Abstract
Search of the protein database with the aflatoxin pathway polyketide synthase (PKS) revealed putative PKSs in the pathogenic fungi Coccidioides immitis and Coccidioides posadasii that could require partnerships with a pair of fatty acid synthase (FAS) subunits for the biosynthesis of fatty acid-polyketide hybrid metabolites. A starter unit:acyl-carrier protein transacylase (SAT) domain was discovered in the nonreducing PKS. This domain is thought to accept the fatty acid product from the FAS to initiate polyketide synthesis. We expressed the C. immitis SAT domain in E. coli and showed that this domain, unlike that from the aflatoxin pathway PKS, transferred octanoyl-CoA four times faster than hexanoyl-CoA. The SAT domain also formed a covalent octanoyl intermediate and transferred this group to a free-standing ACP domain. Our results suggest that C. immitis/posadasii, both human fungal pathogens, contain a FAS/PKS cluster with functional similarity to the aflatoxin cluster found in Aspergillus species. Dissection of the PKS and determination of in vitro SAT domain specificity provides a tool to uncover the growing number of similar sequenced pathways in fungi, and to guide elucidation of the fatty acid-polyketide hybrid metabolites that they produce.
Keywords: fatty acids, polyketides, fungal metabolites, starter unit:acyl-carrier protein transacylase, aflatoxin
1. Introduction
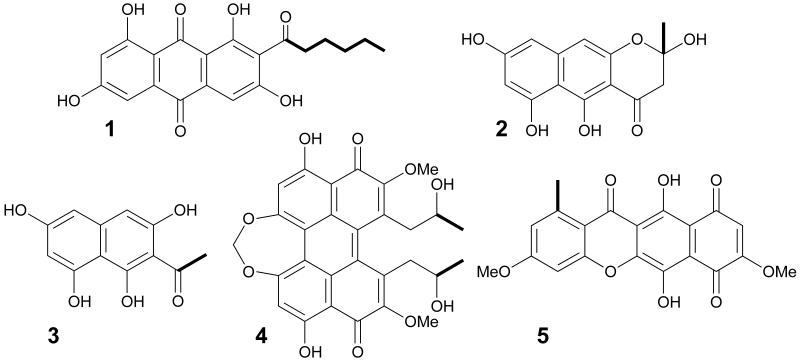
Many apparent secondary metabolic gene clusters are emerging from genome sequencing projects. Among these, fungi have genes predicted to encode as many as 30 polyketide synthases (PKSs) where only few have been correlated to characterized natural products [1]. PKSs use chemistry similar to fatty acid synthases (FASs); however, variable starter unit specificity and differing extents of reduction of the growing polyketide chain underlie the enormous biosynthetic diversity of their products [2]. In addition, PKSs often associate with other PKSs, non-ribosomal peptide synthetases (NRPSs), or FASs to further enhance the structural complexity of products that can arise. FAS and PKS enzymes harbor acyl-carrier proteins (ACPs) that bear the elongating intermediates through the catalytic cycle maintaining high effective substrate concentrations. In some ascomycetes, specialized FASs are required for the production of medium chain length fatty acids that initiate PKSs in the biosynthesis of fatty acid-polyketide hybrid metabolites such as the aflatoxins [3], sterigmatocystin [4], and dothistromin [5]. A starter unit:ACP transacylase (SAT) domain discovered in these accompanying PKSs was demonstrated to selectively accept a C6-fatty acyl group and transfer it to the PKS ACP domain to initiate synthesis of the aflatoxin precursor, norsolorinic acid (1) [6] (Figure 1). Subsequently, SAT domains were found to be widespread based on examination of genome/protein databases. Most are predicted to have high selectivity for acetyl-CoA when not associated with a FAS. This correlation was demonstrated for PKSs involved in the biosynthesis of naphthopyrone YWA1 (2), acetyl-1,3,6,8-tetrahydroxynaphthalene (3), cercosporin (4), and bikaverin (5) [7].
Figure 1. Selected fungal polyketide metabolites.
Structures include norsolorinic acid (1), naphthopyrone YWA1 (2), acetyl-1,3,6,8-tetrahydroxynaphthalene (3), cercosporin (4), and bikaverin (5). Hexanoyl or acetyl starter units are bolded.
The clustering of genes involved in the production of secondary metabolites in fungi and bacteria has allowed the isolation of most, if not all, of the genes that encode enzymes necessary for product formation, which has helped accelerate the functional assignments of gene products. With many fungal genome projects completed and many more underway, establishing the functions of the increasing number of secondary metabolic pathways is a growing need, and only part of the wider challenge in chemistry and biology of associating function to newly discovered gene products [8]. In this paper we describe an approach applicable to fungal “nonreducing” iterative PKSs involving the dissection, expression, and in vitro assay of substrate preference by SAT domains that initiate individual PKS pathways. Such an analysis has allowed us to predict the existence of a secondary metabolic FAS/PKS pathway in the human pathogenic fungi Coccidioides immitis and Coccidioides posadasii. That is, the SAT substrate preference determined experimentally in vitro could provide a method to predict unknown or “cryptic” toxin-producing polyketide biosynthetic pathways discovered in sequence-based searches.
2. Materials and Methods
2.1. Cloning of the C. immitis SAT domain
The three exon templates that constitute the SAT coding sequence were amplified in primary PCR reactions using primers based upon the C. immitis sequence. The highly similar C. posadasii NRRL C735 gDNA was used as a template for PCR. Primer pairs included SAT-immitis-5.1 (NcoI) / SAT-immitis-3.1, SAT-immitis-5.2 / SAT-immitis-3.2, and SAT-immitis-3.3 (NotI) / SAT-immitis-5.3 (Table 1). The full-length cDNA fragment was amplified in a secondary PCR using the three primary products and the end primers, SAT-immitis-5.1 / SAT-immitis-3.3. The full-length product was digested with NcoI and NotI and inserted into the corresponding sites in pET28a (Novagen, Madison, WI) creating the C-terminal His6x-tag fusion plasmid, pECp-SAT. The plasmid was sequenced and the resulting encoded protein for the constructed SAT domain was identical to the available C. immitis protein sequence.
Table 1.
Oligonucleotide cloning sequences.
| Name | Sequence (5′-3′) |
|---|---|
| SAT-immitis-5.1 | GAATTACCATGGCTCAGGACATGCAGGTCTTC |
| SAT-immitis-3.1 | CTCCTCATAATGATTAATAAAGCAGGCCAGCTGCGTC |
| SAT-immitis-5.2 | GGCCTGCTTTATTAATCATTATGAGGAGCCCGGACGT |
| SAT-immitis-3.2 | GCACGGGACCGGCGTTACGCTATCAAACTCTACAAGAGCG |
| SAT-immitis-5.3 | TTTGATAGCGTAACGCCGGTCCCGTGCAAGCTAC |
| SAT-immitis-3.3 | GAATTAGCGGCCGCTCCCACGGGTACGGTGTCGA |
2.2 SAT domain production
The Coccidioides SAT domain was expressed after IPTG induction in Rosetta2(DE3) E. coli cells (Novagen) harboring pECp-SAT. Cultures were initially grown in 100 mL of 2×YT medium supplemented with 25 μg/mL kanamycin and 25 μg/mL chloramphenicol at 30 °C. When the OD600 reached 0.6, IPTG was added (1 mM), and incubation was continued for approximately 16 h at 20 °C. Cells were harvested by centrifugation at 4080 × g for 20 min and resuspended in 5 mL of resuspension buffer (50 mM potassium phosphate pH 7.6, 10% glycerol, 300 mM NaCl, 10 mM imidazole, and 1 mg/mL lysozyme). After incubation on ice for 30 min, the resuspension was disrupted by sonication, and cleared by centrifugation for 30 min at 27000 × g. The His6x-tagged protein was bound to 0.5 mL nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA) and eluted with an imidazole step-gradient according to the manufacturer’s instructions. The purified protein was dialyzed against 100 mM potassium phosphate pH 7.0, and enzyme concentrations were determined in triplicate using the Bradford assay (Bio-RAD, Hercules, CA) with bovine albumin as a standard.
2.3 Chromatographic transacylase assay
The chain length specificity of the Coccidioides SAT domain was determined using 1 mM acyl-CoAs (Sigma-Aldrich, St. Louis, MO) and 0.5 μM enzyme in 100 mM potassium phosphate pH 7.0 as previously described [6]. For octanoyl-CoA the protein concentration was lowered to 0.1 μM to reduce the extent of reaction. Time point samples were flash frozen in liquid nitrogen and reactants/products were quantified using HPLC.
2.4 Synthesis of [1-14C]-octanoyl-CoA
The CoA free-acid and the [1-14C]-octanoic acid sodium salt (53 mCi/mmol) were purchased from Sigma-Aldrich. Octanoyl-CoA was prepared using a described procedure for the synthesis of acyl-CoA thioesters [9], and the product was purified by HPLC.
2.5 Purification of ACPPksA
The holo-ACP monodomain from PksA in Aspergillus parasiticus was expressed in BL21(DE3) (Novagen), purified using Ni NTA-Sepharose chromatography, and activated with the promiscuous phosphopantetheinyltransferase, Svp [10], from Streptomyces verticillus as previously described [6].
2.6 Radiochemical Transacylase Assay
20 μM of enzyme(s) were reacted for either 5 or 10 min with 200 μM [1-14C]-octanoyl-CoA in 100 mM potassium phosphate pH 7.0. Reactions included ACPPksA, SAT, and ACPPksA + SAT. The reactions were quenched in 5× SDS buffer [11], and immediately separated over a 12% SDS-PAGE gel. The PAGE gel was dried between Cellophane sheets and exposed to BioMax XAR film (Eastman Kodak, Rochester, NY).
3. Results and Discussion
The fungal Type I FASs dedicated to secondary metabolic pathways share higher sequence similarity to themselves than to FASs involved in primary metabolism, resulting in phylogenetic clustering of each group and further assistance in assigning their roles (data not shown). The secondary metabolic FAS and PKS genes in aflatoxin-producing strains are located in a 75 kb region [12], and these three proteins form a complex [13]. Except for a few known cases, such as dothistromin biosynthesis [5], the FAS and PKS genes of secondary metabolism are in close proximity, making it possible to predict that the starter unit for polyketide initiation is a synthesized fatty acid rather than acetate (acetyl-CoA), which is readily available in the cell and needs no specialized synthetic apparatus. Of the cases presently characterized, the encoded PKSs associated with FASs are nonreducing and synthesize highly oxidized polyketide metabolites. Most such PKSs include the following domains: SAT, ketosynthase (KS), malonyl-CoA:ACP transacylase (MAT), putative product template (PT, hypothesized to facilitate cyclization chemistry of a hypothetical poly-β-keto intermediate [14]), ACP, and thioesterase/Claisen cyclase (TE/CLC). Kroken et al used alignment of the KS domains from a large group of fungal PKSs to chart their genealogy [15]. They proposed that these nonreducing PKS genes evolved by gaining TE/CLC function while losing reductive function (β-dehydrase, enoyl-reductase, and β-ketoreductase) from their “reducing” PKS ancestors. During this evolutionary transformation, the nonreducing family of PKS genes acquired the SAT domain, which is intimately involved in chain length selection from the associated FAS. For phylogenetic analysis, the sequences of SAT domains, therefore, should be considered in order to gain insight into FAS/PKS partnerships.
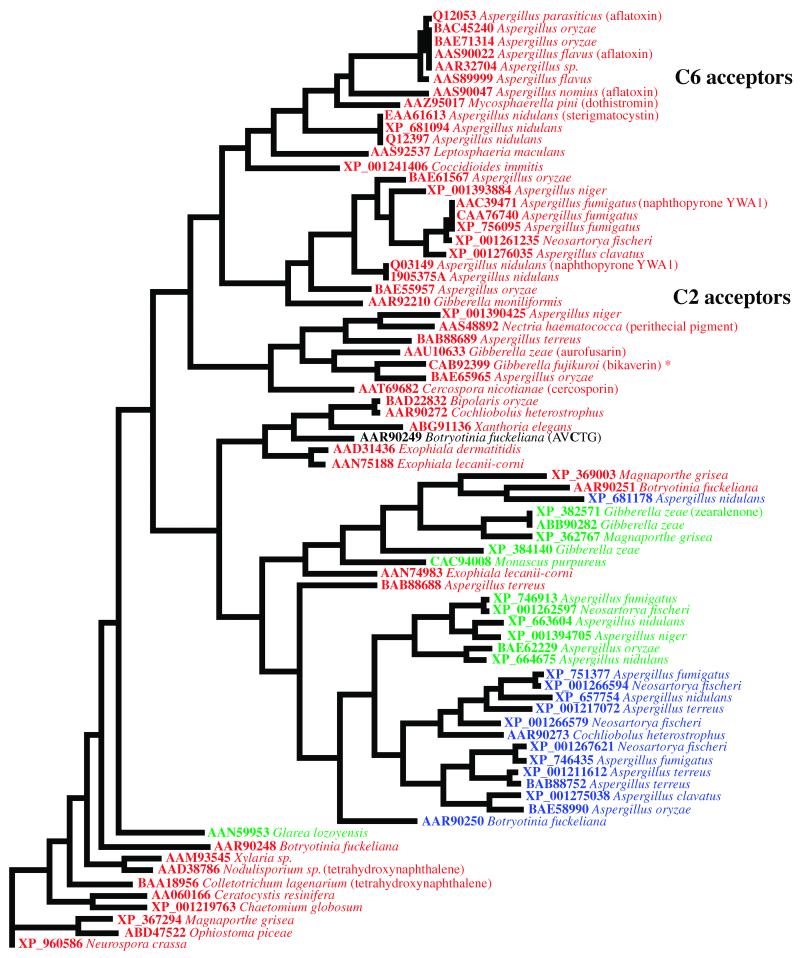
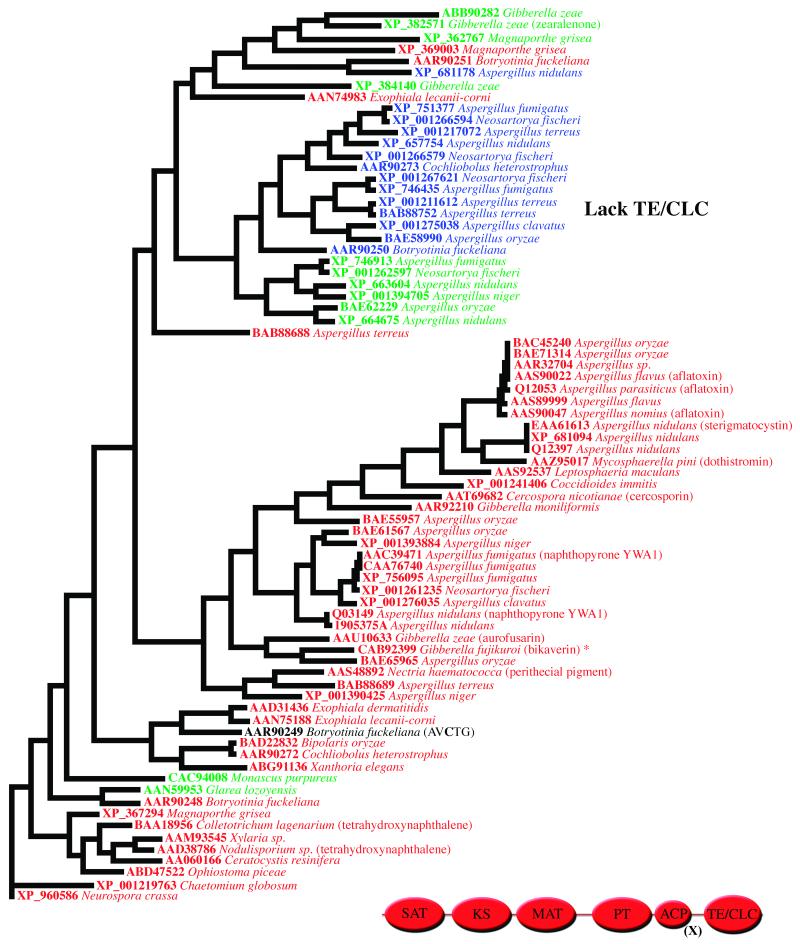
To estimate SAT domain evolution within the nonreducing PKS family, the norsolorinic acid-producing PKS, PksA, from A. parasiticus was used as the query sequence in a Blastp search of the non-redundant protein sequence database (http://www.ncbi.nlm.nih.gov/). Seventy-six sequences (cut-off E-value = 0.0) were aligned in Clustal X. In addition to a full-length protein alignment, the first 350 amino acids (approximate size of SAT domains) were separately aligned. Phylogenetic trees were built in PAUP and rooted with the outgroup Neurospora crassa PKS sequence (XP_960586) to demonstrate cladal organization as well as to estimate possible ancestral origins. Forty-nine of the protein seqeuences harbored the expected GXCXG active-site motif in the SAT domain (Figure 2). This active site Cys tethers the starter unit during PKS chain initiation [6]. Twelve sequences contained a GXSXG motif, which presumably substitutes oxyester chemistry more akin to that of the well-studied acyl transacylases known in fatty acid and polyketide biosynthesis. Fourteen sequences had a GXGXG motif. These SAT domains may be either inactive, but have been maintained for structural reasons, or have an alternative, as yet unknown function. Alternatively, in such PKSs, the MAT domain could account for both starter and extender unit loading [7,16]. Consequently, evolutionary pressure to maintain an active SAT domain would be lost. In the full-length alignment, a group of sequences lack the TE/CLC domain and fall into one clade (Figure 3). These still contain the SAT domain with a GXSXG or GXGXG rather than a GXCXG motif. One of the PKSs in this clade (XP_382571) has been demonstrated to be involved in zearalenone biosynthesis [17]. The presumed SAT domain (GXSXG) could be involved in accepting a partially reduced polyketide starter unit from an associated PKS for additional chain extension leading to zearalenone [18].
Figure 2. Phylogenetic divergence of SAT domains.
Sequences are listed by their GenBank accession numbers and fungal hosts. PKSs giving rise to known compounds are indicated, and the known hexanoyl acceptors cluster separately from the known acetyl acceptors. Closely-grouping sequences could have the same starter unit specificity. Putative active site motifs are color-coded: Red, GXCXG; green, GXSXG; and blue, GXGXG. (*) indicates a revision in the PKS4 sequence [7].
Figure 3. Phylogenetic alignment of full-length nonreducing PKSs.
The tetrahydroxynaphthalene PKSs, PKS1 orthologs, are found closest to the ancestral outgroup Neurospora crassa sequence (XP_960586). Many of the PKSs with SAT sequences containing a GXSXG or GXGXG motif lack the C-terminal TE/CLC domain. The domain organization of nonreducing PKSs is shown. (x) = 1, 2, or 3 ACP domains. The Coccidioides immitis PKS (XP_001241406) contains 2 ACPs.
The PKS involved in melanin production, PKS1 (BAA18956/AAD38786 [19]), is closest to that of the ancestral outgroup Neurospora sequence, supporting the contention that genes coding for other PKSs evolved from gene duplication events of pks1 ancestors. Moreover, SAT domains from species with PKSs that are known hexanoyl acceptors (involved in aflatoxin, sterigmatocystin, and dothistromin biosynthesis) cluster separately from the known acetyl acceptors (involved in naphthopyrone YWA1, 1,3,6,8-tetrahydroxynaphthalene, cercosporin, and bikaverin biosynthesis) further supporting the prediction that such PKSs form complexes with secondary metabolic FASs. Phylogenetic analysis, therefore, is a first step in predicting the initial functionality of some of the myriad PKSs discovered in genomes of ascomycete fungi.
Phylogenetic analysis of SAT domains provides insight into starter unit selection among unknown and cryptic pathways. Among these overall sequence differences, several distinct variations arise when aligning the known hexanoyl acceptors versus the known acetyl acceptors (data not shown). One difference stands out among these. It was proposed that the Phe residue directly adjacent (upstream) to the active site His in the Streptomyces coelicolor MAT catalytic diad was a “selectivity filter” [20]. All of the known hexanoyl acceptors listed in Figure 2 contain a conserved Ala in this position. The known acetyl acceptors [7], however, have either a bulky Phe or Tyr. Consequently, SAT domains might also use a similar steric approach to “selecting” the appropriate acyl chain length. If this analysis is correct, this observed amino acid pattern can be used to assist in predicting the polyketide starter unit, as well as to assess evolutionary relationships among PKS families.
To analyze FAS/PKS partnerships in this manner, a predicted PKS in Coccidioides immitis/posadasii was examined. The primary pathogenic fungi C. immitis and C. posadasii are the causative agents of coccidioidomycosis in humans [21]. Although polyketide products such as melanin and pigments have a role in fungal pathogenesis and virulence [22], a specific polyketide metabolite associated with toxicity and virulence in Coccidioides species has not been identified to date. Of the predicted twelve PKSs in the C. immitis genome (Coccidioides immitis Sequencing Project. Broad Institute of Harvard and MIT, http://www.broad.mit.edu) one (XP_001241406) is in the same clade as the norsolorinic acid synthase PKS, PksA (48.7/63.9/9.9 % id/sim/gap, global alignment), in the phylogenetic trees (Figures 2 and 3). The protein also contains a similarly small Ser residue in the potential SAT “selectivity filter” position. This Coccidioides PKS is located in the genome next to homologs of the norsolorinic acid hexanoyl synthase subunits HexA (XP_001241401, 41.0/57.3/13.0 % id/sim/gap) and HexB (XP_001241402, 39.6/55.9/9.7) from A. parasiticus supporting the notion that a similar fatty acid starter unit is used to initiate polyketide biosynthesis. Homologs to the zinc cluster binuclear transcription factor AflR (XP_001241399 [23]) and the protein that regularly accompanies AflR, AflS (Broad: between sequence 08562 and 08563), are also present. Both positively regulate aflatoxin biosynthesis in aflatoxin-producing species [24]. Their presence in this cluster, hints that the cluster is active. Additionally, an efflux pump (XP_001241400) similar to AflT found in the aflatoxin pathway for possible toxin transport is located within the cluster. This gene in C. posadasii was shown to be upregulated during the early parasitic growth phase compared to that of the latent parasitic growth phase, suggesting that the metabolite produced and secreted could play a role in establishing infection [25]. Identifiable transposable elements are suggestive of the cluster’s nomadic origin. In sum, the sequence information support the view that a FAS pair is encoded to synthesize a medium chain fatty acid, as in the case of NorS, and the PKS SAT domain would accept this medium chain product. To test this proposal, exons 1, 2, and 3 coding for the C. immitis SAT domain (XP_001241406) were amplified in a 3-part overlap extension PCR and inserted into pET28a for production of the C-terminal His6x-tag fusion protein in Rosetta2(DE3) E. coli cells to determine starter unit specificity for this unknown PKS pathway. The protein was purified using Ni NTA-Sepharose chromatography for in vitro specificity studies. Our results confirmed that the C. immitis/posadasii PKS SAT domain is able to accept a fatty acid product, and interestingly, this SAT domain preferentially accepted octanoyl-CoA as the starter unit with a 4-fold preference to hexanoyl-CoA (Figure 4A). The dissected domain also was able to covalently accept and transfer a radioactive octanoyl-group from [1-14C]-octanoyl-CoA to the PksA holo-ACP monodomain further demonstrating its role in starter unit initiation (Figure 4B). Importantly, the ability to interact with a non-cognate ACP suggests domain flexibility in carrier protein recognition, but would likely have an even higher affinity for one or both of the two cognate ACPs found in the Coccidioides PKS. This flexibility reduces the barrier for evolutionary SAT domain acquisition and poses a possible route for engineering metabolites with alternative starter units.
Figure 4.
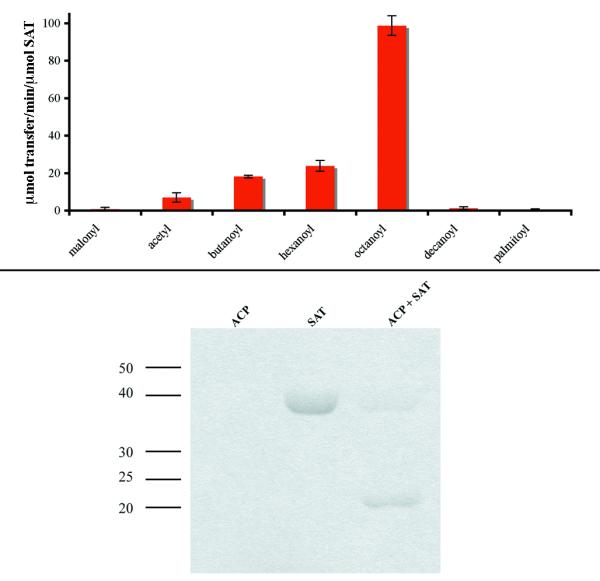
A: Coccidioides immitis SAT domain specificity. Rates are reported in μmol transfer/min/μmol SAT: malonyl, 0.8 ± 0.9; acetyl, 7.0 ± 2.5; butanoyl, 18.2 ± 0.7; hexanoyl, 23.9 ± 2.9; octanoyl, 98.8 ± 5.2; decanoyl, 1.3 ± 0.7; and palmitoyl, 0.3 ± 0.7. B: Radiochemical transacylase assay. The Coccidioides SAT monodomain covalently accepts a 14C-labelled octanoyl-group and transfers it to the holo-ACP monodomain from PksA.
4. Conclusion
The Coccidioides FAS/PKS cluster clearly encodes enzymes that make an as yet uncharacterized metabolite(s), likely with an octanoyl starter unit. Identification of starter unit specificity in SAT domains can guide radiotracer experiments to direct the isolation of unknown fungal aromatic polyketides. For example, the N-acetyl cysteamine thioester of hexanoic acid strongly incorporated into aflatoxin intermediates in whole-cell feeding experiments [26]. Elucidation of the metabolite(s) produced by this probable gene cluster could assist in determining the virulence and toxicity of this infectious fungus. The Coccidioides cluster exemplified here is one of many gene clusters of unknown biosynthetic function now emerging in sequence-based searches. In addition to phylogenetic clustering of the fungal FAS subunits, analogous clustering of the accompanying PKS or, more specifically, the SAT domain corroborates the pivotal role of this catalytic domain in the selection of medium chain fatty acids rather than simple acetate starters. The convergence of FAS and PKS sequence analyses coupled with in vitro specificity studies provides an initial predictive approach to uncovering the structures of unknown FAS/PKS hybrid metabolites in fungi to close the gap between sequence and function.
Acknowledgements
We thank Drs. C.-Y. Hung and G. T. Cole at the University of Texas, San Antonio for providing C. posadasii NRRL C735 gDNA. We also thank Dr. J. D. Galpin for synthesizing [1-14C]-octanoyl-CoA. This work was supported by the National Institutes of Health Grant ES 001670 and the United States Department of Agriculture Grant USDA-SCA-58-6435-6-020.
Dedication Dedicated to the memory of A. Ian Scott, distinguished mentor and friend
Contributor Information
Jason M. Crawford, Department of Chemistry Johns Hopkins University 3400 North Charles Street, Baltimore, MD 21218
Anna L. Vagstad, Department of Chemistry Johns Hopkins University 3400 North Charles Street, Baltimore, MD 21218
Kenneth C. Ehrlich, Southern Regional Research Center United States Department of Agriculture 1100 RE Lee Boulevard, New Orleans, LA 70124
Craig A. Townsend, Department of Chemistry Johns Hopkins University 3400 North Charles Street, Baltimore, MD 21218.
References
- [1].Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- [2].Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- [3].Hitchman T, Schmidt E, Trail F, Rarick M, Linz J, Townsend CA. Bioorg. Chem. 2001;29:293–307. doi: 10.1006/bioo.2001.1216. [DOI] [PubMed] [Google Scholar]
- [4].Brown DW, Adams TH, Keller NP. Proc. Natl. Acad. Sci. USA. 1996;93:14873–14877. doi: 10.1073/pnas.93.25.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bradshaw RE, Jin H, Morgan BS, Schwelm A, Teddy OR, Young CA, Zhang S. Mycopathologia. 2006;161:283–294. doi: 10.1007/s11046-006-0240-5. [DOI] [PubMed] [Google Scholar]
- [6].Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA. Proc. Natl. Acad. Sci. USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Crawford JM, Vagstad AL, Whitworth KP, Ehrlich KC, Townsend CA. manuscript in preparation. [DOI] [PMC free article] [PubMed]
- [8].Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mangroo D, Gerber G. Biochem. Cell. Biol. 1989;68:308–312. [Google Scholar]
- [10].Sanchez C, Du L, Edwards DJ, Toney MD, Shen B. Chem. Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- [11].Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- [12].Ehrlich KC, Yu J, Cotty PJ. Appl. Microbiol. 2005;99:518–527. doi: 10.1111/j.1365-2672.2005.02637.x. [DOI] [PubMed] [Google Scholar]
- [13].Watanabe C, Townsend CA. Chem. Biol. 2002;9:981–988. doi: 10.1016/s1074-5521(02)00213-2. [DOI] [PubMed] [Google Scholar]
- [14].Udwary D, Merski M, Townsend CA. J. Mol. Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Proc. Natl. Acad. Sci. USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ma SM, Tang Y. Febs. J. 2007;274:2854–2864. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- [17].Gaffoor I, Trail F. Appl. Environ. Microbiol. 2006;72:1793–1799. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cox RJ. Org. Biomol. Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- [19].Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y. Biosci. Biotechnol. Biochem. 1999;63:1445–1452. doi: 10.1271/bbb.63.1445. [DOI] [PubMed] [Google Scholar]
- [20].Keatinge-Clay AT, Shelat AA, Savage DF, Tsai SC, Miercke LJ, O’Connell JD, Khosla C, Stroud RM. Structure. (Camb) 2003;11:147–154. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]
- [21].Stevens D. New. Eng. J. Med. 1995;332:1077–1082. doi: 10.1056/NEJM199504203321607. [DOI] [PubMed] [Google Scholar]
- [22].Nosanchuk JD, Yu JJ, Hung CY, Casadevall A, Cole GT. Fungal. Genet. Biol. 2007;44:517–520. doi: 10.1016/j.fgb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- [23].Chang PK, Ehrlich KC, Yu J, Bhatnagar D, Cleveland TE. Appl. Environ. Microbiol. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW. Appl. Environ. Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Johannesson H, Kasuga T, Schaller RA, Good B, Gardner MJ, Townsend JP, Cole GT, Taylor JW. Fungal Genet. Biol. 2006;43:545–559. doi: 10.1016/j.fgb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- [26].Brobst SW, Townsend CA. Can. J. Chem. 1994;72:200–207. [Google Scholar]