Abstract
Xenograft transplantation of human tumor cells into immunodeficient mice is an important method to clarify the roles of specific molecules or chemicals in vivo. Recently, this method has been reported as a definitive examination to identify tumor stem cells. In this study, the authors compared the morphology and the quality and quantity of ribonucleic acid (RNA) and protein in paraffin-embedded tissues of nude mice implanted with human uterine cervical cancer cells, followed by fixation with commonly used fixatives, including 4% paraformaldehyde (PFA), 10% neutral buffered formalin (NBF), 20% NBF, and 99% ethanol (EtOH). The quality of the isolated RNA from PFA- and NBF-fixed paraffin-embedded tissues was high, while EtOH-fixed tissues showed degradation of RNA. NBF-fixed tissues showed excellent quality of morphology, but EtOH-fixed tissues showed contraction of cells. Immunohistochemical results showed differences depending on fixations. The 99% EtOH-fixed samples showed decreases of Ki-67 and VEGF-A immunoreactivities, but improved cytokeratin immunoreactivity. This study indicated that formalin fixation is better than alcohol fixation for RNA preservation in paraffin-embedded cancer cell implantation models. Immunohistochemical results differed markedly depending on fixation materials and antibodies; therefore, suitable fixations are needed to quantify and compare the results of immunohistochemical staining on cancer cell implanted nude mice tissues.
Keywords: formalin, paraformaldehyde, ethanol, subcutaneously implanted tumor, PCR, immunohistochemistry
Paraffin-embedded tissues, after various fixations have been performed, are commonly used for histological analysis and pathological diagnosis worldwide, and they are suitable for storage for long periods. For the molecular biological analysis of human tissues, the fixed paraffin-embedded tissues are one of the most valuable resources. A number of reports have indicated that formalin-fixed paraffin-embedded tissue can be used for the analysis of ribonucleic acid (RNA) and protein expressions, in addition to morphological analysis (Chung et al. 2008; Lehmann and Kreipe 2001; Matsuda et al. 2010; von Smolinski et al. 2005). The extracted RNA from formalin-fixed paraffin-embedded tissue is used for quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) (Castiglione et al. 2007), DNA array technologies (Lehmann and Kreipe 2001), and mRNA copy number analysis (von Smolinski et al. 2005). Protein expression in formalin-fixed paraffin-embedded tissue has been analyzed by immunohistochemistry, western blot analysis and mass spectrometry (Crockett et al. 2005; Ostasiewicz et al. 2010; Scicchitano et al. 2009). Gene and protein expression profiles of formalin-fixed paraffin-embedded tissue can provide insights into molecular mechanisms of diseases, but there are difficulties because of the marked degradation and cross-linking of biomolecules by fixation. Therefore, despite advances in molecular technologies, the quality of RNA and protein from fixed paraffin-embedded tissue remains variable.
The processing parameters that affect the quality of RNAs and proteins in fixed paraffin-embedded tissues have been established. There are three important factors that are involved in RNA and protein preservation: methods of fixations, time before fixation, and length of fixation (Gruber et al. 1994; Pikkarainen et al. 2010). The most widely used fixative to preserve human tissue is 10% neutral buffered formalin (NBF), also known as 3.7% formaldehyde solution. Formalin fixation is a complex process in which formaldehyde forms covalent bands and produces protein–protein and protein–nucleic acid cross-links (Fraenkel-Conrat and Olcott 1948; Shi et al. 1991). Thus, this fixation may affect the preservative conditions of RNAs and proteins owing to alteration of their structures (Allen et al. 2004). Other fixatives including coagulant fixatives such as alcohol, alcohol-based fixatives, and acetone have been reported to be superior to formaldehyde with respect to retaining RNAs and proteins, but to be inferior in morphological quality to formaldehyde (Arnold et al. 1996; Su et al. 2004). There have been no reports on excellent fixative methods for the preservation of morphology, RNAs, and proteins; therefore, researchers use different fixatives at present.
Xenograft transplantation of human tumor cells into immunodeficient mice has been widely used in the field of cancer research, and it is known to be an important method to clarify the effects of specific molecules or chemicals on tumor cells. It is incredibly effective to assess morphological changes, as well as RNA and protein expression, with xenograft tumor tissues. Xenograft tumor tissues can provide a lot of information, such as localization, alteration of quantity, and relation to morphological changes of several proteins and RNAs. Recent studies have shown that a xenograft transplantation model is the most reliable method to identify cancer stem cells in various kinds of cancer cells. It is controversial which fixation is best for RNA and protein expression analysis, as well as morphological analysis.
In this study, we compared the morphology and the quality and quantity of RNA and protein in xenograft tissues fixed with commonly used fixatives, including 4% paraformaldehyde (PFA), 10% NBF, 20% NBF, and 99% ethanol (EtOH) and then paraffin-embedded.
Materials and Methods
Materials
The following were purchased: RNAlater, High Capacity cDNA Reverse Transcription Kit, TaqMan Gene Expression Assays for β-actin (Hs99999903_m1), and TaqMan Fast Universal PCR Master Mix from Applied Biosystems, Inc. (Carlsbad, CA); formaldehyde and paraformaldehyde from Wako Pure Chemical Industries, Ltd. (Osaka, Japan); 99% EtOH from Japan Alcohol Trading Co., Ltd. (Tokyo, Japan); rabbit polyclonal anti-vascular endothelial growth factor-A (VEGF-A) antibody from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); mouse monoclonal anti-AE-1/AE-3 antibody, anti-Ki67 antibody, and anti-E-cadherin antibody from Dako Cytomation (Kyoto, Japan); mouse monoclonal anti-human leukocyte antigen (HLA) class I-A, B, C antibody from Hokudo Co., Ltd. (Sapporo, Japan); Histofine Simple Stain MAX PO (R) or (M) kits from Nichirei (Tokyo, Japan); and New Silane II slide glass and malinol mounting medium from Mutoh Chemical Co., Ltd. (Tokyo, Japan). All other chemicals and reagents were purchased from Sigma-Aldrich Corporation (St. Louis, MO).
Subcutaneous Tumor Tissues
Human uterine cervical squamous cell carcinoma cell line, CaSki cells, were obtained from RIKEN BioResource Center (Ibaraki, Japan) and were grown in RPMI-1640 medium containing 10% heat-inactivated fetal bovine serum (FBS) at 37C under a humidified 5% CO2 atmosphere. Six-week-old male nude mice (N = 2, BALB/cA Jcl-nu/nu; CLEA Japan Inc., Tokyo, Japan) were subcutaneously injected in both sides of their back with 3 × 106 CaSki cells/mouse. After 4 weeks, mice were euthanized and tumors were excised. Animal experiments were carried out according to the institutional animal care guidelines of the Nippon Medical School Animal Ethical Committee.
Tissue Fixation
The fresh tumor specimens were cut into 2 pieces, macroscopically (Fig. 1), and the long axes of all tissue pieces were less than 5 mm. To eliminate contamination, a new surgical blade was used for each specimen. Portions from one half were fixed in one of the following fixatives: 4% PFA, 10% NBF, 20% NBF, or 99% EtOH for 24 hr. The 4% PFA was prepared 1 day before use. Fixed tissues were paraffin-embedded routinely with Vacuum Rotary VRX-23 (Sakura Finetek Japan Co., Ltd., Tokyo, Japan).
Figure 1.
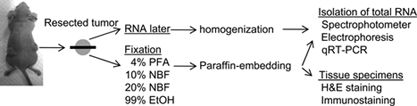
Tissue fixations and subsequent procedures. Nude mice were subcutaneously injected with CaSki cells. After 4 weeks, half of the tumor was immersed in ribonucleic acid (RNA)later, and total RNA was isolated. Portions from the other tumor half were fixed in the respective fixatives, paraffin-embedded, and used for RNA isolation and histological examinations.
RNA Isolation
Paraffin-embedded tissues were sliced using a microtome at a thickness of 10 µm. Paraffin was removed by xylene treatment, and then tissues were washed with EtOH twice to remove xylene. Then, the tissues were treated with proteinase K at 37C overnight. After centrifugation, the supernatant was processed with silica-based spin column to obtain purified total RNA. On the other hand, the samples treated with RNAlater were cut into small pieces and homogenized. Total RNA from all samples was extracted and purified with FastPure RNA Kit. The quantity of RNA was measured using a spectrophotometer (Gene Spec V, Hitachi High-Tech Fielding Corporation, Tokyo, Japan), quantifying single-strand nucleic acid. RNAs were quantified using a spectrophotometer at an OD of 260 nm, and A260/A280 ratios >1.8 were considered to indicate high purity. RNA fragmentation state was evaluated by electrophoresis using the Agilent RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA) following the manufacturer’s protocol.
qRT-PCR
All RNA samples (1 µg) were reverse-transcribed to cDNA using High Capacity cDNA Reverse Transcription Kit following the manufacturer’s protocol. qRT-PCR for housekeeping genes, β-actin, were performed with the StepOnePlus real-time PCR system (Applied Biosystems, Inc., Carlsbad, CA) using specific primers and a TaqMan probe. The amplified size of the β-actin was 171 bp and corresponded to nucleotides 71 to 219 (NM_001101.3). PCR was carried out in 20 µl reaction mixtures containing 10 µl of 2 × TaqMan Fast Universal PCR Master Mix, 2 µl of template cDNA, and 1 µl of TaqMan Gene Expression Assays. Cycling conditions were 20 sec at 95C and then 40 cycles of 1 sec at 95C followed by 20 sec at 60C. Gene expression levels were measured in triplicate.
Morphological Analysis
Paraffin-embedded sections (3 µm) of each fixation protocol were subjected to hematoxylin and eosin (H&E) staining. The morphological changes of H&E-stained tissue with each fixation were analyzed at magnification ×200. The H&E staining was evaluated independently by two of the authors (TF and YM).
Immunohistochemistry
Paraffin-embedded sections (3 µm) were subjected to immunostaining using a Histofine Simple Stain MAX PO (R) or (M) kit. After deparaffinization, the tissue sections for AE-1/AE-3, Ki-67, and E-cadherin immunostaining were preheated in 10 mM citrate buffer (pH 6.0) for 15 min at 121C in an autoclave oven. Then, endogenous peroxidase activity was blocked by incubation for 30 min with 0.3% hydrogen peroxide in methanol. The tissue sections were then incubated with the anti-AE-1/AE-3 antibody (1:200 in dilution), anti-Ki-67 antibody (1:100 in dilution), anti-HLA-class I antibody (1:1000 in dilution), anti-E-cadherin antibody (1:200 in dilution), and anti-VEGF-A antibody (1:200 in dilution) in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) overnight at 4C. Bound antibodies were detected with the Simple Stain MAX PO (R) or (M) reagent, using diaminobenzidine tetrahydrochloride as the substrate. The sections were then counterstained with Mayer’s hematoxylin. Negative control tissue sections were prepared by omitting the primary antibody. As for the evaluation of immunostaining, intensity and proportion of positively stained cancer cells were analyzed at magnification ×200 by two of the authors (TF and YM). As for the evaluation of immunostaining for Ki-67, eight images of ×200 magnification were obtained and then Ki-67-positive nuclei were counted using WinROOF version 6.1.3 (Mitani Corporation, Tokyo, Japan) (Hatanaka et al. 2003).
Statistical Analysis
Quantitative data are presented as mean ± SE values, and were assessed using the Tukey-Kramer test. P<0.05 was taken as statistically significant. Computations were performed using the Stat View J version 5.0 software package (SAS Institute, Inc., Cary, NC).
Results
Quantitative and Qualitative Analyses of Total RNA
RNA was isolated from RNAlater samples and paraffin-embedded samples fixed with 4% PFA, 10% NBF, 20% NBF, or 99% EtOH. The absorbance of each sample was measured, and the quantity of RNA yield was determined (Table 1). A sufficient volume of total RNA for the following experiments was obtained from all samples. OD 260/280 ratio of all samples was more than 1.8; therefore, all samples were considered to be of high purity. Furthermore, the quality of RNA was determined by gel electrophoresis (Fig. 2A). Total RNA from samples treated with RNAlater, exhibited two clear bands at 1800 nucleotides (nts) corresponding to 18S rRNA and 2800 nts corresponding to 28S rRNA. 18S rRNA was detected at high levels in the samples of 4% PFA and 20% NBF, but it was weak in the 10% NBF sample and undetectable in the 99% EtOH sample. In all paraffin-embedded samples, 28S rRNA was undetectable. In contrast to the RNAlater sample, paraffin-embedded samples exhibited many small-size bands (<200 nts). The total RNA sample fixed by 99% EtOH exhibited only small size bands, which were considered as degraded RNAs.
Table 1.
The Quantity and Quality of Total RNA From Paraffin-Embedded Tissues
| RNAlater | 4% PFA | 10% NBF | 20% NBF | 99% EtOH | |
|---|---|---|---|---|---|
| Yield (µg/10 slides) | 323.2 µg/ml | 10.5 | 2.9 | 2.5 | 21.0 |
| 260/280 ratio | 2.10 | 2.05 | 2.02 | 2.08 | 1.99 |
Enough total RNA was obtained from all tissue samples. 260/280 ratio of all samples was >1.8, which were considered to be of high purity. RNA, ribonucleic acid; PFA, paraformaldehyde; NBF, neutral buffered formalin; EtOH, ethanol.
Figure 2.
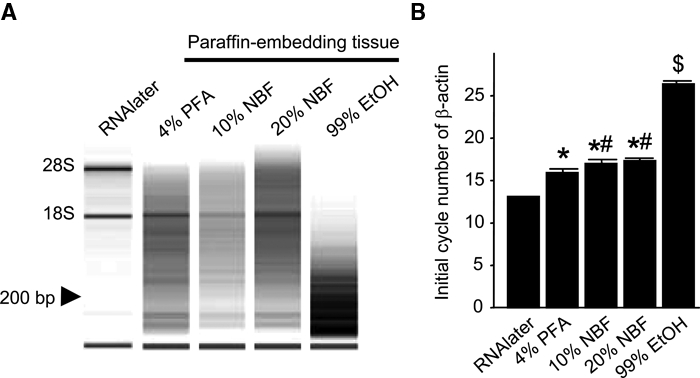
Analysis of quantity and quality of ribonucleic acid (RNA). (A) Levels of RNA degradation were evaluated by electrophoresis. RNAlater samples exhibited clear bands corresponding to 18S rRNA and 28S rRNA, while the samples from 4% paraformaldehyde (PFA), 10% PFA, and 20% neutral buffered formalin (NBF) showed no 28S rRNA band. Neither band was detected in the 99% EtOH sample. (B) Initial cycle number of β-actin in qRT-PCR. The RNAlater sample exhibited the most rapid amplification and the 99% ethanol (EtOH) sample exhibited the slowest amplification for β-actin mRNA (B).
* P<0.05 versus 4% PFA sample.
# P<0.05 versus RNAlater sample.
$ P<0.05 versus RNAlater, 4% PFA, 10%, and 20% NBF.
To further examine the state of preservation of RNA, qRT-PCR amplification plot of β-actin mRNA was measured in RNAlater, 4% PFA, 10% NBF, 20% NBF, and 99% EtOH samples. RNA-specific signals were detectable in all 5 sample types. We analyzed the initial cycle number of qRT-PCR amplification plot of b-actin (Fig. 2B), because the initial cycle number of qRT-PCR amplification plot reflects the total amount of target cDNA. A small initial cycle number indicates a large amount of target RNA and the RNAlater sample showed the smallest number of initial amplified cycles in β-actin mRNA. Among the paraffin-embedded samples, the 4% PFA sample showed the shortest initial amplified cycles, whereas the 99% EtOH sample showed the largest number of initial amplified cycles in β-actin mRNA (Fig. 2B).
Morphological Analysis
Morphological changes with each fixation were determined by H&E staining. The 10% NBF sample showed similar morphology to the 20% NBF samples (Figs. 3B and 3C). Compared with the 20% NBF sample, the 4% PFA sample showed a distinct intercellular space, and an increase of nuclear staining (Fig. 3A). The 99% EtOH sample showed a distinct intercellular space (Fig. 3D), eosinophilic changes of cytoplasm, and a decrease of nuclear staining. In addition, the 99% EtOH sample exhibited contraction of the cells; therefore, cells looked smaller than those in other fixations.
Figure 3.

Morphological and immunohistochemical analysis. The resected tumors were fixed in 4% paraformaldehyde (PFA), 10% neutral buffered formalin (NBF), 20% NBF, or 99% ethanol (EtOH), and stained with hematoxylin and eosin (A–D). These tissue sections were immunostained with anti-VEGF-A (E–H), anti-AE-1/AE-3 (I–L), anti-E-cadherin (M–P), or anti-Ki-67 (Q–T) antibodies. Bars A to D = 50 µm, and E to T = 100 µm.
Preservation of Antigen in the Tissues
To analyze the preservation of antigen in paraffin-embedded tissue, we performed immunostaining for the proteins located in cytoplasm (VEGF-A and AE-1/AE-3), cell membrane (E-cadherin and HLA class I), and nuclei (Ki-67). In VEGF-A, similar immunoreactivities in the cytoplasm of cancer cells were observed in the 4% PFA, 10% NBF, and 20% NBF samples (Figs. 3E, 3F, and 3G). The 99% EtOH sample showed a marked decrease of VEGF-A immunoreactivity (Fig. 3H). By contrast, immunostaining for AE-1/AE-3 showed strong expression in the 99% EtOH sample compared with those in the 4% PFA, 10% NBF, and 20% NBF samples (Figs. 3I, 3J, 3K, and 3L). Immunostaining for E-cadherin showed similar expressions in the membrane of cancer cells in the 4% PFA, 10% NBF, and 20% NBF samples (Figs. 3M, 3N, and 3O), while the 99% EtOH sample showed a decrease of intensity of E-cadherin expression (Fig. 3P). In Ki-67, the 4% PFA sample exhibited slightly high expression of Ki-67 in the nuclei of cancer cells compared with the 10% and 20% NBF samples (Figs. 3Q, 3R, and 3S). Similar expression patterns of Ki-67 in the 10% NBF and 20% NBF samples (Figs. 3R and 3S) were observed. The 99% EtOH sample showed a marked decrease of Ki-67 expression (Fig. 3T). Furthermore, the number of Ki-67-positive cells was counted using image analyzing software WinROOF (Fig. 4). Ki-67-positive cells were 2.5-fold greater in number in 4% PFA samples than in 20% NBF samples. The numbers of Ki-67-positive cells were decreased in the 10% NBF and 99% EtOH samples, and Ki-67 was observed in approximately 6% of cells in the samples fixed with 99% EtOH compared with the level with 4% PFA. Tables 2 and 3 show summaries of the results of intensity and proportion of positive cells in immunohistochemical analyses for Ki-67, VEGF-A, AE-1/AE-3, E-cadherin, and HLA class I.
Figure 4.

Number of Ki-67-positive cells. The number of Ki-67-positive cells was counted using image analyzing software. The 4% paraformaldehyde (PFA) sample had the highest number of Ki-67-positive cells, and the 99% ethanol (EtOH) sample had the lowest number of Ki-67-positive cells.
Table 2.
Intensity of Immunohistochemical Analysis
| 4% PFA | 10% NBF | 20% NBF | 99% EtOH | |
|---|---|---|---|---|
| Ki-67 | 2+ | + | + | ± |
| VEGF-A | + | + | + | - |
| AE-1/AE-3 | + | + | + | 2+ |
| E-cadherin | 2+ | 2+ | 2+ | ± |
| HLA class I | 2+ | + | 2+ | + |
Intensity of immunostaining for Ki-67, VEGF-A, and E-cadherin was markedly decreased in 99% EtOH sample. Intensity of -; negative, ±; weak, +; moderate, 2+; strong. PFA, paraformaldehyde; NBF, neutral buffered formalin; EtOH, ethanol.
Table 3.
Proportion of Positive Cells of Immunohistochemical Analysis
| 4% PFA | 10% NBF | 20% NBF | 99% EtOH | |
|---|---|---|---|---|
| Ki-67 | 2+ | 2+ | 2+ | - |
| VEGF-A | 2+ | 3+ | 3+ | - |
| AE-1/AE-3 | 3+ | 3+ | 3+ | 3+ |
| E-cadherin | 3+ | 3+ | 3+ | 3+ |
| HLA class I | 3+ | 3+ | 3+ | 3+ |
Proportion of positive cells of immunostaining for Ki-67 and VEGF-A was markedly decreased in 99% EtOH sample. -; 0-25%, +; 25-50%, 2+; 50-75%, 3+ 75-100%. PFA, paraformaldehyde; NBF, neutral buffered formalin; EtOH, ethanol.
Discussion
In the present study, the quality of all of the isolated RNA from PFA, NBF, and RNAlater tissues was high, and they could be analyzed by qRT-PCR. Among the formaldehyde fixations, mRNA was most preserved in 4% PFA. However, 99% EtOH tissue showed marked degradation of RNA. These results indicate that formaldehyde fixation is better than alcohol fixation to preserve RNA, and 4% PFA is best for RNA preservation in all fixation methods examined. The reason for the difference between formaldehyde fixations may be that the purity of formaldehyde freshly prepared from PFA is superior to that of commercial stock solutions. Furthermore, the present study showed that the initial cycle number of β-actin, a housekeeping gene, markedly altered with different fixations. The results of the qRT-PCR of β-actin mRNA indicate a similar tendency to those of total RNA in the fixations. The current results differ from those of previous studies reporting that better RNA preservation in tissues can be achieved by using alcohol as a fixative (Ben-Ezra et al. 1991; Goldsworthy et al. 1999; Su et al. 2004). In the present study, we analyzed the effects of various fixatives using a consistent fixation time of 24 hr, and this fixation time with alcohol might have had a deleterious effect on RNA preservation.
In morphological analysis, samples fixed with 4% PFA showed a distinct intercellular space, but sufficient quality was retained for morphological analysis. Ten percent and 20% NBF-fixed tissues showed excellent quality of morphology. Ninety-nine percent EtOH-fixed tissues showed contraction of cells because of dehydration changes. Therefore, 99% EtOH was inferior to formalin-based fixatives in morphological analysis, as previously reported (Su et al. 2004).
Immunohistochemical results showed marked differences depending on fixations. The 99% EtOH-fixed samples showed marked decreases of Ki-67 and VEGF-A immunoreactivities compared with those of formalin-fixed samples. On the other hand, immunoreactivities of HLA class I and E-cadherin showed no changes between the fixations. Immunostaining for AE-1/AE-3 showed the highest expression in the 99% EtOH sample, which is consistent with previous reports that coagulant fixatives improved cytokeratin immunoreactivity (Arnold et al. 1996; Battifora and Kopinski 1986). The present study also indicated that differences of immunoreactivity depended on fixatives, but did not depend on antigen localization. Differences of masks of antigens or changes of epitopes because of fixations might cause alterations of immunoreactivity.
The results of Ki-67 staining with a microscope differed from the results obtained using automatic counting with image-analysis software. The image-analysis software automatically counts the nuclei of cells that have an intensity that exceeds the predetermined threshold level. Therefore, as Table 2 shows, the Ki-67 staining intensity might strongly affect outcomes with software analysis of Ki-67-positive cells. The number of Ki-67-positive cells, as determined using the software, markedly differed between the fixations, and 4% PFA showed the highest number of Ki-67-positive cells. Ki-67-positive index has been widely used in pathological diagnosis and cancer research to determine the proliferative activity or malignant potential of the tumors (Hendricks and Wilkinson 1994; Spyratos et al. 2002). Therefore, the results emphasize the need for careful standardization of immunocytochemical methods, particularly when performing quantitative analysis.
Xenograft transplantation of human tumor cells into immunodeficient mice has been widely used because it is known to be an effective method to clarify the effects of specific molecules or chemicals on tumor cells. Recently, this method has been reported as a definitive examination to identify tumor stem cells in various kinds of tumors. It is controversial which fixation is best for analysis; therefore, researchers usually use various fixations (Aikawa et al. 2008; Arumugam et al. 2005; Sasaki et al. 2008). The present study indicated that it is important to select proper fixations and fixation methods for assessment of expression levels of proteins and RNAs. Furthermore, xenograft transplantation of human cancer cell lines into nude mice is useful to assess proper fixation materials and fixation methods because the model permits us to examine the expression levels of proteins and RNAs in human cancer cells, which originate from single clones of cancer and show a uniform pattern.
In conclusion, the present study indicated that formalin fixation is better than alcohol fixation for RNA preservation in paraffin-embedded cancer cell implantation models. Immunohistochemical results differed markedly depending on fixation materials and antibodies; therefore, suitable fixations and fixation methods are needed to quantify and compare the results of immunohistochemistry for cancer cell-implanted nude mice tissues.
Acknowledgments
The authors thank Mr. Satoshi Takeshima (Mitani Corporation, Tokyo, Japan) for excellent technical assistance. Authors YM and TF are equally contributed to this study.
Footnotes
The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
The authors disclosed receipt of the following financial support for the research and/or authorship of this article: This work was supported by a Grant-in-Aid for Scientific Research (C, No. #22591531) to TI, and Grants-in-Aid for Young Scientists (A, No. #22689038) to YM and (B, No. #22790675) to TY, from the Japan Society for the Promotion of Science, a grant provided by Pancreas Research Foundation of Japan, a grant provided by the Ichiro Kanehara Foundation, and a grant provided by Daiwa Securities Health Foundation to YM.
References
- Aikawa T, Whipple CA, Lopez ME, Gunn J, Young A, Lander AD, Korc M. 2008. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. J Clin Invest. 118:89-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GM, Middleton J, Katrak PH, Lord SR, Gandevia SC. 2004. Prediction of voluntary activation, strength and endurance of elbow flexors in postpolio patients. Muscle Nerve. 30:172-181 [DOI] [PubMed] [Google Scholar]
- Arnold MM, Srivastava S, Fredenburgh J, Stockard CR, Myers RB, Grizzle WE. 1996. Effects of fixation and tissue processing on immunohistochemical demonstration of specific antigens. Biotech Histochem. 71:224-230 [DOI] [PubMed] [Google Scholar]
- Arumugam T, Simeone DM, Van Golen K, Logsdon CD. 2005. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res. 11:5356-5364 [DOI] [PubMed] [Google Scholar]
- Battifora H, Kopinski M. 1986. The influence of protease digestion and duration of fixation on the immunostaining of keratins. A comparison of formalin and ethanol fixation. J Histochem Cytochem. 34:1095-1100 [DOI] [PubMed] [Google Scholar]
- Ben-Ezra J, Johnson DA, Rossi J, Cook N, Wu A. 1991. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J Histochem Cytochem. 39:351-354 [DOI] [PubMed] [Google Scholar]
- Castiglione F, Degl’Innocenti DR, Taddei A, Garbini F, Buccoliero AM, Raspollini MR, Pepi M, Paglierani M, Asirelli G, Freschi G, Bechi P, Taddei GL. 2007. Real-time PCR analysis of RNA extracted from formalin-fixed and paraffin-embeded tissues: effects of the fixation on outcome reliability. Appl Immunohistochem Mol Morphol. 15:338-342 [DOI] [PubMed] [Google Scholar]
- Chung JY, Braunschweig T, Williams R, Guerrero N, Hoffmann KM, Kwon M, Song YK, Libutti SK, Hewitt SM. 2008. Factors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem. 56:1033-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. 2005. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 85:1405-1415 [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Olcott HS. 1948. The reaction of formaldehyde with proteins; cross-linking between amino and primary amide or guanidyl groups. J Am Chem Soc. 70:2673-2684 [DOI] [PubMed] [Google Scholar]
- Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR. 1999. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 25:86-91 [PubMed] [Google Scholar]
- Gruber AD, Moennig V, Hewicker-Trautwein M, Trautwein G. 1994. Effect of formalin fixation and long-term storage on the detectability of bovine viral-diarrhoea-virus (BVDV) RNA in archival brain tissue using polymerase chain reaction. Zentralbl Veterinarmed B. 41:654-661 [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Hashizume K, Nitta K, Kato T, Itoh I, Tani Y. 2003. Cytometrical image analysis for immunohistochemical hormone receptor status in breast carcinomas. Pathol Int. 53:693-699 [DOI] [PubMed] [Google Scholar]
- Hendricks JB, Wilkinson EJ. 1994. Quality control considerations for Ki-67 detection and quantitation in paraffin-embedded tissue. J Cell Biochem Suppl. 19:105-110 [PubMed] [Google Scholar]
- Lehmann U, Kreipe H. 2001. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 25:409-418 [DOI] [PubMed] [Google Scholar]
- Matsuda KM, Chung JY, Hewitt SM. 2010. Histo-proteomic profiling of formalin-fixed, paraffin-embedded tissue. Expert Rev Proteomics. 7:227-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostasiewicz P, Zielinska DF, Mann M, Wisniewski JR. 2010. Proteome, phosphoproteome, and n-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 9:3688-3700 [DOI] [PubMed] [Google Scholar]
- Pikkarainen M, Martikainen P, Alafuzoff I. 2010. The effect of prolonged fixation time on immunohistochemical staining of common neurodegenerative disease markers. J Neuropathol Exp Neurol. 69:40-52 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Miura K, Horii A, Kaneko N, Fujibuchi W, Kiseleva L, Gu Z, Murata Y, Karasawa H, Mizoi T, Kobayashi T, Kinouchi M, Ohnuma S, Yazaki N, Unno M, Sasaki I. 2008. Orthotopic implantation mouse model and cDNA microarray analysis indicates several genes potentially involved in lymph node metastasis of colorectal cancer. Cancer Sci. 99:711-719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scicchitano MS, Dalmas DA, Boyce RW, Thomas HC, Frazier KS. 2009. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J Histochem Cytochem. 57:849-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SR, Key ME, Kalra KL. 1991. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 39:741-748 [DOI] [PubMed] [Google Scholar]
- Spyratos F, Ferrero-Pous M, Trassard M, Hacene K, Phillips E, Tubiana-Hulin M, Le Doussal V. 2002. Correlation between MIB-1 and other proliferation markers: clinical implications of the MIB-1 cutoff value. Cancer. 94:2151-2159 [DOI] [PubMed] [Google Scholar]
- Su JM, Perlaky L, Li XN, Leung HC, Antalffy B, Armstrong D, Lau CC. 2004. Comparison of ethanol versus formalin fixation on preservation of histology and RNA in laser capture microdissected brain tissues. Brain Pathol. 14:175-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Smolinski D, Leverkoehne I, von Samson-Himmelstjerna G, Gruber AD. 2005. Impact of formalin-fixation and paraffin-embedding on the ratio between mRNA copy numbers of differently expressed genes. Histochem Cell Biol. 124:177-188 [DOI] [PubMed] [Google Scholar]


