Abstract
DNA melting under torsion plays an important role in a wide variety of cellular processes. In the present Letter, we have investigated DNA melting at the single-molecule level using an angular optical trap. By directly measuring force, extension, torque, and angle of DNA, we determined the structural and elastic parameters of torsionally melted DNA. Our data reveal that under moderate forces, the melted DNA assumes a left-handed structure as opposed to an open bubble conformation and is highly torsionally compliant. We have also discovered that at low forces melted DNA properties are highly dependent on DNA sequence. These results provide a more comprehensive picture of the global DNA force-torque phase diagram.
Negative DNA supercoiling is known to play an important role throughout the cell cycle. Circular DNA, as seen in plasmids and genomic DNA of prokaryotes, is maintained in an underwound state [1], while negative supercoiling in eukaryotic genomes is predominantly constrained within nucleosomes [2]. Negative torsion affects cellular processes through changes in DNA conformation, such as plectoneme formation [3], cruciform extrusion from palindromic sequences [4], and conversion of B-DNA regions into unusual structures such as Z- or H-DNA [2]. Most importantly, underwinding can promote local melting of the DNA [5] which is a requirement for the initiation of replication [6] and transcription [7]. A common feature of many prokaryotic and eukaryotic replication origins is the so called DNA unwinding element (DUE), an easily denaturing segment of DNA [8]. Since the sequences of DUEs are generally not conserved, it is believed that proteins may recognize general structural features of the unpaired DNA strands, rather than a particular sequence [9].
In order to better understand the regulatory mechanisms of torsionally melted DNA, it is important to know its structural and elastic properties. Early insights were provided by experiments on circular plasmid DNA [10] which yielded estimates of persistence length and torsional modulus of small regions of strand-separated DNA. In the last two decades, single-molecule manipulation techniques have become powerful tools to study DNA mechanics [11-18]. Although these methods have provided insights for B-DNA, torsionally melted DNA has not yet been fully examined and is still poorly understood. Even the structure of melted DNA is controversial as it is unclear whether melted DNA takes on the form of two parallel single strands [13] or a left-handed helix [14, 15]. Elastic parameters of melted DNA have only been roughly estimated by examining the coexistence state of B-DNA and melted DNA [19]. To date, there has been no direct and complete description of the tensile and torsional elasticity of melted DNA, and even less is known about the sequence dependence of the elastic properties of melted DNA.
In the current Letter, we employed our angular optical trap [16, 17, 20-22] to carry out a comprehensive study of torsionally underwound DNA. Direct torque detection is essential to our approach, as it allows unambiguous identification of the phase boundaries [17, 23] as well as direct measurements of torsional elastic parameters. Our experiments allow for the full characterization of both the torsional and tensile properties of melted DNA.
Angular trapping experiments were performed in a configuration identical to that described previously [16, 17, 21]. All experiments were performed in phosphate buffered saline (PBS) with 150 mM NaCl at 23±1 °C. Three different DNA constructs with distinct sequences were used: Sequence A (2.2 kb, 51% GC), Sequence B (1.8 kb, 52% GC), and Sequence C (1.6 kb, 64% GC) [24].
We first carried out experiments on Sequence A under a range of constant forces (Fig. 1). As DNA was continuously twisted, both the torque and the extension were directly monitored. The degree of supercoiling σ is defined as the number of turns added to dsDNA divided by the number of naturally occurring helical turns in the given molecule. The experiment began with the DNA in a slightly overwound state, as evidenced by a torque plateau [15-17, 25] at the far right of Fig. 1. Rotation of the DNA in the underwinding direction resulted first in the disappearance of non-B form structures such as plectonemic loops or supercoiled P-DNA, and the subsequent twisting of B-DNA where torque was proportional to the number of turns [15, 16]. The onset of a DNA torsional melting transition occurred when the torque reached approximately –10 pN nm, a value that compares well with previous work [15, 25, 26]. A large number of turns had to be applied before the molecule completed the transition to a fully melted state. The end of the torsional melting transition was signaled by a decrease in the torque (corresponding to an increase in magnitude) and a concomitant shortening in extension. Interestingly, upon further unwinding, the torque plateaued again, indicating another phase transition, while the extension continued to decrease, albeit at a slower rate.
FIG. 1.
Torque (a) and extension (b) traces for Sequence A, averaged over multiple single-molecule experiments. Dashed lines indicate phase boundaries for measurements at 24 pN.
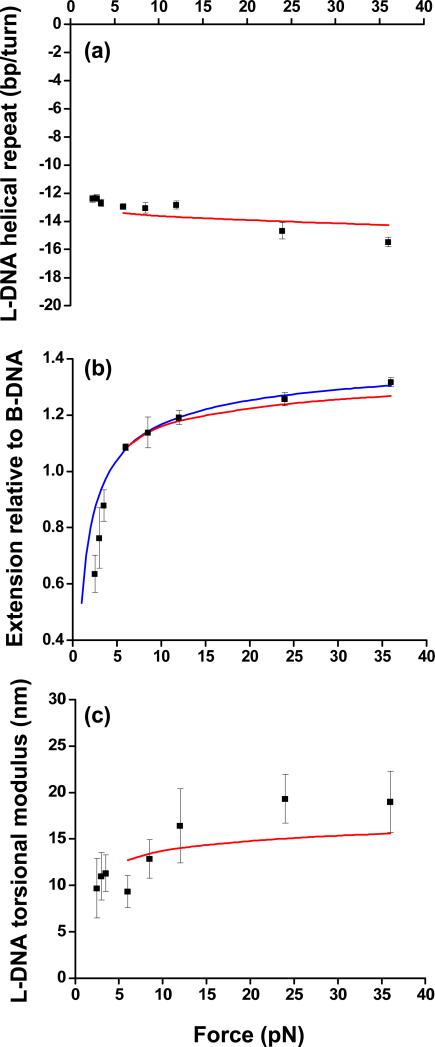
Several parameters can be determined from the data taken in the fully melted state. First is the natural twist of torsionally melted DNA. The degree of supercoiling at the end of the melting transition σend was slightly force dependent [Fig. 2(a)], with an average value of approximately −1.8; the corresponding twist is therefore ≈ −13 bp/turn, comparable to but larger in magnitude than previous indirect estimates of −9 bp/turn [14, 19]. This negative helicity indicates that the melted DNA takes on a left-handed structure, and we will therefore follow the notation of Ref. [15] to designate such a form of torsionally melted DNA as L-DNA. A previous single-molecule study [13] employed glyoxal, which chemically reacts with unpaired DNA bases, to infer σend ~ –1. The discrepancy may lie in the difference in experimental conditions, such as the significantly lower salt concentration of Ref. [13] which may have disfavored the left-handed wrapping that we observe.
FIG. 2.
Force dependence of elastic parameters of L-DNA. Measurements (black squares) are compared with theory discussed in the main text (red [light gray] lines). The extension data were also fit by the wormlike chain model (blue [dark gray] line).
In addition to helicity, several key elastic parameters of L-DNA may also be determined. For this analysis, data below 5 pN were excluded since secondary structures within L-DNA likely existed at low forces (see below). A fit of L-DNA extension at the end of the melting transition versus force to the wormlike chain model [Fig. 2(b)] yielded a persistence length of ~ 3 nm, much smaller than that of B-DNA, and a contour length of 0.48 nm/bp, 40% longer than B-DNA. Because of significant interdependence of the parameters from the full fit we were not able to reliably determine the L-DNA stretch modulus. The torsional modulus C was calculated from the slope of the torque versus linking number data: , where LL is the contour length of L-DNA and Lk0 is the linking number in relaxed B-DNA, calculated as number of basepairs divided by the helical pitch [Fig. 2(c)]. The torsional modulus for L-DNA was found to be ~ 20 nm at high force, much smaller than that of B-DNA (~ 100 nm) [15, 16, 25].
It is informative to compare our measured elastic parameters of torsionally melted DNA with those of alternate structures that can form upon negative supercoiling (Table 1). Unlike a simple “DNA bubble,” L-DNA possesses a well-defined helicity and is significantly torsionally stiffer. On the other hand, it is easily bent, in contrast to highly rigid base-paired Z-DNA. In addition, based on recent data [28], the torque during the B- to Z-DNA transition is estimated to be –5 pN nm, a factor of 2 smaller in magnitude from the measured average melting torque (–10 pN nm). Based on these differences, we conclude that torsionally melted DNA under substantial tension (> 5 pN) likely consists predominantly of a single form (L-DNA), while other sequence-dependent structures (such as Z-DNA) make only a minor contribution to the overall content of underwound DNA.
Table 1.
Comparison of main elastic parameters of L-DNA with those of B-DNA, DNA “bubble,” and Z-DNA.
Recently a phenomenological model of DNA phase transitions under force and torque has been proposed [23], specifically focusing on supercoiling and melting transitions. The theory has been shown to agree well with our previous experiments on positively supercoiled DNA [16]. Here we examine the applicability of the theory to the melting transition, using a similar framework with small modifications, as described below (see also [24]).
For a pure state (i) of DNA structure, the free energy may be expressed using a harmonic approximation:
| (1) |
where εi is an energy offset and σ0, i is the relaxed degree of supercoiling for the state. Assuming the DNA in each state behaves as a semiflexible polymer with twist compliance, stretching energies gi (f) and torsional coefficients ci (f) as a function of force f can be approximated [23, 29]:
| (2) |
| (3) |
where Ai is persistence length, Ci is torsional modulus, Ki is stretch modulus (assumed to be infinity for L-DNA), bi is contour length ratio to that of B-DNA, and ω0 is the natural twist of B-DNA. Following Ref. [23], we analytically expressed the linking numbers at the onset and the end of a transition to a new phase as well as the extension and torque throughout this process [24]. We then performed a global fit of the theory to our data for forces above 5 pN with elastic parameters of L-DNA as fit parameters and those of B-DNA as known parameters. Fitting agreed reasonably well with the experimental data (red lines in Fig. 2 and Fig. S1), yielding elastic parameters of L-DNA in good agreement with those obtained above, as shown in Table S1.
Both thermal- and torsion-induced DNA melting transitions have been shown to occur in a sequence-dependent manner [30-32]. However, little is known about the effects of DNA sequence on the elasticity of melted DNA. We investigated this on the single-molecule level by repeating the experiments described above using two additional constructs, Sequence B and Sequence C, that were derived from two entirely different plasmids and differ from the original Sequence A in GC content and overall sequence.
Surprisingly, while the torque signal during the melting transition was only marginally affected by sequence or GC content (data not shown), the extension change varied considerably in a force-dependent manner. Figure 3(a) depicts individual traces for each construct at a low (3.5 pN) and a high (12 pN) force for the three sequences. Figure 3(b) presents the reversibility data for Sequence B. Extension traces for the different sequences coincide at high forces, and unwinding and rewinding traces are fully reversible, indicating quasiequilibrium measurements. In contrast, significant differences in extension traces were found at low forces, even for sequences with nearly identical overall GC-content. In addition, the extension traces for unwinding and rewinding no longer coincided and thus the process was no longer reversible, indicating the breakdown of the quasiequilibrium condition. This trend became more prominent as force decreased. Based on these observations, we speculate that this sequence-dependent behavior at low forces may be attributable to the formation of secondary structures in L-DNA.
FIG. 3.
Sequence dependence of melted DNA. Representative extension traces of single DNA molecules are shown for different DNA sequences. (a) Sequence dependence. Extension behavior is identical for all three sequences at 12 pN (upper traces) and differs at 3.5 pN (lower traces). (b) Reversibility illustrated using Sequence B. Extension behavior is reversible at high force, and shows hysteresis at low force.
In summary, our single-molecule experiments suggest a structural and mechanical model of torsionally melted DNA under tension > 5 pN. It is a left-handed structure (L-DNA) with a helicity of ~ −13 bp/turn. Its contour length is ~40% longer than B-DNA. L-DNA is quite flexible in response to bending, with a persistence length of ~3 nm, but has a relatively large effective torsional modulus of ~20 nm (at high force). We find that the B-DNA to L-DNA transition and L-DNA unwinding can be well described by a simple phase-transition model. At lower forces (< 5 pN), L-DNA adopts a more compacted configuration whose properties depend strongly on the DNA sequence. This behavior could be attributed to the formation of secondary structures in L-DNA. Indeed, the plateaulike force-extension curves (Fig. S2) are reminiscent of those for ssDNA [33] and P-DNA [13], which form hairpins and supercoiled P-DNA respectively.
Our data on phase transitions in underwound DNA are summarized in Fig. S3, and present a complexity exceeding that found by earlier studies [14, 15]. The range of forces explored here is well within that exerted by DNA-based motor proteins [34, 35], some of which are also known to generate negative supercoiling. It is therefore possible for L-DNA to occur in vivo, where it may serve as a recognition target for regulatory factors. In addition to a detailed study of L-DNA, we have also discovered another DNA phase which appears upon further underwinding of L-DNA. The nature of this phase transition is outside the scope of the present work and requires further investigation. The data may be interpreted as the formation of supercoiled L-DNA (by analogy with supercoiled B-DNA). However, the rate of extension decrease during that phase transition is slower than during L-DNA twisting, and opposite to expectations based on existing data for supercoil formation. An alternative hypothesis can be formulated, based on an early work on DNA torsional melting [13], where it was found that upon overwinding at sufficiently high force DNA bases flip to the outside of the phosphate backbone, forming a new structure named PDNA. Molecular simulations by the same group have shown that a similar structure can be formed upon underwinding. It might thus be possible that the new phase we observed is akin to left-handed P-DNA — “LP-DNA.”
Our current work also extends theoretical understanding of torsionally melted DNA, from previous analytical [23, 36] and computational [37, 38] studies. Further research is needed to elucidate the precise effect sequence has on the structure and properties of L-DNA, as torsionally induced in vivo melting tends to be localized to specific sequences. Additionally, a comparative study between P-DNA and LP-DNA can undoubtedly add more colors to the rich palette of the physics of DNA phase transitions.
We would like to thank J. Sethna for stimulating theoretical discussions, Y.-F. Chang for the gift of pCR-PepA plasmid, Y.-C. Chang and Y.-F. Chang for the help with construct design, J. Cheng for inspiring discussions, and members of the Wang lab for critical reading of the manuscript. We acknowledge support of the NIH (Grants No. GM059849 to M.D.W. and No. U54CA143869-01 to J.F.M.), the NSF (Grants No. MCB-0820293 to M.D.W., No. DMR-0715099 and No. MCB-1022117 to J.F.M.), and the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust (J.F.M.).
Supplementary Material
References
- 1.David MMC, Nelson L. Lehninger Principles of Biochemistry. W. H. Freeman and Company; New York, NY: 2005. [Google Scholar]
- 2.Kouzine F, Levens D. Front Biosci. 2007;12:4409. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 3.Vinograd J, et al. Proc Natl Acad Sci U S A. 1965;53:1104. [Google Scholar]
- 4.Liu Y, West SC. Nat Rev Mol Cell Biol. 2004;5:937. doi: 10.1038/nrm1502. [DOI] [PubMed] [Google Scholar]
- 5.Jacob RJ, Lebowitz J, Printz MP. Nucleic Acids Res. 1974;1:549. doi: 10.1093/nar/1.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker TA, et al. Cell. 1986;45:53. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- 7.Siebenlist U. Nature. 1979;279:651. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- 8.Umek RM, Kowalski D. Cell. 1988;52:559. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis ML. DNA Replication in Eukaryotic Cells. Cold Springs Harbor Press; Cold Spring Harbor, NY: 1996. p. 1058. [Google Scholar]
- 10.Kahn JD, Yun E, Crothers DM. Nature. 1994;368:163. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang MD, et al. Biophys J. 1997;72:1335. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strick TR, et al. Science. 1996;271:1835. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 13.Allemand JF, et al. Proc Natl Acad Sci U S A. 1998;95:14152. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leger JF, et al. Physical Review Letters. 1999;83:1066. [Google Scholar]
- 15.Bryant Z, et al. Nature. 2003;424:338. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 16.Forth S, et al. Phys Rev Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheinin MY, Wang MD. Phys Chem Chem Phys. 2009;11:4800. doi: 10.1039/b901646e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celedon A, et al. Nano Lett. 2009;9:1720. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkar A, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;63:051903. doi: 10.1103/PhysRevE.63.051903. [DOI] [PubMed] [Google Scholar]
- 20.La Porta A, Wang MD. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 21.Deufel C, et al. Nat Methods. 2007;4:223. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 22.Inman J, Forth S, Wang MD. Opt Lett. 2010;35:2949. doi: 10.1364/OL.35.002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marko JF. Physical Review E. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 24.see Supplemental Material at http://link.aps.org/supplemental/10.1103/PhysRevLett.107.108102 for supplemental experimental methods, theoretical fitting results, and DNA force-torque phase diagram.
- 25.Lipfert J, et al. Nat Methods. 2010;7:977. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 26.Strick TR, et al. Biophys J. 1998;74:2016. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas TJ, Bloomfield VA. Nucleic Acids Res. 1983;11:1919. doi: 10.1093/nar/11.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee M, Kim SH, Hong SC. Proc Natl Acad Sci U S A. 2010;107:4985. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moroz JD, Nelson P. Proc Natl Acad Sci U S A. 1997;94:14418. doi: 10.1073/pnas.94.26.14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalski D, Eddy MJ. EMBO J. 1989;8:4335. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strick TR, Croquette V, Bensimon D. Proc Natl Acad Sci U S A. 1998;95:10579. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Nucleic Acids Res. 2006;34:564. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessinges MN, et al. Phys Rev Lett. 2002;89:248102. doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- 34.Wang MD, et al. Science. 1998;282:902. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 35.Smith DE, et al. Nature. 2001;413:748. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 36.Benham CJ. Proc Natl Acad Sci U S A. 1979;76:3870. doi: 10.1073/pnas.76.8.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wereszczynski J, Andricioaei I. Proc Natl Acad Sci U S A. 2006;103:16200. doi: 10.1073/pnas.0603850103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randall GL, Zechiedrich L, Pettitt BM. Nucleic Acids Res. 2009;37:5568. doi: 10.1093/nar/gkp556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.