SUMMARY
We describe the results of a genome-wide analysis of human cells that suggests that most protein-coding genes, including most genes thought to be transcriptionally inactive, experience transcription initiation. We found that nucleosomes with H3K4me3 and H3K9,14Ac modifications, together with RNA polymerase II, occupy the promoters of most protein-coding genes in human embryonic stem cells. Only a subset of these genes produce detectable full-length transcripts and are occupied by nucleosomes with H3K36me3 modifications, a hallmark of elongation. The other genes experience transcription initiation but show no evidence of elongation, suggesting that they are predominantly regulated at post-initiation steps. Genes encoding most developmental regulators fall into this group. Our results also identify a class of genes that are excluded from experiencing transcription initiation, at which mechanisms that prevent initiation must predominate. These observations extend to differentiated cells, suggesting that transcription initiation at most genes is a general phenomenon in human cells.
INTRODUCTION
Studies of individual genes have revealed that transcriptional regulation can occur at many levels, but these regulatory mechanisms fall into two general models (Ptashne, 1986; Krumm et al., 1993; Kuras and Struhl, 1999; Saunders et al., 2006). In one model, transcription is regulated primarily at the initiation step, when DNA-binding transcription factors recruit the transcriptional machinery. In the other, the key regulatory mechanisms occur subsequent to initiation and involve transcript elongation or stability. These models are not mutually exclusive but make different predictions about the chromatin state of the genome, which can be influenced by transcription.
Transcription initiation and elongation are both associated with specific chemical modifications of the histone components of nucleosomes that reside in the promoter regions and transcribed portions of active protein-coding genes (Jenuwein and Allis, 2001; Turner, 2002; Sims et al., 2004). Among the modifications associated with transcription initiation, the histone H3 lysine 4 trimethyl (H3K4me3) mark occurs in nucleosomes found in the promoter regions of actively transcribed genes (Bernstein et al., 2002; Santos-Rosa et al., 2002; Ng et al., 2003; Schneider et al., 2004; Schubeler et al., 2004; Pokholok et al., 2005). The histone methyltransferase (HMT) responsible for H3K4me3 modification in yeast, the Set1 complex, is recruited to the 5′ end of actively transcribed genes by interacting with the RNA polymerase II (Pol II)-associated PAF complex (Miller et al., 2001; Krogan et al., 2003a; Laribee et al., 2005). Recruitment of mammalian homologs of the Set1 complex to promoters may occur by a similar mechanism (Tenney and Shilatifard, 2005). Acetylation of histone H3 lysine 9 and 14 (H3K9,14Ac) also occurs in promoter-associated nucleosomes at actively transcribed genes (Liang et al., 2004; Schubeler et al., 2004; Bernstein et al., 2005; Pokholok et al., 2005). This is catalyzed by several promoter-associated histone acetyltransferases (HATs), including Gcn5, TAF1, and p300/CBP (Sterner and Berger, 2000; Strahl and Allis, 2000).
Among the modifications associated with transcription elongation, the histone H3 lysine 36 trimethyl (H3K36me3) modification occurs at nucleosomes in the 3′ portion of the transcribed region of actively transcribed genes (Strahl et al., 2002; Bannister et al., 2005; Pokholok et al., 2005). This may be a consequence of recruitment of the HMT responsible for this histone modification by the elongating RNA polymerase (Li et al., 2002, 2003; Xiao et al., 2003).
The relationship between transcriptional activity and chromatin state in embryonic stem (ES) cells is of particular interest. ES cells are uniquely capable of differentiating into almost any cell type (Mayhall et al., 2004; Pera and Trounson, 2004). ES cell chromatin proteins have been reported to be in a hyperdynamic state (Meshorer et al., 2006). Recent studies have shown that a small set of ES cell genes that are transcriptionally inactive but poised for expression contain histones whose modifications are characteristic of both active and inactive genes (Bernstein et al., 2006). However, histone modifications have yet to be studied throughout the genome of ES cells.
We report here that the promoter-proximal nucleosomes of at least 75% of all protein-coding genes in human ES cells are trimethylated at histone H3K4. This is surprising because this particular histone modification was thought to be associated with only actively transcribed genes. Further study revealed that transcription initiation and associated histone modifications occur at most promoters in both ES and differentiated cell types. Thus, most genes that have been considered inactive because they produce no detectable transcript nonetheless experience transcription initiation. This initiation is likely responsible for the modified nucleosome landmark we observe at most promoters.
RESULTS
Most Promoters in Embryonic Stem Cells Have Nucleosomes with H3K4me3
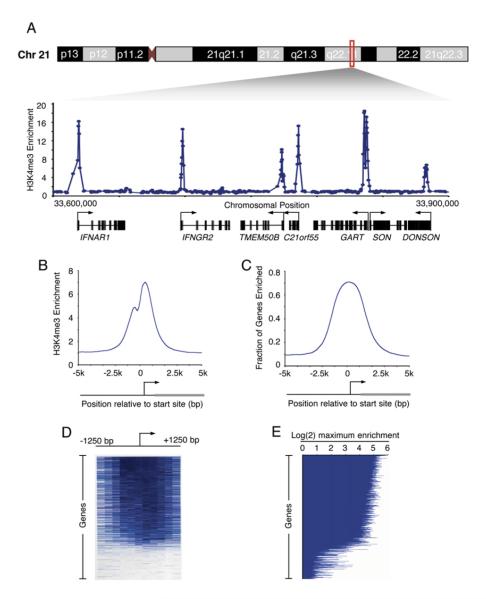
We used chromatin immunoprecipitation coupled to DNA microarray analysis (ChIP-chip) (Boyer et al., 2005; Lee et al., 2006) to determine how nucleosomes with H3K4me3 are distributed across the entire genome in human embryonic stem (hES) cells (Figure 1; see also the Supplemental Experimental Procedures in the Supplemental Data available with this article online). Previous studies had suggested that H3K4me3-modified nucleosomes occur near the transcription start sites of actively transcribed genes (Bernstein et al., 2002; Santos-Rosa et al., 2002; Ng et al., 2003; Schneider et al., 2004; Schubeler et al., 2004; Pokholok et al., 2005). As expected, we found H3K4me3 enriched at sites of transcription initiation (Figure 1A; Table S1). Nearly all sites of H3K4me3 enrichment were within 1 kb of known or predicted transcript start sites, with maximal enrichment immediately downstream of the start site (Figure 1B; Supplemental Experimental Procedures).
Figure 1. H3K4me3-Modified Nucleosomes Are Found at Sites of Transcription Initiation.
(A) Genomic DNA segments enriched for H3K4me3-modified histones in hES cells were identified with ChIP-chip using DNA microarrays tiling the nonrepeat human genome. Enrichment ratios for H3K4me3 across 300 kb of chromosome 21 are shown. Start sites and direction of transcription are indicated by arrows.
(B) Composite H3K4me3 enrichment profile for all known genes. The start site and direction of transcription of the average gene are indicated by an arrow. Details of composite profile generation are described in Supplemental Experimental Procedures.
(C) Fraction of genes enriched for H3K4me3 (enrichment ratio > 2) was calculated for all known promoters. All genes were aligned according to the position of their transcription start sites, and the fraction of genes with enriched H3K4me3 binding was then calculated in a sliding 200 bp window.
(D) Single-probe enrichment ratios of H3K4me3 across the 2.5 kb region surrounding the transcription start site (~10 probes per promoter). Enriched probes (blue) and unenriched probes (white) are shown for each gene. Individual promoters are represented top to bottom and are ordered by average binding ratio within a 1 kb window surrounding the transcription start site.
(E) H3K4me3 maximum enrichment (blue horizontal line) within 1 kb of the transcription start site for each gene represented on the whole-genome array. Genes are ordered as in (D).
Previous studies have estimated that only 30%–45% of genes have detectable mature transcripts (Sato et al., 2003; Brandenberger et al., 2004a; Su et al., 2004), so we were surprised to find that 74% of all annotated promoters were enriched for H3K4 methylation in hES cells at a high confidence level (Figure 1C; Table S2). Using slightly less stringent criteria (2-fold enrichment; see Supplemental Experimental Procedures), we found that 79% of all protein-coding genes have promoter-proximal nucleosomes enriched for H3K4me3 modification (Figures 1D and 1E; Table S3). This was confirmed using a smaller array that tiles all known human promoter regions (Supplemental Experimental Procedures; Table S4). These results led us to believe that a large fraction of genes for which no transcript has been detected nonetheless have promoter-proximal nucleosomes enriched for H3K4me3 modification.
H3K4me3 Is Enriched at Active and Inactive Genes throughout the Genome
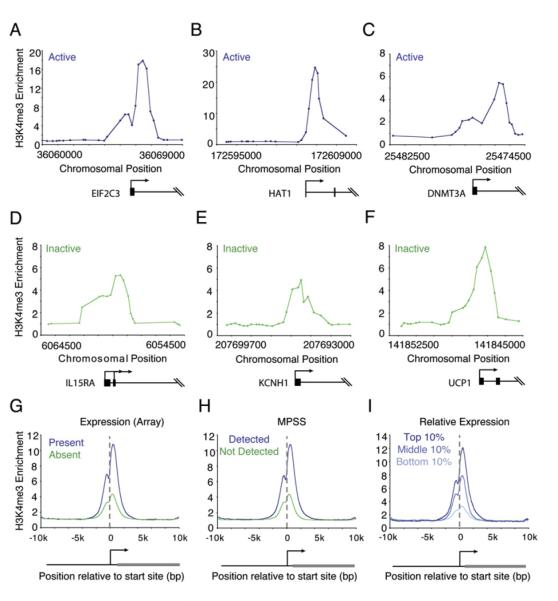
Since H3K4me3 enrichment occurs at a surprisingly large fraction of known and predicted transcription start sites, we reexamined how H3K4me3 methylation is related to transcription. As expected, nucleosomes with H3K4me3 occurred immediately downstream of the transcription start site of actively transcribed genes (Figures 2A–2C) (Bernstein et al., 2002; Santos-Rosa et al., 2002; Ng et al., 2003; Schneider et al., 2004; Schubeler et al., 2004; Pokholok et al., 2005). Nucleosomes with H3K4me3 also occurred immediately downstream of the transcription start site of many genes that are transcriptionally inactive in ES cells. For example, cytokine receptors (IFNAR1), potassium channels expressed in neural tissue (KCNH1), and genes essential for adipose tissue function (UCP1) were all found to be occupied by nucleosomes modified at H3K4me3 (Figures 2D–2F). Transcripts for these genes have not been detected in ES cells based on DNA microarray and massively parallel signature sequencing (MPSS) data (Sato et al., 2003; Abeyta et al., 2004; Brandenberger et al., 2004a; Wei et al., 2005). The signals observed for histone H3K4me3 were typically lower (about 3-fold) at the inactive genes than at active genes but were substantially above background and located at the same position relative to the transcription start site.
Figure 2. H3K4me3-Modified Nucleosomes Are Enriched at Both Active and Inactive Promoters.
(A–F) Genomic DNA segments enriched for H3K4me3-modified histones were identified with ChIP-chip using DNA microarrays spanning the entire nonrepeat portion of the human genome. Enrichment ratios are shown for H3K4me3 at active genes (mRNA transcript called “present” in Affymetrix expression data and detected by massively parallel signature sequencing [MPSS] data) and inactive genes (mRNA transcript called “absent” in Affymetrix expression data and not detected by MPSS data). Gene start and direction of transcription are indicated by arrows.
(G–I) Composite H3K4me3 enrichment profiles for genes called present or absent for mRNA transcript using Affymetrix data (G) (Sato et al., 2003; Abeyta et al., 2004); genes detected or not detected by MPSS (H) (Brandenberger et al., 2004a; Wei et al., 2005); and genes in the top 10%, middle 10%, or bottom 10% by relative expression (I) (Su et al., 2004).
To investigate the relationship between gene transcription and H3K4me3 modification genome-wide, we used three approaches. First, we identified genes whose transcripts in ES cells were consistently called “present” or “absent” in Affymetrix expression data (Sato et al., 2003; Abeyta et al., 2004) and averaged enrichment signals for these genes to create composite histone H3K4me3 profiles for the present and absent genes (Figure 2G). The composite profiles show that, for both gene classes, histone H3K4me3 signals span the 3 kb region surrounding the transcript start site and peak just downstream of the transcript start site. The level of the peak signal for H3K4me3 in the absent class of genes was about 1/3 of that for the present class. Second, we identified genes whose transcripts in ES cells were detected or not detected based on MPSS data (Brandenberger et al., 2004a; Wei et al., 2005) and created a composite histone H3K4me3 profile for the two classes (Figure 2H); the results were very similar to those obtained with microarray expression data. Finally, we used a compendium of microarray expression data for 79 different human cells and tissues to identify genes whose ES cell transcript expression levels were in the top 10%, middle 10%, and bottom 10% relative to all other cells (Su et al., 2004) and created a composite histone H3K4me3 profile for the three classes (Figure 2I). The results confirm that there is a positive correlation between gene transcript levels and histone H3K4me3 signal yet show that histone H3K4me3 is observed even at the promoters of genes for which there is no evidence of transcription in ES cells.
To ensure that the observation that histone H3K4me3 occupies most promoters in human ES cells was not an experimental or analytical artifact, we conducted a series of control experiments. A ChIP-chip experiment with nonspecific IgG produced only background signals that did not show a general enrichment of promoter regions (Figure S4; Supplemental Experimental Procedures). A ChIP-chip experiment with antibody against the transcription factor E2F4, with strength of signal comparable to H3K4me3, showed enrichment of DNA sequences at <10% of genes (Supplemental Experimental Procedures). Finally, an experiment with antibody specific for the histone H3K36me3 modification, which is associated with transcription elongation, showed enrichment of DNA sequences well downstream of the transcription start site at genes that produce RNA transcripts in ES cells (see below).
Evidence for Transcription Initiation at Most Genes
We considered the possibilities that the presence of histone H3K4me3 at most promoters is either due to transcription initiation events that occur at many more genes than suggested by measurements of mature transcripts or is instead independent of transcription. The former model predicts that additional histone modifications associated with transcription initiation, as well as the initiating form of Pol II itself, can be captured at the promoters of more protein-coding genes than are actively transcribed based on RNA transcript data (Sato et al., 2003; Brandenberger et al., 2004b; Su et al., 2004), while the latter would expect no such correlation.
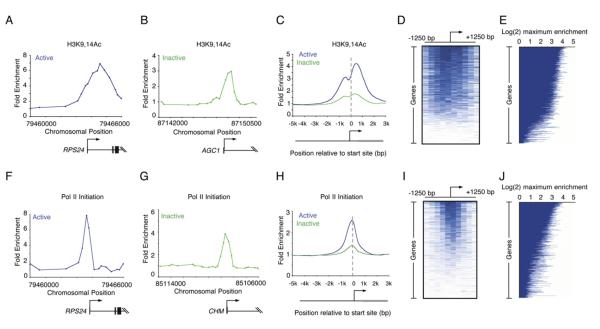
We carried out ChIP-chip experiments in human ES cells using arrays tiling 21,223 known transcription start sites with an antibody specific for H3K9,14Ac, a pair of histone acetylation modifications associated with transcription initiation (Figures 3A–3E). Acetylation of histone H3K9,14 is critical for the recruitment of transcription factor IID (TFIID), an initiating step in transcription (Agalioti et al., 2002), and should therefore be associated with genes that have recently initiated transcription. Nucleosomes with histone H3K9,14Ac were enriched at the promoters of nearly 70% of genes, including both transcriptionally active and inactive genes (Table S2). Nearly all of the promoters (>95%) acetylated on H3K9 and H3K14 were also enriched for H3K4me3.
Figure 3. Enrichment of H3K9,14Ac and Pol II at Inactive Genes.
(A and B) Enrichment ratios are shown for H3K9,14Ac, which is associated with initiation. The start site and direction of transcription are indicated by an arrow.
(C) Composite enrichment profiles for H3K9,14Ac at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G).
(D) Single-probe enrichment ratios of H3K9,14Ac across the average 2.5 kb region surrounding the transcription start site (~10 probes per promoter). Enriched probes (blue) and unenriched probes (white) are shown for each gene. Individual promoters are represented top to bottom and are ordered by average binding ratio within 1 kb surrounding the transcription start site (as in Figure 1D).
(E) H3K9,14Ac maximum enrichment (blue horizontal line) within 1 kb of the transcription start site for each gene represented on the human promoter array. Genes are ordered as in (D).
(F and G) Enrichment ratios are shown for initiating Pol II (antibody 8WG16).
(H) Composite enrichment profiles for initiating Pol II at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G).
(I) Single-probe enrichment ratios of initiating Pol II across the average 2.5 kb region surrounding the transcription start site (~10 probes per promoter). Enriched probes (blue) and unenriched probes (white) are shown for each gene. Individual promoters are represented top to bottom and are ordered by average binding ratio within 1 kb surrounding the transcription start site (as in Figure 1D).
(J) Initiating Pol II maximum enrichment (blue horizontal line) within 1 kb of the transcription start site for each gene represented on the human promoter array. Genes are ordered as in (I).
We then carried out ChIP-chip experiments in human ES cells using an antibody directed against the initiating form of Pol II (8WG16). The initiating form of Pol II occupied promoters both at genes that produce detectable transcripts and at genes that do not, albeit at lower levels (Figures 3F–3J; Table S2). We compared the set of genes whose promoters were occupied by Pol II with those occupied by histone H3K4me3. As expected, most promoters (98%) occupied by Pol II were also occupied by histone H3K4me3, whereas Pol II occupied few genes (2.3%) that lack histone H3K4me3. The fraction of all promoters enriched for the initiating form of Pol II (52% based on a 2-fold enrichment ratio) was somewhat smaller than that occupied by nucleosomes with H3K4me3 modification (79%) or by nucleosomes with H3K9,14Ac modifications (70%). This may be due to greater longevity of the nucleosome modifications than Pol II at these promoters or due to the higher signal produced by ChIP with histone antibodies than ChIP with RNA polymerase antibodies (Supplemental Results and Discussion). Nevertheless, our evidence suggests that the majority of genes experience transcription initiation in human ES cells.
Evidence for Initiation without Transcript Completion at Many Genes
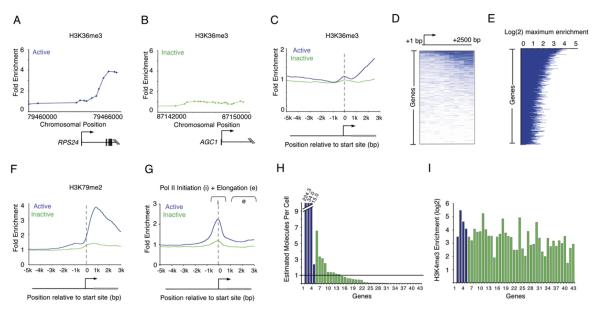
If the promoters of most genes in human ES cells experience transcription initiation but only a portion of these genes produce complete transcripts, then we would expect to find nucleosome modifications associated with elongation only at those genes that produce detectable transcripts. H3K36me3 modification has been associated with elongating Pol II and is enriched within the body of transcriptionally active genes (Krogan et al., 2003b; Xiao et al., 2003; Pokholok et al., 2005). Indeed, we found that H3K36me3 occurs almost exclusively downstream of promoters that produce detectable transcripts in ES cells (Figures 4A–4C). Additionally, the total number of genes enriched for H3K36me3 is significantly lower than the number enriched for modifications associated with transcription initiation (Figures 4D and 4E). Histone H3 lysine 79 dimethyl (H3K79me2) modification has also been shown to be associated with active transcription (Schubeler et al., 2004; Morillon et al., 2005; Pokholok et al., 2005). The ChIP-chip results for H3K79me2 were similar to those for H3K36me3 (Figure 4F). Furthermore, we performed ChIP-chip experiments with antibodies that recognize both initiating and elongating Pol II and found that elongating Pol II signals occur almost exclusively downstream of promoters that produce detectable transcripts (Figure 4G). These data suggest that while transcription initiation occurs at both active and inactive genes, elongation is strongly correlated only with the set of genes that are active based on transcript detection.
Figure 4. Events Associated with Transcriptional Elongation Occur at a Minority of Genes.
(A and B) Enrichment ratios are shown for H3K36me3, which is associated with elongation. The start site and direction of transcription are indicated by an arrow.
(C) Composite enrichment profiles for H3K36me3 at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G). The start site and direction of transcription of the average gene are indicated by an arrow.
(D) Single-probe enrichment ratios of H3K36me3 across the average 2.5 kb region downstream of the transcription start site (~10 probes per promoter). Enriched probes (blue) and unenriched probes (white) are shown for each gene. Individual genes are represented top to bottom and are ordered by average binding ratio across the entire region.
(E) H3K36me3 maximum enrichment (blue horizontal line) between 1 kb and 3 kb of the transcription start site for each gene represented on the human promoter array. Genes are ordered as in (D).
(F) Composite enrichment profiles for H3K79me2 at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G).
(G) Composite enrichment profiles for Pol II (4H8) at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G). Both the initiating and elongating forms of Pol II are recognized by this antibody. The area from 1 kb to 3 kb downstream of the promoter (designated e) was used to determine genes enriched. The start site and direction of transcription of the average gene are indicated by an arrow.
(H) Quantitative RT-PCR analysis of total RNA extracted from hES cells. Estimated transcripts per cell were obtained by dividing the number of molecules of cDNA, as determined by relative quantitation using the standard curve in Figure S5, by the number of cells used for the amplification. Positive control probes (active genes, blue bars) and test sample probes (Inactive genes, green bars) are as follows: (1) No probe, (2) RPLPO, (3) HMGB2, (4) CCNB1, (5) EIF2C3, (6) HOP, (7) GUCY1A3,(8) KITLG, (9) SOX9, (10) PLD1, (11) INCENP, (12) ERCC4, (13) CSF1, (14) HOXB1, (15) GPR143, (16) DSC3, (17) NFKBIL2, (18) ANG, (19) LEFTY1, (20) LNPEP, (21) TCF21, (22) RAP1GA1,(23) KLF2, (24) KCNJ3, (25) TNFSF7, (26) PRRG2, (27) FOXG1B, (28) DLX5, (29) KCNH1, (30) ACADL, (31) CXCL2, (32) HOXD12, (33) ONECUT1, (34) LYST, (35) PCSK2, (36) AVPR1B, (37) SMP3, (38) EPHX2, (39) UCP1, (40) AGC1, (41) ERAF, (42) HBA1, (43) MLNR. Measured values for the genes can be found in Table S6. Broken axis is indicated by white line.
(I) H3K4me3 maximum enrichment (within 1 kb of the transcription start site) for each gene represented in (H).
We tested the possibility that many inactive genes occupied by histone H3K4me3 actually produce full-length transcripts that were not detected previously by either MPSS or microarray methods (Figures 4H–4I). Using quantitative RT-PCR, we assayed transcripts for 38 genes enriched in H3K4me3 but for which transcripts had not been detected previously (Figure 4H, green bars) and, as a positive control, 4 genes selected at random from the list of genes enriched in H3K4me3 for which transcripts had been previously detected (Figure 4H, blue bars). Standard curves derived from known quantities of three independent cDNAs were used to estimate the number of transcripts per cell (Figure S5). While the randomly selected positive control genes had transcript levels averaging 69 molecules per cell, the genes in the inactive set had transcript levels averaging 0.7 molecules per cell. The levels of transcripts in the positive control group ranged from 2 to 224 molecules per cell, whereas the inactive set had 0.0002 to 6.5 molecules per cell. These data show that some of the genes occupied by histone H3K4me3 and whose transcripts had not been detected previously by either MPSS or microarray methods are indeed transcribed, but that little or no transcript accumulates for most of these genes. Interestingly, the levels of histone H3K4me3, the modification associated with transcription initiation, were similar for the genes in both the active and inactive classes (Figure 4I). Thus, there is a class of genes for which there is ample evidence of transcription initiation (promoter occupancy by histone H3K4me3, histone H3K9,14Ac, and initiating Pol II) but for which there is little evidence of transcript elongation (histone H3K36me3, histone H3K79me2, and elongating Pol II) or transcript accumulation (RT-PCR).
For genes whose promoters were enriched in H3K4me3 but that did not accumulate detectable amounts of mRNA (Figure 4H), we attempted to detect RNA molecules originating from the extreme 5′ end of the gene using real-time qPCR. We designed probes to sequences within the 5′-most 70 nucleotides of genes expressed at <1 transcript per cell. The results demonstrate that RNA species containing sequences at the extreme 5′ end can be detected for essentially all genes tested (Figure S6). We conclude that many genes that do not produce full-length mRNAs nonetheless experience transcript initiation.
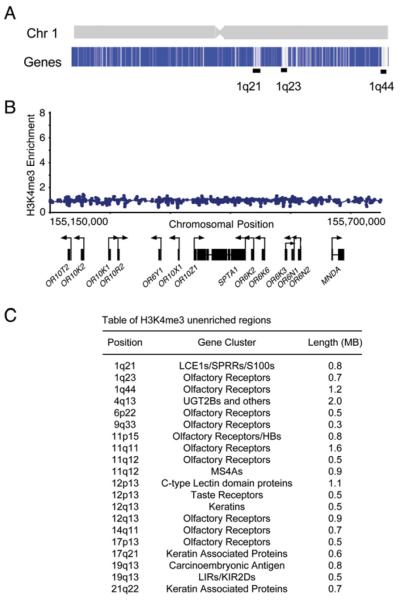
Genes Not Enriched for H3K4me3 Occur in Clusters
We searched for features common to the genes that lacked nucleosomes enriched with histone H3K4me3. Nearly half of the loci lacking histone H3K4me3 contained clusters of ≥3 homologous genes that encode conserved proteins including olfactory receptors, taste receptors, keratins, apolipoproteins, interleukins, and leukocyte antigens (Figure 5). In some instances, gene clusters lacking H3K4me3 enrichment extended across dozens of genes covering Mb regions of the genome (Figures 5A and 5B). For example, only 4 of the 309 olfactory genes represented on the DNA microarray showed >2-fold H3K4me3 enrichment (Table S3). For these clusters of olfactory receptor genes, there is evidence that a single enhancer controls selective transcription initiation at a single gene within the cluster (Lomvardas et al., 2006). It is possible, then, that the loci containing clustered homologous genes lacking histone H3K4me3 are generally regulated by enhancer regions that impose stricter control of initiation than typically observed at other genes.
Figure 5. Genes Not Enriched for H3K4me3 Are Found in Clusters along the Genome.
(A) Top shows schematic representation of chromosome 1; bottom shows maximum enrichment ratios of H3K4me3 within 1 kb of the transcription start site for each gene of chromosome 1. Genes are shaded blue (H3K4me3 positive) to white (H3K4me3 unenriched). Large clusters of genes not enriched for H3K4me3 are indicated by black bars.
(B) Enlarged view of a portion of the 1q23 band of chromosome 1 containing a cluster of olfactory genes devoid of H3K4me3 enrichment. Ratios for H3K4me3 are shown across 550 kb spanning 11 olfactory receptors.
(C) Table of H3K4me3 unenriched regions containing at least ten consecutive genes.
Transcription Initiation at Inactive Genes in Differentiated Cells
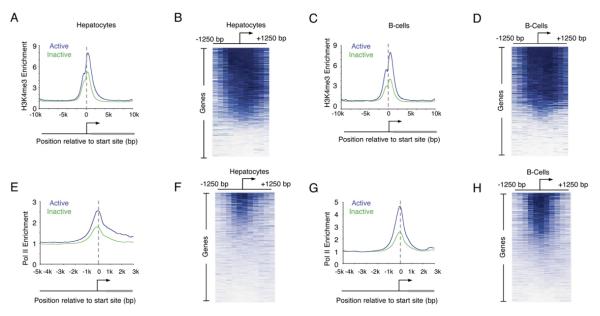
ES cells possess unique properties, including pluripotency, self-renewal, and hyperdynamic chromatin (Meshorer and Misteli, 2006). Numerous reports have suggested that silent genes in ES cells are especially poised to become active upon differentiation (Azuara et al., 2006; Bernstein et al., 2006; Lee et al., 2006). We therefore considered the possibility that widespread transcription initiation at inactive genes is limited to ES cells. To test this, we profiled histone H3K4me3 modification across the genome in primary hepatocytes and B cells (Figure 6; Tables S7 and S8). The results show that the histone H3K4me3 modification is evident both at genes that produce detectable transcripts and at those that do not (Figures 6A and 6C). The promoters of 78% of genes in hepatocytes and 75% of genes in B cells contain nucleosomes with at least a 2-fold enrichment of histone H3K4me3 (Table S3). We conclude that nucleosomes with histone H3K4me3 occupy the majority of all active and repressed genes in both ES cells and differentiated cells of multiple lineages.
Figure 6. Enrichment of H3K4me3 at Inactive Genes Occurs in Differentiated Cells.
(A, C, E, and G) Composite enrichment profiles for H3K4me3 (A and C) and Pol II (E and G) in primary human hepatocytes (A and E) and the REH line of pro-B cells (C and G) at genes called present or absent for mRNA transcript using microarray data as in Figure 2G.
(B, D, F, and H) Enriched probe (blue) and unenriched probe (white) profiles for H3K4me3 (B and D) and Pol II (F and H) are shown for each gene in hepatocytes (B and F) and B cells (D and H). Individual genes are represented top to bottom and are ordered by average binding ratio within 1 kb surrounding the transcription start site as in Figure 1D.
If the presence of nucleosomes with H3K4me3 at the promoters of inactive genes is associated with transcription initiation in these differentiated cells, we would expect to detect the initiating form of Pol II at many of those promoters. Indeed, we found that the initiating form of Pol II is enriched at 42% and 36% of inactive promoters in B cells and hepatocytes, respectively (Figures 6E–6H).
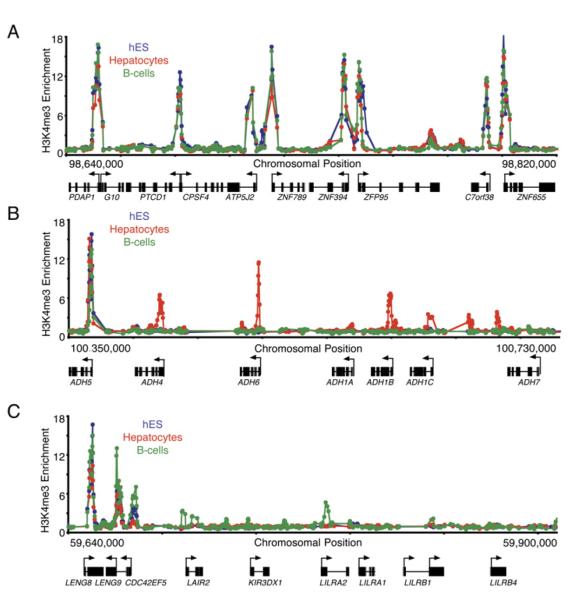
Cell-Type-Specific Differences in H3K4me3
The majority of all genes contain H3K4me3-modified nucleosomes in both ES and differentiated cells, but many promoters do show cell-type-specific differences in H3K4 trimethylation. The promoters of about 25% of genes show differential H3K4 methylation in the three cell types we examined (Figure 7; Supplemental Results and Discussion). These genes frequently have cell-type-specific expression patterns and cell-type-specific function. For example, alcohol dehydrogenase genes do not contain H3K4me3-modified nucleosomes in either ES cells or B cells but do contain methylated nucleosomes in hepatocytes. Only in B cells are the leukocyte receptor cluster genes occupied by H3K4me3 nucleosomes and transcribed (Figure 7C). This cell type specificity of H3K4 methylation suggests that transcription initiation is tightly regulated at this subset of genes and supports our hypothesis that initiation is tightly regulated at many loci containing clustered homologous genes (Figure 5).
Figure 7. Tissue-Specific Methylation of H3K4 at Gene Clusters.
(A) Enrichment ratios for H3K4me3 from hES cells (blue), primary human hepatocytes (red), and the REH line of pro-B cells (green) across 180 kb of chromosome 7 are shown. Start sites and direction of transcription are indicated by arrows.
(B) Enrichment ratios for H3K4me3 from hES cells (blue), primary human hepatocytes (red), and the REH line of pro-B cells (green) across 380 kb of chromosome 4 spanning the alcohol dehydrogenase cluster are shown. ADH1A is specifically expressed in fetal liver, and ADH7 is expressed in gastric tissue. Constitutively H3K4me3-modified ADH5 is expressed in all samples tested.
(C) Enrichment ratios for H3K4me3 from hES cells (blue), primary human hepatocytes (red), and the REH line of pro-B cells (green) across 260 kb of chromosome 19 spanning the leukocyte receptor cluster are shown.
DISCUSSION
Our results suggest that most protein-coding genes in human cells, including most genes thought to be transcriptionally inactive, experience the hallmarks of transcription initiation. We found that nucleosomes with H3K4me3 and H3K9,14Ac modifications, together with the initiating form of Pol II, occupy the promoters of approximately 75% of protein-coding genes in human embryonic stem cells, but only about half of these produce detectable transcripts. Evidence for transcription initiation without transcript accumulation was also found for a similar fraction of genes in hepatocytes and B cells. These results suggest that protein-coding genes fall into three groups of regulatory behavior (Figure S7). The actively transcribed genes are occupied by nucleosomes with histone modifications that are hallmarks of both initiation and elongation, and these generally produce detectable transcripts. A second group experiences transcription initiation without evidence of transcript elongation or accumulation. Within this population of genes, which includes most genes encoding developmental regulators, regulation of events subsequent to transcription initiation must play important roles in preventing production or accumulation of transcripts. The third group consists of genes that are excluded from experiencing transcription initiation, where mechanisms that prevent transcription initiation must predominate.
Previous studies have emphasized that nucleosomes with histone H3K4me3 are associated with actively transcribed genes in various eukaryotes (Bernstein et al., 2002; Santos-Rosa et al., 2002; Ng et al., 2003; Schneider et al., 2004; Schubeler et al., 2004; Pokholok et al., 2005). Similarly, histone acetylation has been associated with actively transcribed genes (Sterner and Berger, 2000). The genome-wide results from human cells described here confirm that nucleosomes with H3K4me3 and H3K9,14Ac are present at actively transcribed genes and show that the relative levels of these modifications increase as transcript levels increase (Figure 2; Figure 3; Figure S6). Among the genes that are actively transcribed in ES cells based on MPSS and microarray data (Sato et al., 2003; Abeyta et al., 2004; Brandenberger et al., 2004a; Wei et al., 2005), 90% contain significantly enriched levels of H3K4me3 and H3K9,14Ac, with the highest levels of enrichment found at the promoters of genes that are most highly expressed (Figure 2I and data not shown). Our results also show that nucleosomes with histone H3K4me3 and H3K9,14Ac occupy promoter regions, with maximum occupancy just downstream of the transcription start site (Figure 1; Figure 2; Figure 3;Figure 6).
A striking result from this study is that among the 55% of genes that are transcriptionally inactive by either Affymetrix or MPSS data in ES cells (Table S5; Sato et al., 2003; Abeyta et al., 2004; Brandenberger et al., 2004a; Wei et al., 2005), at least half contain significantly enriched levels of H3K4me3 and H3K9,14Ac at their promoters but show no evidence of the elongation-associated H3K36me3 or H3K79me2 modifications (Table S2). Several previous studies have noted the presence of H3K4me3 at certain inactive genes. Nucleosomes with H3K4me3 have been reported at inactive genes within the β-globin locus (Schneider et al., 2004), X-linked genes (Brinkman et al., 2006), certain transcriptional regulatory genes (Azuara et al., 2006; Bernstein et al., 2006; Brinkman et al., 2006), and other genes (Kim et al., 2005). Because H3K4me3 was assayed at no more than 2% of all genes in these studies, it was not evident that occupancy of promoters of transcriptionally inactive genes by nucleosomes with H3K4me3 is a frequent and global phenomenon. A recent genome-wide study of H3K4me3 in T cells did note that 60% of genes were enriched for H3K4me3 but concluded that the modification was associated with actively transcribed genes and certain classes of genes such as those that are rapidly inducible (Roh et al., 2006). Our results show that nucleosomes with H3K4me3 are associated with the promoters of more than half of the transcriptionally inactive genes in three cell types, that these inactive genes are not limited to those poised for activation (Table S2; Supplemental Results and Discussion), and that this association is likely due to transcription initiation without transcript accumulation.
The co-occupancy of Pol II with H3K4me3- and H3K9,14Ac-modified nucleosomes at genes without detectable levels of transcription strongly suggests that a large fraction of human genes experience transcription initiation without transcript completion. Indeed, previous studies have also found Pol II localized to inactive genes (Soutoglou and Talianidis, 2002; Radonjic et al., 2005). We do not detect Pol II at all promoters occupied by H3K4me3 nucleosomes, and although this may simply be due to less robust signals for Pol II and the noise associated with microarray-based methods, it is also possible that some promoters acquire H3K4me3 nucleosomes in a manner that does not depend on transcription initiation. Evidence in budding yeast indicates that H3K4me3 modification occurs subsequent to Pol II recruitment and Ser5 phosphorylation of the Pol II C-terminal domain (Santos-Rosa et al., 2002; Krogan et al., 2003a; Ng et al., 2003; Pokholok et al., 2005). Similar studies in mammalian cells show that components of H3K4 methyltransferase complexes interact with the Ser5-phosphorylated form of Pol II, indicating that transcription initiation coincides with H3K4me3 deposition (Hughes et al., 2004). Because multiple H3K4 methylases exist in mammalian cells, it is not clear whether there are multiple mechanisms involved in their recruitment to promoters, and thus it is possible that some promoters acquire H3K4me3 nucleosomes in a manner that does not depend on transcription initiation.
There are at least two general mechanisms that may be responsible for transcription initiation without transcript accumulation. In one, the transcription apparatus is recruited to promoters and initiates transcription but is prevented from efficient transcript completion. This may be due to transcriptional pausing, poor processivity, or abortive initiation, all of which have been described as mechanisms that prevent initiating polymerase from elongating efficiently through specific genes (Conaway et al., 2000; Dvir, 2002; Sims et al., 2004; Saunders et al., 2006). A stable, paused Pol II molecule is found just downstream of the transcription initiation site at the Hsp70 gene in Drosophila (Lis, 1998) and it has been estimated that as many as 20% of Drosophila genes experience some level of transcriptional pause (Law et al., 1998). Paused RNA polymerases have also been observed at mammalian c-Myc, N-myc, and c-Fos promoters (Saunders et al., 2006). Poorly processive RNA polymerase molecules can initiate transcription at the HIV long terminal repeat but do not complete transcription in the absence of the TAT transcriptional activator (Laspia et al., 1993; Wei et al., 1998; Parada and Roeder, 1999). Abortive initiation, which involves premature termination of transcription in promoter regions, is best understood in prokaryotes but has also been described in eukaryotes (Dvir, 2002). Transcriptional pausing, poor processivity, or abortive initiation may all contribute to the observations described here. We do not detect Pol II molecules at all promoters that are occupied by nucleosomes with histone H3K4me3, so it is unlikely that a paused Pol II molecule is stably associated with the promoter regions of all of these inactive genes. Instead, our evidence is consistent with a less stable association of Pol II at promoters of genes that experience transcription initiation without transcript accumulation.
A second general mechanism that may be responsible for transcription initiation without transcript accumulation is posttranscriptional degradation. It is possible that pre-mRNA molecules are actually transcribed from the inactive genes but are rapidly degraded or accumulate at very low levels that are difficult to detect. Some transcripts may be produced and rapidly degraded as part of the process of gene silencing, as has been demonstrated for pericentromeric heterochromatin in Schizosaccharomyces pombe (Kato et al., 2005; Buhler et al., 2006). It is also possible that some low-abundance transcripts are targeted for degradation by miRNAs; indeed, mRNA species that are not detected in specific tissues are more likely to constain target sequences for tissue-specific miRNAs, which may help control any “leaky expression” (Farh et al., 2005). If very low levels of transcripts are produced from the inactive genes, it may be difficult to detect the mRNA or nucleosomes enriched for H3K36me3. Experiments with genome-wide tiling arrays show evidence for small amounts of transcript at almost all genes in numerous cell types (Cheng et al., 2005). While we were able to detect some amount of full-length transcript for almost all the inactive genes we tested using RT-PCR, the majority of these likely exist at levels much lower than one transcript per cell (Figure 4H).
While the majority of genes have promoters enriched for nucleosomes with histone H3K4me3, one class of genes provides a notable exception. Clusters of highly related genes often do not have nucleosomes with H3K4me3 and H3K9,14Ac modifications, nor are they occupied by the initiating form of Pol II (Figure 5; Figure 7). Many of the clustered genes lacking H3K4me3 are expressed in a tissue-specific fashion (Su et al., 2004; Table S3; Supplemental Results and Discussion). The most numerous members of this class of genes are the olfactory receptors, in which there is evidence that a single enhancer controls selective transcription initiation at only one of many such genes (Lomvardas et al., 2006). The fact that the clusters of olfactory genes do not experience transcription initiation and do not contain nucleosomes enriched in H3K4me3 further supports the view that the presence of H3K4me3 is a hallmark of transcription initiation at most genes.
In summary, we have found that transcription initiation and histone H3K4me3 modification occur at the promoters of most protein-coding genes in human cells. This may serve to create a chromatin structure at transcription initiation sites that is more accessible to transcription factors and signaling kinases than at much of the rest of the genome, perhaps providing landmarks that produce a much smaller search space for these regulatory molecules. Such landmarks might facilitate reprogramming of gene expression during differentiation.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Human H9 ES cells (WiCell) were cultured as described (Boyer et al., 2005). Hepatocytes were obtained directly from perfused human liver at the University of Pittsburgh through the Liver Tissue Procurement and Distribution System (S. Strom). REH cells (ATCC) were cultured in RPMI 1640 media containing 10% FBS. Cells were crosslinked as described (Boyer et al., 2005).
Chromatin Immunoprecipitation and DNA Microarray Analysis
ChIP was combined with DNA microarray analysis as described (Boyer et al., 2005). The antibodies used here were specific for H3K4me3 (Abcam ab8580), hypophosphorylated RNA polymerase II (8WG16), H3K9,14Ac (Upstate 06-599), H3K36me3 (Abcam ab9050), H3K79me2 (Abcam ab3594), total histone H3 (Abcam ab1791), E2F4 (Santa Cruz Biotech sc-1082), and nonspecific IgG (Santa Cruz Biotech, sc-2043). The design of the oligo-based arrays, which were manufactured by Agilent Technologies, is described in detail in Supplemental Experimental Procedures. In addition to simple ratio measurements mentioned in the text, a whole-chip error model was used to calculate confidence values from the enrichment ratio and signal intensity of each probe (probe p value) and of each set of three neighboring probes (probe-set p value). Probe sets with significant probe-set p values (p < 0.001) and significant individual probe p values were judged to be bound at high confidence (see Supplemental Experimental Procedures for additional information). Bound regions were assigned to an Entrez gene ID if they were within 1 kb of the transcription start site from one of four genomic databases, RefSeq, MGC, Ensembl, or UCSC Known Gene, where the transcript had been assigned to the Entrez gene (transcript data downloaded from UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway, using NCBI build 35). Enrichment ratio plots were expressed as a sliding average of three neighboring probes.
Gene Expression Analysis
Gene expression data were collated from H1 ES cells (Sato et al., 2003); H9, HSF1, and HSF6 ES cells (Abeyta et al., 2004); and 79 differentiated human cell and tissue types (Su et al., 2004) and were analyzed as described in detail in Supplemental Experimental Procedures and in Lee et al. (2006). These data are summarized in Table S5. Absent/present (AP) calls for hepatocytes and REH cells were derived from liver and CD19+ peripheral blood samples from Su et al. (2004).
Quantitative RT-PCR
Standard curves were produced for three independent cDNAs (ANG, GPR143, and KCNJ3; OriGene Technologies) using 100 ng–0.0001 pg of vector subjected to quantitative real-time PCR in duplicate (TaqMan predeveloped gene expression assays; Applied Biosystems). The standard curves were then averaged to produce a composite standard curve (cycle threshold [Ct] versus molecules present) to which all test measurements were compared (Figure S5). 6 × 106 hES cells cultured as above were enriched from feeder murine embryonic fibroblasts by trypsinization. RNA was extracted from hES cells by the TRIzol method and precipitation (Invitrogen). Total RNA was reverse transcribed by means of the Invitrogen SuperScript III First-Strand Synthesis System using both oligo(dT) and random hexamer primers to produce cDNA. cDNA was amplified using TaqMan predeveloped gene expression assays as described by the manufacturer in an Applied Biosciences 7500 Real-Time PCR Thermocycler in duplicate. cDNA abundance was determined by measuring the point during cycling when amplification could first be detected (auto Ct value determined by Applied Biosciences Prism 7500 software) rather than the endpoint of the 40-cycle reaction. The measured Ct value was used to calculate the estimated transcripts present in the test sample using relative quantitation to the composite standard curve. Genes determined to be active or inactive by Affymetrix and MPSS methods were measured using unique TaqMan predeveloped gene expression assays for each species.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Mitalipova for ES cell expertise; R. Kumar, E. Jacobsen, S. Johnstone, R. Jenner, S. McCuine, E. Herbolsheimer, and the Whitehead Institute Center for Microarray Technology for technical assistance; D. Odom and S. Strom for hepatocytes; and D. Gifford, J. Lis, J. Wysocka, T. Lee, and the Young lab for helpful discussions. This work was supported by NIH grant HG002668. R.A.Y. consults for Agilent Technologies.
Footnotes
Raw Data Further information, methods, and raw data can be found at http://web.wi.mit.edu/young/hES_chromatin/.
Supplemental Data Supplemental Data include Supplemental Experimental Procedures, Supplemental Results and Discussion, Supplemental References, ten figures, and eight tables and can be found with this article online at http://www.cell.com/cgi/content/full/130/1/77/DC1/.
Accession Numbers All microarray data discussed herein are available at ArrayExpress under the accession designation E-TABM-277.
REFERENCES
- Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum. Mol. Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and trimethyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R, Khrebtukova I, Thies RS, Miura T, Jingli C, Puri R, Vasicek T, Lebkowski J, Rao M. MPSS profiling of human embryonic stem cells. BMC Dev. Biol. 2004a;4:10. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R, Wei H, Zhang S, Lei S, Murage J, Fisk GJ, Li Y, Xu C, Fang R, Guegler K, et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004b;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Roelofsen T, Pennings SW, Martens JH, Jenuwein T, Stunnenberg HG. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS toa nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by Pol II. Trends Biochem. Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- Dvir A. Promoter escape by Pol II. Biochim. Biophys. Acta. 2002;1577:208–223. doi: 10.1016/s0167-4781(02)00453-0. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. Pol II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003a;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by Pol II. Mol. Cell. Biol. 2003b;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Groudine M. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays. 1993;15:659–665. doi: 10.1002/bies.950151005. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Wendel P, Mathews MB. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J. Mol. Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- Law A, Hirayoshi K, O’Brien T, Lis JT. Direct cloning of DNA that interacts in vivo with a specific protein: application to Pol II and sites of pausing in Drosophila. Nucleic Acids Res. 1998;26:919–924. doi: 10.1093/nar/26.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of Pol II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with Pol II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Mayhall EA, Paffett-Lugassy N, Zon LI. The clinical potential of stem cells. Curr. Opin. Cell Biol. 2004;16:713–720. doi: 10.1016/j.ceb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. A novel Pol II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Trounson AO. Human embryonic stem cells: prospects for development. Development. 2004;131:5515–5525. doi: 10.1242/dev.01451. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A Genetic Switch. Cell Press and Blackwell Scientific Publications; Cambridge, MA, USA: 1986. [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal Pol II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by Pol II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J. Cell. Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of Pol II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.