Abstract
HLA-B27 is associated with spontaneous viral clearance in hepatitis C virus (HCV) infection. Viral escape within the immunodominant HLA-B27 restricted HCV-specific CD8+ T cell epitope NS5B2841-2849 (ARMILMTHF) has been shown to be limited by viral fitness costs as well as broad T cell cross-recognition, suggesting a potential mechanism of protection by HLA-B27. Here, we studied the subdominant HLA-B27 restricted epitope NS5B2936-2944 (GRAAICGKY) in order to further define the mechanisms of protection by HLA-B27. We identified a unique pattern of escape mutations within this epitope in a large cohort of HCV genotype 1a infected patients. The predominant escape mutations represented conservative substitutions at the main HLA-B27 anchor residue or a T cell receptor contact site, neither of which impaired viral replication capacity as assessed in a subgenomic HCV replicon system. In contrast, however, in a subset of HLA-B27+ subjects rare escape mutations arose at the HLA-B27 anchor residue R2937, which nearly abolished viral replication. Notably, these rare mutations only occurred in conjunction with the selection of two equally rare, and structurally proximal, upstream mutations. Co-expression of these upstream mutations with the rare escape mutations dramatically restored viral replication capacity from <5% to ≥70% of wild-type levels.
Conclusion
The selection of rare CTL escape mutations in this HLA-B27 restricted epitope dramatically impairs viral replicative fitness unless properly compensated. These data support a role for the targeting of highly-constrained regions by HLA-B27 in its ability to assert immune control of HCV and other highly variable pathogens.
Keywords: Hepatitis C virus, T cell response, viral escape, HLA-B27, viral fitness
The HLA class I allele B27 has a protective role in both human immunodeficiency virus (HIV) and hepatitis C virus (HCV) infections (1–2), two of the most highly variable pathogens for which effective vaccines are still lacking. In HIV infection, HLA-B27 is associated with low viral loads, slow CD4+ decline, and the delayed onset of AIDS (1). Protection by HLA-B27 has been linked to the immunodominant targeting of a highly conserved CD8+ epitope in Gag which requires a complicated pathway of viral escape to evade this response. Escape from this B27-KK10 response requires a multitude of mutations. First, an L268M mutation inside the epitope impairs T cell recognition and also impairs dendritic cell (DC) function through increased interaction with an inhibitory receptor (3). A subsequent R264K mutation at the main HLA-B27 binding anchor of the epitope, which achieves more effective CTL escape by abolishing epitope presentation, typically occurs much later in infection and is associated with disease progression (3). The late appearance of the main R264K escape mutation can be attributed to its substantial impact on viral replicative fitness when expressed alone, and thus it is commonly associated in vivo with a S173A compensatory mutation located 90 amino acids upstream of the epitope (3–5). Thus, the ability of HLA-B27 to impart substantial and durable control of HIV is due in part to the requirement for a compensatory mutation to enable effective CTL escape from this dominant immune response.
In HCV genotype 1 infection, the majority of HLA-B27+ individuals are able to clear the virus spontaneously (2, 6). We have previously described several HLA-B27 restricted HCV-specific CD8+ T cell epitopes (7). One of these epitopes, NS5B2841-2849 (ARMILMTHF), is particularly immunodominant since it is recognized in nearly all HLA-B27+ individuals with resolved HCV genotype 1 infection. In addition, nearly all HLA-B27+ patients who progress to chronic HCV genotype 1 infection display viral escape mutations in this epitope, suggesting that this response exerts significant selection pressure (7). Notably, this immunodominant HLA-B27 restricted CD8+ epitope is only present in HCV genotype 1 but absent from the other HCV genotypes, possibly explaining why HLA-B27+ individuals are not protected from chronic non-genotype 1 HCV infection (8). Importantly, viral escape within this epitope does not occur easily. Mutations experimentally introduced in replicon constructs at the HLA-B27 binding anchor residues of the epitope exhibit high fitness costs in vitro, and have therefore not been found in viral sequences derived from patient specimens. In contrast, while mutations at T cell receptor contact residues within the epitope are tolerated by the virus, broad cross-recognition of these escape variants requires multiple ‘clusters’ of these mutations to achieve efficient escape from this CD8+ T cell response (9). The requirement for clustered mutations to escape from dominant HLA-B27 restricted CD8+ T cell responses in both HIV and HCV infections suggests that HLA-B27 may be achieving immune control of these highly variable pathogens through the targeting of critical regions of these viruses in which viral escape is impaired.
To further explore this hypothesis, here we have analyzed the patterns of viral escape in a second HLA-B27 restricted CD8 epitope NS5B2936-2944 (GRAAICGKY). This CD8+ T cell response also selects for viral escape mutations in the majority of patients with chronic HCV genotype 1a infection. We identified a subset of escape mutations within the epitope that nearly abolished viral replication, and thus required additional, compensatory mutations located upstream of the epitope. This was in striking similarity to the escape pathways described for the immunodominant HLA-B27 restricted KK10 epitope in HIV Gag (3–4), providing unique insight into the mechanisms by which some HLA alleles may be mediating their protective effect over HCV and other highly variable pathogens.
Experimental procedures
Patients
Viral sequences from 404 mostly (>90%) treatment-naïve patients with chronic HCV genotype 1a infection were obtained previously (10)(Kuntzen et al., manuscript in preparation). HLA class I typing was performed using standard molecular typing. 19 of the 404 patients were positive for HLA-B27 (supplementary table S1). In addition, PBMCs were obtained from 14 HLA-B27+ patients with chronic HCV genotype 1a infection (10 patients from the above mentioned cohort and 4 patients enrolled at the Freiburg University Medical Center) as well as from 2 HLA-B27+ patients with acute-resolving genotype 1a infection enrolled at the Freiburg University Medical Center. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the institutional review boards at Massachusetts General Hospital, Boston, and the University of Freiburg, respectively. Written informed consent was obtained from each study participant.
Plasmid construction, electroporation, transient-replication assays, NS5B structure analyses, cellular T cell assays and HLA-B2705 peptide binding assay
These methods are described in the supplementary methods section.
Statistics
Frequencies of substitutions in HLA-B27+ and HLA-B27- patients (Table 1) were compared using two-tailed Fisher’s exact test (http://www.graphpad.com/quickcalcs). Replicative capacities of mutants and wild-type (Fig. 2 and 4, supplementary Fig. S1) were compared using paired t test (GraphPad Prism 5.0). P values <0.05 were considered significant.
Table 1.
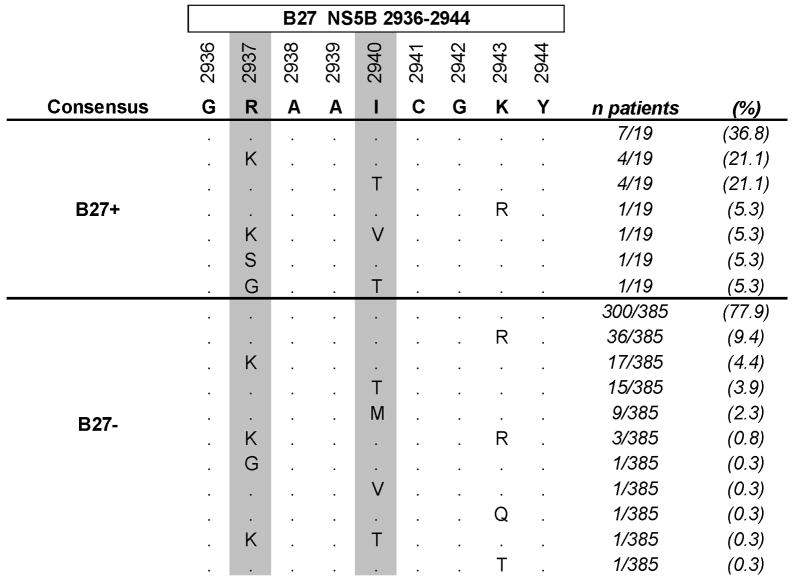
Amnio acid substitutions compared to consensus at Positions 2875, 2881, 2937, and 2940 in HLA-B27 positive and negative patients with chronic HCV genotype 1a infection.
| Residue | Substitution | B27+ (n=19) | B27− (n=385) | P* |
|---|---|---|---|---|
| E2875 | Q | 0 (0.0%) | 4 (1.0%) | 1.0000 |
| K | 3 (15.8%) | 1 (0.3%) | 0.0003 | |
| any | 3 (15.8%) | 5 (1.3%) | 0.0043 | |
| P2881 | L | 4 (21.1%) | 24 (6.2%) | 0.0347 |
| Q | 1 (5.3%) | 5 (1.3%) | 0.2524 | |
| S | 0 (0.0%) | 2 (0.5%) | 1.0000 | |
| any | 5 (26.3%) | 31 (8.1%) | 0.0183 | |
| R2937 | K | 5 (26.3%) | 21 (5.5%) | 0.0047 |
| G | 1 (5.3%) | 1 (0.3%) | 0.0920 | |
| S | 1 (5.3%) | 0 (0.0%) | 0.0470 | |
| any | 7 (36.8%) | 22 (5.7%) | 0.0001 | |
| I2940 | T | 5 (26.3%) | 16 (4.2%) | 0.0017 |
| M | 0 (0.0%) | 9 (2.3%) | 1.0000 | |
| V | 1 (5.3%) | 1 (0.3%) | 0.0920 | |
| any | 6 (31.6%) | 26 (6.8%) | 0.0020 |
P value: two-tailed Fisher’s exact test comparing frequencies in B27+ versus HLA-B27− patients.
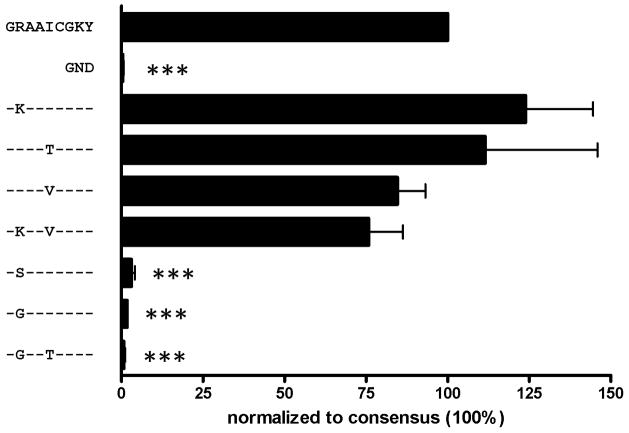
Fig. 2. Rare escape mutations at position two of the HLA-B27 restricted epitope NS5B2936-2944 nearly abolish viral replication.
A wild-type subgenomic replicon, a replication deficient variant (GND), and several variants containing escape mutations within the HLA-B27 restricted epitope NS5B2936-2944 were tested for replication capacity. Replication of the wild-type was set 100%. Means and standard errors of three individual experiments are shown. P values comparing replication of mutants and wild-type were calculated by paired t test and are indicated as follows: * P<0.05, ** P<0.005, and *** P<0.0005.
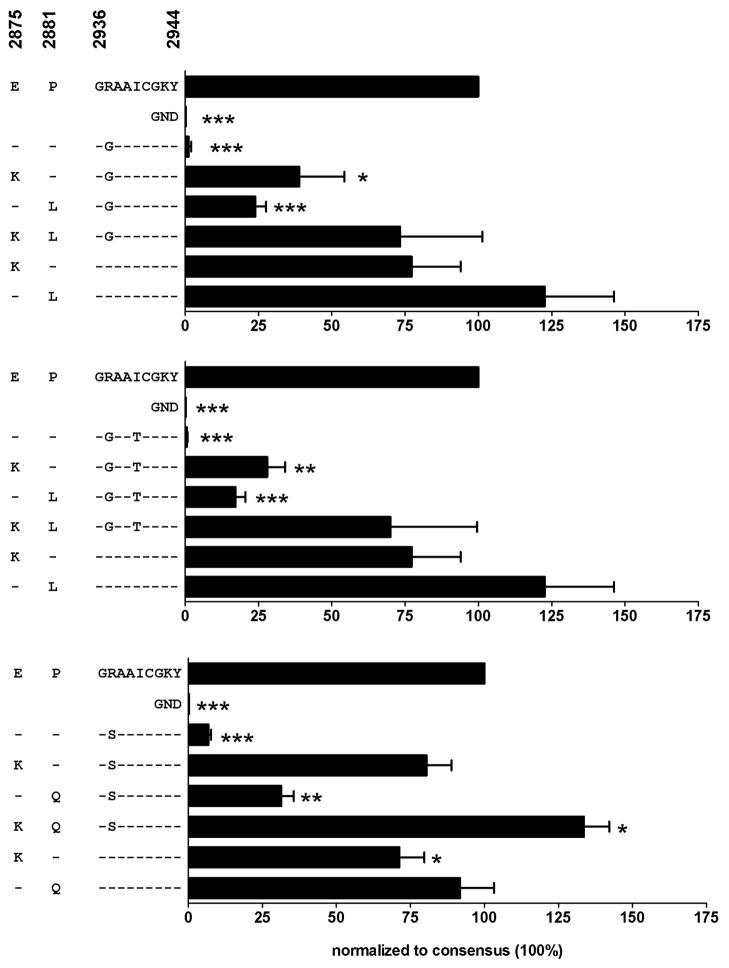
Fig. 4. Viral replication capacity of the rare mutants R2937S, R2937G, and R2937G/I2940T can be restored by mutations E2875K and P2881L/Q.
Replication levels of the wild-type subgenomic replicon, a replication deficient mutant (GND), mutants containing the rare escape mutations alone (R2937G, upper panel; R2937G/I2940T, middle panel; R2937S, lower panel), or in combination with mutations at position 2875 and/or 2881 were compared. Replication of the wild-type was set 100%. Means and standard errors of three individual experiments are shown. P values comparing replication of mutants and wild-type were calculated by paired t test and are indicated as follows: * P<0.05, ** P<0.005, and *** P<0.0005.
Results
CTL escape in the B27-GRAAICGKY epitope typically occurs through conservative mutations
In a large cohort analysis of 404 full-length HCV genotype 1a sequences, we identified that viral escape in the HLA-B27 restricted CD8+ T cell epitope NS5B2936-2944 (GRAAICGKY) was comprised of a unique array of CTL escape mutations. In the 19 HLA-B27+ individuals the predominant escape mutations within this epitope arose at either the main B27 anchor residue (R2937K) or at a T cell receptor contact site (I2940T) (Fig. 1). In each case, these mutations represented conservative substitutions with the swapping of positively charged residues at R2937K, and the substitution of uncharged residues at I2940T. Notably, however, in two B27+ subjects more rare escape mutations were observed, with R2937 replaced by the uncharged amino acids serine (R2937S) or glycine (R2937G) (Fig. 1). In addition, in one B27-positive subject the I2940 residue was replaced by a rare valine (I2940V) substitution in conjunction with the R2937K mutation (Fig. 1). Due to the more frequent development of the R2937K and I2940T mutations we hypothesized that these more conservative substitutions might only minimally impact viral replication (viral fitness), while the more rare mutations might be associated with higher fitness costs.
Fig. 1. HLA-B27 driven sequence variation in the HLA-B27 restricted epitope NS5B2936-2944.
The HCV genotype 1a consensus sequence, the autologous viral sequence in 19 HLA-B27+ patients with chronic HCV genotype 1a infection, and the autologous viral sequence in 385 HLA-B27 negative patients with HCV genotype 1a infection are shown. Positions with HLA-B27 associated sequence polymorphisms (‘footprints’, compare table 1) are shown with a grey background.
Common escape mutations within the B27-GRAAICGKY epitope have no effect on viral replication
To examine the impact of these CTL escape mutations on the virus we constructed subgenomic HCV replicons bearing the different viral escape mutations. Replicative capacities of the mutants were then measured in a transient in-vitro replication system in hepatoma cells using the luciferase reporter gene as a marker for HCV replication. As predicted, no significant differences in the replication capacity of either of the constructs containing the frequent mutations R2937K or I2940T were observed as compared to the parental ‘wild-type’ construct (Fig. 2). In fact, in this in vitro assay, the two variants even exhibited a slight, though not statistically significant, increase in replication. This unimpaired replication is in agreement with the observation that both mutations are more frequently observed in B27-negative subjects, with R2937K being the dominant viral species in 21/385 B27-negative patients (5.5%) and I2940T representing the dominant viral species in 16/385 B27-negative patients (4.2%) (Table 1).
Rare escape mutations within the B27-GRAAICGKY epitope substantially impair viral replication
In contrast to the common R2937K HLA-B27 anchor binding mutation, both of the rare R2937S and R2937G mutations nearly completely abolished viral replication (Fig. 2), including the R2937G mutation in conjunction with I2940T as observed in one patient (Fig. 1). The greater impact of these two rare escape mutations on viral replication is also in agreement with the absence of these mutations in HLA-B27 negative individuals; indeed, the R2937S mutation was not found in any of the 385 B27-negative patients in the cohort, while the R2937G mutation was only observed in a single B27-negative patient (0.3%; Table 1). In addition, the I2940V mutation, alone or in combination with mutation R2937K, was also found to impair viral replication, resulting in reductions in replication of 15% and 24%, respectively, although these results did not reach statistical significance. Considering that the I2940V substitution is only rarely observed in B27-negative patients (1/385 patients, 0.3%; Table 1) suggests that even this moderate reduction in replication capacity may have a significant effect on the virus in vivo analogous to the impact of the M184V drug-resistance mutation in HIV (11).
Mutations at position E2875 and P2881 co-evolve with rare escape mutations at positions R2937 and I2940
The observation that mutations R2937S and R2937G nearly completely impair viral replication was surprising given their presence in subjects demonstrating significant viral loads (e.g., the B27-positive patient with mutations R2937G/I2940T had a viral load of 33.9 × 106 IU/ml, and the B27-negative patient with mutation R2937G had a viral load of 22.5 × 106 IU/ml). We therefore hypothesized that these mutations might occur in conjunction with one or several additional mutations that function as compensatory mutations to alleviate their high costs to viral fitness. In screening our cohort of 404 HCV genotype 1a infected patients for additional substitutions co-evolving with the rare HLA-B27 associated mutations R2937G or R2937S throughout the HCV polyprotein, we found co-evolution (defined as a mutation present in at least 2 of the three patients with the R2937G or R2937S mutation but not frequently occurring in other patients) at only two amino acid residues, E2875 and P2881 (Fig. 3). These two residues do neither overlap with previously reported B27-restricted epitopes nor with predicted B27-restricted epitopes (T cell epitope prediction algorithm: www.iedb.org).
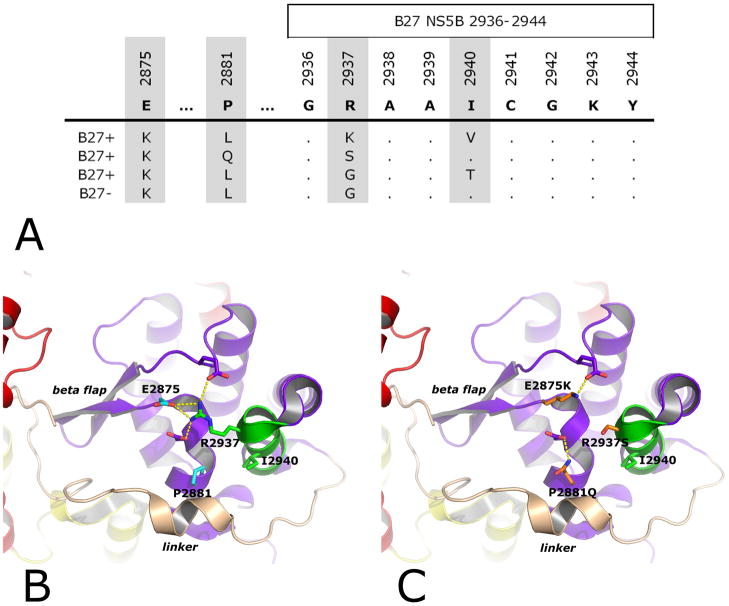
Fig. 3. Rare mutations R2937S, R2937G (with or without I2940T), and R2937K/I2940V within the HLAB27 restricted epitope NS5B2936-2944 only occur in concert with an equally rare cluster of two mutations 71 and 65 amino acids upstream of the epitope which restore a network of interactions in the NS5B thumb domain.
A: Autologous viral sequences are shown for all patients that harbor the E2875K mutation (4/404 patients in the cohort; compare table 1). All four patients also display one of the rare mutations at position 2937 (R2937S, R2937G, or R2940K/I2940V). In addition, all four patients display a mutation at position 2881 (P2881L or P2881Q).
B: Close-up of the epitope region in the three-dimensional structure of genotype 1a NS5B. NS5B is displayed as ribbons and colored by domains (red: fingers; yellow: palm; purple: thumb). The epitope is shown in green with the sites of rare escape mutations R2937 and I2940 displayed as sticks, colored by atom type (blue: nitrogens; green: carbons) and labeled. The sites of compensatory mutations P2881 and E2875 are similarly displayed (red: oxygens; cyan: carbons) as well as two other acidic amino acids (purple: carbons) also in direct interaction (dotted yellow lines) with R2937. The thumb’s ‘beta-flap’ is labeled in italics as well as the linker (in light brown) C-terminal to the thumb. C: Same view as in B, but with mutations R2937S, E2875K and P2881Q modeled into the structure (orange: carbons). E2875K compensates for the negative charge cluster at the base of the beta-flap that would be left by R2937S alone, restoring in part an orientation of the beta-flap that allows proper interaction with the linker. P2881Q completes the remodeling of the network of interactions between thumb and linker.
Residue E2875 was found to be highly conserved, with 396 of 404 patients (98.0%) displaying the consensus residue at this position (Table 1). Only 8 patients carried sequence variations at this position: 4 B27-negative patients (0.8%) exhibited an E2875Q substitution, while this substitution did not occur in HLA-B27+ patients (not significant). Conversely, however, a substitution of the negatively charged amino acid glutamate by the positively charged amino acid lysine (E2875K) occurred in three of the 19 HLA-B27+ patients (15.8%), notably more frequent than in the 0.3% (1/385) of B27-negative subjects (P=0.0003, Table 1). Strikingly, as shown in Figure 3A, all four E2875K substitutions occurred together with one of the rare mutations at position R2937 (R2937S or R2937G) or I2940 (I2940V). The co-expression of these very rare mutations indicated that E2875K may indeed be required for the development of these rare escape mutations, functioning to compensate for their impacts on viral replication. This hypothesis is confirmed by structural analysis of genotype 1a NS5B. As shown in Figure 3B, R2937’s positively charged side chain inserts between three negatively charged side chains, of which the central one is that of E2875. Mutating R2937 to either S (Figure 3C) or G (not shown) leaves a cluster of three negative charges, an unfavorable configuration that is compensated by the single E2875K mutation (Figure 3C). Still, these changes would affect the conformation of the so-called ‘beta-flap’, a flexible beta-structure in NS5B’s ‘thumb’ domain at the base of which are the three negatively charged residues (Figure 3B). The beta-flap is one of two main anchors of the ‘linker’, a structural element whose movement has been proposed to be critical for NS5B’s enzymatic activity (12). The other anchor is made by the outer faces of two helices in the thumb, harboring residue 2881 and the NS5B2937-3944 epitope, respectively. P2881 in particular is sandwiched between two hydrophobic residues of the linker, an interaction that is changed by mutation to the bulkier L (not shown) or to the more polar Q (Figure 3C). Substitutions at position 2881 are more commonly observed than at position 2875, with 36 of 404 patients (8.9%) showing a substitution of the consensus residue proline (P) (Table 1), however, this substitution is also associated with HLA-B27 (5/19 HLA-B27+ patients [26.3%] compared to 31/385 HLA-B27-patients [8.1%], P=0.0183). Notably, three out of the 5 substitutions in B27-positive patients occurred in those three patients with rare mutations at positions R2937 or I2940 (Figure 3A).
Mutations at positions E2875 and P2881 restore the fitness defects of rare escape mutations in the B27-GRAAICGKY epitope
In order to directly test whether the observed mutations at positions E2875 and P2881 can compensate for the impact on viral replication of the rare mutations at positions R2937 or I2940, we co-expressed the different mutations in the subgenomic replicon system. As shown in Figure 4, co-expression of mutations R2937G, R2937G/I2940T, or R2937S with mutation E2875K led to a marked, though not complete restoration of viral replication to levels of 39%, 28%, and 80% compared to wild-type, respectively. The same was true for co-expression of the three respective R2937 mutations with mutations at P2881 (P2881L or P2881Q, respectively), resulting in viral replication levels of 17–31% compared to wild-type. Only the combined co-expression of the position R2937 mutations together with both candidate compensatory mutations (E2875K and P2881L/Q) led to a nearly complete restoration of viral replication, with replication levels of approximately 70% in the case of mutations R2937G and R2937G/I2940T and ≥100% in the case of mutation R2937S (Fig. 4). These results show clearly that the HLA-B27 associated substitutions at positions E2875 and P2881 are indeed compensatory mutations for the strong replicative defects caused by the substitution of arginine at R2937 by Glycine or Serine. When tested alone, the E2875K and P2881Q mutations were associated with a slightly reduced replication level (~77% and 92% of wild-type, respectively) as compared to wild-type, explaining why these substitutions occur rarely in B27-negative patients (1/385 patients [0.3%] and 5/385 patients [1.3%], respectively, Table 1). The subgenomic replicon carrying the P2881L mutation alone, however, replicated at levels that even exceed the wild-type replication level. This finding may at least partially be due to the in vitro system used here. However, it is in agreement with the observation that the P2881L mutation is rather frequently observed in B27-negative patients in vivo (24/385 patients [6.2%], Table 1).
Escape mutations associated with a high viral fitness cost may be required for successful escape from the B27-GRAAICGKY response in some patients
Considering that the rare mutations R2937S, R2937G, and R2937G/I2940T require a complicated pathway of compensatory mutations, it is surprising that these mutations nevertheless occur in a subset of HLA-B27-positive patients. This finding led us to the hypothesis that the ability of epitope-specific CD8+ T cell responses to recognize the common escape mutations in the B27-GRAAICGKY epitope in some subjects may function to select for these rare and highly deleterious mutations. To better address this issue, we first determined the HLA-B27 binding capacity of the different epitope peptide variants. Indeed, a gradual effect of the different amino acid substitutions was observed (Table 2). Substitutions I2940T and I2940V did not show a substantial effect on HLA-B27 binding, consistent with the location of residue I2940 in the core region of the epitope (amino acid position 5), which likely is a T cell receptor contact site rather than an HLA-B27 binding anchor. The frequently observed R2937K substitution still showed some level of HLA-B27 binding, albeit at a strongly reduced level (IC50 1893nM compared to 31nM observed for the wild-type peptide). The fact that this variant still showed some degree of HLA-B27 binding is in accordance with the notion that lysine (K) has been found in a small portion of natural HLA-B27 ligands (13), and also with our previous finding that an arginine to lysine substitution in another HCV-specific HLA-B27 restricted CD8+ T cell epitope had no effect on HLA-B27 binding and only a minor effect on T cell recognition (9). Peptides with the combined R2937K/I2940V mutations or the R2937S mutation showed only borderline HLA-B27 binding capacity (IC50 4668 and 4815nM, respectively), while peptides with the R2937G substitution showed no HLA-B27 binding at all (IC50 >75,000 nM). These results suggest that some of the epitope-specific escape mutations may still be able to be presented and cross-recognized by specific CD8+ T cells.
Table 2.
Binding of epitope peptide variants to HLA-B*2705
| Peptide | IC50 [nM] |
|---|---|
| GRAAICGKY | 31 |
| -K------- | 1893 |
| ----T---- | 122 |
| ----V---- | 32 |
| -K--V---- | 4668 |
| -S------- | 4815 |
| -G------- | >75000 |
| -G--T---- | >75000 |
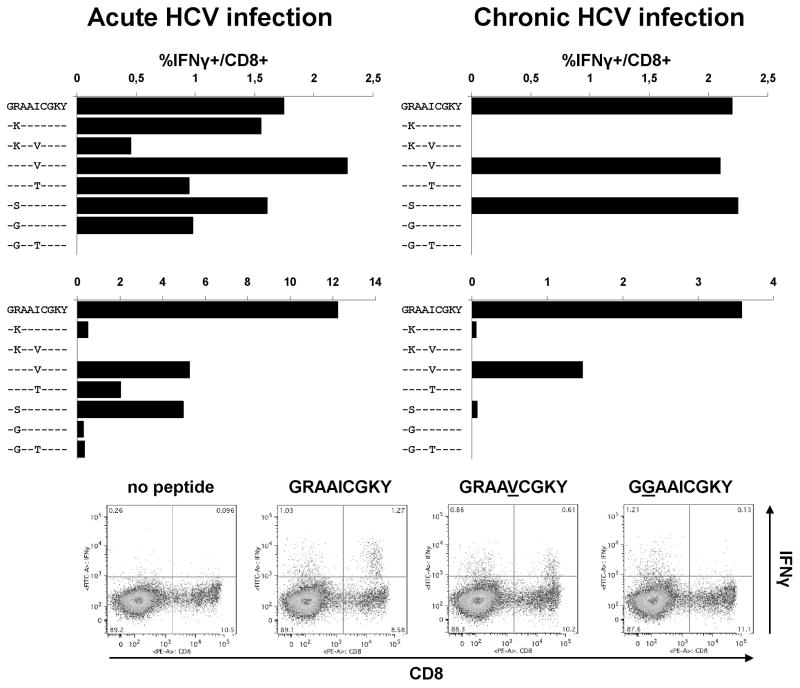
To determine whether variant-specific CD8+ T cell responses are capable of recognizing some of these escape mutations, we tested two B27-positive patients with an acute-resolving course of HCV genotype 1a infection and 12 B27-positive patients with a chronic HCV genotype 1a infection for responses against this epitope. We could establish peptide-specific CD8+ T cell lines from both patients with acute-resolving infection as well as from two out of the twelve patients with chronic infection. Of note, this low rate of detection in chronic HCV infection is similar to what has been observed for other dominant HCV-specific CD8+ T cell responses (14). Next, the cross-recognition of different peptide variants by the wild-type specific T cell line was tested. As shown in Fig. 5, one of the two patients with acute-resolving infection showed a broad cross-recognition of the different peptide variants including the common R2937K and I2940T mutations. Here, only the R2937G/I2940T double mutant led to a complete loss of cross-recognition. In the second acute-resolving patient a rather narrow pattern of cross-recognition was observed with only the R2937S, I2940V, and I2940T variant peptides showing a relevant level of cross-recognition. Finally, in both of the patients with chronic infection a similarly narrow level of cross-recognition was observed (Fig. 5). In sum, these experiments suggest that in some patients a broad degree of peptide variant cross-recognition may drive the selection of rare and highly deleterious mutations such as the R2937G/I2940T double variant, while in other patients, weak T cell responses with a narrow degree of peptide variant cross-recognition select for frequent and well tolerated escape mutations or even allow replication of the wild type sequence.
Fig. 5. NS5B2936-2944 specific CD8+ T cell responses show varying degrees of variant cross-recognition in individual patients.
Epitope-specific T cell lines were derived from two HLA-B27+ patients with acute-resolving HCV genotype 1a infection (left panels) and from two HLA-B27+ patients with chronic HCV genotype 1a infection (right panels) and tested for interferon-γ production in response to wild-type peptide and variant peptides. Representative dot blots from the second patient with acute-resolving infection are shown below.
Discussion
HLA-B27 is associated with a high rate of spontaneous viral clearance in HCV genotype 1 infection (2, 8). We have previously suggested that the immunodominant HLA-B27 restricted HCV-specific CD8+ epitope NS5B2841-2849 (ARMILMTHF) might have an important role in this protective effect (7, 9) due to the fact that viral escape from this epitope requires the evolution of several combined mutations within the epitope. In that epitope, since mutations at the main HLA-B27 binding anchors cannot occur due to viral fitness costs, mutations at T cell contact residues are required for viral escape (9). However, due to a broad cross-recognition of these escape mutations, such TCR escape mutations need to occur in clusters in order to efficiently escape from this dominant CD8+ T cell response.
Here, we studied viral evolution in a second B27-restricted HCV-specific CD8+ T cell epitope in NS5B (NS5B2936-3944, GRAAICGKY). In this epitope region, viral sequence variations were strongly enriched in HLA-B27+ patients with chronic HCV genotype 1a infection as compared to HLA-B27 negative patients. Although cross-sectional data from chronically infected patient cannot formally prove viral evolution, these data strongly indicate that these mutations are indeed HLA-B27 driven viral escape mutations. Indeed, two different patterns of viral escape mutations were observed in patients chronically infected with HCV genotype 1a. In the majority of patients, viral escape was mediated by a conservative mutation either at the main HLA-B27 binding anchor at position 2 (arginine to lysine) or at a T cell receptor contact residue at position 5 (isoleucine to threonine). These mutations were also observed in a longitudinal sequence analysis of two HLA-B27+ patients with acute infection (supplementary table S2), further supporting the notion that the sequence variations observed in chronically infected patients indeed reflect viral escape mutations. Importantly, these frequent mutations had no relevant effect on viral replication as assessed in the subgenomic replicon model. In contrast, in a small subset of HLA-B27+ patients, non-conservative mutations occurred at the main HLA-B27 binding anchor at position 2, replacing the long, positively charged residue arginine by the short, uncharged residue glycine or serine. These mutations led to a nearly abolished viral replication capability. The molecular basis for this effect is readily found by structural analysis of genotype 1a NS5B: These mutations of R2937 (position 2) disrupt a network of charge interactions at the base of NS5B’s beta-flap, preventing correct interactions between the thumb domain and the linker element. These interactions have been proposed to be involved in the rate-limiting early steps of RNA synthesis by NS5B (12). Indeed, modulation of the interactions between thumb and linker through mutation of R2937 was recently found to be involved in replicative properties of strain JFH1 NS5B (15). Accordingly, detrimental escape mutations R2937S or R2937G could only co-evolve in the genotype 1a context in concert with two additional, compensatory mutations located 61 and 55 amino acids upstream of the epitope. The former compensatory mutation E2875K is expected to restore charge complementarity at the base of the beta-flap, while the latter P2881L/Q would additively enhance interaction with the linker at its other binding site in the thumb. While these observations provide an attractive explanation for the need of compensatory mutations in the presence of the R2937S and R2937G mutation, it is important to point out that conserved RNA structures in this region may also provide further functional restrictions on the evolution of viral escape mutations, as has been reported for the 3′ end of the RNA coding for NS5B (16). Evolution of certain amino acid mutations could further be complicated by the need to change two nucleotides from the consensus sequence. Importantly, mutation R2937S can be obtained by a single nucleotide change. The same is true for the R2937G mutation; however, the HLA-B27+ individual displaying this mutation had two nucleotide changes, while the HLA-B27 negative patient with the R2937G mutation had a single nucleotide change (supplementary table S3).
The finding that the two compensatory mutations E2875 and P2881 could restore the viral replication capacity from <5% to 70%-100% of wild-type level allows two important conclusions. First, it may help to explain why associations between certain protective HLA alleles, CD8+ T cell responses or viral escape mutations and HCV viral loads, at least in the chronic phase, are hard to identify. Indeed, the high replication rate as well as the high mutation rate of HCV might allow the selection of compensatory mutations and restore replication levels similar to wild-type even in the presence of mutations that are highly detrimental if occurring alone. In agreement with this finding, the B27+ patient who displayed the rare R2937G/I2940T mutation in concert with the compensatory mutations E2875K and P2881L mutation even had an extraordinary high viral load (supplementary table S1). Second, this finding points out that a modest viral fitness cost, e.g. a reduction of replication levels by 30–50% as observed in previous studies (9, 17), may still have a substantial effect in vivo. Indeed, this effect may be analogous to the benefit derived by maintaining HIV-positive patients on 3TC therapy despite the presence of the M184V 3TC-resistance mutation because of the negative impact of M184V on viral replication (11). This is further supported by the notion that mutations that were associated with a modest viral fitness cost in the in vitro replicon assay (e.g., E2875K, P2881Q, and I2940V) were observed very rarely (0.3%, 1.3%, and 0.3%, respectively) in B27-negative patients (Table 1). In contrast, mutations that had no effect on viral replication levels (or replicated even at >100% compared to wild-type in the in vitro system) occurred more frequently in B27-negative individuals (e.g., P2881L, R2937K, and I2940T), although they were detected significantly more frequently in B27-positive patients. These findings are in line with the observation in chimpanzees that escape mutations with a low impact on viral replication are fixed over time, while escape mutations with a more dramatic effect on viral replication are not sustained and also revert in an in vitro model system (18). It is tempting to speculate that the well replicating P2881L mutation may occur in a minor quasispecies in those patients that select for one of the rare, detrimental escape mutations, enabling sufficient replication until the additional E2875K mutations can evolve and to evade the CD8+ T cell response.
Some of the effects observed in the subgenomic HCV replicon system might be due to the specific sequence of the replicon used. Here, we used a genotype 1b (strain con1) based replicon with NS5B from genotype 1a (strain H77). In order to exclude that the observed effects were due to the genotype 1b proteins, we confirmed the observations made in this hybrid replicon in a replicon that only contained genotype 1a (strain H77) sequences. Importantly, similar effects of these mutations were observed in the genotype 1a replicon (supplementary figure S1). It is also noteworthy that previous studies have identified mutations that facilitate replication in the replicon system, while exhibiting a negative effect on viral replication e.g. in the chimpanzee model (19). Thus, it is important to interpret data obtained from the in vitro system in the context of data available in vivo. As described above, the viral fitness data observed for individual mutations in vitro corresponded well to their frequency observed in patients in vivo, indicating that they were not due to cell-culture specific phenomena.
With the majority of HLA-B27+ patients spontaneously clearing HCV genotype 1 infection (2, 8), these data may help to explain the protective effect of HLA-B27 in HCV infection. We have previously shown that a dominant response against the HLA-B27 restricted CD8+ T cell epitope NS5B2841-2849 (ARMILMTHF) contributes to protection by HLA-B27, and could further demonstrate that a broad cross-recognition of viral escape mutations, as well as limitations of viral escape through fitness costs, are determinants of this protection (9). The requirement of clustered escape mutations in that epitope results in a complicated pathway of escape to evade the immune response. During acute infection, this difficulty in rapidly achieving effective escape may provide the epitope-specific CD8+ T cell response sufficient time to clear the virus before effective escape mutations can develop. In the present study, we reveal an equally complicated pathway of viral escape in a second neighboring HLA-B27 restricted CD8+ T cell epitope that requires the evolution of a network of two compensatory mutations in at least a subset of patients. This requirement for complex escape pathways for two epitopes restricted by HLA-B27 would hamper the development of effective escape mutations to both of these responses, and thus prevent a major determinant of persistent infection (20–22).
Of note, mutations that compensate for viral fitness costs associated with viral escape mutations have not been described previously for HCV, but have now been observed in the HLA-B27 restricted epitope described here, in an HLA-B57 restricted epitope (Oniangue-Ndza et al., personal communication), as well as in an HLA-A3 restricted epitope (23). Since all three HLA alleles have been described to be associated with spontaneous HCV clearance (2, 24), the requirement for compensatory mutations due to the high viral fitness costs of specific escape mutations may be a common characteristic of protective HLA alleles in HCV infection. As noted earlier, a similar requirement for compensatory mutations has been made for the immunodominant HLA-B27-restricted KK10 epitope in HIV Gag (3–4), as well as for the HLA B57-restricted KF11 epitope in Gag (25), each of which are believed to contribute importantly to the immune control of HIV associated with each of these alleles.
In sum, our data suggest that CD8+ T cell responses targeting the HLA-B27 restricted epitope NS5B2936-2944 (GRAAICGKY) are uniquely capable of driving the selection of rare and highly deleterious mutations in some HLA-B27-positive patients. These escape mutations require the evolution of a network of compensatory mutations located outside the epitope in order to restore viral replication. This complicated pathway of viral escape may significantly contribute to viral control in the acute phase of infection and thus contribute to protection by HLA-B27 observed in HCV infection.
Supplementary Material
Acknowledgments
We thank all participating patients, clinical collaborators, and collaborators at the Broad Institute of MIT and Harvard who conducted the full-genome virus sequencing (10). In addition, we thank Ralf Bartenschlager (University of Heidelberg) for providing plasmid I341PILuc_NS3_3ET as well as Huh7-Lunet cells, and Charles Rice (Rockefeller University) for providing plasmid H77 (H/FL).
Financial support: This project was funded in part with Federal funds from the National Institute of Allergy and Infectious Disease (NIAID) under grant R01-AI067926 (TMA) U19 AI082630 (TMA, GL), with funds from the Deutsche Forschungsgemeinschaft (Emmy Noether-Programm, NE 1567/1-1 to CNH; KU 2250/1-1 to TK; FOR1202 to VL), with funds from the Bundesministerium für Bildung und Forschung (BMBF 01EO0803 to CNH), with funds from the French National Agency for Research on AIDS and Viral Hepatitis (ANRS, grant AO 2010/CSS4 to SB), and with funds from Association pour la recherche sur le cancer (ARC, postdoctoral fellowship to CCS).
Abbreviations
- AIDS
acquired immunodeficiency syndrome
- CTL
cytotoxic T lymphocyte
- DC
dendritic cell
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- HLA
human leukocyte antigen
- NS
non-structural protein
- PBMC
peripheral blood mononuclear cell
- RdRp
RNA-dependent RNA polymerase
Contributor Information
Christoph Neumann-Haefelin, Email: christoph.neumann-haefelin@uniklinik-freiburg.de.
Cesar Oniangue-Ndza, Email: coniangue-ndza@partners.org.
Thomas Kuntzen, Email: t.kuntzen@gmx.de.
Julia Schmidt, Email: julia.schmidt@uniklinik-freiburg.de.
Katja Nitschke, Email: katja.nitschke@uniklinik-freiburg.de.
John Sidney, Email: jsidney@liai.org.
Célia Caillet-Saguy, Email: celia.caillet@vms.cnrs-gif.fr.
Marco Binder, Email: marco_binder@med.uni-heidelberg.de.
Nadine Kersting, Email: nadine.kersting@uniklinik-freiburg.de.
Michael W. Kemper, Email: earthmk@umich.edu.
Karen A. Power, Email: kapower@partners.org.
Susan Ingber, Email: susanzellsingber@gmail.com.
Laura L. Reyor, Email: lreyor@partners.org.
Kelsey Hills-Evans, Email: khills-evans@partners.org.
Arthur Y. Kim, Email: akim1@partners.org.
Georg M. Lauer, Email: glauer@helix.mgh.harvard.edu.
Volker Lohmann, Email: volker_lohmann@med.uni-heidelberg.de.
Alessandro Sette, Email: alex@liai.org.
Matthew R. Henn, Email: mhenn@broadinstitute.org.
Stéphane Bressanelli, Email: stephane.bressanelli@vms.cnrs-gif.fr.
Robert Thimme, Email: robert.thimme@uniklinik-freiburg.de.
Todd M. Allen, Email: tallen2@partners.org.
References
- 1.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 3.McMichael AJ. Triple bypass: complicated paths to HIV escape. J Exp Med. 2007;204:2785–2788. doi: 10.1084/jem.20072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, et al. Structural and functional constraints limit options for CTL escape in the Immunodominant HLA-B27 restricted epitope in HIV-1 capsid. J Virol. 2008;82:5594–5605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Streeck H, Li B, Poon AF, Schneidewind A, Gladden AD, Power KA, et al. Immune-driven recombination and loss of control after HIV superinfection. J Exp Med. 2008;205:1789–1796. doi: 10.1084/jem.20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hraber P, Kuiken C, Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 7.Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 8.Neumann-Haefelin C, Timm J, Schmidt J, Kersting N, Fitzmaurice K, Oniangue-Ndza C, et al. Protective effect of human leukocyte antigen B27 in hepatitis C virus infection requires the presence of a genotype-specific immunodominant CD8+ T-cell epitope. Hepatology. 2010;51:54–62. doi: 10.1002/hep.23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dazert E, Neumann-Haefelin C, Bressanelli S, Fitzmaurice K, Kort J, Timm J, et al. Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J Clin Invest. 2009;119:376–386. doi: 10.1172/JCI36587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuntzen T, Timm J, Berical A, Lennon N, Berlin AM, Young SK, et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 2008;48:1769–1778. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wainberg MA. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev Anti Infect Ther. 2004;2:147–151. doi: 10.1586/14787210.2.1.147. [DOI] [PubMed] [Google Scholar]
- 12.Harrus D, Ahmed-El-Sayed N, Simister PC, Miller S, Triconnet M, Hagedorn CH, et al. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J Biol Chem. 2010;285:32906–32918. doi: 10.1074/jbc.M110.151316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez de Castro JA, Alvarez I, Marcilla M, Paradela A, Ramos M, Sesma L, et al. HLA-B27: a registry of constitutive peptide ligands. Tissue Antigens. 2004;63:424–445. doi: 10.1111/j.0001-2815.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- 14.Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127:924–936. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt M, Scrima N, Radujkovic D, Caillet-Saguy C, Simister PC, Friebe P, et al. A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strain JFH1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture across Genotypes. J Virol. 2011;85:2565–2581. doi: 10.1128/JVI.02177-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salloum S, Oniangue-Ndza C, Neumann-Haefelin C, Hudson L, Giugliano S, Aus dem Siepen M, et al. Escape from HLA-B*08 restricted CD8 T cells by hepatitis C virus is associated with fitness costs. J Virol. 2008 doi: 10.1128/JVI.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uebelhoer L, Han JH, Callendret B, Mateu G, Shoukry NH, Hanson HL, et al. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog. 2008;4:e1000143. doi: 10.1371/journal.ppat.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci U S A. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox AL, Mosbruger T, Mao Q, Liu Z, Wang XH, Yang HC, et al. Cellular immune selection with hepatitis C virus persistence in humans. J Exp Med. 2005;201:1741–1752. doi: 10.1084/jem.20050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tester I, Smyk-Pearson S, Wang P, Wertheimer A, Yao E, Lewinsohn DM, et al. Immune evasion versus recovery after acute hepatitis C virus infection from a shared source. J Exp Med. 2005;201:1725–1731. doi: 10.1084/jem.20042284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker CM. Adaptive immunity to the hepatitis C virus. Adv Virus Res. 2010;78:43–86. doi: 10.1016/B978-0-12-385032-4.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merani S, Petrovic D, James I, Chopra A, Cooper D, Freitas E, et al. Effect of Immune Pressure on Hepatitis C Virus Evolution: Insights From a Single-Source Outbreak. Hepatology. 2011;53:396–405. doi: 10.1002/hep.24076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuniholm MH, Kovacs A, Gao X, Xue X, Marti D, Thio CL, et al. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology. 2010;51:1514–1522. doi: 10.1002/hep.23515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.