liver. Int J Exp Pathol. 2004;85:249–256. doi: 10.1111/j.0959-9673.2004.00393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, Huber-Lang M, Kumar-Sinha C, Barrette TR, Shankar-Sinha S, Sarma VJ, Padgaonkar VA, Ward PA. Molecular signatures of sepsis: Multiorgan gene expression profiles of systemic inflammation. Am J Pathol. 2001;159(4):1199–1209. doi: 10.1016/S0002-9440(10)62505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy KD, Lorenz K, Hahn E, Christiansen B, Hofmann C, Gamelli RL. Down-regulation of tissue specific tumor necrosis factor-α in the liver and lung after burn injury and endotoxemia. J Trauma. 1997;42:169–176. doi: 10.1097/00005373-199702000-00001. [DOI] [PubMed] [Google Scholar]
- Cobb JP, O’Keefe GE. Injury research in the genomic era. Lancet. 2004;363(9426):2076–2083. doi: 10.1016/S0140-6736(04)16460-X. [DOI] [PubMed] [Google Scholar]
- Dahn MS, Mitchell RA, Berberoglu ED, Lange P. The isolated perfused rat liver as an experimental model. Am Surg. 1992;58(9):521–525. discussion 526. [PubMed] [Google Scholar]
- Dahn MS, Mitchell RA, Lange MP, Smith S, Jacobs LA. Hepatic metabolic response to injury and sepsis. Surgery. 1995;117(5):520–530. doi: 10.1016/s0039-6060(05)80251-x. [DOI] [PubMed] [Google Scholar]
- Dasu MR, Cobb JP, Laramie JM, Chung TP, Spies M, Barrow RE. Gene expression profiles of livers from thermally injured rats. Gene. 2004;327(1):51–60. doi: 10.1016/j.gene.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Demling RH, Seigne P. Metabolic management of patients with severe burns. World J Surg. 2000;24(6):673–680. doi: 10.1007/s002689910109. [DOI] [PubMed] [Google Scholar]
- Des Rosiers C, Lloyd S, Comte B, Chatham JC. A critical perspective of the use of (13)C-isotopomer analysis by GCMS and NMR as applied to cardiac metabolism. Metab Eng. 2004;6:44–58. doi: 10.1016/j.ymben.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Deutschman CS, Maio A, Clemens MG. Sepsis-induced attenuation of glucagon and 8-BrcAMP modulation of the phosphoenolpyruvate carboxykinase gene. Am J Physiol. 1995;269:R584–R591. doi: 10.1152/ajpregu.1995.269.3.R584. [DOI] [PubMed] [Google Scholar]
- Duan X, Yarmush DM, Jayaraman A, Yarmush ML. Dispensable role for interferon-gamma in the burn-induced acute phase response: A proteomic analysis. Proteomics. 2004;4(6):1830–1839. doi: 10.1002/pmic.200300696. [DOI] [PubMed] [Google Scholar]
- Fitzwater J, Purdue GF, Hunt JL, O’Keefe GE. The risk factors and time course of sepsis and organ dysfunction after burn trauma. J Trauma. 2003;54(5):959–966. doi: 10.1097/01.TA.0000029382.26295.AB. [DOI] [PubMed] [Google Scholar]
- Hampton WA, Townsend MC, Hybron DM, Shirmer WJ, Fry DE. Effective hepatic blood flow and hepatic bioenergy status in murine peritonitis. J Surg Res. 1987;42:33–38. doi: 10.1016/0022-4804(87)90061-8. [DOI] [PubMed] [Google Scholar]
- Hasselgren PO, James JH, Fischer JE. Inhibited muscle amino acid uptake in sepsis. Ann Surg. 1986;203:360–365. doi: 10.1097/00000658-198604000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerstein MK, Murphy E. Stable isotope-mass spectrometric measurements of molecular fluxes in vivo: Emerging applications in drug development. Curr Opin Mol Ther. 2004;6:249–264. [PubMed] [Google Scholar]
- Herndon DN, Tompkins RG. Support of the metabolic response to burn injury. Lancet. 2004;363(9424):1895–1902. doi: 10.1016/S0140-6736(04)16360-5. [DOI] [PubMed] [Google Scholar]
- Hsieh YC, Hsu C, Yang RC, Lee PY, Hsu HK, Sun YM. Isolation of bona fide differentially expressed genes in the 18-hour sepsis liver by suppression subtractive hybridization. Shock. 2004;21:549–555. doi: 10.1097/01.shk.0000126148.83935.6a. [DOI] [PubMed] [Google Scholar]
- Hua Q, Yang C, Oshima T, Mori H, Shimizu K. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl Environ Microbiol. 2004;70(4):2354–2366. doi: 10.1128/AEM.70.4.2354-2366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson SM, Wallin R, Hall TR. Identification of mitochondrial branched chain aminotransferase and its isoforms in rat tissues. J Biol Chem. 1992;267(22):15681–15686. [PubMed] [Google Scholar]
- Jayaraman A, Yarmush ML, Roth CM. Dynamics of gene expression in rat hepatocytes under stress. Metab Eng. 2000;2(3):239–251. doi: 10.1006/mben.2000.0153. [DOI] [PubMed] [Google Scholar]
- Jeng JC, Jablonski K, Bridgeman A, Jordan MH. Serum lactate, not base deficit, rapidly predicts survival after major burns. Burns. 2002;28(2):161–166. doi: 10.1016/s0305-4179(01)00098-5. [DOI] [PubMed] [Google Scholar]
- Jin ES, Jones JG, Merritt M, Burgess SC, Malloy CR, Sherry AD. Glucose production, gluconeogenesis, and hepatic tricarboxylic acid cycle fluxes measured by nuclear magnetic resonance analysis of a single glucose derivative. Anal Biochem. 2004;327:149–155. doi: 10.1016/j.ab.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Kromer JO, Sorgenfrei O, Klopprogge K, Heinzle E, Wittmann C. Indepth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J Bacteriol. 2004;186(6):1769–1784. doi: 10.1128/JB.186.6.1769-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Berthiaume F, Stephanopoulos GN, Yarmush ML. Metabolic flux analysis: A powerful tool for monitoring tissue function. Tissue Eng. 1999;5:347–368. doi: 10.1089/ten.1999.5.347. [DOI] [PubMed] [Google Scholar]
- Lee K, Berthiaume F, Stephanopoulos GN, Yarmush DM, Yarmush ML. Metabolic flux analysis of postburn hepatic hypermetabolism. Metab Eng. 2000;2(4):312–327. doi: 10.1006/mben.2000.0160. [DOI] [PubMed] [Google Scholar]
- Lee K, Berthiaume F, Stephanopoulos GN, Yarmush ML. Profiling of dynamic changes in hypermetabolic livers. Biotechnol Bioeng. 2003;83(4):400–415. doi: 10.1002/bit.10682. [DOI] [PubMed] [Google Scholar]
- Lohmann R, Souba WW, Zakrzewski K, Bode BP. Stimulation of rat hepatic amino acid transport by burn injury. Metabolism. 1998;47:608–616. doi: 10.1016/s0026-0495(98)90248-7. [DOI] [PubMed] [Google Scholar]
- Maitra SR, Wang SJ, Brathwaite CEM, El-Maghrabi MR. Alterations in glucose 6-phoaphatase gene expression in sepsis. J Trauma. 2000;49:38–42. doi: 10.1097/00005373-200007000-00005. [DOI] [PubMed] [Google Scholar]
- Marin S, Lee WN, Bassilian S, Lim S, Boros LG, Centelles JJ, FernAndez-Novell JM, Guinovart JJ, Cascante M. Dynamic profiling of the glucose metabolic network in fasted rat hepatocytes using [1,2-13C2]glucose. Biochem J. 2004;381:287–294. doi: 10.1042/BJ20031737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Surmacz CA. Liver perfusion: An in vitro technique for the study of intracellular protein turnover and its regulation in vivo. Proc Nutr Soc. 1984;43(2):161–177. doi: 10.1079/pns19840039. [DOI] [PubMed] [Google Scholar]
- Nguyen A, Yaffe MB. Proteomics and systems biology approaches to signal transduction in sepsis. Crit Care Med. 2003;31(Suppl 1):S1–S6. doi: 10.1097/00003246-200301001-00001. [DOI] [PubMed] [Google Scholar]
- Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. J Biol Chem. 2002;277(15):13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- Otero-Anton E, Gonzalez-Quintela A, Lopez-Soto A, Lopez-Ben S, Llovo J, Perez LF. Cecal ligation and puncture as a model of sepsis in the rat: Influence of the puncture size on mortality, bacteremia, endotoxemia and tumor necrosis factor alpha levels. Eur Surg Res. 2001;33(2):77–79. doi: 10.1159/000049698. [DOI] [PubMed] [Google Scholar]
- Pawlick TM, Lohmann R, Souba WW, Bode BP. Hepatic glutamine transporter activation in burn injury: Role of amoni acids and phos-phatidylinositol-3-kinase. Am J Physiol. 2000;278:G532–G541. doi: 10.1152/ajpgi.2000.278.4.G532. [DOI] [PubMed] [Google Scholar]
- Segre D, Zucker J, Katz J, Lin X, D’Haeseleer P, Rindone WP, Kharchenko P, Nguyen DH, Wright MA, Church GM. From annotated genomes to metabolic flux models and kinetic parameter fitting. Omics. 2003;7(3):301–316. doi: 10.1089/153623103322452413. [DOI] [PubMed] [Google Scholar]
- Sheridan RL, Ryan CM, Yin LM, Hurley J, Tompkins RG. Death in the burn unit: Sterile multiple organ failure. Burns. 1998;24(4):307–311. doi: 10.1016/s0305-4179(97)00062-4. [DOI] [PubMed] [Google Scholar]
- Shinnick FL, Harper AE. Branched-chain amino acid oxidation by isolated rat tissue preparations. Biochim Biophys Acta. 1976;437(2):477–486. doi: 10.1016/0304-4165(76)90016-7. [DOI] [PubMed] [Google Scholar]
- Tredget EE, Yu YM. The metabolic effects of thermal injury. World J Surg. 1992;16(1):68–79. doi: 10.1007/BF02067117. [DOI] [PubMed] [Google Scholar]
- Tummala SB, Junne SG, Paredes CJ, Papoutsakis ET. Transcriptional analysis of product-concentration driven changes in cellular programs of recombinant Clostridium acetobutylicum strains. Biotechnol Bioeng. 2003;84(7):842–854. doi: 10.1002/bit.10851. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula M, Berthiaume F, Jayaraman A, Yarmush ML. Expression profiling analysis of the metabolic and inflammatory changes following burn injury in rats. Physiol Genomics. 2004;18(1):87–98. doi: 10.1152/physiolgenomics.00189.2003. [DOI] [PubMed] [Google Scholar]
- von Allmen D, Hasselgren PO, Fischer JE. Hepatic protein synthesis in a modified septic rat model. J Surg Res. 1990;48(5):476–480. doi: 10.1016/0022-4804(90)90016-u. [DOI] [PubMed] [Google Scholar]
- Wong MS, Raab RM, Rigoutsos I, Stephanopoulos GN, Kelleher JK. Metabolic and transcriptional patterns accompanying glutamine depletion and repletion in mouse hepatoma cells: A model for physiological regulatory networks. Physiol Genomics. 2004;16(2):247–255. doi: 10.1152/physiolgenomics.00088.2003. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Yu YM, Zupke C, Yarmush DM, Berthiaume F, Tompkins RG, Yarmush ML. Effect of burn injury on glucose and nitrogen metabolism in the liver: Preliminary studies in a perfused liver system. Surgery. 1997;121(3):295–303. doi: 10.1016/s0039-6060(97)90358-5. [DOI] [PubMed] [Google Scholar]
- Yang D, Brunengraber H. Glutamate, a window on liver intermediary metabolism. J Nutr. 2000;130:991S–994S. doi: 10.1093/jn/130.4.991S. [DOI] [PubMed] [Google Scholar]
- Yarmush ML, Banta S. Metabolic engineering: Advances in modeling and intervention in health and disease. Annu Rev Biomed Eng. 2003;5:349–381. doi: 10.1146/annurev.bioeng.5.031003.163247. [DOI] [PubMed] [Google Scholar]
- Yarmush ML, Berthiaume F. Metabolic engineering and human disease. Nature Biotechnol. 1997;15(6):525–528. doi: 10.1038/nbt0697-525. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Banta S, Berthiaume F, Nagrath D, Tompkins RG, Yarmush ML. Evolution of intrahepatic carbon, nitrogen, and energy metabolism in a D-galactosamine-induced rat liver failure model. Metab Eng. 2005;7(2):88–103. doi: 10.1016/j.ymben.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Zupke C, Sinskey AJ, Stephanopoulos G. Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995;44:27–36. doi: 10.1007/BF00164476. [DOI] [PubMed] [Google Scholar]