Abstract
SIRT1 is the closest mammalian homologue of yeast SIR2, an important ageing regulator that prolongs lifespan in response to caloric restriction. Despite its importance, the mechanisms that regulate SIRT1 activity are unclear. Our study identifies a novel post-translational modification of SIRT1, namely sumoylation at Lys 734. In vitro sumoylation of SIRT1 increased its deacetylase activity. Conversely, mutation of SIRT1 at Lys 734 or desumoylation by SENP1, a nuclear desumoylase, reduced its deacetylase activity. Stress-inducing agents promoted the association of SIRT1 with SENP1 and cells depleted of SENP1 (but not of SENP1 and SIRT1) were more resistant to stress-induced apoptosis than control cells. We suggest that stress-inducing agents counteract the anti-apoptotic activity of SIRT1 by recruiting SENP1 to SIRT1, which results in the desumoylation and inactivation of SIRT1 and the consequent acetylation and activation of apoptotic proteins.
Sirtuins (SIRTs) are mammalian NAD+-dependent histone deacetylases (HDACs)1. Of the seven SIRTs, SIRT1 is the closest homologue of yeast SIR2 (ref. 2, 3), which has important roles in diverse cellular processes, including transcriptional silencing4, rDNA recombination5, glucose metabolism and energy homeostasis6, DNA repair and cell survival7–9. Because of its dependency on NAD+, the activity of SIRT1 is regulated by the NAD+:NADH ratio and, is therefore sensitive to the status of redox and cellular metabolism. Similar to SIR2 in lower organisms, SIRT1 is potentially a nutrient sensor that regulates the lifespan of mammals in response to caloric restriction or nutrient starvation9–13. By deacetylating and inactivating apoptotic proteins (such as the p53 tumour suppressor14–17), SIRT1 also protects cancer cells from apoptosis induced by DNA damage. The expression of SIRT1 is known to be controlled at both transcriptional and post-transcriptional levels, but post-translational mechanisms that regulate SIRT1 activity have yet to be defined.
Sumoylation is a reversible post-translational modification in which proteins termed small ubiquitin-related modifiers (SUMOs) are covalently linked to lysine residues of target proteins. Similar to ubiquitination, sumoylation is catalysed by a three-step enzymatic reaction involving an E1 activating enzyme, an E2 conjugating enzyme and E3 ligases. The reverse reaction is catalysed by SENP desumoylases, a family of SUMO specific isopeptidases18. In contrast to polyubiquitination through Lys 48 of ubiquitin, sumoylation does not signal protein degradation; instead, it may be involved in transcriptional silencing19,20, genomic stabilization20 and the stress response21,22.
SIRT1 and sumoylation are both implicated in cellular responses to genotoxic stress. SIRT1 deacetylates protein substrates with established roles in the DNA-damage response. These proteins include p53 (refs 14–16), FOXO factors23–25 and Ku70 (ref. 9), and deacetylation of p53 reduces both its transcriptional and apoptotic activities. Evidence is also accumulating that SUMOs control pathways important for the surveillance of genome integrity; for example, proteins that maintain genomic stability by functioning at the interface between DNA replication, recombination and repair processes undergo sumoylation20,26. The functional similarity between SIRT1 and SUMOs prompted us to address the possibility that SIRT1 is sumoylated. Here, we show that SIRT1 is indeed sumoylated, and that its desumoylation by SENP1 regulates cellular response to genotoxic stress.
Results
Sumoylation of human SIRT1 on Lys 734 increases its activity
To determine whether SIRT1 is sumoylated, extracts of DU145 cells were subjected to reciprocal coprecipitation analyses with antibodies against Sumo-1 and SIRT1. Anti-SIRT1 coprecipitated endogenous Sumo-1 (Fig. 1a). Conversely, anti-Sumo-1 coprecipitated endogenous SIRT1 (Fig. 1b). An unrelated antibody (anti-Flag) did not precipitate either protein. As expected, sumoylation increased the molecular mass of SIRT1, as evident from reduced mobility on SDS–PAGE. Indicative of covalent linkage, nickel beads precipitated SIRT1 from denatured extracts of cells expressing His–Sumo-1 (pulldown assays; Fig. 1c). SIRT1 was also sumoylated in in vitro sumoylation reactions containing E1 and E2 enzymes (Fig. 1d).
Figure 1.
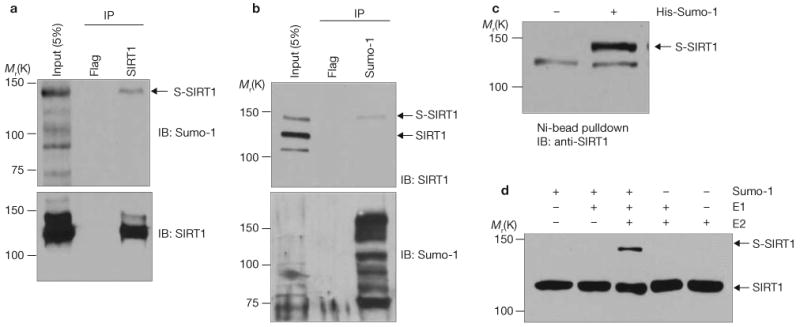
SIRT1 is sumoylated in vivo and in vitro. (a) DU145 cell extracts were immunoprecipitated (IP) with antibodies against Flag or SIRT1 and immunoblotted (IB) with antibodies against Sumo-1 or SIRT1, as indicated. S-SIRT1, sumoylated SIRT1. (b) Cell extracts were immunoprecipitated with antibodies against Flag or Sumo-1 and immunoblotted with antibodies against SIRT1 or Sumo-1, as indicated. Note that the antibody against SIRT1 detected three proteins in the input lane. This antibody is known to detect a non-specific protein with an Mr of approximately 100K in western blots. (c) Cells expressing His–Sumo-1 were extracted under denaturing conditions. His–Sumo-1 conjugates were isolated with Ni-NTA agarose and immunoblotted with antibodies against SIRT1. The error bars represent s.d. (d) Sumoylation reactions using recombinant human SIRT1 from bacteria were carried out in the presence of the indicated components. Reaction mixtures were immunoblotted with antibodies against SIRT1. Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
The consensus sumoylation site sequence is Ψ-Lys-X-Glu/Asp, where Ψ is a large hydrophobic amino acid (such as isoleucine or valine) and X is any amino acid19. As determined with SUMOplot(tm) analyses program (http://www.abgent.com/subplot.html), and using the web-based sumoylation prediction software SUMOplot™, human SIRT1 contains two potential sumoylation sites in the carboxy-terminal region, Lys 610 and Lys 734 (scores are 0.5 and 0.93, respectively; Fig. 2a). Substitution of Lys 610 with Arg (SIRT1K610R) did not prevent the sumoylation of SIRT1, as monitored by its coprecipitation with His–Sumo-1, which was expressed in cells and isolated with nickel beads (Fig. 2b). SIRT1, however, was not sumoylated when mutated at Lys 734 (SIRT1K734R) or when mutated at both sites (SIRT1DM). Deletion of the 50 C-terminal amino acids of SIRT1 (either wild-type or mutated at Lys 610; SIRT1ΔCT) also attenuated SIRT1 sumoylation. The lack of sumoylation of SIRT1K734R is not due to its aberrant localization to cytoplasm, as both wild-type SIRT1 and SIRT1K734R were predominantly detected in the nucleus (Fig. 2c).
Figure 2.
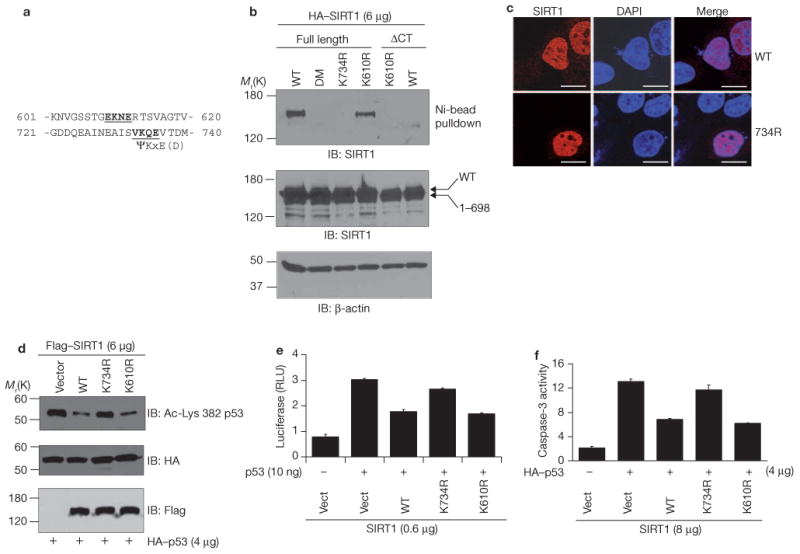
Mutation of the sumoylation sites of SIRT1 inhibits its function. (a) Putative sumoylation sites in the C-terminus of human SIRT1. (b) H1299 cells were cotransfected with His-Sumo-1 and either wild-type (WT) HA-SIRT1 or HA-SIRT1 lacking C-terminal 50 amino acids (HA-SIRT1ΔCT). Additional mutants included in the analyses are SIRT1 mutated at Lys 734 (SIRT1K734R), at Lys 610 (SIRT1K610R) or both (SIRT1DM). Sumoylated SIRT1 was isolated by Ni-NTA agarose and detected by immunoblotting with antibodies against HA. Cell extracts were immunoblotted with antibodies against HA or β-actin (loading control). (c) Cells were transfected with wild-type SIRT1 or SIRT1K734R and the cellular distribution of SIRT1 was determined by fluorescence staining and confocal imaging. For detection of all cells, cells were stained with DAPI. The scale bars represent 20 μm. (d) Cells were cotransfected with HA-p53 and either wild-type Flag-SIRT1, Flag–SIRT1K743R or Flag–SIRT1K610R. Anti-HA immunoprecipitates were immunoblotted with antibodies against HA (total p53) or against p53 acetylated at Lys 382 (a SIRT1 deacetylation site). Cell extracts were immunoblotted with antibodies against Flag. (e) p53-null MEFs were transfected with p21–Luc and either control vector (Vect), Flag–SIRT1, Flag–SIRT1K610R or Flag–SIRT1K734R, and luciferase assays were performed. Each data point is the average of triplicates (n = 3) from a representative experiment. The error bars represent s.d. (f) H1299 cells were transfected with HA–p53 together with wild-type Flag–SIRT1, Flag–SIRT1K734R or Flag–SIRT1K610R, and caspase-3 activity was determined. Each data point is the average of triplicates (n = 3) from a representative experiment. The error bars represent s.d. Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
Importantly, mutation of SIRT1 at Lys 734 reduced its ability to deacetylate and inhibit the activity of p53. H1299 cells were transfected with HA–p53 and either wild-type SIRT1, SIRT1K610R or SIRT1K734R, and HA immunoprecipitates were immunoblotted with an antibody that recognizes p53 acetylated at Lys 382, the site of SIRT1 deacetylation14–17. To monitor p53 activity, cells were either cotransfected with a p53-driven luciferase reporter construct or assayed for caspase-3 activity. Wild-type SIRT1 and SIRT1K610R mutant reduced amounts of acetylated p53 (Fig. 2d) and both the transcriptional (Fig. 2e) and apoptotic (Fig. 2f) activities of p53, whereas SIRT1K734R had little effect. Thus, mutation of the SIRT1 sumoylation site affects the acetylation status of proteins in addition to p53. Collectively, the data in Figs 1 and 2 identify Lys 734 as the major sumoylation site of SIRT1 and show that sumoylation of SIRT1 is functionally relevant.
To determine whether sumoylation regulates the intrinsic deacetylase activity of SIRT1, recombinant SIRT1 was sumoylated in vitro and its deacetylase activity was measured using an in vitro fluorometric assay with a p53 peptide acetylated at Lys 382 as the substrate27. In vitro sumoylation of wild-type SIRT1 increased its deacetylase activity (Fig. 3a, b). As expected, SIRT1K734R was not sumoylated in vitro, and incubation of SIRT1K734R with Sumo-1 did not change its deacetylase activity, which was lower than that of wild-type SIRT1 after sumoylation. In addition to p53, cellular levels of histone H4 acetylation at Lys 16 were decreased by the wild-type SIRT1 and SIRT1K610R, but not by SIRT1K734R (Fig. 3c), indicating that the effect of sumoylation is not restricted to the ability of SIRT1 to deacetylate p53, but instead reflects a decrease in the intrinsic deacetylase activity of SIRT1.
Figure 3.
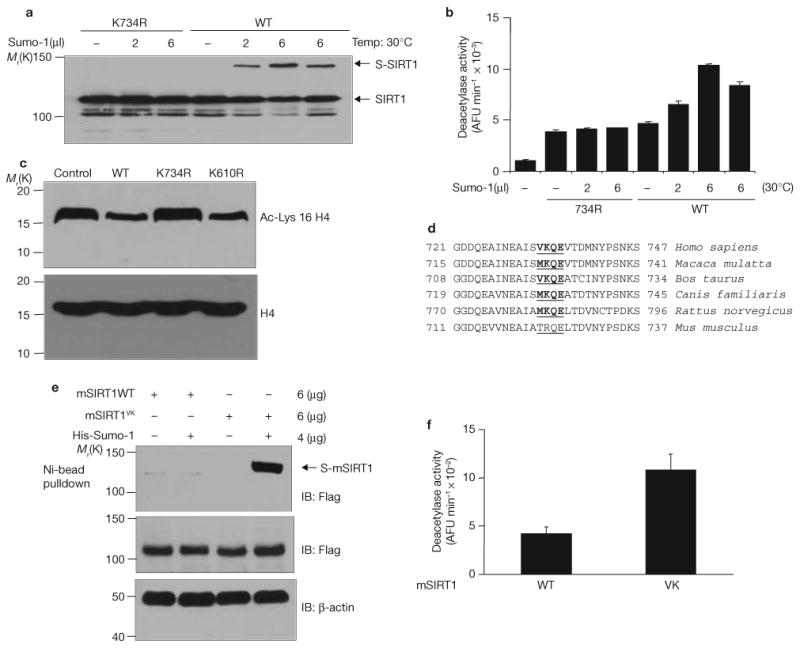
Sumoylation of SIRT1 at Lys 734 increases its intrinsic deacetylase activity. (a) Wild-type SIRT1 and K734R mutant were produced and purified in a cell-free system as His-tagged protein. In vitro sumoylation of SIRT1 was performed in the presence of indicated amounts of Sumo-1 and detected by immunoblotting of half of the reaction mixture with antibodies against SIRT1. (b) The other half of the reaction mixture described in a was assayed for deacetylase activity using a fluorometric assay system. Each data point is the average of triplicate (n = 3) samples analysed in parallel. The error bars represent s.d. AFU, arbitrary fluorescence unit. (c) Cells were transfected with wild-type Flag–SIRT1, Flag–SIRT1K734R or Flag–SIRT1K610R. Cell extracts were immunoblotted with antibodies against acetyl-Lys 16 of histone H4 or against histone H4. (d) Sequence alignment of the C-terminus of mammalian SIRT1 proteins. The four amino acids in comparison are underlined. The sumoylation consensus sites are shown in bold. (e) Cells were transfected with His–Sumo-1 together with Flag-tagged wild-type or mutant (VK) mouse SIRT1 (mSIRT1) in which Thr 722 and Arg6 723 were mutated to valine and lysine, respectively. His–Sumo-1 conjugated SIRT1 was isolated by nickel beads and detected by immunoblotting with antibodies against Flag. (f) Cells were transfected with Flag-tagged wild-type or mutant mouse SIRT1. Flag–SIRT1 was purified from cell extracts on M2-conjugated affinity columns and its deacetylase activity determined in vitro using a fluorometric assay system. Each data point is the average of triplicate samples (n = 3) analysed in parallel. The error bars represent s.d. Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
Sequence comparison reveals that the Lys 734 sumoylation motif is evolutionarily conserved from rat to human (Fig. 3d). Mouse SIRT1 contains an argine residue instead of a lysine reside at the same position. Mutation of threonine to arginine and valine to lysine in mouse SIRT1 created a functional sumoylation site (Fig. 3e) and increased its deacetylase activity (Fig. 3f). This finding suggests that SIRT1 sumoylation at Lys 734 may allow SIRT1 to operate at increased levels, when needed, in mammals with large body sizes and long lifespan. Although speculative, it is possible that SIRT1 sumoylation is one of the critical features accounting for some of the phenotypic or metabolic differences between mice and humans.
SENP1 is a SIRT1 desumoylase
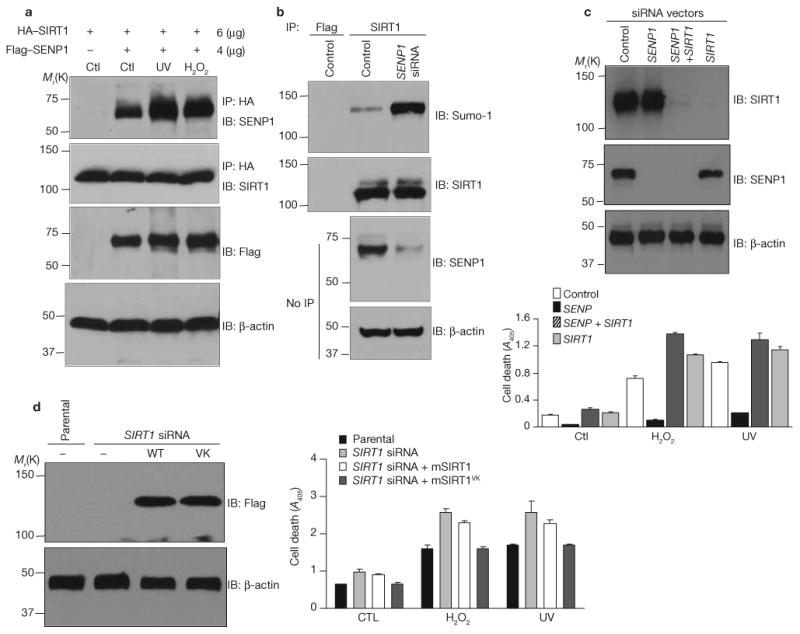
As a potential in vivo modulator of SIRT1 sumoylation, we examined SENP1—a SUMO-specific protease that desumoylates proteins such as HDAC1 (ref. 28). Reciprocal immunoprecipitation and immunoblot analyses demonstrated interaction of both ectopic (Fig. 4a) and endogenous (Fig. 4b) SIRT1 and SENP1. Consistent with their interaction, SENP1 and SIRT1 colocalized in the nucleus, as shown by confocal microscopy (Fig. 4c). To assess the effects of SENP1 on SIRT1 sumoylation, H1299 cells were transfected with HA–SIRT1, His–Sumo-1, and either wild-type or mutant (inactive) Flag–SENP1, and amounts of His–Sumo-1-conjugated HA–SIRT1 were determined by pulldown assays with nickel beads. As shown in Fig. 4d, much less sumoylated SIRT1 was observed in cells expressing wild-type SENP1 than in cells expressing Flag vector alone or inactive SENP1.
Figure 4.
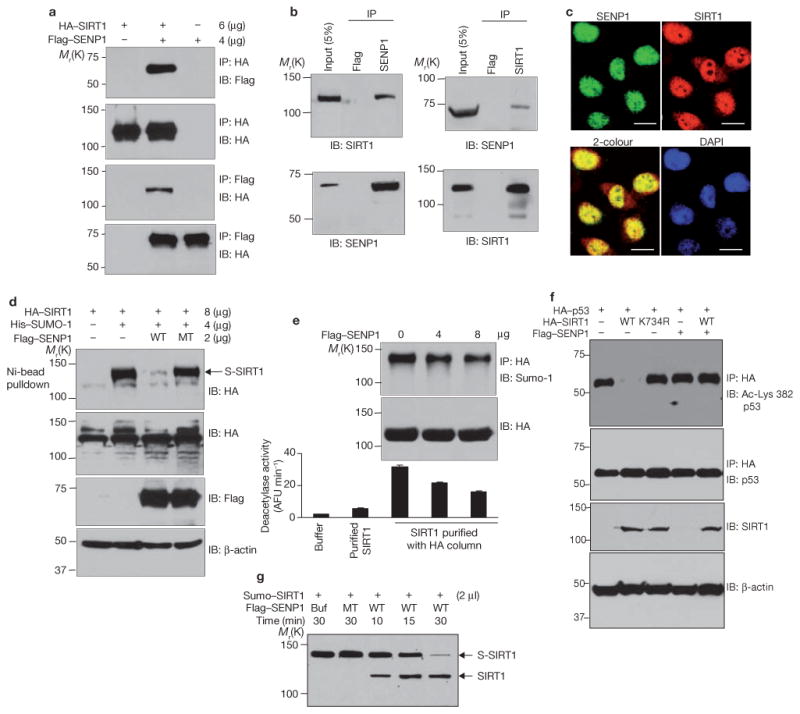
Desumoylation of SIRT1 by SENP1 reduces its ability to deacetylate p53. (a) H1299 cells were cotransfected with HA–SIRT1 and Flag–SENP1. HA and Flag immunoprecipitates were immunoblotted with antibodies against Flag or HA. (b) Extracts of H1299 cells were immunoprecipitated with antibodies against Flag, SENP1 or SIRT1 and immunoblotted with antibodies as indicated. (c) H1299 cells were fixed and immunostained with antibodies against SENP1 or SIRT1. Colocalization was detected by confocal fluorescence microscopy. The scale bars represent 20 μm. (d) Cells were cotransfected with HA–SIRT1, His–Sumo-1 and Flag–SENP1, as indicated. Sumoylated HA–SIRT1 was isolated by Ni-NTA agarose and detected by western blotting with antibodies against HA. Amounts of SIRT1 (anti-HA), wild-type and mutant (MT) SENP1 (anti-Flag), and β-actin were determined by western blotting. (e) Cells were cotransfected with HA–SIRT1 and SENP1. HA–SIRT1 was purified from cell extracts on an anti-HA conjugated affinity column, and its activity was determined by a fluorometric assay system. Commercially obtained recombinant SIRT1 protein is included in the assay as a reference (lane 2). Each data point is the average of triplicate samples (n = 3) analysed in parallel. The error bars represent s.d. (f) Cells were cotransfected with HA–p53, Flag–SENP1 and wild-type Flag–SIRT1 or Flag–SIRT1K734R. Anti-HA immunoprecipitates were immunoblotted with antibodies against p53 (total p53) or with antibodies against p53 acetylated at Lys 382. Cell extracts were immunoblotted with antibodies against HA or β-actin. (g) SIRT1 conjugated to His–SUMO-1 was isolated from HEK 293 cells using the μMACS His-tagged protein isolation kit and incubated with wild-type and inactive mutant SENP1 produced in vitro. Sumoylated and desumoylated SIRT1 were detected by immunoblotting with antibodies against SIRT1. Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
Consistent with the need for sumoylation of SIRT1 for deacetylase activity, ectopic expression of SENP1 (and consequent desumoylation of SIRT1) reduced SIRT1 activity (Fig. 4e). In this experiment, H1299 cells were cotransfected with HA–SIRT1 and various amounts of Flag– SENP1. HA–SIRT1 was isolated with anti-HA beads and activity was determined by an in vitro fluorometric assay27. As amounts of transfected Flag–SENP1 increased, acetylation of the p53 peptide decreased as did HA–SIRT1 sumoylation. H1299 cells were cotransfected with Flag– SENP1, HA–p53 and Flag–SIRT1 and amounts of acetylated HA–p53 were determined by western blotting (Fig. 4f). Flag–SIRT1 deacetylated HA–p53 in the absence but not the presence, of Flag–SENP1. Wild-type SENP1 also desumoylated Sumo–SIRT1 in vitro whereas inactive SIRT1 did not (Fig. 4g).
SENP1 promotes stress-induced apoptosis by interacting with and desumoylating SIRT1
Desumoylation and consequent inactivation of SIRT1 may be required for or contribute to apoptosis. We tested this hypothesis using two stress-inducing apoptotic agents, UV radiation and hydrogen peroxide. Both agents reduced the sumoylation of ectopic (Fig. 5a) and endogenous (Fig. 5b) SIRT1 as demonstrated by pulldown assays with nickel beads and by reciprocal precipitation assays respectively. Consistent with reduced sumoylation, SIRT1 was less active in cells exposed to UV radiation or hydrogen peroxide than in control cells (Fig. 5c). More importantly, ectopic expression of wild-type SIRT1 partially protected H1299 cells from apoptosis induced by UV radiation and hydrogen peroxide, whereas the sumoylation-deficient mutant (SIRT1K734R) did not (Fig. 5d). This finding suggests that sumoylation of SIRT1 at Lys 734 is a molecular switch that tips the balance from survival to death when DNA is damaged.
Figure 5.
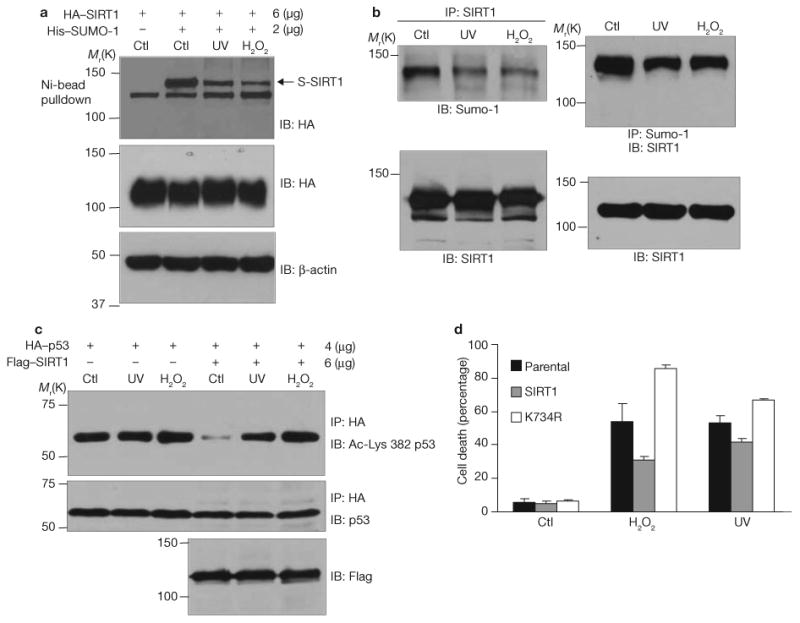
SIRT1 desumoylation by UV radiation and hydrogen peroxide contributes to stress-induced apoptosis. (a) H1299 cells expressing HA–SIRT1 and His–Sumo-1 were treated with 0.5 mM hydrogen peroxide for 6 h or irradiated with 0.3 J UV. Sumoylated SIRT1 was isolated by Ni-NTA agarose and detected by immunoblotting with antibodies against HA. Cell extracts were immunoblotted with antibodies against HA and β-actin. (b) Cells were treated as in a and the level of endogenous SIRT1 sumoylation was determined by reciprocal coimmunoprecipitations. (c) Cells were cotransfected with HA–p53 and Flag–SIRT1 and treated with UV radiation or hydrogen peroxide. HA immunoprecipitates were immunoblotted with antibodies against p53 or p53 acetylated at Lys 382. Cellular extracts were immunoblotted with antibodies against Flag. (d) H1299 cells stably transfected with either wild-type Flag–SIRT1 or Flag–SIRT1K743R were treated with UV radiation or hydrogen peroxide. The percentage of apoptotic cells was determined by monitoring DNA content by flow cytometry. Each data point is the average of triplicate samples (n = 3) analysed in parallel. The error bars represent s.d. Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
Additional studies show that stress-inducing agents inhibit the sumoylation of SIRT1 by promoting its interaction with SENP1. Antibodies against HA–SIRT1 coprecipitated more Flag–SENP1 from cells treated with UV radiation or hydrogen peroxide than from control cells (Fig. 6a). Amounts of total HA–SIRT1 were similar in all conditions as were amounts of total Flag–SENP1.
Figure 6.
SENP1 interacts with and desumoylates SIRT1 and promotes stress-induced apoptosis. (a) Cells were cotransfected with HA–SIRT1 and Flag–SENP1 and treated with UV radiation or hydrogen peroxide. HA immunoprecipitates were immunoblotted with antibodies against SENP1 or SIRT1. Cell extracts were immunoblotted with antibody against Flag or β-actin. (b) SENP1 siRNA was stably transfected into H1299 cells. Endogenous SENP1 expression, and SIRT1 expression and sumoylation, were determined by immunoblotting. (c) H1299 cells stably expressing control vector, SENP1 siRNA, SIRT1 siRNA or both were treated with UV radiation or hydrogen peroxide. Amounts of the indicated proteins were determined by western blotting. Cell apoptosis was determined by measuring DNA fragmentation by ELISA. Each data point is the average of triplicate samples (n = 3) analysed in parallel. The error bars represent s.d. (d) H1299 cells stably expressing SIRT1 siRNA were transfected with control plasmid or expression vectors for Flag-tagged wild-type or mutant mouse SIRT1 (mSIRT1). The expression of mouse SIRT1 and β-actin was determined by immunolotting. Cellular apoptosis was determined by measuring DNA fragmentation by ELISA. Each data point is the average of triplicate samples (n = 3) analysed in parallel. The error bars represent s.d. Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
To determine whether SENP1 abundance modulates stress-induced apoptosis, H1299 cells were depleted of SENP1 by stable expression of SENP1 short interfering RNA (siRNA; Fig. 6b). Reductions in amounts of SENP1 were accompanied by increases in amounts of sumoylated SIRT1. Importantly, cells depleted of SENP1 were much less susceptible to UV radiation- and hydrogen peroxide-induced cell death than control cells (Fig. 6c). Simultaneous silencing of SIRT1 reversed the effect of SENP1 depletion on cell survival.
Silencing of SIRT1 by siRNA modestly increased the sensitivity of H1299 cells to apoptosis induction by UV radiation and hydrogen peroxide (Fig. 6c, d). Coexpression of mouse SIRT1 with the artificial sumoylation site prevented this increase, whereas wild-type mouse SIRT1 did not. Previous studies show that siRNA against human SIRT1 does not deplete mouse SIRT1 (ref. 29). Taken together, the data in Figs. 5 and 6 show that SENP1 antagonizes cell survival and that it requires SIRT1 to do so. These data strongly suggest that SIRT1 inactivation by SENP1-mediated desumoylation decreases resistance of UV radiation- and hydrogen peroxide-treated cells to apoptosis.
Members of the p53 family are SIRT1 target proteins in stress-induced apoptosis
As shown above, wild-type SIRT1 inhibits the acetylation of p53, the activity of a p53-driven reporter gene and p53-mediated caspase activation, whereas a sumoylation-deficient SIRT1 mutant (SIRT1K734R) does not. The studies suggest that SIRT1 protects cells from apoptosis by inactivating p53 and that stress induces apoptosis, at least in part, by counteracting the effects of SIRT1 on p53 – that is, by eliciting SIRT1 desumoylation and consequent p53 activation. In support of this hypothesis, p53-intact HCT116 colon cancer cells were more sensitive to UV radiation-induced apoptosis than were their p53-depleted counterparts, and the expression of wild-type SIRT1 but not SIRT1K734R reduced the sensitivity of p53-intact cells to UV radiation (Fig. 7a). Western-blot analyses show comparable expression of wild-type SIRT1 and SIRT1K734R, thus indicating that their different effects are not because of their differential expression in these cells.
Figure 7.
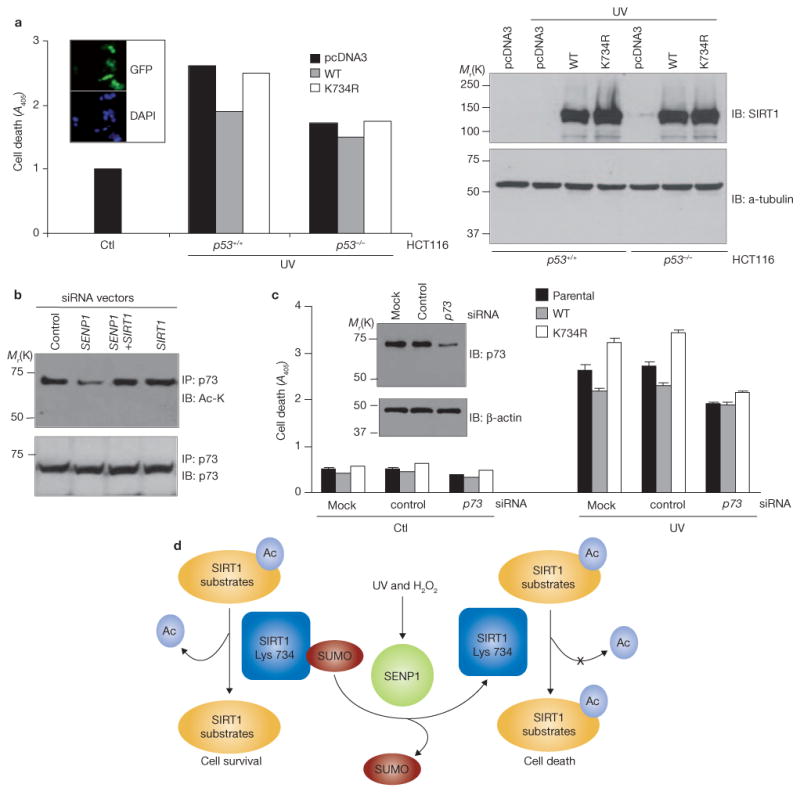
p53 and p73 mediate the effect of SIRT1 desumoylation on the cellular response to genotoxic stress. (a) Parental (p53+/+) or p53-deficient (p53−/−) HCT116 cells were transfected with control vector, HA–SIRT1 or HA–SIRT1K743R and treated with UV radiation for 6 h. Apoptosis was determined by ELISA. Each data point is the average of duplicate samples (n = 2) analysed in parallel and repeated once. HA–SIRT1 and α-tubulin expression was determined by immunoblotting with antibodies against HA and α-tubulin. The insert shows representative HCT116 cells transfected with GFP and stained with DAPI to illustrate transfection efficiency. (b) Extracts of H1299 cells stably expressing the indicated siRNAs were immunoprecipitated with anti-p73, followed by immunoblotting with antibodies against p73 or acetyl-lysine (Ac-Lys). (c) H1299 cells stably expressing wild-type or mutant SIRT1 were transfected with synthetic scrambled (control) or p73 siRNA oligonucleotides and exposed to UV radiation. Apoptosis induced by UV radiation was determined by ELISA. Each data point is the average of triplicate samples (n = 3) analysed in parallel. The insert shows the efficiency and specificity of the synthetic siRNA oligonucleotides, as determined by immunoblotting of H1299 extracts with antibodies against p73 and β-actin. The error bars denote s.d. (d) Schematic representation of a model of how SIRT1 sumoylation regulates its deacetylase activity and determines cellular response to stress and DNA damage (see text for details). Uncropped images of the scans are shown in the Supplementary Information, Fig. S1.
H1299 cells are p53-null and wild-type SIRT1 protects them from apoptosis, whereas the SIRT1K734R mutant does not (Fig. 5c). This finding suggests that SIRT1 has targets that regulate apoptosis in addition to p53. The p53-related protein p73 seems to be such a target. Silencing of SENP1 in H1299 cells reduced amounts of acetylated p73 in the absence but not presence of SIRT1 siRNA (Fig. 7b). H1299 cells depleted of p73 were less sensitive to UV radiation-induced apoptosis than were control cells regardless of whether they expressed wild-type SIRT1, SIRT1K734R or vector alone (Fig. 7c). These data tentatively place p73 downstream of SIRT1 in the pathway in which UV radiation-induced apoptosis promotes SIRT1 desumoylation and H1299 apoptosis. It is important to note that p73 or p53 depletion only modestly changed cellular sensitivity to UV radiation. Other SIRT1 substrates, such as FOXO factors23, Ku70 (ref. 30) or E2F1 (ref. 29), may also mediate the inhibitory effects of SENP1 depletion on UV radiation-induced cell death.
Discussion
Based on our results, we propose SIRT1 sumoylation as a molecular switch that determines the fate of cells when DNA damage occurs (Fig. 7d). In the absence of acute DNA damage, SIRT1 is sumoylated and active, and suppresses the activity of its apoptotic substrates by deacetylation. In response to stress, SIRT1 associates with SENP1 and becomes desumoylated and less active. As a result, pro-apoptotic SIRT1 substrates become active and cause cell death.
Our data show that SENP1 associates with SIRT1 in cells exposed to UV radiation and hydrogen peroxide, and inhibits the deacetylase activity of SIRT1 through desumoylation. Our data do not exclude the possibility that SENP1 also regulates SIRT1 function independently of desumoylation; for example, SENP1 may prevent SIRT1 from interacting with its substrates or other proteins. Our preliminary analyses detected complex formation between desumoylase-inactive SENP1 and the sumoylation-deficient SIRT1K734R mutant, similarly to their wild-type counterparts (data not shown). In addition, the effect of SENP1 depletion on stress-induced cell death may also be mediated by SIRT1-independent mechanisms involving p53 or its ubiquitin ligase MDM2. SENP1 desumoylates p53 (ref. 31) and MDM2 (ref. 31), and UV radiation decreases MDM2 sumoylation through decreased binding of the E2 conjugating enzyme Ubc9 (ref. 32). Because both sumoylation and acetylation occur on lysine residues, p53 desumoylation by SENP1 may enhance p53 acetylation without the involvement of SIRT1.
SIRT1 promotes the survival of both aging and cancer cells. Its activators27,33 and inhibitors34 are being investigated as potential anti-aging and anti-cancer drugs, respectively. Overexpression of SIRT1 increases cellular resistance to ionizing radiation and cisplatin7,35. Thus, our finding that sumoylation regulates SIRT1 activity by its sumoylation is clinically relevant and agents that regulate SIRT1 sumoylation are potential novel targets for intervention on aging and tumorigenesis.
Methods
Cell lines, antibodies and plasmids
H1299 non-small cell lung cancer cells, p53-intact and p53-deficient HCT116 colon cancer cells, DU145 prostate cancer cells and p53-null mouse embryonic fibroblasts (MEFs) were maintained in DMEM medium with 10% FBS. Antibodies against HA (Covance, Berkeley, CA), Flag (Sigma, St Louis, MO), SUMO-1 (Zymed Laboratories, San Francisco, CA), SIRT1 (Santa Cruz Biotechnology, Santa Cruz, CA), p53 (DO-1; BD Biosciences, San Jose, CA), acetyl-Lys 382 p53 (Trevigen, Gaithersburg, MD), and SENP1 (Abgent, San Diego, CA) were all obtained from commercial sources. Except where indicated, all expression vectors encode human proteins. Expression vectors for HA–SIRT1 (ref. 25), Flag–SIRT1 (ref. 25), mouse Flag–SIRT1 (ref. 14), SIRT1 siRNA29, Flag–SENP1 (refs 28, 36), SENP1 siRNA28,36, His–SUMO-1 and HA-SUMO-1 (ref. 31)31 have been described previously. SIRT1ΔCT was generated by inserting the BamH1–EcoR1 fragment of human SIRT1 cDNA into the pcDNA3–HA vector. For SIRT1 expression in Escherichia coli, SIRT1 cDNA was inserted into the BamH1–Not1 sites of the pET-30a vector. SIRT1 mutants were generated by site-directed mutagenesis using the Quick Change kit from Stratagene (La Jolla, CA). The sequences of the mutagenesis primers were: 5′ -TAATGAAGCTATATCTGTGAGACAGGAAGTAAC- 3′ for Lys 734; 5′-TGG TTCTAGTACTGGGGAGAGAAATGAAAGAACT- 3′ for Lys 610; and 5′-GTT GTT AAT GAA GCT ATA GCT GTA AAA CAG GAA TTG ACA G- 3′ for mouse SIRT1VK.
Immunological assays
Whole-cell extracts were prepared by sonication of cells in a buffer containing 20 mM Tris–HCl at pH 7.5, 0.5% NP-40, 150 mM NaCl, 3 mM EDTA, 3 mM EGTA, 10 μg ml−1 aprotinin, 10 μg ml−1 leupeptin, 10 mM benzamidine and 1 mM phenylmethylsulfonyl fluoride. The extracts were incubated with 4 μg antibody for 4 h at 4 °C and subsequently with protein G agarose beads for an additional 4 h. After washing five times with lysis buffer, the immunoprecipitates were heated in SDS–PAGE sample buffer. Proteins in the supernatant were resolved on 8% SDS–PAGE gels and transferred to nitrocellulose membranes. The membranes were probed with antibodies as indicated and proteins recognized by the antibodies were visualized by enhanced chemiluminesence. To detect p53 acetylation at Lys 382, approximately 1 × 106 H1299 cells were plated on 100-mm dishes. Twenty-four hours later, the cells were transfected with 4 μg HA–p53 and additional plasmids as indicated. Eighteen hours post-transfection, cells were treated with 0.5 mM H2O2 or exposed to 0.3 J UV radiation (UV crosslinker, Stratagene) if necessary. Cells were harvested 6 h later, and amounts of acetylated p53 were determined by immunoprecipitation or immunoblotting of cell extracts with antibody to acetyl-Lys 382 p53.
To detect SIRT1 sumoylation, H1299 or DU145 cells were transfected with 4 μg Sumo-1 and SIRT1 plasmids 24 h after plating on 100-mm dishes. If needed, cells were then treated with 0.5 mM H2O2 or exposed to 0.3 J UV radiation 18 h post transfection and harvested 6 h later for immunoprecipation and immunoblotting analysis.
For immunofluorescence analyses, H1299 cells transfected with wild-type SIRT1 and SIRT1K734R on coverslips were washed three times with PBS and fixed in 2% paraformaldehyde for 15 min at room temperature. After additional washing with PBS, fixed cells were made permeable with a solution containing 1% Triton X-100 and 1% BSA. Permeabilized cells were incubated with mouse anti-SIRT1 or rabbit anti-SENP1 antibodies for 2 h at room temperature followed by incubation with goat anti-mouse IgG conjugated with Alexa Fluor 594 (red) or anti-rabbit IgG conjugated with fluorescein isothiocyanate (FITC, green; Molecular Probes, Carlsbad, CA) for 1 h at room temperature. Cells were then washed three times in PBS and stained with DAPI in anti-fade mounting medium. Fluorescent microscopic images were obtained with a Leica confocal laser scanning microscope or a Leitz Orthoplan 2 microscope.
In vitro sumoylation, deacetylase and desumoylation assays
To measure SIRT1 sumoylation in vitro, His–SIRT1 was produced using the E. coli T7 S30 Extract System (Promega, Madison, WI) and purified with the μMACX His-tagged protein isolation kit (Miltenyi Biotec Inc, Auburn, CA) following the manufacturer's protocols. In brief, after five high-stringency washes with buffer containing 50 mM Tris at pH 8.0, 500 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS, and rinsing in 20 mM Tris at pH 8.0, His–SIRT1 was eluted from the beads with the buffer for the native condition and dialysed against sumoylation buffer containing 20 mM HEPES at pH 7.5, 5 mM MgCl2, 2 mM ATP. Purified SIRT1 protein (1 μg) was added to a 20 μl reaction in an in vitro sumoylation system (LAE Biotech International, Rockville, MD) containing different sumoylation components as indicated. Reactions were carried out at 37 °C for 2 h, or as indicated. Reaction mixtures were boiled in SDS sample buffer and separated by SDS-PAGE and SIRT1 protein was detected by immunoblotting with an anti-SIRT1 antibody.
For deacetylase assays, the SIRT1 sumoylation reactions were dialysed against deacetylase assay buffer containing 50 mM Tris at pH. 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2 and 1 mg ml−1 BSA and deacetylase activity was determined using the SIRT1 fluorimetric drug discovery kit (AK-555; BIOMOL International, Plymouth Meeting, PA) following the manufacturer's protocol. In brief, sumoylated SIRT1 was incubated in the assay buffer with the acetylated p53 peptide Arg-His-His-Lys (ε-acetyl) and NAD+ at 37 °C. After 30 min, the deacetylase reactions were stopped by adding Developer II. The deacetylation of the substrate sensitizes it to Developer II which then generates a fluorophore. The fluorophore was then excited with 360 nm light and the emission light at 460 nm detected on a multi-detection microplate reader (Synergy HT; Bio-Tek Instruments, Winoosky, VT).
To assay the deacetylase activity of SIRT1 isolated from cells, HA-tagged human SIRT1 or Flag-tagged mouse SIRT1 was transfected into H1299 cells and purified on an anti-HA or M2 antibody-conjugated affinity column. Cell lysates were loaded onto an anti-HA or M2 affinity column equilibrated with a buffer containing 20 mM Tris–HCl at pH 7.5, 100 mM NaCl, 0.1 mM EDTA, and the columns were washed three times with buffer containing 20 mM Tris–Hcl at pH 7.5 and 1 M NaCl. Proteins were eluted in equilibration buffer containing 1 mg ml−1 HA or 100 μg ml−1 Flag peptide. The deacetylase activity of the eluted SIRT1 proteins was assayed using the fluorimetric drug discovery kit.
For in vitro desumoylation, HA–SIRT1 conjugated to His–Sumo-1 was purified from HEK293 cells expressing HA–SIRT1 and His–Sumo-1 using the μMACX His-tagged protein isolation kit. After five high stringency washes of the beads, the sumoylated SIRT1 protein was eluted from the beads with the buffer for native conditions and dialysed against desumoylation buffer containing 10 mM Tris–HCl at pH 8.0, 150 mM NaCl and 1 mM DTT. Wild-type and inactive mutant Flag–SENP1 proteins were produced in an in vitro TNT translation system (Promega) and purified using M2 affinity beads. After five high stringency washes, Flag–SENP1 was eluted from the beads with a solution containing 100 μg ml−1 Flag peptide. Eluted proteins were dialysed against desumoylation buffer. SENP1 desumoylation assays were performed as previously described37.
Transfections and reporter assays
p53-null MEFs were plated in 12-well plates in DMEM containing 10% FBS at 1 × 105 cells per well. One day after plating, cells were transfected by Lipofectamine Plus following the protocol provided by Gibco–BRL. Cell lysates were prepared by directly adding lysis buffer (25 mM Tris-phosphate at pH 7.8, 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N′,N′,N′,N′-tetraacetic acid, 10% glycerol and 0.2% Triton X-100) to the cells on ice. Luciferase activity was determined using the dual luciferase assay system from Promega. The reporter activity was normalized to the activity of renilla luciferase to minimize the artifacts caused by variation transfection efficiency.
Ni-bead pulldown assays
To detect SIRT1 conjugates with His–SUMO-1, transfected cells were divided into two aliquots. One aliquot was immunoblotted to determine the level of protein expression. The second aliquot was lysed under denaturing conditions in a buffer containing 10 mM Tris–HCl at pH 8.0, 6 M guanidinium–HCl, 100 mM Na2HPO4−NaH2PO4, 5 mM imidazole and 10 mM β-mercaptoethanol, and incubated with Ni2+-NTA beads (Qiagen, Valencia, CA) for 4 h at room temperature. The beads were washed with lysis buffer followed by two sequential washes with a buffer containing 10 mM Tris–HCl at pH 8.0, 8 M urea, 100 mM Na2PO4/−NaH2PO4 and 10 mM β-mercaptoethanol, and a buffer containing 10 mM Tris–HCl at pH 6.3, 8 M urea, 100 mM Na2PO4−NaH2PO4 and 10 mM β-mercaptoethanol. His–Sumo-1 conjugated SIRT1 was eluted from the beads with a buffer containing 150 mM Tris–HCl at pH 6.7, 200 mM imidazole, 30% glycerol, 0.72 M β-mercaptoethanol and 5% SDS. The eluted proteins were immunoblotted with anti-Flag or HA antibodies.
Apoptotic assays
To measure p53-induced caspase-3 activation, cells were permeabilized, fixed and stained for active caspase-3 with FITC-conjugated anti-caspase-3 antibody. The percentage of apoptotic cells was determined by flow cytometry according to the instructions of the active caspase-3 apoptosis kit I (BD Biosciences).
To measure cell death, cells (1 × 106 per 100-mm dish) were stained with propidium iodide, and DNA content was determined by flow cytometry on a Becton-Dickinson FACScan. As alternative approaches, apoptosis was also analysed by the cell death detection ELISAPLUS (Roche, Indianapolis, IN), according to the manufacturer's protocol. The assay system quantifies the amount of histone-coupled DNA fragments by measuring absorbance at 405 nm.
To measure the effect of SIRT1 and SENP1 siRNA on UV radiation- and H2O2-induced cell death, H1299 cells were transfected with SENP1 siRNA28 in pSuppressorNeo vector (IMGENEX Corporation, San Diego, CA) and selected against G418. SIRT1 siRNA was delivered into the cells by retroviral infection followed by selection with puromycin, as previously described29. SMART pool siRNA specific to p73 and scrambled controls were synthesized by Dharmacon and transfected into cells by lipofectamine 2000 following manufacturer's protocols. After treatment with UV radiation and H2O2, cell apoptosis were analysed by cell death detection ELISAPLUS.
Supplementary Material
Acknowledgments
The authors thank E. T. Yeh at the M. D Anderson Cancer Center for SENP1 and SENP1 siRNA vectors. FACS analyses were performed in the Flow Cytometry core facility at H. Lee Moffitt Cancer Center and Research Institute. The work was supported by the Public Health Service grant CA93666 (W.B) and a DOD Prostate Cancer grant DAMD17-02-1-0140 (W.B).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
Author Contributions: Y.Y. designed (with W.B.) and performed (with W.F.) the included studies. J.C., X.Z. and K.B. contributed scientifically to the revision. The manuscript was written by W.B., edited by N.O. and S.V.N. and read by all authors. The senior author (W.B.) designed the project, helped with the analyses and the organization of the data, and provided the financial support. All authors have discussed the data and had scientific input into the manuscript.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 2.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 3.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 6.Haigis MC, Guarente LP. Mammalian sirtuins — emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita N, et al. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells. 2005;10:321–332. doi: 10.1111/j.1365-2443.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- 8.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2α, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 9.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 10.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 11.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 13.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 15.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 17.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 19.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 21.Manza LL, et al. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 22.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 23.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 24.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moschos SJ, Mo YY. Role of SUMO/Ubc9 in DNA damage repair and tumorigenesis. J Mol Histol. 2006;37:309–319. doi: 10.1007/s10735-006-9030-0. [DOI] [PubMed] [Google Scholar]
- 27.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 28.Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol Cell Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nature Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 30.Cohen HY, et al. Acetylation of the C-terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- 32.Buschmann T, Lerner D, Lee CG, Ronai Z. The Mdm-2 amino terminus is required for Mdm2 binding and SUMO-1 conjugation by the E2 SUMO-1 conjugating enzyme Ubc9. J Biol Chem. 2001;276:40389–40395. doi: 10.1074/jbc.M103786200. [DOI] [PubMed] [Google Scholar]
- 33.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 34.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 35.Chu F, Chou PM, Zheng X, Mirkin BL, Rebbaa A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005;65:10183–10187. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- 36.Cheng J, Perkins ND, Yeh ET. Differential regulation of c-Jun-dependent transcription by SUMO-specific proteases. J Biol Chem. 2005;280:14492–14498. doi: 10.1074/jbc.M412185200. [DOI] [PubMed] [Google Scholar]
- 37.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


