Abstract
Pluripotency is a developmental ground state that can be recreated by direct reprogramming. Establishment of pluripotency is crucially dependent on the homeodomain-containing transcription factor Nanog. Compared with other pluripotency-associated genes, however, Nanog shows relatively low sequence conservation. Here, we investigated whether Nanog orthologs have the capacity to orchestrate establishment of pluripotency in Nanog–/– somatic cells. Mammalian, avian and teleost orthologs of Nanog enabled efficient reprogramming to full pluripotency, despite sharing as little as 13% sequence identity with mouse Nanog. Nanog orthologs supported self-renewal of pluripotent cells in the absence of leukemia inhibitory factor, and directly regulated mouse Nanog target genes. Related homeodomain transcription factors showed no reprogramming activity. Nanog is distinguished by the presence of two unique residues in the DNA recognition helix of its homeodomain, and mutations in these positions impaired reprogramming. On the basis of genome analysis and homeodomain identity, we propose that Nanog is a vertebrate innovation, which shared an ancestor with the Bsx gene family prior to the vertebrate radiation. However, cephalochordate Bsx did not have the capacity to replace mouse Nanog in reprogramming. Surprisingly, the Nanog homeodomain, a short sequence that contains the only recognizable conservation between Nanog orthologs, was sufficient to induce naive pluripotency in Nanog–/– somatic cells. This shows that control of the pluripotent state resides within a unique DNA-binding domain, which appeared at least 450 million years ago in a common ancestor of vertebrates. Our results support the hypothesis that naive pluripotency is a generic feature of vertebrate development.
Keywords: Nanog, Homeodomain, Induced pluripotency
INTRODUCTION
Pluripotent stem cells from mice and humans differ in important biological and molecular aspects, including culture requirements (Evans and Kaufman, 1981; Thomson et al., 1998; Ying et al., 2008), gene expression (Ginis et al., 2004; Richards et al., 2004) and X chromosome status (Maherali et al., 2007; Shen et al., 2008; Tchieu et al., 2010). It was initially thought that these differences reflected variation between species. However, stem cell lines derived from the mouse post-implantation epiblast (EpiSCs) were found to have properties similar to human embryonic stem (ES) cells (Brons et al., 2007; Tesar et al., 2007). This suggested that differences between mouse and human ES cells may reflect a developmental distinction between naive and primed pluripotent states, rather than species-specific differences (Nichols and Smith, 2009; Rossant, 2008). Upon expression of defined factors and manipulation of the culture environment, EpiSCs can be converted to naive pluripotency (Greber et al., 2010; Guo et al., 2009; Hanna et al., 2009a). A similar strategy was recently shown to convert human ES cells into a cellular state more akin to the naive pluripotent state observed in mice (Hanna et al., 2010). This has raised the intriguing possibility that naive pluripotency may be a generic feature of mammalian development.
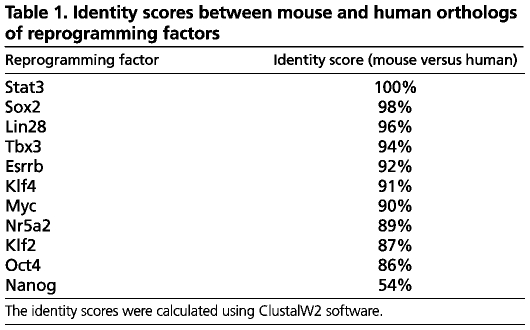
One approach to this issue is to consider whether molecular determinants of naive pluripotency, specifically components of its core transcriptional circuitry, are conserved between species. Most genes associated with pluripotency and reprogramming are highly conserved between eutherian mammals (Table 1). However, mouse and human Nanog orthologs share only 54% sequence identity, far below the average of 92% for other pluripotency-associated and reprogramming factors (Table 1). Nanog is also poorly conserved relative to the global average of 85% sequence identity between mouse and human proteins (Makalowski et al., 1996). This is remarkable, as Nanog occupies a central position in the transcriptional network controlling ES cell pluripotency (Ivanova et al., 2006; Loh et al., 2006; Niwa, 2007; Wang et al., 2006). Genetic studies in the early mouse embryo have shown that Nanog is required for the isolation of ES cells (Mitsui et al., 2003), and for the establishment of the naive pluripotent epiblast (Silva et al., 2009). Selection for activation of the endogenous Nanog locus allows the isolation of fully reprogrammed induced pluripotent stem cell (iPS) cells (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). Endogenous Nanog is not expressed in highly proliferative and transgene-dependent transduced somatic cells (pre-iPS), but is upregulated during the transition to full pluripotency (Mikkelsen et al., 2008; Silva et al., 2008; Sridharan et al., 2009; Theunissen et al., 2011). Constitutive expression of Nanog accelerates reprogramming and enables induced pluripotency in conditions that do not support the self-renewal of ES cells (Hanna et al., 2009b; Theunissen et al., 2011). Without Nanog, somatic cell reprogramming does not progress to full pluripotency in chemically defined conditions that support naive pluripotency (Silva et al., 2009). Thus, Nanog can be seen as a molecular switch that controls the establishment of pluripotency during embryogenesis and reprogramming (Theunissen and Silva, 2011).
Table 1.
Identity scores between mouse and human orthologs of reprogramming factors
Previous studies suggested that Nanog has only limited, if any, functional conservation. Human Nanog had a significantly reduced capacity to support mouse ES cell self-renewal in the absence of leukemia inhibitory factor (LIF) (Chambers et al., 2003). Although recognizing some targets of mouse Nanog, urodele Nanog had no activity in this self-renewal assay (Dixon et al., 2010). In addition, non-eutherian orthologs of Nanog lack the tryptophan repeat (WR) domain that was previously shown to be important for Nanog dimerization and for interactions with other pluripotency regulators (Mullin et al., 2008; Wang et al., 2008). However, these studies did not interrogate the capacity to establish naive pluripotency, the process for which Nanog is genetically indispensable in the mouse. In fact, once pluripotency is established, Nanog can be permanently deleted without eliminating the self-renewal or developmental potential of pluripotent stem cells (Chambers et al., 2007; Silva et al., 2009). Furthermore, these self-renewal experiments were performed in the presence of endogenous mouse Nanog (mNanog), which raises the issue of whether the observed phenotypes can be solely attributed to ectopically expressed Nanog orthologs. To uncover the full extent of Nanog functional conservation, we interrogated the capacity of Nanog orthologs to establish pluripotency in the complete absence of endogenous mouse Nanog.
MATERIALS AND METHODS
Cells
Nanog–/– neural stem (NS) cells and mouse embryonic fibroblasts (MEFs) were derived as previously described (Silva et al., 2009) from E13.5 mouse chimeras and were purified by two rounds of flow cytometry for constitutive GFP expression. Nanog–/– somatic cells were transduced with pMXs-based retroviral reprogramming factors (Silva et al., 2008; Takahashi and Yamanaka, 2006). Cultures were changed into ES cell medium (serum/LIF) at day 3 post-transduction, and re-plated onto feeders at day 5 to expand Nanog–/– pre-iPS cells. Transfections of Nanog ortholog transgenes were performed in Nanog–/– pre-iPS cells to avoid variability due to differences in viral titers between experiments. The introduction of Nanog before or after retroviral infection does not affect the outcome of this reprogramming assay (Silva et al., 2009). Nanog–/– pre-iPS cells were nucleofected (Amaxa) with 1 μg of PB-CAG-loxP-Transgene-loxP-PGK-Hygromycin plus 2 μg PBase expression vector, pCAGPBase (Silva et al., 2009). Selection was applied to transfectants for at least 10 days and stable transgene expression was confirmed by qRT-PCR. Stable pre-iPS cell transfectants (1×105) were seeded in a six-well plate on a fibroblast feeder layer in serum/LIF medium. After 2 days, medium was switched to 2i/LIF. Geneticin (100-400 μg/ml) selection for activation of a neomycin transgene under the endogenous Nanog regulatory elements was applied after 6 days of 2i/LIF induction to eliminate background pre-iPS cells (Chambers et al., 2007). No iPS cells emerged in empty vector transfectants whether or not geneticin was applied. Alkaline phosphatase (AP) staining was performed after 10 days of 2i/LIF treatment. EpiSCs derived from E5.5 Oct4GiP epiblast were transfected using LipofectamineTM 2000 (Invitrogen) with 1 μg of B-CAG-loxP-Transgene-loxP-PGK-Hygromycin plus 2 μg PBase expression vector, pCAGPBase (Guo et al., 2009). Stable EpiSC transfectants were seeded in a six-well plate in EpiSC medium. After 2 days medium was switched as indicated. Puromycin (1 μg/ml) was applied after 6 days of 2i/LIF induction to select for expression of the Oct4GiP reporter transgene. All reprogramming experiments in this paper were repeated two to four times. LIF-independent self-renewal was assessed in iPS–/– cells before and after tamoxifen-induced Cre-excision of the PB transgene, and in E14Tg2A ES cells stably transfected with empty, mouse Nanog, chick Nanog and zebrafish Nanog PB transgenes. Six-hundred cells were plated into 6 wells in ES cell medium containing 10% FCS minus LIF. AP staining was performed after 7 days. Details of cDNA sequences used in this study can be found in Fig. S8 (supplementary material).
Culture media
Pre-iPS cells were cultured on a fibroblast feeder layer in GMEM containing 10% FCS, 1×NEAA, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol and 2 mM L-glutamine, supplemented with LIF (complete medium). Reprogramming experiments were performed in N2B27 medium (Stem Cell Sciences, SCS-SF-NB-02) supplemented with LIF and 2i inhibitors (Ying et al., 2008), CHIR99021 (3 μM) and PD0325901 (1 μM). For expansion of established iPS cell lines, 2i/LIF was added in N2B27 or knockout serum replacement (KSR) medium. Basal KSR medium is GMEM containing 10% KSR (Invitrogen, 10828-028), 1% FCS, 1×NEAA, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol and 2 mM L-glutamine. NS cells were maintained in NDiff basal RHB-A (Stem Cell Sciences, SCS-SF-NB-01) supplemented with 10 ng/ml of both EGF and FGF2. EpiSCs were cultured in activin A (20 ng/ml) and Fgf2 (12 ng/ml) in N2B27 medium on fibronectin-coated plates.
Blastocyst injection and morula aggregation
iPS–/– cells were treated for 48 hours with 500 nM 4-hydroxy-tamoxifen (4OHT) for Rosa26-CreERT2-induced transgene excision prior to blastocyst injection. Chimaeras were generated by microinjection using host blastocysts of C57BL/6 strain. At least one-third of littermates showed coat color chimerism after every round of blastocyst injection using iPS–/– cells generated with human Nanog, chick Nanog, zebrafish Nanog, mouse Nanog homeodomain (HD) or zebrafish Nanog HD. We did not assess germline transmission in chimeric animals generated using iPS–/– cells as Nanog is required for germ cell development (Chambers et al., 2007). The capacity to contribute to the germ lineage was assessed at E12.5 in wild-type EpiSC-derived iPS cells generated with zebrafish Nanog. These cells contain an Oct4-GFP reporter transgene. Prior to morula aggregation, the loxP-flanked zebrafish Nanog transgene was excised by 4OHT induction of a stably transfected Cre-ERT2 plasmid.
Immunofluorescence and RNA FISH
Cells were cultured overnight on glass slides and fixed directly in 4% PFA, followed by permeabilization in 0.5% Triton X-100. Mouse monoclonal anti-FLAG M2 (1:500) from Sigma (F1804) was used as primary antibody. Subsequently, a goat anti-mouse secondary antibody (1:1000) from Molecular Probes was applied. RNA FISH was carried out as described previously (Heard et al., 2001). The probe was prepared by labeling plasmid DNA containing a mouse Xist exon 1 sequence.
Bisulfite sequencing
Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen). Bisulfite treatment was performed using the EpiTect Bisulfite Kit (Qiagen). Amplified products were cloned into pCR2.1-TOPO (Invitrogen). Randomly selected clones were sequenced and analyzed using Quantification Tool for Methylation Analysis (QUMA, http://quma.cdb.riken.jp/).
Chromatin immunoprecipitation
ChIP-IT Express (Active Motif) was used according to supplier’s recommendations. Cells were crosslinked using 1% formaldehyde for 10 minutes at room temperature. Formaldehyde was quenched by a 5-minute incubation with glycine, cells were rinsed twice with ice-cold PBS, collected by scraping and pelleted at 573 g for 10 minutes at 4°C. Chromatin was sonicated using a Bioruptor200 (Diagenode) at high frequency on 30 seconds ON/30 seconds OFF cycles for 10 minutes. At least 15 μg of chromatin was incubated with 2 μg of mouse monoclonal anti-FLAG M2 (1:500) from Sigma (F1804) or rabbit polyclonal anti-Nanog (Bethyl Laboratories, A300-397A) for 1 hour at 4°C and subsequently with protein G magnetic beads. Purified DNA and 1% input were analyzed by Taqman qPCR, using fourfold dilutions of the concentrated input for standard curves and triplicates per sample. Occupancy is plotted as fold enrichment after normalization to the input; error bars represent standard deviation of the technical replicates of the qPCR for each experiment.
Microarray
Amplification and labeling of RNA were performed according to the TotalPrep-96 RNA Amplification Kit for the Illumina platform (Ambion). Subsequent hybridization, staining and scanning were performed according to the Whole Genome Gene Expression Direct Hybridization Guide on the MouseWG-6 v2.0 Expression BeadChip (Illumina). Data were loaded into the R package lumi (Du et al., 2008) and then divided into subsets to be analyzed. The data were transformed using Variance Stabilization (VST) (Lin et al., 2008) and normalized using quantile normalization. Comparisons were performed in the R package limma (Smyth, 2004) and the results were corrected using False Discovery Rate (FDR). Our analysis employed a 5% confidence interval. Microarray data are presented as heatmaps that show the correlation between samples (all replicates included). Microarray data has been deposited at Gene Expression Omnibus (Accession Number GSE32715).
RT-PCR and qRT-PCR
Total RNA was extracted using the RNeasy kit (Qiagen) and cDNA generated using Superscript III (Invitrogen). Expression of HD fragments was determined using Taq DNA polymerase (Qiagen) according to manufacturer recommendations and the following thermalcycler settings: 94°C for 3 minutes, 30 cycles of (94°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds) and 72°C for 10 minutes. Relative gene expression levels were determined using the TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and FAM-labeled TaqMan gene expression assays. Average threshold cycles were determined from triplicate reactions and the levels of gene expression were normalized to GAPDH (VIC-labeled endogenous control assay). Relative expression levels of Xist were determined using Fast SYBR Green Master Mix (Applied Biosystems). Mean quantity of expression was determined from triplicate reactions and a standard curve. Expression levels were normalized to GAPDH. Error bars indicate ±1 s.d. qRT-PCR experiments were performed on a StepOnePlus Real Time PCR System (Applied Biosystems). Details of all primers used in this study can be found in Table S1 (supplementary material).
RESULTS
Mammalian orthologs of Nanog establish full pluripotency in Nanog–/– somatic cells
To examine rigorously the ability of Nanog orthologs to establish naive pluripotency, we undertook a genetic complementation experiment (supplementary material Fig. S1A). It has previously been reported that Nanog–/– neural stem (NS) cells infected with retroviral transgenes encoding Oct4, Klf4 and Myc give rise to pre-iPS cells, but do not transit to pluripotency in the presence of small molecule inhibitors of MAP kinase (MEK) and GSK3 (2i) with leukemia inhibitory factor (LIF) (Silva et al., 2009). This is an optimal culture condition not only for the derivation and maintenance of mouse ES cells, but also for promoting induced pluripotency (Silva et al., 2008; Theunissen et al., 2011; Ying et al., 2008). Transfection with a constitutive mouse Nanog (mNanog) transgene enables successful generation of iPS cells from Nanog–/– pre-iPS cells in 2i/LIF conditions (Silva et al., 2009). Thus, Nanog–/– somatic cells provide a genetically controlled system to assess the functional conservation and structural requirements of Nanog in induced pluripotency.
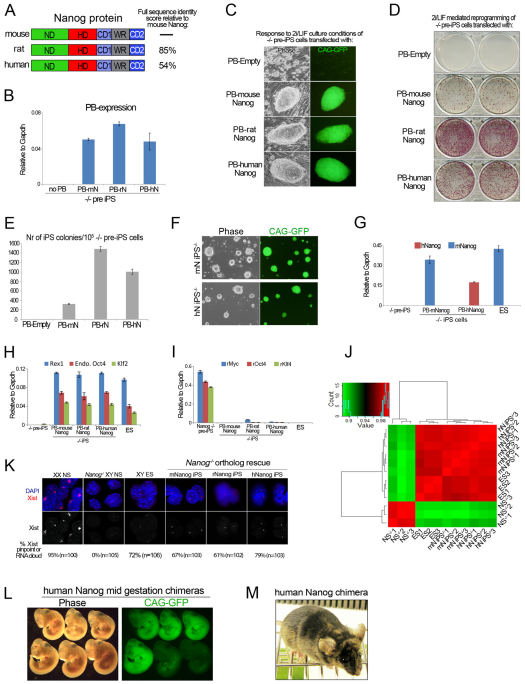
We asked whether the reprogramming potential of Nanog–/– somatic cells could be restored by rat Nanog (rNanog) or human Nanog (hNanog) (Fig. 1A, supplementary material Fig. S1B). Nanog–/– pre-iPS cells were stably transfected with transgenes encoding rNanog or hNanog (Fig. 1B) and medium was switched to 2i/LIF (Fig. 1C). Alkaline phosphatase (AP) staining after 10 days of 2i/LIF culture showed that rNanog enabled efficient reprogramming (Fig. 1D,E). This was expected given the recent derivation of naive pluripotent rat ES cells (Buehr et al., 2008; Li et al., 2008). Interestingly, we also observed efficient iPS cell generation with hNanog (hNanog iPS–/–) (Fig. 1D,E). These showed expression of hNanog but not of mNanog transcript (Fig. 1F,G). Quantitative (q) RT-PCR analysis indicated reactivation of pluripotency-associated genes and silencing of retroviral transgenes (Fig. 1H,I). To ascertain whether reprogramming with hNanog resulted in any global expression changes, we performed microarray analysis. This revealed a close clustering between iPS–/– cells derived with mNanog and hNanog, indicating that hNanog faithfully generated a pluripotent transcriptome (Fig. 1J). Another feature of naive pluripotency is the unique nuclear pattern of Xist RNA, a large non-coding RNA that induces X-chromosome inactivation in female eutherian mammals (Deakin et al., 2009). It has recently been demonstrated that Nanog is directly involved in regulating Xist expression (Navarro et al., 2008). The expected pattern for naive pluripotent cells, an Xist RNA pinpoint signal, was detected in rNanog and hNanog iPS–/– cells (Fig. 1K). As constitutive Nanog expression is likely to interfere with embryonic development, we assessed the contribution to somatic development after tamoxifen-induced Cre excision of the loxP-flanked hNanog transgene. Mid-gestation and adult chimeras were obtained after blastocyst injection (Fig. 1L,M). These results demonstrate that the capacity to induce naive pluripotency is fully conserved in eutherian mammalian orthologs of Nanog. This is compatible with the recent report that a putative naive pluripotent state can be captured in the human system (Hanna et al., 2010).
Fig. 1.
Eutherian mammalian orthologs of Nanog induce naive pluripotency. (A) Structural alignment of eutherian Nanog ortholog proteins. Identity scores are indicated on the right and were calculated using ClustalW2. ND, N-terminal domain; HD, homeodomain; CD1, C-terminal domain 1; WR, tryptophan repeat; CD2, C-terminal domain 2 (see supplementary material Fig. S1B for sequence alignment). (B) qRT-PCR analysis for expression of the piggyBac (PB) sequence in Nanog–/– pre-iPS cells transfected with mouse (m) Nanog (N), rat (r) Nanog or human (h) Nanog PB. (C) Phase contrast and GFP images of emerging Nanog–/– iPS cell colonies. (D) Alkaline phosphatase (AP) staining of emerging Nanog–/– iPS cell colonies. (E) Quantification of the total number of AP-positive iPS cell colonies. Data are mean values. Error bars indicate 1 s.d. (n=2). (F) Phase contrast and GFP images of Nanog–/– iPS. (G) Quantitative RT-PCR (qRT-PCR) analysis confirming the presence of mNanog transcript or hNanog transcript in mNanog iPS–/– cells and hNanog iPS–/– cells, respectively. Data are relative levels. Error bars indicate 1 s.d. (H,I) qRT-PCR analysis for pluripotent gene (H) and retroviral (r) transgene (I) expression in Nanog–/– pre-iPS cells, mNanog iPS–/– cells, rNanog iPS–/– cells, hNanog iPS–/– cells and ES cells. Data are relative levels. Error bars indicate 1 s.d. (J) Correlation heatmap of global gene expression between Nanog–/– NS cells, ES cells, mNanog (mN) iPS–/– cells and hNanog (hN) iPS–/– cells. Three biological replicates were included for each cell line. (K) RNA FISH for Xist in Nanog–/– iPS cells. (L) Phase contrast and fluorescence images of mid-gestation chimeras of hNanog iPS–/– cells following Cre excision of the hNanog transgene. (M) Chimeric adult mouse with hNanog iPS–/– cells. Agouti coat color indicates chimerism.
Vertebrate orthologs of Nanog establish full pluripotency in Nanog–/– somatic cells
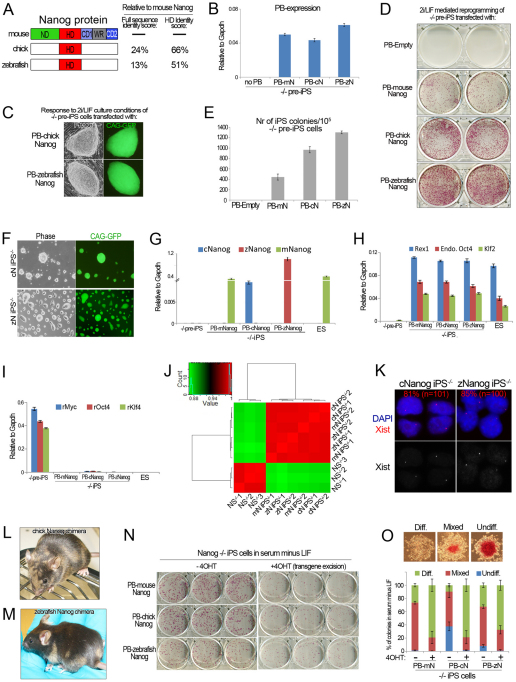
We then examined the functional conservation of Nanog genes isolated from non-eutherian species. Phylogenetics and comparative genomics support full orthology between Actinopterygii and Sarcopterygii Nanog homeobox genes (supplementary material Fig. S2A,B). Chick Nanog (cNanog) and zebrafish Nanog (zNanog; GenBank JN809237) share as little as 13% protein sequence identity with mNanog, and do not contain a WR domain (Fig. 2A). Surprisingly, we found that both cNanog and zNanog had the capacity to replace mNanog in iPS cell generation (Fig. 2B,C), and could do so without compromising reprogramming efficiency (Fig. 2D,E). The resultant iPS cells expressed cNanog or zNanog, respectively, but not mNanog (Fig. 2F,G). Upregulation of endogenous pluripotency genes and silencing of retroviral transgenes was confirmed (Fig. 2H,I). Global gene expression in cNanog and zNanog iPS–/– cells was highly similar to mNanog iPS–/– cells (Fig. 2J). In fact, individual replicates of mNanog iPS–/– samples clustered more closely with cNanog or zNanog iPS–/– samples than with each other. Additionally, the presence of an Xist RNA pinpoint signal indicates that non-mammalian Nanog orthologs acquire the characteristic Xist RNA nuclear pattern of naive pluripotent cells (Fig. 2K). Finally, we performed blastocyst injection after Cre excision of the cNanog or zNanog transgene. In both cases, adult chimeras were obtained (Fig. 2L,M). We conclude that the capacity to induce naive pluripotency in murine cells is functionally and robustly conserved in vertebrate orthologs of Nanog separated by at least 450 million years of evolution.
Fig. 2.
Vertebrate Nanog orthologs induce naive pluripotency. (A) Structural alignment of mNanog with non-mammalian Nanog ortholog proteins. Identity scores between full-length proteins or homeodomain (HD) only are indicated on the right. The identity scores were calculated using ClustalW2. ND, N-terminal domain; CD1, C-terminal domain 1; WR, tryptophan repeat; CD2, C-terminal domain 2. (B) qRT-PCR analysis for expression of the piggyBac (PB) sequence in Nanog–/– pre-iPS cells transfected with mouse (m), chick (c) or zebrafish (z) Nanog (N) PB. Data are relative levels. Error bars indicate 1 s.d. (C) Phase contrast and GFP images of emerging Nanog–/– iPS cell colonies. (D) Alkaline phosphatase (AP) staining of emerging Nanog–/– iPS cell colonies. (E) Quantification of the total number of AP-positive iPS cell colonies. Data are mean values. Error bars indicate 1 s.d. (n=2). (F) Phase contrast and GFP images of Nanog–/– iPS cells generated with cNanog or zNanog. (G) qRT-PCR analysis confirming the presence of cNanog or zNanog transcript in cNanog iPS–/– cells and zNanog iPS–/– cells, respectively. This panel only serves to verify the expression of the intended ortholog. Relative expression levels from different Taqman assays cannot be cross-compared. Quantification of total PB transgene expression in these samples is provided in Fig. 3H. Data are relative levels. Error bars indicate 1 s.d. (H,I) qRT-PCR analysis for pluripotent gene (H) and retroviral (r) transgene (I) expression in Nanog–/– pre-iPS cells, mNanog, cNanog and zNanog iPS–/– cells, and ES cells. Data are relative levels. Error bars indicate 1 s.d. (J) Correlation heatmap of global gene expression between Nanog–/– NS cells, mNanog (mN) iPS–/– cells, cNanog (cN) iPS–/– cells and zNanog (zN) iPS–/– cells. Two biological replicates were included for each cell line. (K) RNA FISH for Xist in Nanog–/– iPS cells. (L,M) Adult chimeric mouse obtained after blastocyst injection of either cNanog iPS–/– cells (L) or zNanog iPS–/– cells (M) following Cre excision of the Nanog transgene. Agouti coat color indicates chimerism. (N) AP staining of mNanog, cNanog and zNanog iPS–/– cells plated at clonal density in serum minus LIF before and after tamoxifen-induced Cre excision of the PB transgene. (O) Quantification of the proportion of undifferentiated, mixed and differentiated colonies in each condition shown in N. Error bars indicate 1 s.d. (n=3). Representative examples of colonies in each category are shown.
To validate these observations independently we tested the capacity of Nanog orthologs to induce naive pluripotency in alternative reprogramming systems. We first confirmed the capacity of zNanog to induce naive pluripotency in an independent Nanog–/– somatic cell type. Nanog–/– MEFs transduced with retroviral transgenes encoding Oct4, Klf4, Myc and Sox2 transited to pluripotency upon transfection with mNanog or zNanog, but not an empty vector transgene (supplementary material Fig. S3A,B). MEF-iPS–/– cells derived with zNanog expressed pluripotency-associated genes and zNanog, but not mNanog (supplementary material Fig. S3C-E). After Cre excision of the zNanog transgene, these MEF-iPS–/– cells contributed to adult mouse development (supplementary material Fig. S3F). Thus, Nanog orthologs enable induction of naive pluripotency in distinct somatic cells and this activity is independent of endogenous mNanog. EpiSCs can be reprogrammed to naive pluripotency by transfection with defined factors such as Nanog (Guo et al., 2009; Silva et al., 2009). We stably transfected constitutive transgenes encoding Nanog orthologs in EpiSCs carrying an Oct4-GFP-ires-puromycinr cassette (Silva et al., 2009). After transfer to 2i/LIF culture conditions, multiple GFP-positive, puromycin-resistant iPS cell colonies emerged from EpiSC lines expressing rNanog, hNanog, cNanog or zNanog (supplementary material Fig. S3G,H). These iPS cells expressed markers of naive pluripotency (supplementary material Fig. S3I,J). No iPS cells were derived in empty vector transfectants, as previously shown for EpiSCs maintained without feeders (Guo et al., 2009; Silva et al., 2009). The ability of Epi-iPS cells derived with zNanog to contribute towards the germ lineage was assessed by morula aggregation after transgene excision. Presence of Oct4-GFP reporter activity in the genital ridge at E12.5 demonstrated proof of germ lineage contribution (supplementary material Fig. S3K). These results show that Nanog orthologs are also sufficient to induce naive pluripotency in EpiSCs.
We recently reported that Nanog induces pluripotency in culture conditions that do not support ES cell self-renewal (Theunissen et al., 2011). To investigate whether a distant ortholog of Nanog could confer the same phenotype, we attempted to reprogram EpiSCs expressing zNanog in serum-free medium supplemented with LIF, but without further additives such as 2i or BMP4. iPS cells were readily obtained in this condition. Upon withdrawal of LIF, these iPS cells could be expanded for at least 10 passages in serum-free medium alone while maintaining pluripotency gene expression (supplementary material Fig. S3L-N). This suggested that vertebrate Nanog orthologs may also have the capacity to sustain self-renewal of pluripotent cells in the absence of LIF, a crucial property of mNanog (Chambers et al., 2003). To investigate this further, we assessed whether vertebrate Nanog orthologs could support self-renewal of iPS–/– cells in serum minus LIF. Both cNanog and zNanog sustained self-renewal at clonal density to a similar extent to mNanog, and this effect was strictly dependent upon the presence of the PB transgenes (Fig. 2N,O). These results were corroborated in ES cells (supplementary material Fig. S3O,P). We conclude that vertebrate Nanog orthologs recapitulate the full repertoire of reprogramming activities attributed to mNanog, and maintain the self-renewal of pluripotent cells without LIF.
Vertebrate orthologs of Nanog directly regulate target genes of mouse Nanog in iPS cells
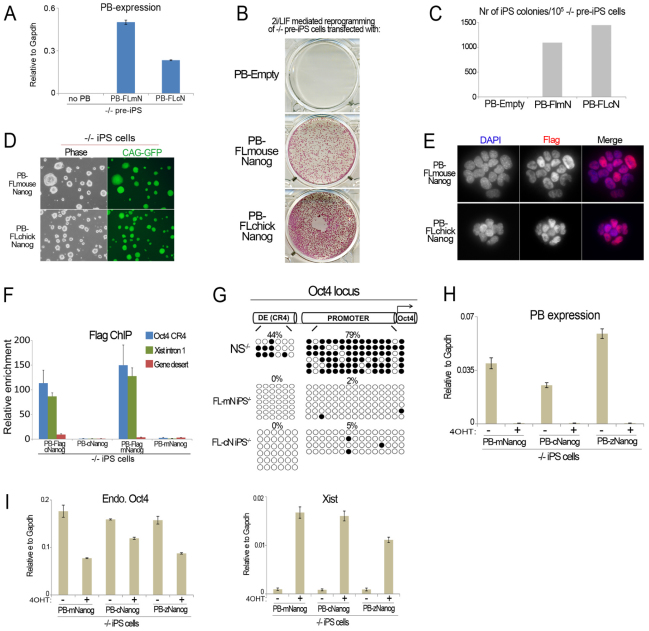
To determine whether non-mammalian Nanog orthologs have the capacity to bind DNA targets of mNanog, we first reprogrammed Nanog–/– somatic cells with constitutive transgenes expressing FLAG-tagged mNanog (FL-mNanog) or FLAG-tagged cNanog (FL-cNanog). Successful generation of iPS–/– cells with FL-mNanog and FL-cNanog demonstrates that the FLAG tag does not impair reprogramming capacity (Fig. 3A-D). Expression of mNanog or cNanog in the respective iPS–/– cell lines was confirmed by qRT-PCR (supplementary material Fig. S4A). iPS–/– cells generated with FL-mNanog and FL-cNanog showed silencing of retroviral transgenes and upregulation of endogenous pluripotency genes (supplementary material Fig. S4B,C). Immunofluorescence analysis indicated nuclear localization of FL-mNanog and FL-cNanog in iPS–/– cells (Fig. 3E). The CR4 element in the Oct4 distal enhancer and Xist intron 1, defined targets of Nanog in ES cells (Loh et al., 2006; Navarro et al., 2008), were enriched after chromatin immunoprecipitation (ChIP) with an anti-FLAG antibody in both FL-mNanog and FL-cNanog iPS–/– cells (Fig. 3F). Bisulfite sequencing of the Oct4 locus in both FL-mNanog and FL-cNanog iPS–/– cells indicated that the loss of DNA methylation marks in the CR4 element and Oct4 promoter (Fig. 3G).
Fig. 3.
Vertebrate Nanog orthologs regulate the expression of mouse Nanog targets in iPS cells. (A) qRT-PCR analysis for expression of piggyBac (PB) transgene sequence in Nanog–/– pre-iPS cells transfected with N-terminal FLAG (FL)-tagged mouse (m) Nanog (N) or FL-chick (c) Nanog PB transgenes. (B) AP staining of Nanog–/– pre-iPS cells cultured in 2i/LIF conditions for 10 days. (C) Quantification of the total number of AP-positive iPS cell colonies at day 10 after medium switch. (D) Phase contrast and GFP images of Nanog–/– iPS cells. (E) Immunofluorescence staining with anti-FLAG antibody of FL-mNanog and FL-cNanog iPS–/– cells. (F) Chromatin immunoprecipitation (ChIP) analysis of Nanog binding at the CR4 element in the Oct4 distal enhancer, Xist intron 1 and a gene desert control region. ChIP was performed using anti-FLAG antibody. Non-tagged iPS cells were used as negative controls. Occupancy is plotted as fold enrichment after normalization to the input, and error bars represent s.d. of the technical replicates of the qPCR for each experiment. (G) Bisulfite sequencing analysis of DNA methylation in the Oct4 distal enhancer and promoter in Nanog–/– NS cells, FL-mNanog and FL-cNanog iPS–/– cells. (H) qRT-PCR analysis for expression of the piggyBac sequence in mNanog, cNanog and zNanog iPS–/– cells before and after tamoxifen (4OHT)-induced Cre excision of the Nanog transgene. 4OHT was applied for 10 days. Data are relative levels. Error bars indicate 1 s.d. (I) qRT-PCR analysis for expression of endogenous Oct4 and Xist in the samples shown in H. Data are relative levels. Error bars indicate 1 s.d.
We then considered whether vertebrate orthologs of Nanog not only bind, but actively regulate, target genes of mNanog in iPS cells. For this purpose, we examined the transcriptional consequences of removing Nanog ortholog transgenes. Cre excision of the mNanog, cNanog or zNanog transgene in iPS–/– cells induced an upregulation of Xist expression of up to 20-fold, and a downregulation of Oct4 expression of up to twofold (Fig. 3H,I). These changes in gene expression cannot be attributed to iPS cell differentiation as this would cause Xist gene silencing in male cells. Instead, the data indicate that vertebrate orthologs of Nanog recapitulate regulatory functions of mNanog in pluripotent cells, including activation of Oct4 and repression of Xist. The latter is remarkable given that Xist is a gene that specifically evolved in eutherian mammals (Deakin et al., 2009). This is of particular note considering that Nanog is required for reactivation of the silent X chromosome in the female naive epiblast (Silva et al., 2009), which is accompanied by downregulation of Xist expression on the paternal X chromosome. These results suggest that vertebrate orthologs of Nanog directly regulate target genes of mNanog in iPS cells.
The capacity to induce naive pluripotency is unique to Nanog and arose in a common ancestor of vertebrates
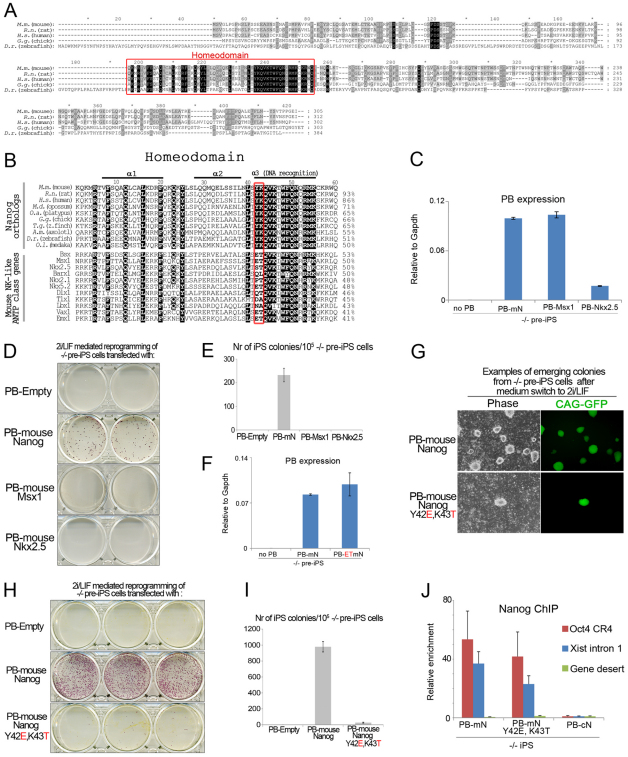
Protein conservation between Nanog orthologs is found only within the homeodomain (HD) (Fig. 4A). However, sequence identity between mNanog and zNanog HD is just 51% (Fig. 4B). This represents a poor degree of conservation as it does not even meet the criteria, i.e. minimum 60% identity, to be assigned to an HD family (Kappen et al., 1993). In fact, sequence identity between the HD of mNanog and either zNanog or mouse NK-like class proteins is similar (Fig. 4B), the latter being the subclass most closely related to Nanog (Chambers et al., 2003). We therefore examined whether Msx1 and Nkx2.5, two distinct NK-like class members, had the ability to restore reprogramming potential in Nanog–/– somatic cells (Fig. 4C, supplementary material Fig. S5A). Neither factor, however, could generate any iPS–/– cells (Fig. 4D,E). In fact, Nkx2.5 expression appeared to be detrimental to cell growth, and we only obtained pre-iPS cells expressing low levels of PB-Nkx2.5
Fig. 4.
Capacity to induce naive pluripotency is unique to Nanog. (A) Sequence alignment of vertebrate Nanog ortholog proteins. Red frame indicates homeodomain (HD). (B) Sequence alignment of the HDs of Nanog orthologs and related mouse NK-like ANTP class proteins. Identical amino acids are highlighted in black. Red frame indicates residues unique to Nanog orthologs. Identity scores relative to mouse Nanog are shown on the right and were calculated using ClustalW2. (C) qRT-PCR analysis for expression of the piggyBac (PB) sequence in Nanog–/– pre-iPS cells transfected with piggyBac (PB) mouse (m) Nanog (N), Msx1 or Nkx2.5. Data are relative levels. Error bars indicate 1 s.d. (D) AP staining of Nanog–/– pre-iPS cells cultured in 2i/LIF conditions for 10 days. (E) Quantification of the total number of AP-positive iPS cell colonies. Error bars indicate 1 s.d. (n=2). (F) qRT-PCR analysis for expression of the PB sequence in Nanog–/– pre-iPS cells transfected with mNanog (mN) or mNanogY42E, K43T (ET-mN). Data are relative levels. Error bars indicate 1 s.d. (G) Phase contrast and GFP images of emerging Nanog–/– iPS cell colonies during 2i/LIF induction of Nanog–/– pre-iPS cells. (H) AP staining of Nanog–/– pre-iPS cells. (I) Quantification of the total number of AP-positive iPS cell colonies. Data are mean values. Error bars indicate 1 s.d. (n=3). (J) ChIP analysis of mNanog and mNanogY42E, K43T binding at the CR4 element in the Oct4 distal enhancer, Xist intron 1 and a gene desert control region. ChIP was performed using an antibody to the N-terminal end of mouse Nanog. Chick (c) Nanog iPS–/– cells were used as negative controls. Occupancy is plotted as fold enrichment after normalization to the input; error bars represent the s.d. of the technical replicates of the qPCR for each experiment.
(Fig. 4C). Thus, the capacity to induce naive pluripotency appears to be unique to Nanog and is not present in related HD-containing transcription factors. Close inspection of HD sequences indicated that Nanog orthologs have two unique residues that are located in the DNA recognition helix: tyrosine (Y) at position 42 and lysine (K) at position 43 (Fig. 4B). Substitution of these residues by glutamic acid (E) and threonine (T), the respective amino acids present in related HD-containing transcription factors such as Bsx and Msx1, significantly impaired the efficiency of reprogramming in both Nanog–/– somatic cells and wild-type EpiSCs (Fig. 4F-I; supplementary material Fig. S3G). This shows an association between one or both of these positions and reprogramming capacity of Nanog. These amino acid substitutions are not predicted to alter the 3D structure of the mNanog HD (supplementary material Fig. S5B). iPS–/– cells derived with mNanogY42E, K43T nonetheless had a pluripotent gene expression profile (supplementary material Fig. S5C). In addition, ChIP analysis with an antibody to the N-terminal domain of mNanog revealed that mNanogY42E, K43T still bound efficiently to the Oct4 distal enhancer and Xist intron 1 (Fig. 4J). This suggests that Y42 and/or K43 affect specificity for other targets in DNA or interactions between Nanog and other proteins. As Y42 and K43 were not absolutely required for the reprogramming activity of Nanog, it is formally possible that the capacity to induce naive pluripotency was acquired in a non-vertebrate precursor of Nanog.
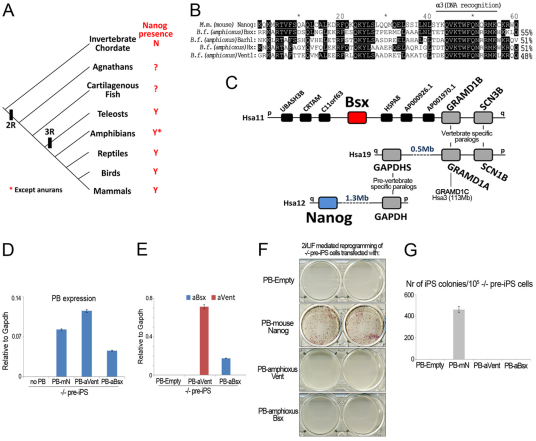
The amphioxus, an invertebrate chordate, does not contain Nanog in its genome, nor are Nanog-like sequences found in available invertebrate genomes, namely protostomes (Fig. 5A). In amphioxus, the gene with the highest HD sequence identity to Nanog is Bsx (Fig. 5B). In addition, we find that two gene families (GRAMD1 and SCN3B) present in the vicinity of the Bsx gene in human chromosome 11 (Hsa11) have duplicated members nearby GAPDHS in Hsa19. GAPDH, an evolutionary relative of GAPDHS, maps upstream close to hNanog in human chromosome 12 (Hsa12) (Fig. 5C). Bsx and Nanog also share an exon boundary within the HD, between amino acid positions 44 and 45. This boundary is only present in a small number of HD-containing transcription factors (data not shown). Taken together, these data suggest that Nanog and Bsx once shared the same genomic location and evolved from the same duplication event in early vertebrate ancestry. However, neither amphioxus Bsx nor amphioxus Vent1, a protein with 48% identity to Nanog HD, had reprogramming activity in Nanog–/– cells (Fig. 5D-G). We then investigated whether the introduction of Y42 and K43, in lieu of E42 and T43, would be sufficient to confer reprogramming activity on either amphioxus Bsx or amphioxus Vent1. However, neither of these mutants generated any iPS cell colonies in Nanog–/– cells (supplementary material Fig. S6). This indicates that Y42 and K43 are not sufficient to confer reprogramming potential on related HD transcription factors, a result verified using mouse Msx1 (supplementary material Fig. S6A-C). We conclude that the capacity to induce naive pluripotency is unique to Nanog, but cannot exclude that it originated in a Bsx-like precursor of Nanog. The appearance of Y42 and K43 may have been a subsequent addition that made Nanog more robust in its function.
Fig. 5.
Nanog is a vertebrate innovation. (A) Nanog homeobox gene family distribution in the chordate phylum (N, not present;?, currently unknown; Y, present). (B) Sequence alignment between the HD of Mus musculus (M.m.) Nanog and the HDs with higher identity score within the Branchiostoma floridae (B.f.) genome. Identical amino acids are highlighted in black. Identity scores were calculated using ClustalW2. (C) Synteny analysis linking Homo sapiens (Hsa) Nanog, located in chromosome 12, to Hsa Bsx, located in chromosome 11. Phylogenetic relationships of the flanking gene families were inferred from ensembl.org. (D) qRT-PCR analysis for expression of the piggyBac (PB) sequence in Nanog–/– pre-iPS cells transfected with mouse Nanog (mN), amphioxus (a) Bsx or amphioxus (a) Vent1. Data are relative levels. Error bars indicate 1 s.d. (E) qRT-PCR analysis confirming presence of aVent1 or aBsx transcript in Nanog–/– pre-iPS cells transfected with the respective transgene. Data are relative levels. Error bars indicate 1 s.d. (F) AP staining of Nanog–/– pre-iPS cells cultured in 2i/LIF conditions for 10 days. (G) Quantification of the total number of AP-positive iPS cell colonies. Data are mean values. Error bars indicate 1 s.d. (n=2).
The Nanog homeodomain is sufficient to induce naive pluripotency
As the HD was the only sequence with a degree of conservation between Nanog orthologs, we tested whether the Nanog HD was sufficient to induce naive pluripotency. Nanog–/– pre-iPS cells were transfected with a constitutive transgene encoding a 70 amino acid sequence that includes the mNanog HD (Fig. 6A). This fragment makes up only 23% of the total protein length of mNanog. Upon transfer to 2i/LIF conditions, iPS–/– cells were generated with this fragment (Fig. 6B-D). mNanog HD iPS–/– cells were readily expanded in culture (Fig. 6E), and expressed mNanog HD, but not the full-length transcript (Fig. 6F). qRT-PCR analysis of mNanog HD-only iPS–/– cells indicated reactivation of pluripotency-associated genes and silencing of retroviral transgenes (Fig. 6G). An Xist RNA pinpoint signal was detected in mNanog HD-only iPS–/– cell lines (Fig. 6H). Demethylation of the Oct4 distal enhancer and promoter regions in mNanog HD iPS–/– cells was confirmed by bisulfite sequencing (Fig. 6I). Finally, we examined the developmental potential of mNanog HD-only iPS–/– cells by performing blastocyst injection after Cre excision of the HD transgene. Contribution of mNanog HD iPS–/– cells to adult chimeras demonstrates that the capacity to induce naive pluripotency resides within the Nanog HD (Fig. 6J). In parallel, we tested reprogramming capacity of a slightly larger (81 amino acid) fragment that includes the zNanog HD in Nanog–/– pre-iPS cells (supplementary material Fig. S7A). This fragment makes up only 21% of the total protein length of zNanog, but still supported production of iPS–/– cells capable of contribution to adult chimeras (supplementary material Fig. S7B-J). These data show that a full-length reprogramming factor can be replaced by a much shorter fragment that includes its active domain.
Fig. 6.
Nanog homeodomain is sufficient to induce naive pluripotency. (A) qRT-PCR analysis for expression of the piggyBac (PB) transgene sequence in Nanog–/– pre-iPS cells transfected with a 70 amino acid fragment that includes the mNanog HD and five flanking amino acids on either side. Data are relative levels. Error bars indicate 1 s.d. (B) Phase contrast and GFP images of emerging Nanog–/– iPS cell colonies. (C) AP staining of Nanog–/– iPS cell colonies. (D) Quantification of the total number of AP-positive iPS cell colonies. Data are mean values. Error bars indicate 1 s.d. (n=2). (E) Phase contrast and GFP images of established Nanog–/– iPS cells. (F) RT-PCR analysis confirming the presence of transcripts corresponding to mNanog HD, but not full-length mNanog, in HD-only iPS–/– cells. (G) qRT-PCR analysis for full-length mNanog, pluripotent gene and retroviral (r) transgene expression in mNanog iPS–/– cells and HD-only iPS–/– cells. Data are relative levels. Error bars indicate 1 s.d. (H) RNA FISH for Xist in PB-mHD Nanog–/– iPS cells. (I) Bisulfite sequencing analysis of DNA methylation in the Oct4 distal enhancer and promoter in Nanog–/– NS cells, mNanog iPS–/– cells and mNanog HD-only iPS–/– cells. (J) Chimeric mice obtained after blastocyst injection of mNanog HD-only iPS–/– cells following Cre excision of the HD transgene. Fluorescence and agouti coat color indicate chimerism.
DISCUSSION
These findings have three main implications. First, our data demonstrate that the capacity of Nanog to establish pluripotency is fully conserved in vertebrates. The extent to which pluripotency is conserved during evolution remains the subject of much speculation. In comparison with other pluripotency genes, Nanog stands out as having relatively low sequence conservation among vertebrates. The absence in non-eutherian Nanog orthologs of a WR domain, which was reported to be crucial for dimerization and protein interactions (Mullin et al., 2008; Wang et al., 2008), further suggested that Nanog is not functionally conserved. In this study, however, we specifically interrogated the capacity of Nanog orthologs to establish pluripotency, the process for which Nanog is genetically essential in the mouse (Silva et al., 2009; Theunissen and Silva, 2011). In addition, we performed our experiments in cells in which both endogenous Nanog alleles were removed, thus ruling out any functional compensation by mNanog. This analysis uncovered an unexpected complete functional conservation between vertebrate orthologs of Nanog sharing as little as 13% sequence identity with mNanog. These results show that an apparent lack of sequence conservation can obscure robust functional conservation. On the basis of genome analysis and functional studies, we propose that Nanog is a vertebrate novelty that evolved from a Bsx-like ancestor after the invertebrate/vertebrate transition. Nanog is distinguished by two unique residues in the DNA recognition helix of its HD. This molecular signature is not found in any available invertebrate genome, and contributes strongly to reprogramming activity. Accordingly, only Nanog orthologs have the capacity to establish pluripotency. It is important to bear in mind that functional complementation experiments in mouse cells can provide only indirect evidence about the role of Nanog in different species. It is possible that Nanog performs a different function in lower vertebrates, which is co-opted during induction of pluripotency. However, the temporal and spatial expression profile of Nanog orthologs in vivo is consistent with the hypothesis that Nanog has a conserved role in specifying pluripotency. Specific expression of Nanog has been reported during the stages corresponding to epiblast formation in chick (Lavial et al., 2007) and axolotl (Dixon et al., 2010). In fish, Nanog is present during the earliest stages of embryonic development and Nanog knockdown produced developmental arrest and embryonic death (Camp et al., 2009). Nanog is absent in anurans, but this may be explained by the evolution of germ plasm, which obviates the need for germ cell specification from a pluripotent epiblast (Johnson et al., 2011). These results raise the possibility that naive pluripotency is a generic feature of vertebrate development. Previous studies reported that tetrapod homologs of Oct4 could support mouse ES cell self-renewal in complementation assays (Morrison and Brickman, 2006; Niwa et al., 2008). A recent in silico analysis suggested that the regions bound by pluripotency factors in mouse ES cells only have limited conservation outside mammals (Fernandez-Tresguerres et al., 2010), but extensive rewiring of the binding sites of functionally conserved transcription factors is not unusual during vertebrate evolution (Schmidt et al., 2010).
Second, by examining sequence identity between functionally conserved Nanog orthologs, we identified the HD as the active module for establishment of pluripotency. This finding has a number of implications. If the primary biological requirement for Nanog resides in a small domain with a limited number of conserved residues then the remainder of the protein would not have been under the same selective constraint. That would not exclude a subsequent refinement in, for example, further mechanisms of its regulation. However, this would have been a secondary event that occurred after vertebrate radiation, i.e. after the separation of the different classes of vertebrates. In the mouse, Nanog expression is rapidly downregulated after formation of the naive pluripotent epiblast and its forced expression blocks ES cell differentiation (Chambers et al., 2003; Ying et al., 2003). Orthologs of Nanog may be subject to different regulatory mechanisms that evolved separately and allow the smooth transition of pluripotent cells into the embryonic lineages. The sufficiency of the Nanog HD for induced pluripotency is also of interest for next-generation reprogramming technologies. A major challenge in the iPS cell field is to develop strategies for the delivery of reprogramming transgenes that do not involve DNA integration events. Promising results have been obtained thus far with the use of synthetic RNAs or recombinant proteins (Kim et al., 2009; Warren et al., 2010; Zhou et al., 2009). Reducing the size of reprogramming factors, which we demonstrate is possible by more than 75% in the case of Nanog, may alleviate the complexity of producing synthetic reprogramming molecules.
Finally, this work changes the way we view the role of Nanog in induced pluripotency. Extensive interactions have been reported between Nanog and other pluripotency factors in ES cells (Liang et al., 2008; Orkin et al., 2008; Wang et al., 2006). Based on these studies, we and others proposed that Nanog contributes to the reprogramming process by coordinating binding of the reprogramming factors to their cognate ES cell targets (Silva et al., 2009; Sridharan et al., 2009; Theunissen and Silva, 2011). However, many of the interactions between Nanog and other pluripotency regulators in ES cells were found to be mediated through the WR domain (Wang et al., 2008), which is absent in non-mammalian orthologs of Nanog. This suggests that Nanog may control the establishment of pluripotency through direct regulation of a select number of target genes or interactions with a subset of its known protein network. The challenge now is to identify those protein interactions and genomic targets that are shared between structurally divergent Nanog orthologs. This may also shed light on the reason why orthologs of Nanog appear to be more efficient than mouse Nanog in certain reprogramming systems. It is possible that the reprogramming activity of mouse Nanog is constrained by repressive interacting proteins, but that the same interactors do not engage in physical interactions with orthologs of Nanog. Alternatively, Nanog orthologs may escape microRNA-mediated negative regulation of mouse Nanog.
In summary, our work offers insights into the evolutionarily origins of pluripotency and how Nanog works during the acquisition of naive pluripotency. Importantly, it also provides a proof-of-principle demonstration that a single protein domain can substitute for a full-length reprogramming factor.
Supplementary Material
Acknowledgments
We thank Austin Smith and Jennifer Nichols for scientific discussions and critical reading of the manuscript. We are grateful to Rachael Walker for flow cytometry, and to William Mansfield and Charles-Etienne Dumeau for blastocyst injections.
Footnotes
Funding
This study was supported by Wellcome Trust Fellowships [WT086692MA and WT079249]. T.W.T is a Wellcome Trust PhD Fellow and J.C.R.S. is a Wellcome Trust Career Development Fellow. Deposited in PMC for immediate release.
Competing interests statement
A patent application has been filed based on this work by Cambridge University Enterprise (inventors J.C.R.S., T.W.T., Y.C. and L.F.C.C.).
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.068775/-/DC1
References
- Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q. L., Smith A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 [DOI] [PubMed] [Google Scholar]
- Camp E., Sanchez-Sanchez A. V., Garcia-Espana A., Desalle R., Odqvist L., O’Connor J. E., Mullor J. L. (2009). Nanog regulates proliferation during early fish development. Stem Cells 27, 2081–2091 [DOI] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- Deakin J. E., Chaumeil J., Hore T. A., Marshall Graves J. A. (2009). Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Res. 17, 671–685 [DOI] [PubMed] [Google Scholar]
- Dixon J. E., Allegrucci C., Redwood C., Kump K., Bian Y., Chatfield J., Chen Y. H., Sottile V., Voss S. R., Alberio R., et al. (2010). Axolotl Nanog activity in mouse embryonic stem cells demonstrates that ground state pluripotency is conserved from urodele amphibians to mammals. Development 137, 2973–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W. A., Lin S. M. (2008). lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 [DOI] [PubMed] [Google Scholar]
- Evans M. J., Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- Fernandez-Tresguerres B., Canon S., Rayon T., Pernaute B., Crespo M., Torroja C., Manzanares M. (2010). Evolution of the mammalian embryonic pluripotency gene regulatory network. Proc. Natl. Acad. Sci. USA 107, 19955–19960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginis I., Luo Y., Miura T., Thies S., Brandenberger R., Gerecht-Nir S., Amit M., Hoke A., Carpenter M. K., Itskovitz-Eldor J., et al. (2004). Differences between human and mouse embryonic stem cells. Dev. Biol. 269, 360–380 [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J. Y., Han D. W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P., et al. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6, 215–226 [DOI] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A. W., Cassady J. P., Staerk J., Carey B. W., Lengner C. J., Foreman R., Love J., et al. (2009a). Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A., Jaenisch R. (2009b). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., Cassady J. P., Muffat J., Carey B. W., Jaenisch R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 107, 9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Rougeulle C., Arnaud D., Avner P., Allis C. D., Spector D. L. (2001). Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107, 727–738 [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. (2006). Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Richardson E., Bachvarova R. F., Crother B. I. (2011). Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction 141, 291–300 [DOI] [PubMed] [Google Scholar]
- Kappen C., Schughart K., Ruddle F. H. (1993). Early evolutionary origin of major homeodomain sequence classes. Genomics 18, 54–70 [DOI] [PubMed] [Google Scholar]
- Kim D., Kim C. H., Moon J. I., Chung Y. G., Chang M. Y., Han B. S., Ko S., Yang E., Cha K. Y., Lanza R., et al. (2009). Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C., Leonard N., De Bolle X., Depiereux E. (2002). ESyPred3D: Prediction of proteins 3D structures. Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
- Lavial F., Acloque H., Bertocchini F., Macleod D. J., Boast S., Bachelard E., Montillet G., Thenot S., Sang H. M., Stern C. D., et al. (2007). The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development 134, 3549–3563 [DOI] [PubMed] [Google Scholar]
- Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R. E., Schulze E. N., Song H., Hsieh C. L., et al. (2008). Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., et al. (2008). Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- Lin S. M., Du P., Huber W., Kibbe W. A. (2008). Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36, e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., et al. (2007). Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70 [DOI] [PubMed] [Google Scholar]
- Makalowski W., Zhang J., Boguski M. S. (1996). Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 6, 846–857 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- Morrison G. M., Brickman J. M. (2006). Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development 133, 2011–2022 [DOI] [PubMed] [Google Scholar]
- Mullin N. P., Yates A., Rowe A. J., Nijmeijer B., Colby D., Barlow P. N., Walkinshaw M. D., Chambers I. (2008). The pluripotency rheostat Nanog functions as a dimer. Biochem. J. 411, 227–231 [DOI] [PubMed] [Google Scholar]
- Navarro P., Chambers I., Karwacki-Neisius V., Chureau C., Morey C., Rougeulle C., Avner P. (2008). Molecular coupling of Xist regulation and pluripotency. Science 321, 1693–1695 [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 [DOI] [PubMed] [Google Scholar]
- Niwa H. (2007). How is pluripotency determined and maintained? Development 134, 635–646 [DOI] [PubMed] [Google Scholar]
- Niwa H., Sekita Y., Tsend-Ayush E., Grutzner F. (2008). Platypus Pou5f1 reveals the first steps in the evolution of trophectoderm differentiation and pluripotency in mammals. Evol. Dev. 10, 671–682 [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Wang J., Kim J., Chu J., Rao S., Theunissen T. W., Shen X., Levasseur D. N. (2008). The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb. Symp. Quant. Biol. 73, 195–202 [DOI] [PubMed] [Google Scholar]
- Richards M., Tan S. P., Tan J. H., Chan W. K., Bongso A. (2004). The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells 22, 51–64 [DOI] [PubMed] [Google Scholar]
- Rossant J. (2008). Stem cells and early lineage development. Cell 132, 527–531 [DOI] [PubMed] [Google Scholar]
- Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C. P., Mackay S., et al. (2010). Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Matsuno Y., Fouse S. D., Rao N., Root S., Xu R., Pellegrini M., Riggs A. D., Fan G. (2008). X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl. Acad. Sci. USA 105, 4709–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. (2009). Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Tchieu J., Kuoy E., Chin M. H., Trinh H., Patterson M., Sherman S. P., Aimiuwu O., Lindgren A., Hakimian S., Zack J. A., et al. (2010). Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- Theunissen T. W., Silva J. C. R. (2011). Switching on pluripotency: a perspective on the biological requirement of Nanog. Philos. Trans. Royal Soc. London Ser. B Biol. Sci. 366, 2222–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T. W., van Oosten A. L., Castelo-Branco G., Hall J., Smith A., Silva J. C. (2011). Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 21, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006). A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- Wang J., Levasseur D. N., Orkin S. H. (2008). Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 105, 6326–6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P. D., Ahfeldt T., Loh Y. H., Li H., Lau F., Ebina W., Mandal P. K., Smith Z. D., Meissner A., et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wu S., Joo J. Y., Zhu S., Han D. W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.