Abstract
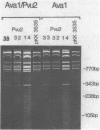
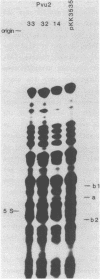
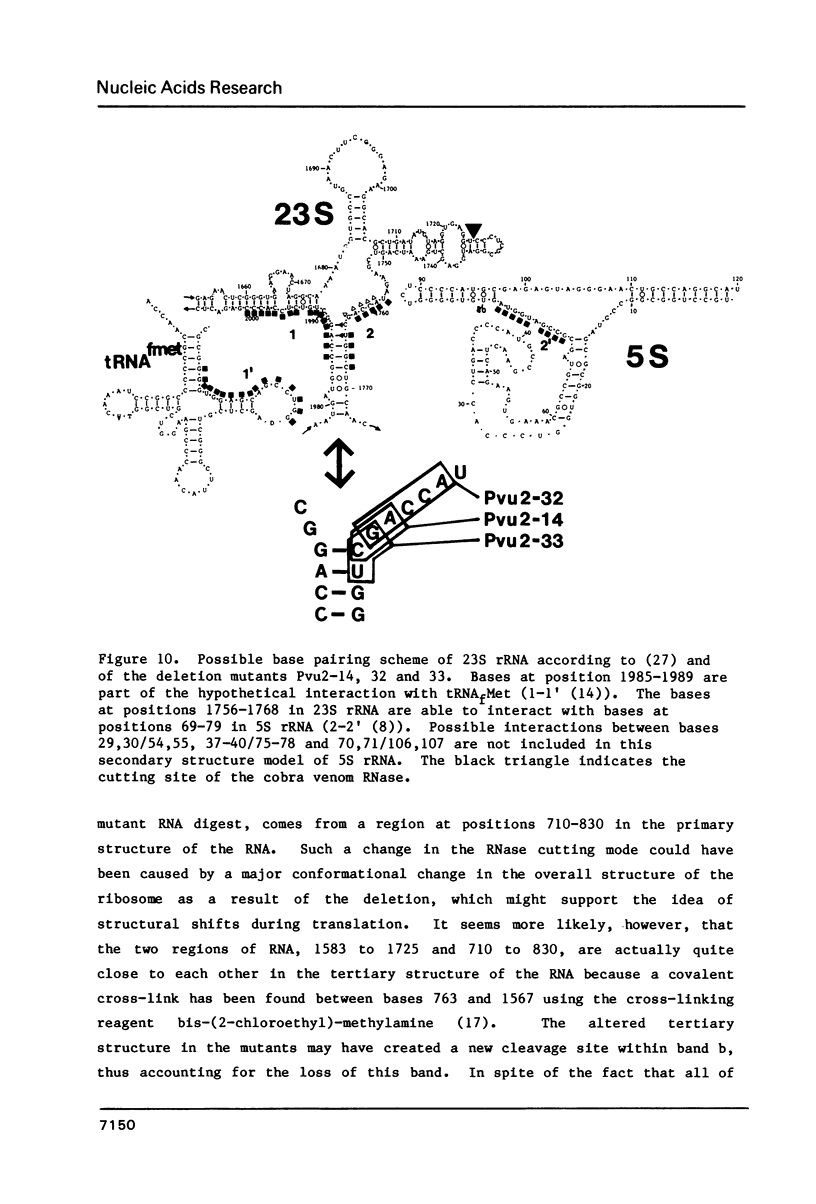
Three different small deletions were produced at a single Pvu 2 restriction site in E. coli 23S rDNA of plasmid pKK 3535 using exonuclease Bal 31. The deletions were located around position 1760 in 23S rRNA and were characterized by DNA sequencing as well as by direct fingerprinting and S1-mapping of the rRNA. Two of the mutant plasmids, Pvu 2-32 and Pvu 2-33, greatly reduced the growth rate of transformed cells while the third mutant, Pvu 2-14 grew as fast as cells containing the wild-type plasmid pKK 3535. All three mutant 23S rRNAs were incorporated into 50S-like particles and were even found in 70S ribosomes and polysomes in vivo. The conformation of mutant 23S rRNA in 50S subunits was probed with a double-strand specific RNase from cobra venom. These analyses revealed changes in the accessibility of cleavage sites near the deletions around position 1760 and in the area around position 800 in all three mutant rRNAs. We suggest, that an altered conformation of the rRNAs at the site of the deletion is responsible for the slow growth of cells containing mutant plasmids Pvu 2-32 and Pvu 2-33.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dingman C. W., Peacock A. C. Electrophoretic characterization of bacterial polyribosomes in agarose-acrylamide composite gels. J Mol Biol. 1969 Apr 14;41(1):139–147. doi: 10.1016/0022-2836(69)90131-4. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Lund E., Kjeldgaard N. O. Some effects of antibiotics on bacterial polyribosomes as studied by gel electrophoresis. J Mol Biol. 1973 Aug 25;78(4):627–636. doi: 10.1016/0022-2836(73)90284-2. [DOI] [PubMed] [Google Scholar]
- Dahlberg J. E., Kintner C., Lund E. Specific binding of tRNAMet to 23S rRNA of Escherichia coli. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1071–1075. doi: 10.1073/pnas.75.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Stark M. J., Dahlberg A. E. Site-directed mutagenesis of ribosomal RNA. Construction and characterization of deletion mutants. J Mol Biol. 1982 Aug 15;159(3):397–416. doi: 10.1016/0022-2836(82)90291-1. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Oostra B. A., Ely B. K., Smith A. E. Deletion loop mutagenesis: a novel method for the construction of point mutations using deletion mutants. Nucleic Acids Res. 1982 Sep 11;10(17):5161–5171. doi: 10.1093/nar/10.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly P., Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983 Nov 11;11(21):7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Experimental determination of interacting sequences in ribosomal RNA. Nature. 1979 Sep 27;281(5729):271–276. doi: 10.1038/281271a0. [DOI] [PubMed] [Google Scholar]
- Stark M. J., Gourse R. L., Dahlberg A. E. Site-directed mutagenesis of ribosomal RNA. Analysis of ribosomal RNA deletion mutants using maxicells. J Mol Biol. 1982 Aug 15;159(3):417–439. doi: 10.1016/0022-2836(82)90292-3. [DOI] [PubMed] [Google Scholar]
- Stiege W., Zwieb C., Brimacombe R. Precise localisation of three intra-RNA cross-links in 23S RNA and one in 5S RNA, induced by treatment of Escherichia coli 50S ribosomal subunits with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1982 Nov 25;10(22):7211–7229. doi: 10.1093/nar/10.22.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Hearst J. E. Structure-function relations in E. coli 16S RNA. Cell. 1983 May;33(1):19–24. doi: 10.1016/0092-8674(83)90330-6. [DOI] [PubMed] [Google Scholar]
- Uchida T., Bonen L., Schaup H. W., Lewis B. J., Zablen L., Woese C. The use of ribonuclease U2 in RNA sequence determination. Some corrections in the catalog of oligomers produced by ribonuclease T1 digestion of Escherichia coli 16S ribosomal RNA. J Mol Evol. 1974 Feb 28;3(1):63–77. doi: 10.1007/BF01795977. [DOI] [PubMed] [Google Scholar]
- Vassilenko S. K., Carbon P., Ebel J. P., Ehresmann C. Topography of 16 S RNA in 30 S subunits and 70 S ribosomes accessibility to cobra venom ribonuclease. J Mol Biol. 1981 Nov 15;152(4):699–721. doi: 10.1016/0022-2836(81)90123-6. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. Micro thin-layer techniques for rapid sequence analysis of 32P-labeled RNA: double digestion and pancreatic ribonuclease analyses. Anal Biochem. 1977 Nov;83(1):228–239. doi: 10.1016/0003-2697(77)90531-0. [DOI] [PubMed] [Google Scholar]
- Weidner H., Yuan R., Crothers D. M. Does 5S RNA function by a switch between two secondary structures? Nature. 1977 Mar 10;266(5598):193–194. doi: 10.1038/266193a0. [DOI] [PubMed] [Google Scholar]