Abstract
Huntington disease (HD) is a neurodegenerative disorder caused by an elongated polyglutamine tract in huntingtin (htt). Htt normally undergoes different posttranslational modifications (PTMs), including phosphorylation, SUMOylation, ubiquitination, acetylation, proteolytic cleavage and palmitoylation. In the presence of the HD mutation, some PTMs are significantly altered and can result in changes in the clinical phenotype. A rate limiting PTM is defined as one which can result in significant effects on the phenotype in animal models. For example the prevention of proteolysis at D586 as well as constitutive phosphorylation at S13 and S16 can obviate the expression of phenotypic features of HD. The enzymes involved in these modifications such as caspase-6, the IκB kinase (IKK) complex and still to be characterized phosphatases therefore represent promising therapeutic targets for HD. Identifying and testing specific modulators of PTMs now constitutes the next big challenge in order to further validate these targets and proceed towards the goal of a mechanism-based treatment for HD.
Huntington disease: one mutation, multiple pathways
Htt is a 350 kDa protein which when mutated by an elongated polyglutamine (polyglutamine) tract (>35) causes HD, which manifests with symptoms that include personality changes, involuntary movement and intellectual decline usually in mid-adulthood (Novak and Tabrizi 2010). On a cellular level, HD is characterized by neuronal dysfunction initially followed by the loss of medium spiny neurons in the striatum and subsequent more wide-spread damage (Albin and others 1990).
Even though its mutant form causes predominantly brain pathology, htt is ubiquitously expressed within and outside the nervous system (Strong and others 1993) and is present in different subcellular compartments including the nucleus, the Golgi complex, mitochondria, microtubules and vesicular structures in neurites and at synapses (Caviston, Ross, Antony, Tokito and Holzbaur 2007; Choo, Johnson, MacDonald, Detloff and Lesort 2004; Hoffner, Kahlem and Djian 2002; Kang and others 2007; McGuire, Rong, Li and Li 2006; Strehlow, Li and Myers 2007). A large variety of htt-interacting proteins have been described with a multitude of functions ranging from endocytosis and vesicle transport to cell signalling, apoptosis and transcriptional regulation. Htt acts as a scaffolding protein bringing together its interaction partners and allowing them to transfer information between cellular compartments (Harjes and Wanker 2003), PTMs are postulated to play a major role in the flexibility of htt’s protein-protein interactions and to influence its subcellular localization.
Htt protein structure and localization of PTMs
The human Htt protein (Fig. 1) has a polymorphic glutamine stretch at the N-terminus, adjacent to a proline-rich region which is an important mediator of protein-protein interactions, may regulate turnover of the htt protein and keep the protein in a non-aggregated state (Bhattacharyya and others 2006; Dehay and Bertolotti 2006; Southwell and others 2008; Zuchner and Brundin 2008). This region is followed by four clusters of HEAT repeats (named for the proteins where they were first described: huntingtin, elongation factor 3, the PR65/A subunit of protein phosphatase 2A, and the lipid kinase TOR), which are also sites of protein-protein interactions (Warby and others 2008). A cytoplasmic retention signal at the N-terminus as well as a nuclear export signal at the C-terminus have been identified, both of which could be modified by PTMs and thus change the subcellular localization of htt (Rockabrand and others 2007; Xia, Lee, Taylor, Vandelft and Truant 2003). Although no classical nuclear localization signal (NLS) has been found in the htt sequence, stretches between amino acids 90-100 and 1188-1204 are suggestive of NLS motifs (Bessert, Gutridge, Dunbar and Carlock 1995; Hackam and others 1998). In addition, htt could be shuttled in and out of the nucleus through its binding partners (Preisinger, Jordan, Kazantsev and Housman 1999).
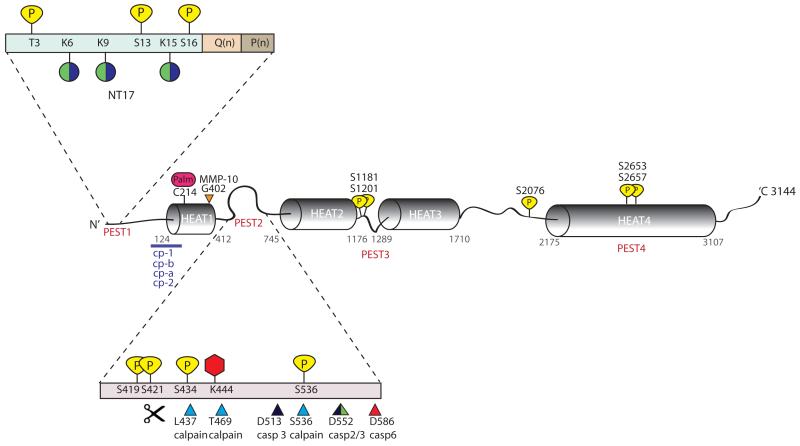
Figure 1. Schematic representation of htt protein and its PTMs.
Predicted HEAT repeats and PEST domains are marked along the length of the htt protein. PEST 1 domain (containing the first N-terminal 17 amino acids (NT17), polyglutamine tract Q(n) and polyproline sequence P(n)) and PEST 2 domain have been magnified to show the large number of clustered post-translational modifications in these regions. Phosphorylation sites at threonine (T) or serine (S) residues are indicated in yellow. Lysines (K) which are modified either by the addition of ubiquitin or sumo-1 are indicated by green and purple coloured circles. Acetylation at lysine (K) 444 is represented as a red hexagon and palmitoylation at cysteine (C) 214 is represented as a pink oval. Protease cleavage sites are marked by triangles; calpain sites are indicated in blue, caspase −2, −3 and −6 sites are indicated by green, black, and red, respectively and matrix metalloprotease-10 (MMP-10) site at 404 is indicated in orange. Additional proteolytic cleavage sites lie at the N-terminus of htt and predicted cleavage regions between residues 81-214 are indicated by a purple line. Cleavage between amino acid 104-114, 205-214, 81-129 and at R167 generate cp-A, cp-B , cp-1 and cp-2 fragments, respectively.
The current structural view of the Htt protein is that of a solenoid with a hydrophobic core consisting of stacked HEAT repeats (Li, Serpell, Carter, Rubinsztein and Huntington 2006). Four PEST domains (regions with predicted proteolytic sites enriched in proline, glutamic acid, serine and threonine) have been reported (Warby and others 2008), three of which are localized between HEAT repeats. Interestingly, most of the reported PTMs have been found in these PEST domains: The N-terminal 17 amino acids adjacent to the polyglutamine tract contain SUMOylation, ubiquitination, phosphorylation, acetylation and proteolytic cleavage sites, and the region between amino acids 412 and 745 contain phosphorylation, acetylation, palmitoylation and protease cleavage sites. Additional phosphorylation sites have been found in close vicinity to PEST domains 3 and 4 as well as in the linker region between HEAT repeats 3 and 4. Proteolysis can have a major impact on the toxicity, aggregation and subcellular localization of mhtt as well as its interaction with binding partners, which has led to the concept that cleavage is not just a mechanism of degradation (DiFiglia and others 1997; Warby and others 2008; Wellington and others 2000).
The importance of PTMs in different polyglutamine diseases- a common theme
The 9 known neurodegenerative polyglutamine disorders (HD, spinal and bulbar muscular atrophy (SMBA), dentatorubro-pallidolysian atrophy (DRPLA), spinocerebellar ataxias (SCAs) 1, 2, 3, 6, 7 and 17) are all characterized by a polyglutamine expansion in the respective disease-causing protein (Pennuto, Palazzolo and Poletti 2009). For most of the polyglutamine containing proteins, PTMs have been reported that often show similar effects on the disease phenotype (Table 1):
Table 1.
PTMs in different polyglutamine diseases and associated disease mechanisms.
| Type of modification |
affected polyglutamine protein |
impact on toxicity |
Disease mechanism |
|---|---|---|---|
| SUMOylation | htt, ataxin-1, atrophin androgen receptor |
↑
↓ |
proteasomal degradation, aggregation, transcriptional dysregulation? |
| ubiquitylation | htt, ataxin-3 | ↓ | proteasomal degradation, aggregation |
| phosphorylation | htt, androgen receptor ataxin-1 |
↓
↑ |
BDNF trafficking, excitotoxicity |
| acetylation | htt, androgen receptor | ↓ | autophagy, transcriptional regulation |
| palmitoylation | htt, androgen receptor | ↓ | aggregation, excitotoxicity? |
| proteolysis | htt, androgen receptor, atrophin, ataxin-3, ataxin-7 |
↑ | excitotoxicity, aggregation |
In five of these disorders, proteolysis leading to the generation of small polyglutamine containing protein fragments is consistently associated with neurotoxicity (Walsh, Storey, Stefani, Kelly and Turnbull 2005). Htt, androgen receptor, atrophin, ataxin-3 and ataxin-7 are for example all substrates for proteases of the caspase family, and cleavage site mutations or the treatment with caspase inhibitors has proven beneficial in different disease model systems (Berke, Schmied, Brunt, Ellerby and Paulson 2004; Colomer Gould and others 2007; Ellerby and others 1999; Garden and others 2002; Graham and others 2006a; Haacke and others 2006; Jung, Xu, Lessing and Bonini 2009; Kobayashi and others 1998; Miyashita, Okamura-Oho, Mito, Nagafuchi and Yamada 1997; Wellington and others 1998; Wellington and others 2000; Young and others 2007). Protein fragments containing the polyglutamine tract are further more prone to misfolding and aggregation, a common feature among the polyglutamine disorders (Ellerby and others 1999; Haacke and others 2006; Li, Chevalier-Larsen, Merry and Diamond 2007; Wellington and others 2000).
Ubiquitination is another mechanism that shows similarities among the different polyglutamine containing proteins. This PTM targets the mutant proteins for proteasomal degradation and reduces polyglutamine toxicity (Jana and others 2005; Matsumoto and others 2004; Tsai, Fishman, Thakor and Oyler 2003). However, for many neurodegenerative diseases the gradual accumulation and aggregation of ubiquitinated polyglutamine proteins can be observed, indicating that the detoxification mechanism has failed (DiFiglia and others 1997).
Other PTMs show differences in their impact on the disease depending on the respective polyglutamine protein (Pennuto, Palazzolo and Poletti 2009). Phosphorylation by Akt can be either beneficial (mhtt, mutant androgen receptor)) or detrimental (mutant ataxin-1) (Chen and others 2003; Emamian and others 2003; Humbert and others 2002; Palazzolo and others 2007). Htt phosphorylation by Akt at S421 has been shown to regulate BDNF vesicle transport, and loss of phosphorylation in HD could therefore contribute to the observed loss of BDNF (Colin and others 2008; Gauthier and others 2004). Furthermore increased BDNF levels can ameliorate many of the disease features in the YAC128 mouse model of HD (Dey and others 2010; Xie, Hayden and Xu in press). The levels of phospho-421 mhtt are also reduced following excitotoxic stimulation in an HD model, linking phosphorylation to excitotoxicity as a disease mechanism (Metzler and others in press). For ataxin-1, constitutive phosphorylation of the wild-type protein surprisingly evokes a phenotype similar to the polyglutamine expanded mutant, indicating that a single PTM is sufficient to turn wild-type ataxin-1 into a toxic protein (Duvick and others 2010).
SUMOylation is detrimental in the context of mhtt and atrophin, but ameliorates mutant androgen receptor toxicity (Chan, Warrick, Andriola, Merry and Bonini 2002; Mukherjee, Thomas, Dadgar, Lieberman and Iniguez-Lluhi 2009; Steffan and others 2004; Terashima, Kawai, Fujitani, Maeda and Yasuda 2002; Ueda and others 2002). In HD, SUMOylation could be responsible for the altered transcriptional regulation through an increase in the nuclear localization of mhtt (Steffan and others 2004). Furthermore, SUMOylation prevents the proteasomal degradation of mhtt and increases its aggregation, an effect that has also been shown for mutant ataxin-1 (Ryu, Cho, Park and Lee do 2010; Steffan and others 2004).
Acetylation has been reported for the androgen receptor and modulates its function in transcription, but seems to have a different function in htt, where it has been associated with the degradation of the mutant protein by autophagy (Fu and others 2000; Jeong and others 2009). Both mhtt and the androgen receptor are furthermore targets for palmitoylation, but a disease-modulating effect of this PTM has only been shown in HD so far (Pedram and others 2007; Yanai and others 2006). Altered palmitoylation could result in the synaptic changes and excitotoxicity observed in HD, and lack of palmitoylation leads to mhtt mislocalization and aggregation (Huang and others 2010; Yanai and others 2006).
Taken together, the similarities in PTMs among different polyglutamine proteins could account for similarities in disease phenotypes such as aggregate formation and transcriptional dysregulation. However the interplay between different PTMs is likely different in each disorder reflecting the importance of protein context with differential effects on cell type toxicity of the polyglutamine tract in each disease.
The study of PTMs- methods and caveats
The importance of single PTMs can be defined by their impact on the disease phenotype in different model systems, particularly in vivo, and both the prevention of caspase cleavage at D586 and constitutive phosphorylation at S13 and S16 have been shown to prevent the phenotype in full-length HD mouse models (Graham and others 2006a; Gu and others 2009). However, in vivo studies of htt PTMs often make use of animal models expressing mhtt with point mutations introduced at the site of the respective PTM, i.e. rendering the protein resistant against cleavage, phosphorylation, palmitoylation or acetylation or introducing an amino acid that mimicks the respective PTM (phosphomimetic mutants). While these experiments can inform us about the impact of PTMs on the disease phenotype, there are several caveats that have to be taken into consideration when interpreting these results. First, mhtt toxicity is highly dependent on the expression level of the protein, with higher expression levels causing more severe symptoms in otherwise identical animal models (Graham and others 2006b). Comparisons between animal models with or without mutations at PTM sites therefore need to be carefully matched for mhtt expression levels to ensure that the observed differences in phenotype are truly a result of changes in PTMs. Second, the possibility needs to be taken into consideration that a point mutation in mhtt could act as a dominant negative, trapping the active enzyme without allowing it to catalyze reactions with other substrates. Although such an effect does argue for the importance of the enzyme in HD, it would overestimate the impact of the PTM on the features and pathogenesis of the disease. Third, htt, and especially mhtt, is a conformationally unstable protein with a high tendency to misfold and aggregate. The introduction of additional mutations could therefore severely impact the folding state and thus influence the toxicity of the protein in a way that is different from the effect of endogenous PTMs. Taken together, it is therefore important to confirm results obtained by the introduction of mutations at PTM sites with other methods, such as the chemical modification of enzymatic activity or direct knockdown/knockout of the respective enzymes.
PTMs of the htt protein: SUMOylation and ubiquitination
SUMOylation and ubiquitination are the covalent attachment of either SUMO-1 (small ubiquitin-like modifier) or ubiquitin molecules to lysine residues. Both modifications are biochemically similar but generally have different functional consequences for the target protein. The SUMOylation and ubiquitination processes involve three enzymatic steps catalyzed by E1 (activating), E2 (carrier) and E3 (ligase) proteins, whereas de-SUMOylation and de-ubiquitination can be performed by SUMO isopeptidases of the SENP protein family and ubiquitin hydrolases, respectively (for reviews, see (Wilkinson, Nakamura and Henley 2010) (Denuc and Marfany 2010; Haas and Broadie 2008)). Historically, ubiquitination has been associated with the targeting of proteins for degradation by the proteasome. However recently a wider range of functions for the ubiquitin modifier has been shown, including DNA repair, endocytosis, nuclear export and the regulation of enzymatic activity, depending on whether the modification involves one or more ubiquitin molecules (mono- or polyubiquitination) and how the ubiquitin molecules in a polyubiquitin chain are linked (Haas and Broadie 2008). SUMOylation on the other hand usually involves the conjugation of a single SUMO moiety and plays a role in genome stability, progression of the cell cycle and subcellular transport, with many nuclear proteins as known targets of SUMO modifications (Wilkinson, Nakamura and Henley 2010). In neurons, SUMO plays a role in the transcriptional regulation of neuronal development and function, mitochondrial dynamics, the modulation of kinase pathways, synaptic function and axonal trafficking (Wilkinson, Nakamura and Henley 2010), whereas ubiquitination plays a critical role at the synapse in the targeted removal of proteins during synaptogenesis, synapse elimination as well as synaptic plasticity and remodeling (Haas and Broadie 2008).
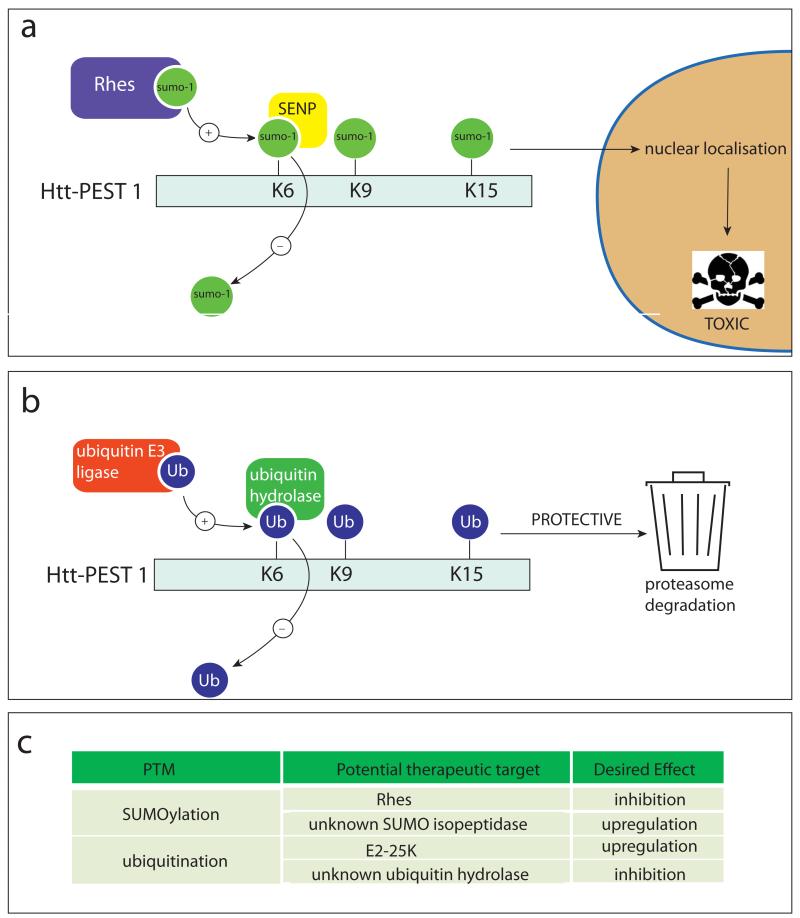
In the htt protein, SUMOylation and ubiquitination compete for the same target lysines at amino acids 6, 9 and 15 (Fig. 2) (Steffan and others 2004). Ubiquitination has been linked to reduced toxicity of mutant htt (mhtt) most likely through increased clearance by the proteasome (Jana and others 2005; Kalchman and others 1996), whereas SUMOylation stabilizes the protein, reduces aggregation and exacerbates transcriptional dysregulation by mhtt as well as its toxicity in a Drosophila model (Steffan and others 2004). The ubiquitin ligase E2-25K has been shown to mediate htt ubiquitination (Kalchman and others 1996), and the E3 ligase Rhes (Ras homolog enriched in striatum) has been implicated in the SUMOylation of mhtt (Subramaniam, Sixt, Barrow and Snyder 2009). Rhes shows significantly higher affinity for the mutant over the wild-type (wt) protein, and could mediate mhtt toxicity by targeting the protein to the nucleus and preventing its ubiquitination and subsequent degradation. Rhes is highly expressed in the striatum, which could contribute to the selective pathology observed in HD (Falk and others 1999). Ubiquitin hydrolases and SUMO isopeptidases are not known so far, making E2-25K and Rhes the only potential therapeutic targets to regulate the SUMO/ubiquitin equilibrium in HD to date.
Figure 2. Enzymes catalyzing the addition and removal of SUMO-1 and ubiquitin from htt.
(a) Sumoylation of htt at lysine’s (K) 6, 9 and 15 is catalysed by the E3 ligase, Rhes. This leads to masking of the cytoplasmic retention signal and subsequent nuclear localisation of mutant htt, which causes cell toxicity by mechanisms including transcriptional dysregulation. The SENP family of enzymes is responsible for the de-SUMOylation of htt. (b) Alternatively lysines 6, 9 and 15 of htt can be ubiquitinated by E3 ligases, including E2-25K, which directs htt for degradation by the proteasome . Removal of ubiquitin from htt is carried out by ubiquitin hydrolases. (c) Therapeutic targets relating to SUMOylation and ubiquitination of htt are indicated.
Phosphorylation
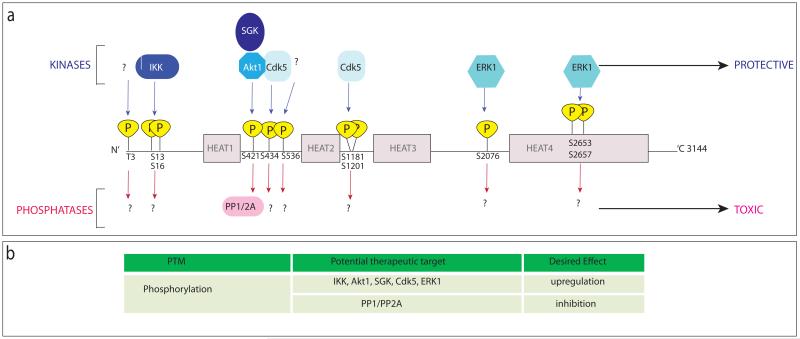
The attachment of phosphate groups to proteins by kinases as well as the inverse reaction catalyzed by phosphatases is an important regulatory step in cellular mechanisms such as protein degradation, signalling, protein-protein interactions or nuclear translocation. The htt protein has several known sites of phosphorylation (Fig. 3): T3, S13 and S16 at the N-terminus (PEST domain 1), S421, S434 and S536 in the second PEST domain, S1181 and S1201 in close vicinity to the third PEST domain, S2076 in the linker region between HEAT repeats 3 and 4 and S2653 and S2657 in PEST domain 4. At all known sites, mhtt is less phosphorylated than the wt protein, and htt phosphorylation is associated with reduced toxicity, although the neuroprotective effect is attributed to different mechanisms depending on the location of the phosphorylation site (see below).
Figure 3. Schematic representation of htt indicating phosphorylation sites and enzymes responsible for phosphorylation and de-phosphorylation.
(a) Phosphorylation of htt by different kinases (indicated above the protein in blue) is protective to the cell, whilst reduced phosphorylation of htt contributes to toxicity. Known phosphatases of htt are indicated below the protein in pink. (b) Potential therapeutic targets relating to phosphorylation of htt are highlighted.
The most N-terminal amino acid subject to phosphorylation is T3, with the lowest levels of phosphorylation found in cells and tissues that are most susceptible to HD pathology (striatal < cortical, neuronal < HeLa cells) (Aiken and others 2009). The level of phosphorylation at T3 is also polyglutamine-length dependent, where a longer polyglutamine stretch is associated with less phosphorylated T3 (Aiken and others 2009). Experiments using phosphomimetic (Asp) and phosphorylation resistant (Ala) mutants in cell culture and Drosophila models further show that T3 phosphorylation is associated with enhanced mhtt aggregation, whereas both phosphomimetic and non-phosphorylatable T3 mutations reduced mhtt toxicity (Aiken and others 2009). This could be due to a secondary modification at the same or an adjacent residue that is prevented by changing Thr to either Asp or Ala, or due to intrinsic toxicity of the Thr residue itself.
Residues S13 and S16 are phosphorylated by the IKK (IκB kinase) complex, again with less efficiency for mhtt than the wt protein (Thompson and others 2009). S13 thereby seems to be a direct target of IKK, whereas the phosphorylation of S16 is facilitated by previous phosphorylation of S13. Phosphorylation of wt htt at these sites leads to increased degradation of the protein by both proteasomal and lysosomal pathways, a mechanism which is non-functional if the polyglutamine tract expands (Thompson and others 2009). This effect could be due to an interaction between the phosphorylation and ubiquitination/SUMOylation at residues in close spatial proximity, processes that have been linked to the proteasomal degradation of htt (see above and (Kalchman and others 1996; Steffan and others 2004)).
Additional evidence for the neuroprotective function of htt phosphorylation comes from recently generated mouse lines expressing S13 and S16 phosphomimetic or non-phosphorylatable mhtt (Gu and others 2009). This study shows that introducing the two phosphomimetic mutations into full-length mhtt abolishes the motor, psychiatric and neuropathological phenotypes in the BACHD mouse model and results in a dramatic reduction of mhtt aggregates in the mice expressing phosphomimetic mhtt (Gu and others 2009).
All PTMs at the N-terminus of mhtt influence the aggregation behaviour of the protein: SUMOylation and phosphorylation at S13 and S16 reduce inclusion formation, while ubiquitination and phosphorylation at T3 accelerate the aggregation process (Aiken and others 2009; Gu and others 2009; Steffan and others 2004). These findings highlight the importance of the N-terminal domain for the correct folding of the htt protein and show the disconnect between observed toxicity and appearance of large aggregates, which is the subject of ongoing discussions (Arrasate, Mitra, Schweitzer, Segal and Finkbeiner 2004; Slow and others 2005).
In summary, inefficient phosphorylation at the N-terminus in combination with reduced clearance could therefore be responsible for the accumulation and toxicity of mhtt, with S13 and S16 being the dominant phosphorylation sites whose modification has a profound impact on the HD phenotype in vivo. The kinase IKK currently represents the only therapeutic target to modify phosphorylation at the N-terminus, since the respective phosphatase(s) remain unknown.
Among the htt phosphorylation sites identified in the second PEST domain (S421, S434 and S536), S421 is the best characterized. It can be phosphorylated by two kinases that are both activated through the IGF-1 (insulin-like growth factor 1) pathway, Akt and SGK (serum- and glucocorticoid-induced kinase) (Humbert and others 2002; Rangone and others 2004). There are conflicting lines of evidence pointing towards a dysregulation of these kinases in HD: Proteolytic processing of Akt, presumably leading to its inactivation, has been seen in late stage HD patient brain tissues (Humbert and others 2002), whereas increased SGK levels were reported from similar samples (Rangone and others 2004). In mice, the Q111/Q111 knock-in model of HD showed constitutive elevation of Akt activity (Gines, Ivanova, Seong, Saura and MacDonald 2003), whereas no changes were observed in a YAC mouse model of HD (Warby and others 2005). These conflicting data indicate that loss of mhtt phosphorylation is not simply due to a loss in the activity of the respective kinases, and more complicated compensatory mechanisms might be at play.
The regional pattern of phosphorylation at S421 in the brain parallels the pathology of HD, with highest phospho-htt levels in the cerebellum, less in the cortex and least in the striatum (Warby and others 2005), providing further evidence for a neuroprotective role of phosphorylation in HD. Functionally, phosphorylation at S421 has been shown to influence htt’s role in neuronal vesicle transport (Colin and others 2008; Pineda and others 2009; Zala and others 2008), in particular controlling the transport of BDNF vesicles through cortico-striatal projection neurons that is crucial for the survival of striatal neurons (Baquet, Gorski and Jones 2004; Gauthier and others 2004). Although no in vivo studies with phosphomimetic mutants have been done for this PTM, it has been shown that phosphorylation of S421 modifies caspase cleavage of htt and could thus have a profound impact on the pathogenesis of HD (see below, and (Metzler and others in press; Warby and others 2009).
Akt and SGK are the kinases that could be targeted to modulate htt phosphorylation in the second PEST domain, and recently the phosphatases PP1 and PP2A have been identified as their counterparts in htt phosphorylation at S421 (Metzler and others in press). For htt, phosphatases represent a more attractive therapeutic target than kinases, since generally inhibition of enzymatic activity can be achieved more easily than upregulation.
Another kinase that has been found to phosphorylate htt at S434, S1181 and S1201 is Cdk5, and despite its widespread expression in adult tissues, its activating proteins p35/p39 are highly expressed in neurons, leading to high Cdk5 activity in these cells (Tsai, Takahashi, Caviness and Harlow 1993). Interestingly, preventing phosphorylation at S1181 and S1201 in wt htt makes the protein toxic, indicating the importance of these PTMs for htt’s function in pro-survival pathways (Anne, Saudou and Humbert 2007). Phosphorylation at all three Cdk5 sites is decreased upon polyglutamine expansion, and constitutive phosphorylation protects against polyglutamine toxicity (Anne, Saudou and Humbert 2007; Luo, Vacher, Davies and Rubinsztein 2005). Cdk5 activity is increased upon DNA damage in striatal neurons, and htt phosphorylation by this kinase is neuroprotective after irradiation- or oxidative stress-induced DNA damage (Anne, Saudou and Humbert 2007). In HD, the protective role of the Cdk5 pathway may be ineffective due to less efficient phosphorylation of the mhtt protein, or increased oxidative stress and DNA damage in affected tissues (Trushina and McMurray 2007). Further, the phosphorylation site at S434 is in very close proximity to a cluster of proteolytic cleavage sites, which themselves may play a crucial role in the pathogenesis of HD (see below). Phosphorylation at S434 has been shown to reduce htt cleavage by caspase-3 at residue 513 (Luo, Vacher, Davies and Rubinsztein 2005).
Additionally, residues S2076, S2653 and S2657 can be phosphorylated and have been predicted as targets of the ERK-1 kinase (Schilling and others 2006). The same study describes a phosphorylation site around amino acids 533-536, that has the potential to modulate calpain cleavage of htt at S536 (Schilling and others 2006).
Therapeutic targeting of CDK5 or its activating proteins p35/p39, as well as Erk-1 should modulate htt phosphorylation at the C-terminal sites, but the respective phosphatases for these residues have not been determined. Such studies should be able to verify the impact of S434 phosphorylation on caspase-3 cleavage of htt; however, the importance of the more C-terminal phosphorylation sites for htt function and HD pathogenesis is not understood.
Acetylation
Acetylation is a modification that has first been identified in histone proteins as a mechanism of transcriptional regulation and cellular differentiation. In neurons, histone acetylation is involved in learning, memory formation and the expression of behaviour and is modulated by environmental stimuli (Crepaldi and Riccio 2009). In a paradigm of contextual fear conditioning, for example, ERK activation in the CA1 area of the hippocampus led to the acetylation of Histone H3, and histone deacetylase inhibitors enhanced the induction of long term potentiation, indicating that histone acetylation plays a role in the formation of long term memory (Levenson and others 2004).
For non-histone proteins, acetylation is also often associated with their nuclear function, such as the modification of their DNA binding affinity or transcriptional activation (Glozak, Sengupta, Zhang and Seto 2005). However, other roles for acetylation are still being discovered, and emerging new concepts include the promotion of protein-protein interactions as well as the modulation of protein stability and turnover through acetylation (Glozak, Sengupta, Zhang and Seto 2005).
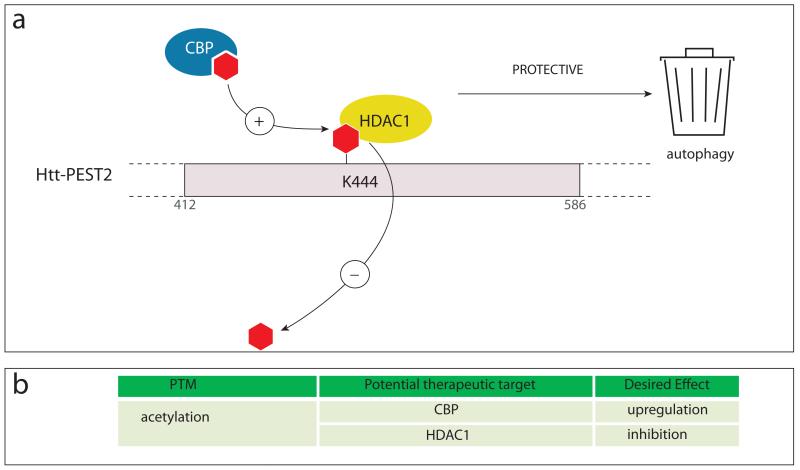
Along these lines, acetylation at K444 (Fig. 4) has recently been identified as a PTM that directly influences HD by facilitating the autophagic clearance of mhtt (Jeong and others 2009). The acetyltransferase mediating the reaction is CBP (CREB-binding protein), and the de-acetylation process is catalyzed by histone deacetylase 1 (HDAC1) (Jeong and others 2009). Interestingly, preferential acetylation of mhtt over the wt protein was observed in cell culture as well as in brains from HD mouse models and patients, and experiments with acetylation-resistant mutants showed that acetylation is correlated with protection from mhtt toxicity. Acetylated mhtt is trafficked to autophagosomes for degradation (Jeong and others 2009), which most likely constitutes the detoxification mechanism. This clearance pathway therefore provides an attractive starting point for the design of therapies that specifically target mhtt without affecting the wt protein. HDAC-1 inhibitors as well as autophagy enhancing agents could be used to promote mhtt clearance, and further elucidation of the degradation mechanism could identify additional enzymes as drug targets. Interestingly, the acetyltransferase CBP is, like htt, a substrate for caspase-6, and the recently reported upregulation of caspase-6 activity in HD could therefore lead to CBP cleavage and its inactivation (Graham and others in press; Rouaux and others 2003). The pan-HDAC inhibitor SAHA (Suberoylanilide hydroxamic acid) has already been shown to improve the phenotype in the R6/2 mouse model, providing evidence for the therapeutic usefulness of this class of compounds (Hockly and others 2003).
Figure 4. Enzymes catalyzing acetylation and de-acetylation of htt.
(a) CREB binding protein (CBP) acetylates htt at lysine 444 facilitating autophagic clearance of the protein. HDAC1 deacetylates htt. (b) Potential therapeutic targets relating to acetylation of htt are highlighted.
An additional acetylation site has been found by mass spectrometry at residue K9. However it does not seem to be acetylated unless the kinase IKK is co-expressed in the same cells. Thus, the importance of K9 acetylation and its presence in vivo is unclear (Thompson and others 2009).
Palmitoylation
The attachment of a palmitoyl group to proteins is a common lipid modification that plays an important role in neuronal development and synaptic plasticity (Fukata and Fukata 2010). Palmitoylation can anchor proteins to the plasma membrane as well as to other membrane compartments in the cell, and since it is a rapidly reversible PTM it can be used to regulate protein trafficking (Fukata and Fukata 2010). In neurons, many pre- and postsynaptic proteins such as PSD95, SNAP25, AMPAR subunits and GLURs are palmitoylated, indicating that this PTM is an important regulator of synaptic transmission (Fukata and Fukata 2010). Palmitoylation can also serve as a signal for the proper folding and assembly of protein complexes, with reduced palmitoylation often leading to the formation of aggregates (Drisdel, Manzana and Green 2004; Rakhilin and others 1999).
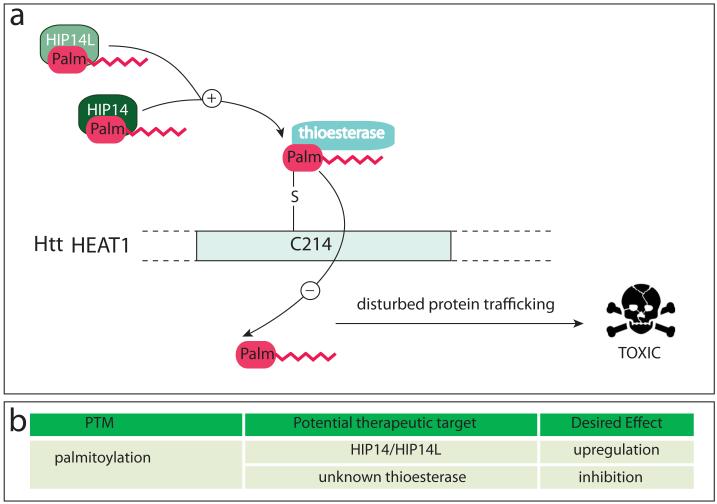
Htt is palmitoylated at C214 by the palmitoyl transferases HIP14 and HIP14L (Fig. 5) (Huang and others 2009; Yanai and others 2006), and a mutation rendering the protein palmitoylation-resistant leads to increased inclusion formation, increased nuclear localization and increased toxicity in neurons (Yanai and others 2006). The interaction of htt with HIP14 is polyglutamine length-dependent, and mhtt exhibits less palmitoylation in an HD mouse model (Yanai and others 2006). As a consequence, reduced palmitoylation could be responsible for the axonal trafficking defects seen in HD, and mhtt that fails to reach its appropriate intracellular destination could end up forming insoluble aggregates (Yanai and others 2006). From a therapeutic point of view, an increase in palmitoylation should be beneficial and could be achieved by either upregulation of HIP14 and HIP14L activity or the inhibition of yet to be defined thioesterases that antagonize htt palmitoylation.
Figure 5. Enzymes catalyzing the palmitoylation and de-palmitoylation of htt.
(a) Protein acyl transferases (PATs) HIP14 and its closely related family member, HIP14L, catalyse the addition of palmitate (palm) to htt at cysteine 214 via a thioester bond . Removal of palmitate is carried out by an unidentified thioesterase. Reduced palmitoylation can result in altered protein trafficking leading to protein aggregation and synaptic dysfunction. (b) Potential therapeutic targets related to palmitoylation of htt are highlighted.
Proteolysis
Htt is subject to cleavage by a variety of different proteases (Fig. 6): Caspase (Goldberg and others 1996; Hermel and others 2004; Wellington and others 1998; Wellington and others 2000) and calpain (Gafni and Ellerby 2002; Gafni and others 2004) cleavage sites as well as sites that could be recognized by aspartyl proteases (Lunkes and others 2002) have been described. Additionally, there are less well defined cleavage fragments that have not been linked to specific proteases but show preferential cleavage in certain brain regions over others (Mende-Mueller, Toneff, Hwang, Chesselet and Hook 2001).
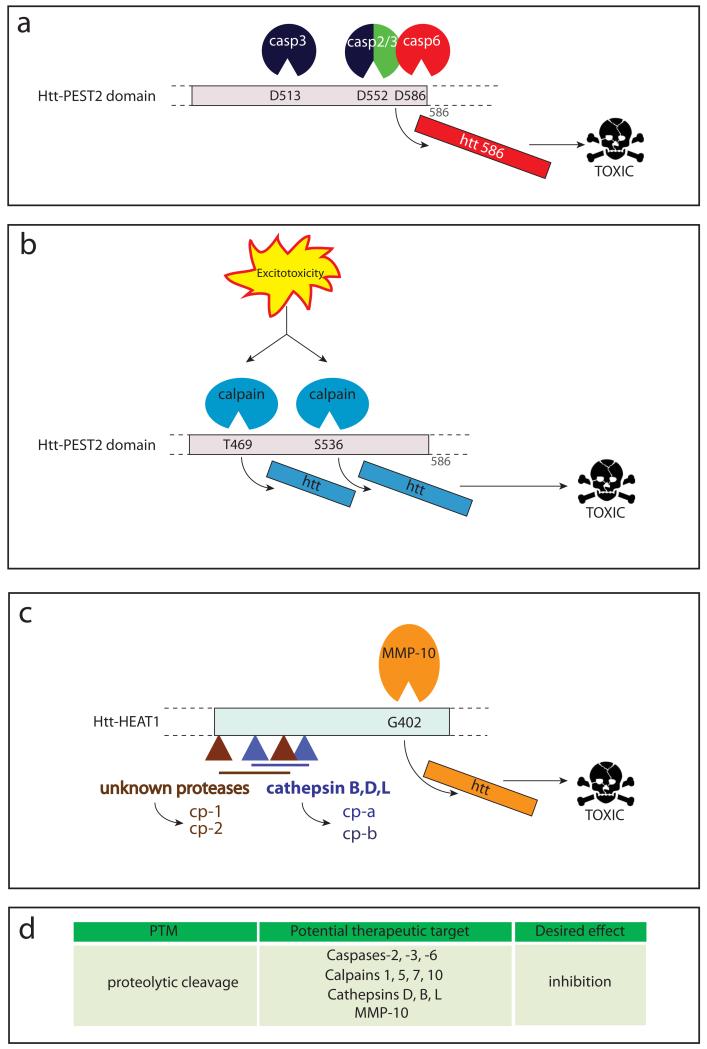
Figure 6. Proteolysis of htt.
Htt is cleaved by a number of (a) caspases , (b) calpains and (c) matrix-metalloprotease-10 (MMP-10), cathepsins and other unidentified proteases generating toxic htt fragments. (d) Potential therapeutic targets relating to huntingtin proteolysis are highlighted.
Caspase cleavage
Proteases of the caspase family are mostly known for their role in programmed cell death (apoptosis), where they are responsible for the systematic destruction of a cell without the generation of an inflammatory response. Caspases dismantle the cell with surgical precision, working on a small number of substrates, often targeting whole protein complexes and generating stable protein fragments that contain single, functional domains (Dix, Simon and Cravatt 2008; Mahrus and others 2008). In addition to their apoptotic function, however, recent evidence suggests the involvement of caspases in non-cell death pathways: In neurons, caspases −3 and −6 have been involved in synaptic plasticity, memory and learning (Li and others 2010). These processes call for tightly controlled, local activation of the proteases in order to prevent activation of the cell death cascade, the disturbance of this delicate balance could lead to neuronal degeneration under pathological conditions (Gray, Mahrus and Wells 2010).
The wt htt protein has an anti-apoptotic function in the adult brain, protecting against ischemic injury as well as against excitotoxicity (Leavitt and others 2006; Zhang and others 2003). This protection is lost and htt becomes proapoptotic upon polyglutamine expansion (Saudou, Finkbeiner, Devys and Greenberg 1998; Zeron and others 2002). Elevated caspase activity has been demonstrated in neuronal cells expressing mhtt as well as in brain samples from mouse models and human HD patients, and the addition of caspase inhibitors reduces mhtt toxicity (Graham and others in press; Hermel and others 2004; Kim and others 1999).
Htt was the first neuronal substrate shown to be proteolyzed by caspases (Goldberg and others 1996). Five sites for caspase cleavage have been predicted in the htt sequence, three of which are cleaved by caspase-3 (D513, D552), caspase-2 (D552) and caspase-6 (D586), respectively (Fig. 6a), whereas D530 and D589 are silent sites (Wellington and others 2000). There is no evidence for preferential cleavage of either mutant or wt htt, but the generation of toxic fragments containing an expanded polyglutamine tract could create a vicious cycle, leading to an increase in caspase activity and even more mhtt cleavage (toxic fragment hypothesis, (Wellington and Hayden 2000)). The age-dependent general increase in caspase-6 activity in the brain could thereby further contribute to this feedback loop and the adult onset of neurodegeneration (Albrecht and others 2007).
The generation of caspase-resistant mhtt constructs has allowed for the dissection of single caspase cleavage events, and cleavage at the 586 site has been identified as a crucial event necessary for the development of HD in a YAC mouse model (Graham and others 2006a; Milnerwood and others 2010). Cleavage-resistant mhtt even seems to have re-acquired some of wt htt’s normal functions, as it is able to protect neurons against excitotoxic injury (Graham and others 2006a). Although this study did not observe any improvement in the HD phenotype when making mhtt resistant to cleavage at amino acids 513 and 552, these proteolysis events take place in vivo and could still play an important role downstream of cleavage at D586 (Hermel and others 2004; Warby and others 2008).
Similar observations have been made in the context of Alzheimer’s disease, where studies on the cleavage of the amyloid precursor protein by caspase-6 showed that inhibition of this cleavage event can prevent the onset of symptoms in a mouse model of Alzheimer’s disease (Galvan and others 2006). Together with the emerging literature on caspase-6 activation in other neurodegenerative diseases (Nikolaev, McLaughlin, O’Leary and Tessier-Lavigne 2009; Park, Grosso, Aubert, Kaplan and Miller 2010; Schoenmann and others 2010), these studies provide compelling evidence for the importance of caspase-6 in neuronal dysfunction and make it a most interesting therapeutic target for HD and probably other neurodegenerative diseases such as Alzheimer’s disease (Lu and others 2000).
Calpain cleavage
Proteases of the calpain family (calpains −1, −5, −7 and −10) have been shown to cleave htt (Fig. 6b), generating small N-terminal fragments that, like the 586 fragment, accumulate in the nucleus where they co-localize with active calpains (Gafni and others 2004). Two calpain cleavage sites have been identified at amino acids 469 and 536, with mhtt being cleaved more readily than the wt protein. Inhibiting cleavage at these sites reduces toxicity in cell culture (Gafni and Ellerby 2002; Gafni and others 2004). Increased calpain activity has been reported in brains of HD mouse models and human patients, and could result from increased Ca2+ influx into neurons after excitotoxic stimuli (Gafni and Ellerby 2002; Gafni and others 2004). Calpain inhibitors could be useful therapeutic agents for HD and ameliorate the neuronal damage caused by excitotoxicity.
Other proteolytic events
A recent study describes the involvement of matrix metalloproteinases (MMPs), in particular MMP-10 in htt proteolysis (Fig. 6c) (Miller and others 2010). The cleavage site has been mapped to amino acid 402 (in PEST domain 2), and prevention of this proteolytic event by knocking down MMP-10 rescued striatal neurons expressing mhtt from cell death. Elevated MMP levels have also been found in HD mouse models, indicating that these proteases might play a role in the generation of toxic mhtt fragments in vivo (Miller and others 2010).
Aspartyl proteases, in particular the lysosomal proteases cathepsin D, B and L also play a role in htt proteolysis (Kim and others 2006). They are likely responsible for the generation of N-terminal fragments termed cp-A and cp-B (cleavage product A and B, Fig. 6c) that are a major component of nuclear and cytoplasmic inclusions, respectively (Lunkes and others 2002). The cleavage site leading to the generation of cp-A has been narrowed down to the region between amino acids 104-114, whereas the site responsible for cp-B cleavage lies between amino acids 205-214 (Kim and others 2006; Lunkes and others 2002). The expression of mhtt or proteasome inhibition can both increase the activity of cathepsins, suggesting that an age-dependent decrease in proteasome activity could lead to increased proteolysis of mhtt and account for the slow accumulation of N-terminal mhtt fragments in HD.
At the N-terminus of htt, additional cleavage events have been reported that generate small protein fragments (Fig. 6c): Cleavage between residues 81 and 129 generates the cp-1 fragment, and proteolysis at R167 gives rise to a fragment termed cp-2 (Ratovitski and others 2009; Ratovitski and others 2007). The cp-2 fragment can be generated from the 586 fragment (Ratovitski and others 2009), suggesting that the small N-terminal htt fragments are the final product of multiple, sequential proteolysis steps catalyzed by different proteases. However, the time course and natural history of different cleavage events and their relation to each other is not known, precluding their evaluation as therapeutic targets.
A recent study describes the effort to map proteolytic cleavage sites in both wt and mutant htt, using a panel of antibodies spanning the whole protein and lysates from different brain regions of an HD mouse model (Landles and others 2010). A total of 14 fragments of different sizes was detected, with at least four of them potentially terminating at caspase or calpain cleavage sites. The cleavage pattern was consistent throughout the brain, although the relative band intensities varied among brain regions, in agreement with another study that reports enhanced levels of htt fragments in striatum compared to cortical tissues from HD patients (Landles and others 2010; Mende-Mueller, Toneff, Hwang, Chesselet and Hook 2001). A quantification of fragment accumulation over time showed that while aging mice exhibit lower levels of soluble mhtt fragments in the cytoplasm, a small increase of soluble mhtt as well as the appearance of aggregates in the nucleus was detected, indicating that mhtt fragments accumulate in the nucleus where they make up the insoluble inclusions seen in end-stage HD brains (Landles and others 2010).
Interactions between PTMs in the PEST1 domain
Many of the PTMs described above are arranged in clusters in the htt protein sequence (Fig. 1), giving rise to the question whether the single modifications interact and regulate each other. This concept is not unique to htt, as the formation of a complex interaction network between PTMs has been shown for other proteins. PTMs of the p53 protein, for example, have been studied extensively and they provide a fine-tuning mechanism that enables cells to regulate the stability and and activity of this tumor suppressor. More than 36 different amino acids within p53 have been shown to be subject to PTMs, phosphorylation, ubiquitination, acetylation, methylation, sumoylation, neddylation, glycosylation and ribosylation sites have been identified in numerous studies (reviewed in (Kruse and Gu 2009)). Ubiquitination has been shown to compete with acetylation for the same residues in p53, and acetylation dominates over ubiquitination under stress conditions, thereby stabilizing the protein and allowing for its activation (Kruse and Gu 2009; Yang and Seto 2008).
This mechanism is reminiscent of the competition between SUMOylation and ubiquitination for the same residues in the htt protein, with SUMOylation preventing proteasomal degradation and causing htt accumulation as well as increasing transcriptional repression by the protein (Fig. 7) (Steffan and others 2004). A very similar scenario has also been reported for another polyglutamine expansion disorder, spinal and bulbar muscular atrophy (SMBA). The androgen receptor (AR), which in its mutated form causes SMBA, can be ubiquitinated and SUMOylated at the same lysine residues, with SUMOylation leading to AR-dependent repression of transcription (Nishida and Yasuda 2002; Poukka, Karvonen, Janne and Palvimo 2000). In HD and potentially also in other polyglutamine expansion disorders, the beneficial effect of ubiquitination and subsequent degradation of the mutant protein can thus be overridden by SUMOylation, which increases neurodegeneration by exacerbating nuclear toxicity.
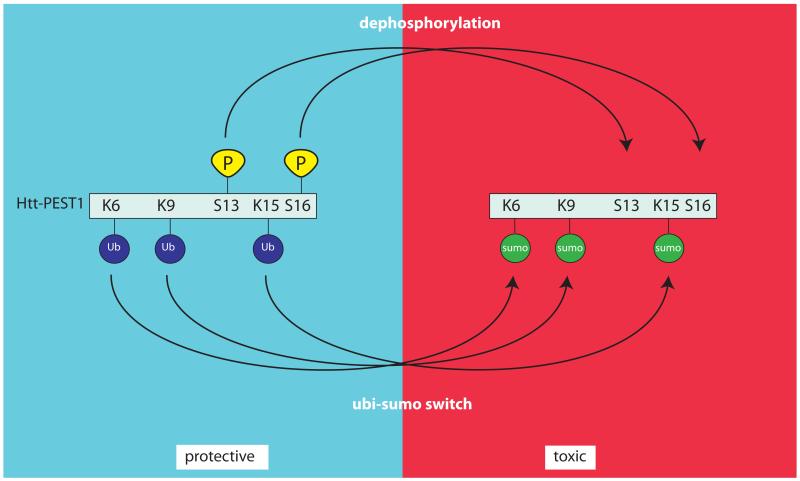
Figure 7. Competition between ubiquitination and sumoylation of htt.
Phosphorylation of htt at S13 and S16 results in its polyubiquitination at lysines K6, K9 and K15 (left-handside of diagram). Absence of phosphorylation at these sites results in sumoylation of htt at lysines K6, K9 and K15 and toxic consequences, as observed in HD mouse models (right-handside of diagram).
Phosphorylation adds another layer of complexity to the PTM network. In p53, phosphorylation at Ser residues adjacent to the acetylation sites is used to promote the interaction with the acetyl transferase p300 and activates p53 in response to DNA damage (Yang and Seto 2008). At the Htt N-terminus, a similar regulation of the ubiquitination/SUMOylation equilibrium by phosphorylation has been shown: Preventing phosphorylation at S13 reduces the polyubiquitination of mhtt and increases its poly-SUMOylation (Thompson and others 2009), in agreement with the neuroprotective role of S13 phosphorylation (Gu and others 2009). These findings put phosphorylation at S13 upstream of other N-terminal PTMs, however it still remains unclear if modifications in the first 17 amino acids interact with PTMs at other residues including those in PEST domain 2.
PTM interactions in PEST domain 2
The second PEST domain is dominated by proteolytic cleavage sites and residues at which phosphorylation and palmitoylation occur (Fig. 1). It is interesting to note that proteolytic cleavage by caspases is known to be regulated by phosphorylation in the vicinity of the cleavage site in many different proteins: Caspase cleavage of the BH-3 protein Bid, the tumor suppressor PTEN, phospholipase C-γ1 and of IκBα are all prevented by phosphorylation. However examples of increased caspase cleavage after adjacent phosphorylation have also been reported (Kurokawa and Kornbluth 2009).
In Htt, phosphorylation at S421 has hereby been shown to reduce htt cleavage at D586 and decrease the accumulation of nuclear fragments, in agreement with the protective role of phosphorylation and the detrimental effects of proteolysis (Warby and others 2009). Similarly, also phosphorylation at S434 reduces caspase cleavage of htt, although in this case only proteolysis at D513 and D552 was assessed (Luo, Vacher, Davies and Rubinsztein 2005). A potential mechanism linking caspase cleavage and phosphorylation in the second PEST domain is excitotoxicity (Fig. 8). Phosphorylation at S421 is reduced in primary neurons and a YAC HD mouse model after excitotoxic stimuli, and blocking dephosphorylation protects YAC primary neurons from excitotoxic death (Metzler and others in press; Metzler and others 2007). At the same time, mice expressing mhtt resistant to caspase-6 cleavage (C6R mice) are protected from excitotoxicity, suggesting a role for the mhtt 586 fragment in NMDAR excitotoxicity (Graham and others 2006a; Milnerwood and others 2010).
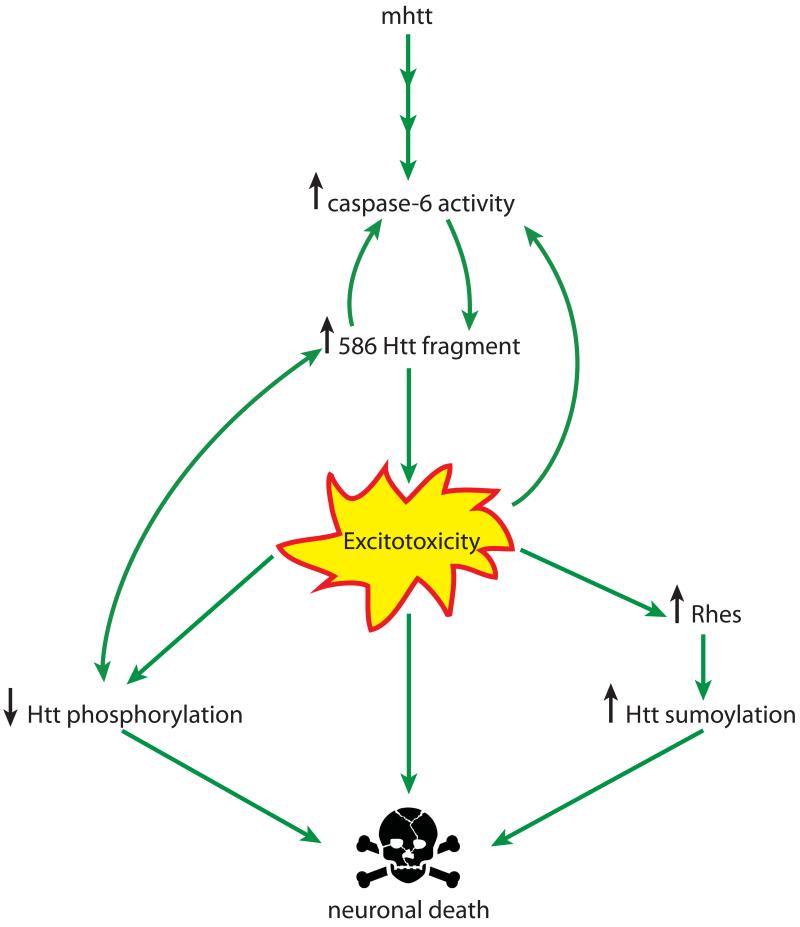
Figure 8. Interplay of PTMs of htt in disease pathology.
Excitotoxicity links reduced phosphorylation, caspase-6 cleavage and increased SUMOylation of mutant htt in the pathogenesis of Huntington disease.
Taking these observations together a feedback loop may be envisaged, with mhtt cleavage resulting in toxic fragments that amplify caspase-6 activity and increase the susceptibility to excitotoxicity, which in turn decreases phosphorylation at S421 and enhances the generation of more htt cleavage fragments.
Interactions spanning the PEST1 and PEST2 domains
An excitotoxic mechanism also links N-terminal SUMOylation of mhtt with its cleavage by caspase-6: Increased extrasynaptic NMDAR activity in an HD model requires the cleavage of mhtt at D586 (Milnerwood and others 2010) and leads to increased levels in Rhes (Okamoto and others 2009), the E3 ligase responsible for the SUMOylation of htt (Subramaniam, Sixt, Barrow and Snyder 2009). Both SUMOylation and cleavage have been shown to increase the toxicity of mhtt (Steffan and others 2004; Warby and others 2008), which is reversed upon blockage of extrasynaptic NMDAR stimulation with low-dose memantine treatment (Okamoto and others 2009). Through the increased influx of calcium ions upon NMDAR stimulation, excitotoxicity also leads to the activation of calpains, a mechanism which is thought to stimulate cleavage of htt by these proteases (Gafni and Ellerby 2002; Gafni and others 2004). Although the precise sequence of events is unclear, cleavage of mhtt at D586 seems to be upstream, since mice expressing caspase-6 resistant mhtt are protected from excitotoxicity (Graham and others 2006a), whereas calpains would be activated further downstream in the pathological cascade.
Another example of an interplay between SUMOylation, phosphorylation and caspase cleavage has recently been published for the protein SATB1 (special AT-rich sequence-binding protein 1): In this case, SUMOylation promotes caspase-6 mediated cleavage of SATB1, but phosphorylation at T188 prevents the interaction of SATB1 with its SUMO E3 ligase PIAS, thus inhibiting its SUMOylation and subsequent caspase-6 cleavage (Tan, Song, Chen and Durrin 2010), indicating that the interplay between phosphorylation, SUMOylation and caspase-6 cleavage might be a more widespread regulatory mechanism in the nucleus.
Htt modifying enzymes as therapeutic targets
Studies on PTMs in the htt protein have identified a number of potential therapeutic targets: SUMO and ubiquitin ligases, kinases, proteases, palmitoyl transferases, acetyl transferases as well as SUMO- and ubiquitin hydrolases, phosphatases, thioesterases and deacetylases are involved, and many of these enzymes are known for htt (Table 2). Knowledge of the enzymes could then theoretically allow for the design or identification of inhibiting or activating compounds. It is generally easier to inhibit than to increase activity, and inhibitors of proteases, phosphatases, deacetylases, SUMO ligases, ubiquitin hydrolases or thioesterases might represent tractable approaches for therapy development. While target selectivity is a desirable feature to avoid potential off-target side-effects, most approved drugs do not demonstrate exquisite selectivity. Imatinib for example was originally developed as an inhibitor of the tyrosine kinase c-ABL and later found to also inhibit c-kit and PDGFR (Landry and Gies 2008). Drug discovery efforts furthermore need to evaluate the druggability of the intended target, meaning the likelihood that drug binding pockets are available in the protein structure and the intended alteration of activity can be achieved by small drug-like molecules (Keller, Pichota and Yin 2006; Schmidtke and Barril 2010). Computational methods for the estimation of target druggability therefore rely on the 3D structure of the protein, which is unfortunately not always known (Keller, Pichota and Yin 2006). The recently published crystal structure of active caspase-6, for example, will be helpful in the design and evaluation of inhibitor compounds for this enzyme (Wang and others 2010).
Table 2.
Enzymes responsible for catalyzing PTMs of htt
| Modification | Amino acid | Responsible enzyme |
Altered activity in HD models |
Antagonisti c enzyme, altered in HD? |
|---|---|---|---|---|
| SUMOylation | K6, K9, K15 | Rhes | increased | |
| ubiquitination | K6, K9, K15 | Ubiquitin E3 ligase eg. E2-25K |
reduced | |
| phosphorylati on |
T3 | ? | reduced | |
| S13,S16 | IKK | reduced | ||
| S421 | Akt1, SGK | unresolve d |
PPT1 / PP2A, reduced |
|
| S434 , S1181, S1201 | Cdk5 | reduced | ||
| S2076, S2653, S2657 |
ERK1 | reduced | ||
| acetylation | K444 | CBP | unknown | HDAC1 |
| cleavage | D513 | Caspase-3 | no | |
| D552 | Caspase-2/3 | no | ||
| D586 | Caspase-6 | increased | ||
| T469, S536 | Calpain 1,5,7,10 | increased | ||
| 104-114, 205-214 | Cathepsin D,B,L | increased | ||
| 81-129, R167 | Undefined proteases |
unknown | ||
| G402 | MMP-10 | increased | ||
| palmitoylatio n |
C214 | HIP14 | reduced |
Even if efficient and selective agonists or antagonists for htt modifying enzymes were available, however, the downstream effects of such a pharmacological intervention on other target proteins have to be considered. The most attractive therapeutic targets are therefore those enzymes, whose activity is generally increased or decreased in the disease state, since enzymatic activity towards all target proteins can be modified by compounds targeting the active site or allosteric sites modulating overall activity. An example for a target with deregulated activity is caspase-6, which has similar affinities for wt and mhtt but shows increased activity in human HD brain and in different HD model systems (Graham and others in press; Hermel and others 2004). An inhibitor normalizing these levels could therefore be expected to show protection without the side effect of impairing the physiological cleavage of other substrates. The successful development of kinase inhibitors such as imatinib (i.e. Gleevec) for chronic myelogenous leukemia shows that enzyme-targeted molecular therapy is a viable approach for the treatment of chronic illnesses (Sherbenou and Druker 2007).
For enzymes that do not show a pathological increase or decrease in activity but rather change their affinity for htt depending on the length of the polyglutamine stretch, selective agonists or antagonists would be necessary to attack exosites responsible for htt binding, similar to the thrombin exosites targeted by anticoagulants used for the treatment of thrombosis (Bock, Panizzi and Verhamme 2007). This could prove to be a more difficult approach since for most enzymes htt-binding exosites are not well defined at the moment.
Another approach to identify therapeutic targets is the exploration of upstream regulators of the enzymes ultimately modifying htt. For example, the IGF-1 pathway leading to Akt activation via PI3-kinase and culminating in the phosphorylation of htt (Humbert and others 2002) is already well elucidated. For caspase-6, recent reports point towards an activation mechanism in neurodegeneration that involves death receptor signalling either via death receptor 6 (DR6) or p75NTR, members of the tumor necrosis factor receptor (TNFR) family (Nikolaev, McLaughlin, O’Leary and Tessier-Lavigne 2009; Park, Grosso, Aubert, Kaplan and Miller 2010). For p75NTR, the pathway is thought to involve Rho signalling through ROCK and activation of the c-Jun N-terminal kinase (JNK) (Park, Grosso, Aubert, Kaplan and Miller 2010), whereas signalling downstream of DR6 has not been elucidated. Activation pathways for other enzymes such as the palmitoyl transferases or phosphatases are less well studied (Dalva 2009; Noritake and others 2009) and htt thioesterases are still unknown. Therefore, a better understanding of the events leading to deregulated enzymatic activities and altered PTM of mhtt will likely identify additional drug targets.
In summary, PTMs of mhtt are attractive starting points for therapeutic development. Many of the modifying enzymes are known, and it may be possible to identify and test the respective agonists and antagonists in the appropriate cellular and in vivo disease model systems. Such agents will also provide helpful tools for the elucidation of the timecourse and interdependence of PTMs in order to identify the most tractable and most upstream therapeutic target(s).
Acknowledgments
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR MOP-84438) and the Cure Huntington Disease Initiative (CHDI) ‘TREAT HD’ grant to MRH. DEE is a CIHR fellowship awardee and held a postdoctoral fellowship of the Austrian Science Fund (FWF). MRH is a University Killam Professor and holds a Canada Research Chair in Human Genetics.
References
- Aiken CT, Steffan JS, Guerrero CM, Khashwji H, Lukacsovich T, Simmons D. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J Biol Chem. 2009;284(43):29427–36. doi: 10.1074/jbc.M109.013193. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin RL, Young AB, Penney JB, Handelin B, Balfour R, Anderson KD. Abnormalities of striatal projection neurons and N-methyl-D-aspartate receptors in presymptomatic Huntington’s disease. N Engl J Med. 1990;322(18):1293–8. doi: 10.1056/NEJM199005033221807. others. [DOI] [PubMed] [Google Scholar]
- Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC. Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol. 2007;170(4):1200–9. doi: 10.2353/ajpath.2007.060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J Neurosci. 2007;27(27):7318–28. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24(17):4250–8. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke SJ, Schmied FA, Brunt ER, Ellerby LM, Paulson HL. Caspase-mediated proteolysis of the polyglutamine disease protein ataxin-3. J Neurochem. 2004;89(4):908–18. doi: 10.1111/j.1471-4159.2004.02369.x. [DOI] [PubMed] [Google Scholar]
- Bessert DA, Gutridge KL, Dunbar JC, Carlock LR. The identification of a functional nuclear localization signal in the Huntington disease protein. Brain Res Mol Brain Res. 1995;33(1):165–73. doi: 10.1016/0169-328x(95)00124-b. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006;355(3):524–35. doi: 10.1016/j.jmb.2005.10.053. others. [DOI] [PubMed] [Google Scholar]
- Bock PE, Panizzi P, Verhamme IM. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemost. 2007;5(Suppl 1):81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104(24):10045–50. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum Mol Genet. 2002;11(23):2895–904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113(4):457–68. doi: 10.1016/s0092-8674(03)00349-0. others. [DOI] [PubMed] [Google Scholar]
- Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13(14):1407–20. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- Colin E, Zala D, Liot G, Rangone H, Borrell-Pages M, Li XJ. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27(15):2124–34. doi: 10.1038/emboj.2008.133. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer Gould VF, Goti D, Pearce D, Gonzalez GA, Gao H, Bermudez de Leon M. A mutant ataxin-3 fragment results from processing at a site N-terminal to amino acid 190 in brain of Machado-Joseph disease-like transgenic mice. Neurobiol Dis. 2007;27(3):362–9. doi: 10.1016/j.nbd.2007.06.005. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepaldi L, Riccio A. Chromatin learns to behave. Epigenetics. 2009;4(1):23–6. doi: 10.4161/epi.4.1.7604. [DOI] [PubMed] [Google Scholar]
- Dalva MB. Neuronal activity moves protein palmitoylation into the synapse. J Cell Biol. 2009;186(1):7–9. doi: 10.1083/jcb.200906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Bertolotti A. Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006;281(47):35608–15. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- Denuc A, Marfany G. SUMO and ubiquitin paths converge. Biochem Soc Trans. 2010;38(Pt 1):34–9. doi: 10.1042/BST0380034. [DOI] [PubMed] [Google Scholar]
- Dey ND, Bombard MC, Roland BP, Davidson S, Lu M, Rossignol J. Genetically engineered mesenchymal stem cells reduce behavioral deficits in the YAC 128 mouse model of Huntington’s disease. Behav Brain Res. 2010;214(2):193–200. doi: 10.1016/j.bbr.2010.05.023. others. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–3. doi: 10.1126/science.277.5334.1990. others. [DOI] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134(4):679–91. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel RC, Manzana E, Green WN. The role of palmitoylation in functional expression of nicotinic alpha7 receptors. J Neurosci. 2004;24(46):10502–10. doi: 10.1523/JNEUROSCI.3315-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick LA, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J. SCA1-like Disease in Mice Expressing Wild-Type Ataxin-1 with a Serine to Aspartic Acid Replacement at Residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerby LM, Andrusiak RL, Wellington CL, Hackam AS, Propp SS, Wood JD. Cleavage of atrophin-1 at caspase site aspartic acid 109 modulates cytotoxicity. J Biol Chem. 1999;274(13):8730–6. doi: 10.1074/jbc.274.13.8730. others. [DOI] [PubMed] [Google Scholar]
- Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38(3):375–87. doi: 10.1016/s0896-6273(03)00258-7. others. [DOI] [PubMed] [Google Scholar]
- Falk JD, Vargiu P, Foye PE, Usui H, Perez J, Danielson PE. Rhes: A striatal-specific Ras homolog related to Dexras1. J Neurosci Res. 1999;57(6):782–8. others. [PubMed] [Google Scholar]
- Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275(27):20853–60. doi: 10.1074/jbc.M000660200. others. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci. 2010;11(3):161–75. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- Gafni J, Ellerby LM. Calpain activation in Huntington’s disease. J Neurosci. 2002;22(12):4842–9. doi: 10.1523/JNEUROSCI.22-12-04842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J Biol Chem. 2004;279(19):20211–20. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV, Sitaraman S. Reversal of Alzheimer’s-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc Natl Acad Sci U S A. 2006;103(18):7130–5. doi: 10.1073/pnas.0509695103. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, Libby RT, Fu YH, Kinoshita Y, Huang J, Possin DE. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22(12):4897–905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–38. doi: 10.1016/j.cell.2004.06.018. others. [DOI] [PubMed] [Google Scholar]
- Gines S, Ivanova E, Seong IS, Saura CA, MacDonald ME. Enhanced Akt signaling is an early pro-survival response that reflects N-methyl-D-aspartate receptor activation in Huntington’s disease knock-in striatal cells. J Biol Chem. 2003;278(50):50514–22. doi: 10.1074/jbc.M309348200. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, Graham RK. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13(4):442–9. doi: 10.1038/ng0896-442. others. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA. Cleavage at the 586aa caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci. doi: 10.1523/JNEUROSCI.2071-10.2010. others. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006a;125(6):1179–91. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Graham RK, Slow EJ, Deng Y, Bissada N, Lu G, Pearson J, et al. Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol Dis. 2006b;21(2):444–55. doi: 10.1016/j.nbd.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142(4):637–46. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–40. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haacke A, Broadley SA, Boteva R, Tzvetkov N, Hartl FU, Breuer P. Proteolytic cleavage of polyglutamine-expanded ataxin-3 is critical for aggregation and sequestration of non-expanded ataxin-3. Hum Mol Genet. 2006;15(4):555–68. doi: 10.1093/hmg/ddi472. [DOI] [PubMed] [Google Scholar]
- Haas KF, Broadie K. Roles of ubiquitination at the synapse. Biochim Biophys Acta. 2008;1779(8):495–506. doi: 10.1016/j.bbagrm.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, et al. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol. 1998;141(5):1097–105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28(8):425–33. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, et al. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ. 2004;11(4):424–38. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffner G, Kahlem P, Djian P. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: relevance to Huntington’s disease. J Cell Sci. 2002;115(Pt 5):941–8. doi: 10.1242/jcs.115.5.941. [DOI] [PubMed] [Google Scholar]
- Huang K, Kang MH, Askew C, Kang R, Sanders SS, Wan J, et al. Palmitoylation and function of glial glutamate transporter-1 is reduced in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2010;40(1):207–15. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Huang K, Sanders S, Singaraja R, Orban P, Cijsouw T, Arstikaitis P, et al. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009;23(8):2605–15. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, et al. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2(6):831–7. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, et al. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280(12):11635–40. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr., Mazzulli JR, Cui L, Savas JN, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137(1):60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Xu K, Lessing D, Bonini NM. Preventing Ataxin-3 protein cleavage mitigates degeneration in a Drosophila model of SCA3. Hum Mol Genet. 2009;18(24):4843–52. doi: 10.1093/hmg/ddp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, et al. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271(32):19385–94. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- Kang BS, Ahn JY, Kim MK, Kim HJ, Kang L, Lim HC, et al. Heat shock protein 70 alters the endosome-lysosomal localization of huntingtin. Exp Mol Med. 2007;39(1):38–46. doi: 10.1038/emm.2007.5. [DOI] [PubMed] [Google Scholar]
- Keller TH, Pichota A, Yin Z. A practical view of ‘druggability’. Curr Opin Chem Biol. 2006;10(4):357–61. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Kim M, Lee HS, LaForet G, McIntyre C, Martin EJ, Chang P, et al. Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J Neurosci. 1999;19(3):964–73. doi: 10.1523/JNEUROSCI.19-03-00964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Sapp E, Cuiffo BG, Sobin L, Yoder J, Kegel KB, et al. Lysosomal proteases are involved in generation of N-terminal huntingtin fragments. Neurobiol Dis. 2006;22(2):346–56. doi: 10.1016/j.nbd.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Miwa S, Merry DE, Kume A, Mei L, Doyu M, et al. Caspase-3 cleaves the expanded androgen receptor protein of spinal and bulbar muscular atrophy in a polyglutamine repeat length-dependent manner. Biochem Biophys Res Commun. 1998;252(1):145–50. doi: 10.1006/bbrc.1998.9624. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138(5):838–54. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H, Finkbeiner S, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285(12):8808–23. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry Y, Gies JP. Drugs and their molecular targets: an updated overview. Fundam Clin Pharmacol. 2008;22(1):1–18. doi: 10.1111/j.1472-8206.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- Leavitt BR, van Raamsdonk JM, Shehadeh J, Fernandes H, Murphy Z, Graham RK, et al. Wild-type huntingtin protects neurons from excitotoxicity. J Neurochem. 2006;96(4):1121–9. doi: 10.1111/j.1471-4159.2005.03605.x. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279(39):40545–59. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li M, Chevalier-Larsen ES, Merry DE, Diamond MI. Soluble androgen receptor oligomers underlie pathology in a mouse model of spinobulbar muscular atrophy. J Biol Chem. 2007;282(5):3157–64. doi: 10.1074/jbc.M609972200. [DOI] [PubMed] [Google Scholar]
- Li W, Serpell LC, Carter WJ, Rubinsztein DC, Huntington JA. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J Biol Chem. 2006;281(23):15916–22. doi: 10.1074/jbc.M511007200. [DOI] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia JM, Lo SC, Whitcomb DJ, Jiao S, et al. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell. 2010;141(5):859–71. doi: 10.1016/j.cell.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, et al. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med. 2000;6(4):397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Lindenberg KS, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer GB, et al. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell. 2002;10(2):259–69. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- Luo S, Vacher C, Davies JE, Rubinsztein DC. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J Cell Biol. 2005;169(4):647–56. doi: 10.1083/jcb.200412071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134(5):866–76. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, et al. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23(3):659–69. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire JR, Rong J, Li SH, Li XJ. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281(6):3552–9. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF, Hook VY. Tissue-specific proteolysis of Huntingtin (htt) in human brain: evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum. J Neurosci. 2001;21(6):1830–7. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Gan L, Mazarei G, Graham RK, Liu L, Bissada N, et al. Phosphorylation of Huntingtin at Ser421 in YAC128 Neurons Is Associated with Protection of YAC128 Neurons from NMDA-Mediated Excitotoxicity and Is Modulated by PP1 and PP2A. J Neurosci. doi: 10.1523/JNEUROSCI.1589-10.2010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Gan L, Wong TP, Liu L, Helm J, Georgiou J, et al. NMDA receptor function and NMDA receptor-dependent phosphorylation of huntingtin is altered by the endocytic protein HIP1. J Neurosci. 2007;27(9):2298–308. doi: 10.1523/JNEUROSCI.5175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, Zhang N, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease. Neuron. 2010;67(2):199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65(2):178–90. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Okamura-Oho Y, Mito Y, Nagafuchi S, Yamada M. Dentatorubral pallidoluysian atrophy (DRPLA) protein is cleaved by caspase-3 during apoptosis. J Biol Chem. 1997;272(46):29238–42. doi: 10.1074/jbc.272.46.29238. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Thomas M, Dadgar N, Lieberman AP, Iniguez-Lluhi JA. Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem. 2009;284(32):21296–306. doi: 10.1074/jbc.M109.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457(7232):981–9. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nishida T, Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J Biol Chem. 2002;277(44):41311–7. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. 2009;186(1):147–60. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak MJ, Tabrizi SJ. Huntington’s disease. BMJ. 2010;340:c3109. doi: 10.1136/bmj.c3109. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, et al. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15(12):1407–13. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzolo I, Burnett BG, Young JE, Brenne PL, La Spada AR, Fischbeck KH, et al. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum Mol Genet. 2007;16(13):1593–603. doi: 10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- Park KJ, Grosso CA, Aubert I, Kaplan DR, Miller FD. p75NTR-dependent, myelin-mediated axonal degeneration regulates neural connectivity in the adult brain. Nat Neurosci. 2010;13(5):559–66. doi: 10.1038/nn.2513. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem. 2007;282(31):22278–88. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- Pennuto M, Palazzolo I, Poletti A. Post-translational modifications of expanded polyglutamine proteins: impact on neurotoxicity. Hum Mol Genet. 2009;18(R1):R40–7. doi: 10.1093/hmg/ddn412. [DOI] [PubMed] [Google Scholar]
- Pineda JR, Pardo R, Zala D, Yu H, Humbert S, Saudou F. Genetic and pharmacological inhibition of calcineurin corrects the BDNF transport defect in Huntington’s disease. Mol Brain. 2009;2(1):33. doi: 10.1186/1756-6606-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci U S A. 2000;97(26):14145–50. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger E, Jordan BM, Kazantsev A, Housman D. Evidence for a recruitment and sequestration mechanism in Huntington’s disease. Philos Trans R Soc Lond B Biol Sci. 1999;354(1386):1029–34. doi: 10.1098/rstb.1999.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhilin S, Drisdel RC, Sagher D, McGehee DS, Vallejo Y, Green WN. alpha7 subunits in two different disulfide-bonded conformations. J Cell Biol. 1999;146(1):203–18. doi: 10.1083/jcb.146.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangone H, Poizat G, Troncoso J, Ross CA, MacDonald ME, Saudou F, et al. The serum- and glucocorticoid-induced kinase SGK inhibits mutant huntingtin-induced toxicity by phosphorylating serine 421 of huntingtin. Eur J Neurosci. 2004;19(2):273–9. doi: 10.1111/j.0953-816x.2003.03131.x. [DOI] [PubMed] [Google Scholar]
- Ratovitski T, Gucek M, Jiang H, Chighladze E, Waldron E, D’Ambola J, et al. Mutant huntingtin N-terminal fragments of specific size mediate aggregation and toxicity in neuronal cells. J Biol Chem. 2009;284(16):10855–67. doi: 10.1074/jbc.M804813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski T, Nakamura M, D’Ambola J, Chighladze E, Liang Y, Wang W, et al. N-terminal proteolysis of full-length mutant huntingtin in an inducible PC12 cell model of Huntington’s disease. Cell Cycle. 2007;6(23):2970–81. doi: 10.4161/cc.6.23.4992. [DOI] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16(1):61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22(24):6537–49. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Cho S, Park BC, Lee do H. Oxidative stress-enhanced SUMOylation and aggregation of ataxin-1: Implication of JNK pathway. Biochem Biophys Res Commun. 2010;393(2):280–5. doi: 10.1016/j.bbrc.2010.01.122. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95(1):55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Schilling B, Gafni J, Torcassi C, Cong X, Row RH, LaFevre-Bernt MA, et al. Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem. 2006;281(33):23686–97. doi: 10.1074/jbc.M513507200. [DOI] [PubMed] [Google Scholar]
- Schmidtke P, Barril X. Understanding and predicting druggability. A high-throughput method for detection of drug binding sites. J Med Chem. 2010;53(15):5858–67. doi: 10.1021/jm100574m. [DOI] [PubMed] [Google Scholar]
- Schoenmann Z, Assa-Kunik E, Tiomny S, Minis A, Haklai-Topper L, Arama E, et al. Axonal degeneration is regulated by the apoptotic machinery or a NAD+− sensitive pathway in insects and mammals. J Neurosci. 2010;30(18):6375–86. doi: 10.1523/JNEUROSCI.0922-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherbenou DW, Druker BJ. Applying the discovery of the Philadelphia chromosome. J Clin Invest. 2007;117(8):2067–74. doi: 10.1172/JCI31988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, et al. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci U S A. 2005;102(32):11402–7. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, Khoshnan A, Dunn DE, Bugg CW, Lo DC, Patterson PH. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci. 2008;28(36):9013–20. doi: 10.1523/JNEUROSCI.2747-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304(5667):100–4. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Strehlow AN, Li JZ, Myers RM. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum Mol Genet. 2007;16(4):391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- Strong TV, Tagle DA, Valdes JM, Elmer LW, Boehm K, Swaroop M, et al. Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nat Genet. 1993;5(3):259–65. doi: 10.1038/ng1193-259. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324(5932):1327–30. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JA, Song J, Chen Y, Durrin LK. Phosphorylation-dependent interaction of SATB1 and PIAS1 directs SUMO-regulated caspase cleavage of SATB1. Mol Cell Biol. 2010;30(11):2823–36. doi: 10.1128/MCB.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima T, Kawai H, Fujitani M, Maeda K, Yasuda H. SUMO-1 co-localized with mutant atrophin-1 with expanded polyglutamines accelerates intranuclear aggregation and cell death. Neuroreport. 2002;13(17):2359–64. doi: 10.1097/00001756-200212030-00038. [DOI] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187(7):1083–99. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, McMurray CT. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145(4):1233–48. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- Tsai LH, Takahashi T, Caviness VS, Jr., Harlow E. Activity and expression pattern of cyclin-dependent kinase 5 in the embryonic mouse nervous system. Development. 1993;119(4):1029–40. doi: 10.1242/dev.119.4.1029. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278(24):22044–55. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- Ueda H, Goto J, Hashida H, Lin X, Oyanagi K, Kawano H, et al. Enhanced SUMOylation in polyglutamine diseases. Biochem Biophys Res Commun. 2002;293(1):307–13. doi: 10.1016/S0006-291X(02)00211-5. [DOI] [PubMed] [Google Scholar]
- Walsh R, Storey E, Stefani D, Kelly L, Turnbull V. The roles of proteolysis and nuclear localisation in the toxicity of the polyglutamine diseases. A review. Neurotox Res. 2005;7(1-2):43–57. doi: 10.1007/BF03033775. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, et al. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010 doi: 10.1038/embor.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Chan EY, Metzler M, Gan L, Singaraja RR, Crocker SF, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum Mol Genet. 2005;14(11):1569–77. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet. 2008;17(15):2390–404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- Warby SC, Doty CN, Graham RK, Shively J, Singaraja RR, Hayden MR. Phosphorylation of huntingtin reduces the accumulation of its nuclear fragments. Mol Cell Neurosci. 2009;40(2):121–7. doi: 10.1016/j.mcn.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273(15):9158–67. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Hayden MR. Caspases and neurodegeneration: on the cutting edge of new therapeutic approaches. Clin Genet. 2000;57(1):1–10. doi: 10.1034/j.1399-0004.2000.570101.x. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, et al. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J Biol Chem. 2000;275(26):19831–8. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Nakamura Y, Henley JM. Targets and consequences of protein SUMOylation in neurons. Brain Res Rev. 2010;64(1):195–212. doi: 10.1016/j.brainresrev.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Lee DH, Taylor J, Vandelft M, Truant R. Huntingtin contains a highly conserved nuclear export signal. Hum Mol Genet. 2003;12(12):1393–403. doi: 10.1093/hmg/ddg156. [DOI] [PubMed] [Google Scholar]
- Xie Y, Hayden MR, Xu B. BDNF Overexpression in the Forebrain Rescues Huntington’s Disease Phenotypes in YAC128 Mice. J Neurosci. doi: 10.1523/JNEUROSCI.1637-10.2010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9(6):824–31. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31(4):449–61. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JE, Gouw L, Propp S, Sopher BL, Taylor J, Lin A, et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J Biol Chem. 2007;282(41):30150–60. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- Zala D, Colin E, Rangone H, Liot G, Humbert S, Saudou F. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum Mol Genet. 2008;17(24):3837–46. doi: 10.1093/hmg/ddn281. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, et al. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron. 2002;33(6):849–60. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li M, Drozda M, Chen M, Ren S, Mejia Sanchez RO, et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J Neurochem. 2003;87(1):101–6. doi: 10.1046/j.1471-4159.2003.01980.x. [DOI] [PubMed] [Google Scholar]
- Zuchner T, Brundin P. Mutant huntingtin can paradoxically protect neurons from death. Cell Death Differ. 2008;15(3):435–42. doi: 10.1038/sj.cdd.4402261. [DOI] [PubMed] [Google Scholar]