Abstract
Pregnane X receptor (PXR), acting as a xenobiotic-activated transcription factor, regulates the hepatic metabolism of therapeutics as well as endobiotics such as steroid hormones. Given our finding that PXR activation by rifampicin (RIF) represses the estrogen sulfotransferase (SULT1E1) gene in human primary hepatocytes and hepatocellular carcinoma Huh7 cells, here we have investigated the molecular mechanism of this repression. First the PXR-responsive enhancer was delineated to a 100 bp sequence (−1000/−901), which contains three half sites that constitute the overlapping direct repeat 1 (DR1) and direct repeat 2 (DR2) motifs and two forkhead factor binding sites. siRNA knockdown, chromatin immunoprecipitation and chromatin conformation capture assays were employed to demonstrate that hepatocyte nuclear factor 4α (HNF4α) bound to the PXR-responsive enhancer, and activated the enhancer by looping its position close to the proximal promoter. Upon activation by RIF, PXR indirectly interacted with the enhancer, decreasing the interaction with HNF4α and dissolving the looped SULT1E1 promoter with deacetylation of histone 3. Removal of the DR sites from the enhancer hampers the ability of HNF4α to loop the promoter and that of PXR to repress the promoter activity. Thus, PXR represses human SULT1E1, possibly attenuating the inactivation of estrogen.
INTRODUCTION
Pregnane X receptor (PXR), an orphan member of the nuclear steroid/thyroid receptor super-family, was first characterized as an activator of transcription of the CYP3A4 gene in response to various drugs and steroid metabolites (1). PXR has now been established as a key transcription factor that induces the hepatic capacity for xenobiotic metabolism and excretion by activating a group of genes that encode xenobiotic-metabolizing enzymes and transporters. In addition, PXR has recently been reported to regulate p38 mitogen-activated protein kinase-mediated cell migration signals by activating the GADD45β gene (2). The general mechanism underlying these activations is a direct binding of PXR to the response enhancers of its target genes. PXR has also been demonstrated to repress transcription of hepatic genes such as PCK1, G6Pase and CYP7A1, thereby regulating energy and cholesterol homeostasis (3–5). The repression of these genes appears to be regulated through interactions of PXR with other transcription factors and co-activators such as FOXO1, CREB and PGC1α, but not by direct DNA binding. However, the molecular mechanism by which PXR represses transcription remains elusive at the present time. Utilizing human estrogen sulfotransferase (SULT1E1) gene as a model, here we have investigated the molecular mechanism of PXR-mediated repression of gene expression.
Estrogen levels can be regulated by altering biosynthesis by aromatase, metabolic inactivation by sulfotransferase and re-activation by sulfatase. Although the actions of estrogen and the development of estrogen-related diseases by effects on aromatase are well established, estrogen metabolism is also involved in the regulation of estrogen activity. SULT1E1 exhibits the highest affinity for estrogens among known sulfotransferases and is the primary enzyme responsible for sulfation of β-estradiol and estrone. SULT1E1 is expressed at low levels in the livers of normal mice and is highly expressed in livers of diabetogenic db/db mice (6). Expression of SULT1E1 in adipose tissue of transgenic female mice revealed that SULT1E1 activity regulates fat and glucose homeostasis (7). Sult1e1−/− female mice were reported to develop placental thrombosis and spontaneous fetal loss (8). High estrogen sulfatase (STS) activity has been detected in tumor tissues of breast cancer patients and was correlated with a poor prognosis for breast cancer in women (9,10). Drugs and steroid hormones are known to induce SULT1E1 in mouse liver via activation of nuclear receptors such as constitutive active/androstane receptor (CAR), liver X receptor and glucocorticoid receptor (11–13).
In this present study, given that the SULT1E1 gene was repressed by rifampicin (RIF) in human primary hepatocytes, we utilized human hepatocellular carcinoma Huh7 and HepG2 cells to investigate the molecular mechanism of the PXR-mediated repression of the SULT1E1 gene.
First the repressive activity was delineated to a 100 bp distal enhancer within the SULT1E1 promoter. Subsequently, chromatin immunoprecipitation (ChIP) and chromatin conformation capture (3 C) assays were employed to identify hepatocyte nuclear factor 4α (HNF4α) and its binding site within the 100 bp enhancer as the target of PXR to repress the SULT1E1 gene. Here, we present these experimental considerations to suggest a novel mechanism, in which a drug-activated PXR cross-talks with HNF4α, disrupting an active chromatin structure, and thereby repressing transcription of the SULT1E1 gene.
EXPERIMENTAL PROCEDURES
Materials
RIF, SR12813, cycloheximide (CHX), TCPOBOP, anti-FLAG-M2 HRP-conjugated antibody and FLAG-M2 agarose were purchased from Sigma-Aldrich (St Louis, MO, USA); CITCO from BioMol (Plymouth Meeting, PA, USA); restriction endonucleases and DNA-modifying enzymes from New England Biolabs, Inc. (Beverly, MA, USA); antibody to human PXR from Perseus Proteomics Inc. (Tokyo, Japan); mouse, goat and rabbit normal IgGs, antibodies against HNF3β (M-20), HNF4α (H-171) and β-actin (C4) from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-V5 HRP-conjugated antibody from Invitrogen (Carlsbad, CA, USA); anti-H3K9K14ac (06–599) from Millipore (Billerica, MA, USA); [32P]dATP from GE Healthcare (Piscataway, NJ, USA) and [35S]methionine from PerkinElmer (Waltham, MA, USA). ON-TARGETplus SMART pool HNF4α (catalog number L-003406-00-0005), HNF3β (catalog number L-010089-00-0005), PGC1α (catalog number L-005111-00-0005) and ON-TARGETplus siCONTROL non-targeting pool (catalog number D-001810-10) were purchased from Dharmacon Research (Lafayette, CO, USA). All primers, oligonucleotides and real-time PCR probes used for present experiments are listed in Supplementary Table S1–S4.
Expression plasmids and adenovirus
In all vectors, the prefixes h and m in front of inserts denote human and mouse, respectively. pCR3/hPXR, pCR3/FLAGhPXR, pcDNA3.1/hHNF4α, XREM-3A4-Luc, pCR3/mCART176V, pGL3/1.8 kbp-2B6-Luc, adeno-hPXR and adeno-β-gal have been described previously (4,14–16). cDNA encoding the full-length human HNF3β was amplified with appropriate PCR primers and cloned into pcDNA3.1/V5-His-TOPO (Invitrogen) to yield pcDNA3.1/hHNF3β.
Cloning of human SULT1E1 promoter
The 5.3 and 1.1 kb DNA fragments containing the 5′-flanking region of human SULT1E1 gene were amplified from human genomic DNA (Promega, Madison, WI, USA) using appropriate pairs of PCR primers. Amplified DNA fragments were cloned into KpnI/XhoI-digested pGL3-Basic vector (Promega) to yield pGL3/5.3 kb-hSULT1E1 and pGL3/1.1 kb-hSULT1E1, respectively. The 100-bp enhancer region was amplified from pGL3/5.3 kb-hSULT1E1 using appropriate pairs of PCR primers and cloned into KpnI/XhoI-digested pGL3-Basic vector containing a 160 bp thymidine kinase promoter to yield pGL3/hSULT1E1enhancer-TK (17). Each of the mutant promoter constructs was produced by site-directed mutagenesis using appropriate oligonucleotides. To construct Luc reporter constructs for 3 C assays, a double-stranded oligonucleotide 5′-GATCACTAGCAAAATAGGCTGTCCCCAGA-3′ encoding RV primer 3 was inserted into the BglII-digested site of each Luc reporter constructs, and then the original sequence of RV primer 3 in the pGL3-Basic was deleted by site-directed mutagenesis.
Cell culture, drug treatment, transfection and infection
Human hepatocellular carcinoma Huh7 cells were maintained in minimal essential medium (MEM) supplemented with 10% FBS, 2 mM l-glutamine, penicillin (100 U/ml) and streptomycin (100 µg/ml) in an atmosphere of 5% CO2 at 37°C. For Luc reporter assays, Huh7 cells were transiently transfected with Firefly luciferase reporter construct along with pCR3/hPXR or pCR3/mCART176V, using FuGene 6 (Roche Applied sciences, Indianapolis, IN, USA). pRL-TK (Promega) was co-transfected as control. The final amounts of transfected DNAs were adjusted to constant levels by adding pCR3 (Invitrogen) empty vector. Twenty-four hours after transfection, the cells were treated with a given drug in FBS-free MEM for an additional 24 h and luciferase reporter assays were performed as previously described (3). For adenoviral infection, cells were cultured in MEM medium containing adeno-β-gal or adeno-hPXR at a multiplicity of infection of 10. After incubation for 30 h, these cells were treated with either RIF (10 µM) or SR12813 (3 µM) in FBS-free MEM for a given time. For siRNA knockdown, trypsinized cells were reverse transfected with siRNA (50 pmol) in MEM medium, using Lipofectamine 2000 (Invitrogen). After incubation for 24 h, culture medium was replaced with FBS-free MEM and cells were incubated for another 16 h. For real-time PCR, trypsinized Huh7 cells were reverse transfected with pCR3 or pCR3/mCART176V using FuGene6. After 30 h, cells were treated with TCPOBOP (250 nM) for 24 h. Human primary heptocytes were kindly provided by Dr Stephen S. Ferguson (Life Technologies Corporation, Durham, NC, USA). Hepatocytes were treated with RIF (10 µM) or CITCO (250 nM) in William's E medium supplemented with 2 mM l-glutamine, penicillin (100 U/ml) and streptomycin (100 µg/ml) for 24 h.
Real-time PCR
Real-time PCR was performed using an ABI prism 7700 sequence detection system (Applied Biosystems, Foster City, CA, USA). Assays-on-Demand probes (Applied Biosystems) were used for PCR with the TaqMAN PCR Master Mix (Applied Biosystems). To measure the expression of human STS and CYP7A1 genes, SYBR Green PCR Master Mix (Applied Biosystems) was used with specific primers. The TaqMAN human β-actin probe (Applied Biosystems) was used as an internal control.
Gel shift assays
cDNAs for hHNF4α and hHNF3β in pcDNA3.1-V5-His-TOPO vector were in vitro translated using the TNT-coupled reticulocyte lysate system (Promega) according to the manufacture's instructions. Gel shift assays were performed as described previously (18). For competition or antibody supershift assays, unlabeled oligonucleotide, normal IgG, anti-HNF4α or anti-HNF3β antibodies was added 15 min before adding radioactive oligonucleotides.
GST pull-down assays
Recombinant GST and GST-fusion proteins of hPXR and hHNF4α were expressed in Escherichia coli BL21 (DE3) cells, and were purified through glutathione-Sepharose. A series of cDNAs for hPXR deletions and hHNF4α in pCR3 or pcDNA3.1-V5-His-TOPO vector were in vitro translated in the presence of [35S] methionine using a TNT-coupled reticulocyte lysate system (Promega) according to the manufacture's instructions. GST pull-down assays were carried out as described previously (19).
ChIP assays
ChIP assays were performed using a ChIP assay kit (Millipore) according to the manufacture's instruction. Thirty hours after infection with adenovirus or reverse transfection, cells were treated with RIF in FBS-free MEM for an another 2 or 6 h, and then cross-linked by adding formaldehyde [final 1% (v/v)] for 10 min at room temperature. Cells were lysed in SDS lysis buffer containing protease inhibitor cocktail (Roche Applied Science) and sonicated to shear chromatin to lengths between 200 and 600 bp. After pre-clearing by shaking with Protein A or G agarose/salmon sperm DNA, cell lysates were incubated with 3 µg of antibodies or normal IgG at 4°C for 16 h. After incubation with Protein A or G agarose/salmon sperm DNA, the immunocomplexes were collected by centrifugation. After de-crosslinking and protease-digestion, the immunoprecipitated DNA was purified by following the manufacture's instructions combined with the QIAquick PCR purification kit (Qiagen). The purified DNA was used as a template for semi-quantitative and real-time PCRs with specific primers.
3C assays
3C assays were performed as previously described with minor modifications (20,21). Thirty hours after infection with adenovirus, Huh7 cells were treated with RIF in FBS-free MEM for another 6 h. For 3 C assays with Luc reporter constructs, trypsinized Huh7 cells were reverse transfected with each construct, using FuGene 6. Twenty-four hours after transfection, the culture medium was replaced with FBS-free MEM and the cells were incubated for another 2 h. Cells were cross-linked by adding formaldehyde [final 1% (v/v)] for 10 min at room temperature, and the reaction was stopped by adding glycine (final 125 mM). After washing with ice-cold PBS, cells were incubated in a lysis buffer [10 mM Tris, pH 7.5, 10 mM NaCl, 0.2% (v/v) NP-40] and protease inhibitor cocktail on ice for 15 min, followed by homogenization in a dounce homogenizer. After centrifugation, nuclei were collected as a pellet and suspended in restriction enzyme buffer 2 containing 0.3% (w/v) SDS. After incubation at 37°C for 1 h, Triton X-100 was added to 1.8% (v/v) final concentration, followed by incubation for another 1 h at 37°C. Aliquots of 1.5 × 106 nuclei were incubated with 500 U of BstYI at 37°C for 16 h. The enzyme reaction was terminated by adding SDS to 1.6% (v/v) and incubated for 20 min at 65°C. The digestion efficiency was confirmed by gel electrophoresis on a 2.0%, agarose gel. An aliquot of digested chromatin was diluted with T4 DNA ligase buffer to 1% (v/v) final concentration Triton X-100 and 2.5 ng/μl of DNA and incubated at 37°C for 1 h. The DNAs were ligated by using 1600 cohesive end units of T4 ligase for 4 h at 16°C followed by 30 min at room temperature. Proteinase K, NaCl and EDTA were added to the ligation mixture to final concentrations of 40 µg/ml, 0.2 M and 1 mM, respectively. These mixtures were incubated at 65°C to reverse cross-linking. After 16 h incubation, mixtures were treated with RNase A at 37°C for 30 min, and the DNA was purified by phenol extraction and ethanol precipitation. The purified DNAs were re-suspended in water at 50 ng/µl, and subjected to semi-quantitative and real-time PCR of ligated and control fragments using specific pairs of primers.
Immunoprecipitation assays
Trypsinized cells were reverse transfected with pCR3/FLAGhPXR for 24 h, and were subsequently treated with 0.1% (v/v) DMSO or RIF for another 2 h. Then, cells were lysed in cold immunoprecipitation buffer [1% (v/v) Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EDTA, 1 mM EGTA, 0.2 mM sodium ortho-vanadate, 0.2 mM PMSF, 0.5% (v/v) NP-40 and 0.1% (v/v) DMSO or 20 µM RIF] containing phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich) at 4°C for 25 min, and were centrifuged at 16 000g for 15 min to prepare whole cell lysates. For co-immunoprecipitation assays, FLAG-M2 agarose was incubated with cell lysates at 4°C for 2 h, washed with immunoprecipitation buffer and subjected to Western blotting with anti-FLAG-M2 HRP-conjugated antibody or anti-V5 HRP-conjugated antibody.
Statistical analysis
All data are presented as mean ± SD. Analysis was performed by using Student's t-test. A statistical probability of P < 0.05 was considered significant.
RESULTS
PXR repression of the human SULT1E1 gene
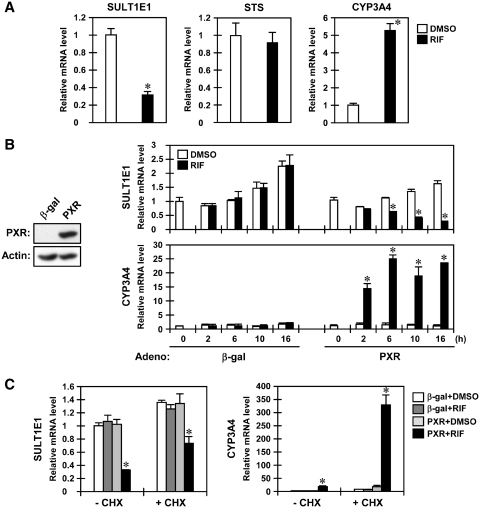
Real-time PCR, performed using RNAs prepared from human primary hepatocytes treated for 24 h with the human PXR activator RIF, determined that SULT1E1 mRNA levels decreased to approximately one-third of those in DMSO-treated hepatocytes after RIF treatment, while STS mRNA showed no change (Figure 1A). The CYP3A4 mRNA was greatly increased as expected. Repression of the SULT1E1 gene by RIF was also observed in human hepatocellular carcinoma Huh7 cells in which PXR was ectopically expressed through infection of an adenovirus. RIF treatment decreased the levels of SULT1E1 mRNA in a time-dependent manner in Huh7 cells infected with adeno-hPXR, but not with adeno-β-gal; the decrease began 6 h after treatment and continued to an ~85% decrease after 16 h (Figure 1B). Levels of STS mRNA, on the other hand, were maintained in Huh7 cells even after RIF treatment (Supplementary Figure S1A). These PXR-mediated expression patterns were conserved in HepG2 cells (Supplementary Figure S1B). Compared to the rapid increase in CYP3A4 mRNA, which began 2 h after RIF treatment and peaked at the 6 h mark, the gradual and slow decrease in SULT1E1 mRNA levels prompted us to examine whether or not protein synthesis was involved in this repression. Co-treatment with cycloheximide that inhibits translation did not block this decrease of SULT1E1 mRNA levels, thus indicating that protein synthesis is not required for PXR to repress transcription of the SULT1E1 gene (Figure 1C). Similarly, cycloheximide did not inhibit RIF induction of the CYP3A4 mRNA.
Figure 1.
PXR represses the SULT1E1 gene in human primary hepatocytes and Huh7 cells. (A) Human primary hepatocytes were treated with RIF for 24 h in William's E medium, from which total RNAs were prepared and subjected to real-time PCR. The levels of the SULT1E1, STS and CYP3A4 mRNAs are expressed by taking those in the DMSO-treated cells as one. *P < 0.05 (DMSO versus RIF). (B and C) After 30 h infection with adeno-β-gal or adeno-hPXR, cells were treated with RIF for indicated time in FBS-free MEM (B) or were pre-treated with CHX (10 µg/ml) for 2 h, and then co-treated with RIF for another 16 h (C). Total RNAs were prepared at the indicated time points and subjected to real-time PCR. The levels of the SULT1E1, STS and CYP3A4 mRNAs are expressed taking those in the DMSO-treated Huh7 cells infected with adeno-β-gal as one. Bars represent the mean ± SD. *P < 0.05 (PXR + DMSO versus PXR + RIF).
The PXR response enhancer within the SULT1E1 gene
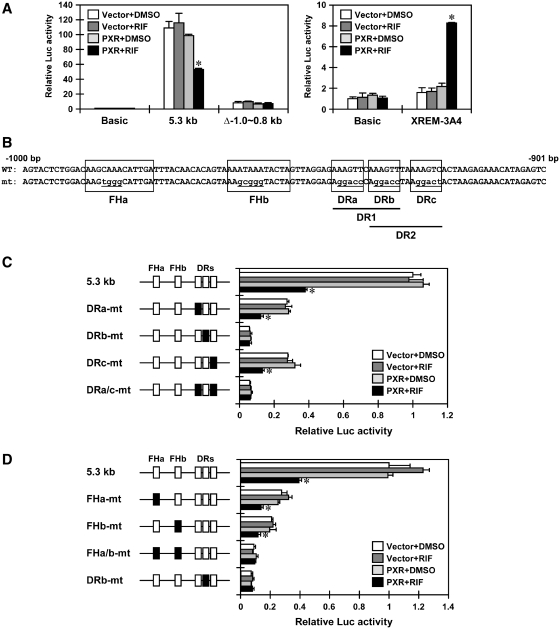
First, we found that the 5.3 kb SULT1E1 promoter-Luc reporter was repressed ~50% by RIF treatment in PXR-expressing Huh7 cells (Figure 2A). Given the fact that the internal deletion (Δ−1.0/−0.8 kb) abolished this repression (Figure 2A), further deletion analysis delineated the PXR-mediated repression activity to a distal 100 bp composite enhancer sequence (−1000/−901 bp) (Figure 2B). This enhancer sequence contains three direct repeat (DR) half sites that constitute the overlapping DR1 and DR2 motifs and two forkhead protein binding sites FHa and FHb. To determine the role of these half sites, they were mutated singularly or simultaneously within the context of the 5.3 kb SULT1E1 promoter. Mutation of DRb (DRb-mt) decreased activity of the promoter and eliminated its ability to respond to PXR, thereby repressing activity of the reporter (Figure 2C). While single mutation of either DRa or DRc fully retained PXR-mediated repression of the promoter, double mutations of both DRa and DRc abolished this repression. Thus, these results indicate that the DRb plays a central role in the repression of the promoter activity. FHa and FHb sites were also mutated to examine their role in repression of activity of the 5.3 kb SULT1E1 promoter; only the double mutation abolished the response of the promoter to PXR (Figure 2D). Taken together, these results lead us to conclude that it is the DRb site that plays the most critical role in the PXR-mediated repression of the the 5.3 kb SULT1E1 promoter, although the two FH sites may also be involved in this repression.
Figure 2.
PXR represses the SULT1E1 promoter via the 100 bp distal enhancer. (A) Huh7 cells were transiently transfected with the pGL3-Basic, pGL3/5.3 kb-hSULT1E1, pGL3/5.3 kb-hSULT1E1del−1.0 ~ −0.8 kb or XREM-CYP3A4-Luc reporter constructs along with pCR3 or pCR3/hPXR for 24 h and then treated with RIF or DMSO for another 24 h in FBS-free MEM. pRL-TK was also included in all transfections as a control. Relative Luc activity was calculated by taking the activity of cells co-transfected with pCR3 and pGL3-Basic and treated with DMSO as one. *P < 0.05 (PXR + DMSO versus PXR + RIF). (B) Schematic diagram of a distal enhancer of the SULT1E1 promoter. (C and D) Huh7 cells were transiently transfected with the pGL3/5.3 kb-hSULT1E1 or its indicated mutant constructs along with pCR3 or pCR3/hPXR for 24 h and then treated with RIF or DMSO for another 24 h in FBS-free MEM. pRL-TK was also included in all transfections as a control. Relative Luc activity was calculated by taking the activity of cells co-transfected with pCR3 and pGL3/5.3 kb-hSULT1E1 and treated with DMSO as one. Bars represent the mean ± SD. *P < 0.05 (PXR + DMSO versus PXR + RIF).
Roles of HNF4α and HNF3β in regulation of the 100 bp enhancer
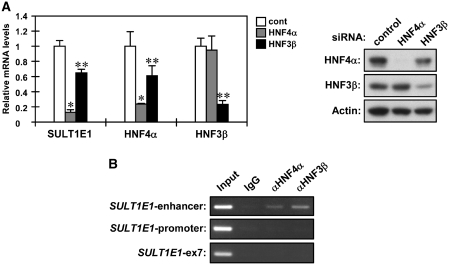
Subsequent to the confirmation by gel shift assays that HNF4α and HNF3β bind to the DR and FH sites, respectively (Supplementary Figure S2), we employed siRNAs to knockdown HNF4α and HNF3β in order to examine their roles in regulating the SULT1E1 gene in Huh7 cells. Both knockdowns were found to decrease SULT1E1 mRNA levels (Figure 3A). Knockdown of HNF4α was the most effective in decreasing SULT1E1 mRNA, and reduced the mRNA levels by 90%. On the other hand, knockdown of HNF3β only reduced levels of SULT1E1 mRNA by ~40%. The HNF3β knockdown also resulted in a small reduction of HNF4α, which is consistent with the fact that HNF3β is one of the transcription factors that activates the HNF4α gene (22). To examine interactions of the 100 bp enhancer with HNF4α and HNF3β, ChIP assays were performed for the SULT1E1 gene in Huh7 cells. Both HNF4α and HNF3β bound to the enhancer region of the SULT1E1 promoter, but not to the proximal promoter region (Figure 3B). These results indicate that HNF4α may play the most critical role in enhancer activation, although HNF3β is also involved in this activation.
Figure 3.
HNF4α is a primary factor regulating the SULT1E1 gene. (A) Huh7 cells were reverse transfected with control, HNF4α or HNF3β siRNAs for 24 h and were subsequently incubated in FBS-free MEM for another 24 h. From these cells, total RNAs and whole cell lysates were prepared and subjected to real-time PCR for SULT1E1, HNF4α and HNF3β mRNAs and western blottings using anti-HNF4α, anti-HNF3β and anti-Actin antibodies. mRNA levels of each gene were expressed by taking those in the Huh7 cells reverse transfected with control siRNA as one. Bars represent the mean ± SD. *P < 0.05 (control siRNA versus HNF4α siRNA). **P < 0.05 (control siRNA versus HNF3β siRNA). (B) Huh7 cells were subjected to ChIP assays using normal IgG, anti-HNF4α antibody or anti-HNF3β antibody as described in ‘Experimental Procedures’ section. From the immunoprecipitated DNA fragments, the enhancer and proximal promoter regions were amplified by PCR using primers specific for the SULT1E1 enhancer and SULT1E1 promoter. A region of exon 7 of the SULT1E1 gene was also amplified as a negative control.
Disruption of the chromatin structure
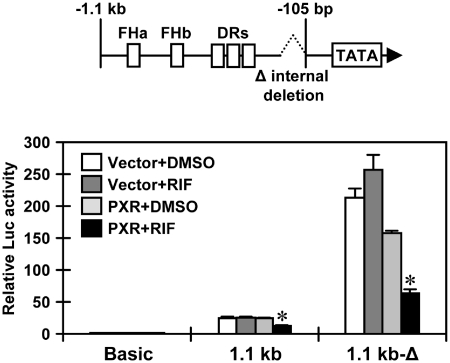
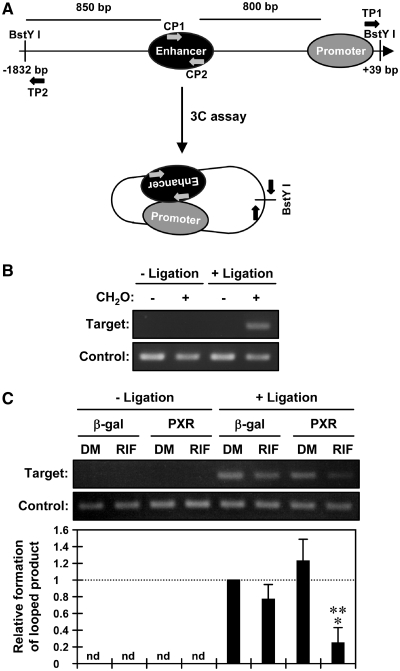
Utilizing deletion promoters, the regions required for repression of activity by PXR were localized in the SULT1E1 promoter. The activity of a −1.1 kb promoter was repressed by PXR, while a −1.1 kb-Δ promoter, which places the enhancer in front of a 105 bp proximal promoter via an internal deletion, greatly enhanced the basal transcription activity and fully retained the repression of activity by PXR (Figure 4). However, the 100 bp enhancer repressed promoter activity when it was placed directly in the front of the TK promoter. Thus, the repressive activity was minimized to the 100 bp enhancer and the −105 bp proximal promoters (Supplementary Figure S3). Noticing that the −1.1 kb-Δ promoter exhibited higher activity than the −1.1 kb promoter, we hypothesized that the SULT1E1 promoter adopts a looped structure in its active conformation. Utilizing the restriction enzyme BstYI to digest the SULT1E1 gene, 3 C assays were designed to detect a 135 bp DNA fragment by PCR amplification upon ligation of the BstYI sites that are located −1832 bp and +39 bp from the transcriptional start site (Figure 5A). Accordingly, this 3 C assay demonstrated that the promoter was looped in its active conformation (Figure 5B). The same 3 C assays were performed on the SULT1E1 gene in PXR-expressing Huh7 cells after RIF treatment; amplification of a 135 bp DNA fragment was detected by both semi-quantitative and real-time PCR. The degree of amplification was strongly attenuated in the presence of PXR after RIF treatment (Figure 5C). Subsequently, ChIP assays were employed on the SULT1E1 gene using an antibody that detects acetylated histone 3 at lysine 9 and lysine 14 (H3K9K14ac); acetylation of histone 3 is known to facilitate activation of gene transcription. H3K9K14ac was reduced on both the enhancer and proximal promoter regions of the SULT1E1 gene in PXR-expressing Huh7 cells after RIF treatment (Figure 6A and B). Thus, the PXR-dependent chromatin structure disruption as observed in our 3 C assays correlated with this deacetylation of histone 3 on the SULT1E1 gene.
Figure 4.
Both the enhancer and proximal promoter are required for the response to PXR. Huh7 cells were transiently transfected with the pGL3-Basic, pGL3/1.1 kb-hSULT1E1 or pGL3/1.1 kb-hSULT1E1Δ along with pCR3 or pCR3/hPXR for 24 h and then treated with RIF or DMSO for another 24 h in FBS-free MEM. pRL-TK was also included in all transfections as a control. Relative Luc activity was calculated by taking the activity of cells co-transfected with pCR3 and pGL3-basic as one. Bars represent the mean ± SD. *P < 0.05 (PXR + DMSO versus PXR + RIF).
Figure 5.
PXR disrupts the enhancer-proximal promoter loop of the SULT1E1 gene. (A) A schematic representation of the 3 C assay of the SULT1E1 gene. Numbers indicates positions relative to the transcriptional start site; arrows indicate positions of PCR primers. (B) After cross-linking by CH2O treatment, nuclei were prepared from Huh7 cells and subjected to 3 C assays as described in ‘Experimental Procedures’ section. From purified DNAs, formation of the 3C ligated product was detected by PCR amplification using TP1 and TP2 primers. Control product was also amplified using CP1 and CP2 primers to verify the quantity and quality of the DNA. (C) Huh7 cells were infected with adeno-β-gal or adeno-hPXR for 30 h, treated with RIF or DMSO (DM) for another 6 h in FBS-free MEM and then subjected to 3 C assays as described in ‘Experimental Procedures’ section. From purified DNAs, formation of the 3 C ligated product was determined by real-time PCR. Values were normalized by amplification of control product and expressed by taking those levels in cells infected with adeno-β-gal as one. Bars represent the mean ± SD. *P < 0.05 (β-gal + RIF versus PXR + RIF); **P < 0.05 (PXR + DMSO versus PXR + RIF).
Figure 6.
PXR induces histone deacetylation of the SULT1E1 gene in Huh7 cells. Huh7 cells were infected with adeno-β-gal or adeno-hPXR for 30 h and treated with RIF or DMSO (DM) for additional 6 h in FBS-free MEM. ChIP assays were performed with normal IgG or an anti-H3K9K14ac antibody using both semi-quantitative PCR (A) and real-time PCR (B). Values are normalized by amplification in sample inputs and expressed by taking those in the DMSO-treated Huh7 cells infected with adeno-β-gal as one. Bars represent the mean ± SD. *P < 0.05 (β-gal + RIF versus PXR + RIF); **P < 0.05 (PXR + DMSO versus PXR + RIF).
HNF4α as a key PXR target for repression of promoter activity
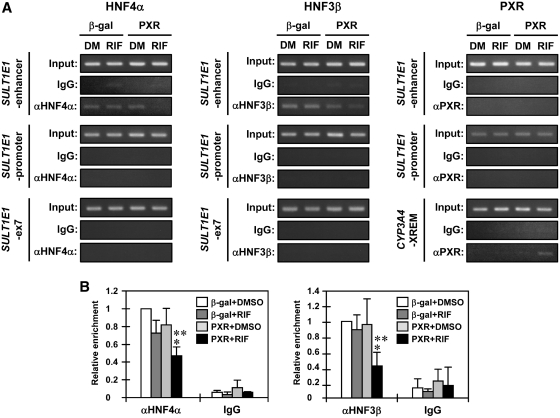
Next, ChIP assays were performed to determine the interactions of HNF4α, HNF3β and PXR with the SULT1E1 promoter. Binding of both HNF4α and HNF3β to the enhancer was attenuated after RIF treatment in PXR-expressing Huh7 cells; no such attenuation was observed with control β-gal-expressing Huh7 cells (Figure 7A and B). HNF4α and HNF3β were not detected on the proximal promoter even after RIF treatment under our assay conditions. There was no PXR binding to either the enhancer or to the proximal promoter before or after RIF treatment (Figure 7A). Thus, PXR appeared to decrease the binding of HNF4α and HNF3β to the enhancer, repressing SULT1E1 promoter activity. To further distinguish the roles of HNF4α and HNF3β, plasmid-based 3 C assays were developed in which the 5.3 kb SULT1E1 promoter plasmid was transfected into Huh7 cells and used as template. BstY1 digestion was designed to produce a 111 bp DNA fragment when the promoter adopted a loop structure (Figure 8A and B). Within the context of the 5.3 kb SULT1E1 promoter, either DRb or both FH sites were mutated and these mutated promoters were subjected to 3 C assays. Mutation of the DRb disrupted the loop structure of the promoter by ~60%, whereas only <10% was decreased after mutation of the FH sites (Figure 8C). This weak contribution by FH sites to the loop structure was consistent with the less effect of HNF3β knockdown on the expression of SULT1E1 gene (Figure 3A).
Figure 7.
PXR prevents HNF4α and HNF3β from binding to the 100 bp enhancer. Huh7 cells were infected with adeno-β-gal or adeno-hPXR for 30 h, treated with RIF or DMSO (DM) for another 2 h in FBS-free MEM and then subjected to ChIP assays using normal IgG, anti-HNF4α antibody, anti-HNF3β antibody or anti-human PXR antibody. (A) From the immunoprecipitated DNA fragments, the SULT1E1 enhancer, SULT1E1 promoter and SULT1E1-ex7 were amplified by PCR. The region containing the DR3 sequence in the CYP3A4 gene was also amplified for CYP3A4-XREM as a positive control for PXR activity. (B) The relative enrichment of the SULT1E1 enhancer in the immunoprecipitated DNA fragments with anti-HNF4α and anti-HNF3β antibodies was determined by real-time PCR. Values are normalized by amplification in sample inputs and expressed by taking those in the DMSO-treated Huh7 cells infected with adeno-β-gal as one. Bars represent the mean ± SD. *P < 0.05 (β-gal + RIF versus PXR + RIF); **P < 0.05 (PXR + DMSO versus PXR + RIF).
Figure 8.
HNF4α determines the chromatin structure of the SULT1E1 promoter. (A) A schematic presentation of ‘plasmid’ 3 C assay of the SULT1E1 gene. Arrows indicate positions of PCR primers. (B) Huh7 cells were reverse transfected with pGL3-BasicVec3C or pGL3/5.3 kb-hSULT1E1Vec3C for 24 h and then treated with DMSO in FBS-free MEM for another 2 h. After cross-linking by CH2O treatment, nuclei were prepared and subjected to ‘plasmid’ 3 C assays as described in ‘Experimental Procedures' section. From purified DNAs, formation of 3 C ligated product was detected by PCR amplification using VecTP1 and VecTP2 primers. Control product was also amplified using VecCP1 and VecCP2 primers to verify the transfection efficiency and quality of the DNAs. (C) Huh7 cells were reverse transfected with pGL3/5.3 kb-hSULT1E1Vec3C, pGL3/5.3 kb-hSULT1E1DRbVec3C or pGL3/5.3 kb-hSULT1E1FHa/bVec3C. The relative looping formation in the purified DNAs was determined by real-time PCR. Values were normalized by amplification of control product and expressed by taking those in cells transfected with pGL3/5.3 kb-hSULT1E1Vec3C as one. Bars represent the mean ± SD. *P < 0.05 (5.3 kb + ligation versus DRb + ligation).
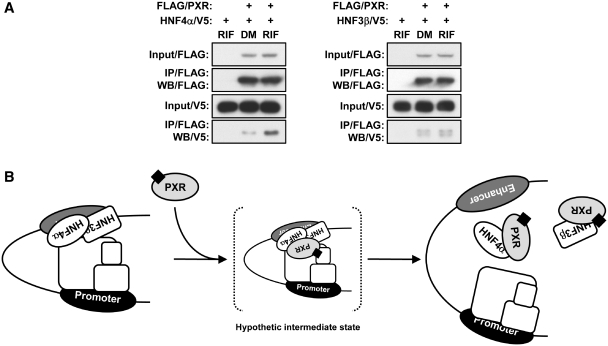
Co-immunoprecipitation assays demonstrated that PXR formed a complex with HNF4α in Huh7 cells, and that formation of this complex formation was increased following RIF treatment (Figure 9A). No such increase was observed with HNF3β. These results, therefore, lead us to conclude that it is the HNF4α that plays the primary role in adopting the SULT1E1 promoter in the active loop formation, and that PXR targets HNF4α to disrupt this loop.
Figure 9.
PXR exhibits a ligand-dependent interaction with HNF4α. (A) Huh7 cells were reverse transfected with indicated expression plasmids for 30 h and then treated with RIF or DMSO (DM) for another 2 h in FBS-free MEM, from which whole cell extracts were prepared and subjected to immunoprecipitation with FLAG-M2 agarose. Immunoprecipitated proteins were detected by western blotting with anti-FLAG-M2 HRP-conjugated antibody or anti-V5 HRP-conjugated antibody. (B) Model of mechanism for the PXR-mediated repression of the SULT1E1 gene.
DISCUSSION
Here, we have identified the upstream enhancer of the SULT1E1 promoter that plays a crucial role in regulating the activity of SULT1E1 transcription. HNF4α binds to the enhancer and acts as a mediator that enables the SULT1E1 promoter to form an active chromatin structure by placing the enhancer close to the proximal promoter, thereby determining the basal rate of transcription. Upon activation by RIF, PXR, through its interactions with HNF4α, prevents the promoter from forming the active chromatin structure, thereby repressing transcription of the SULT1E1 gene. PXR was previously reported to repress the expression of the G6Pase and CYP7A1 genes through its interactions with HNF4α and PGC1α (4,5,19). However, PGC1α does not appear to be involved in the repression of the SULT1E1 gene, since the transcription of the SULT1E1 gene was not affected by siRNA knockdown of PGC1α in Huh7 cells (Supplementary Figure S4). The chromatin structure-mediated repression mechanism for the SULT1E1 gene differs from the previous PGC1α-dependent gene repression by PXR. FOXO1, HNF3β and CREB were also suggested to be PXR targets for the protein–protein interactions, which repress the expression of the G6Pase, CPT1A and HMGCS2 genes, the molecular mechanism for which remains unclear (3,4,23). Thus, SULT1E1 appears to be the first gene whose repression by PXR is now understood at the level of chromatin structure.
PXR interacted with HNF4α in response to RIF activation, suggesting that this interaction can be a critical regulatory step within the underlying mechanism, by which PXR represses the expression of the SULT1E1 gene. Gel Shift assays indicated that PXR does not directly bind to any DNA sequence within the 100 bp enhancer (data not shown). Therefore, if in fact PXR interacted with the 100 bp enhancer, this interaction would have occurred via protein–protein interactions. However, our ChIP assays did not confirm that PXR interacted with either the enhancer or the proximal promoter of the SULT1E1 gene in RIF-treated Huh7 cells. Several reasons for this can be speculated, one of which is that the interaction of PXR with HNF4α on the SULT1E1 gene is transient, so that ability of ChIP assay to detect this transient interaction may be limited (Figure 9B). Although GST pull-down assays determined that PXR and HNF4α bind through their DNA binding domains, gel shift assays found that PXR does not inhibit binding of HNF4α to the overlapping DR1 and DR2 motifs within the 100 bp enhancer (Supplementary Figures S5 and S6). Therefore, PXR does not appear to just simply prevent the binding of HNF4α. In this respect, HNF4α activity can be regulated by post-translational modifications such as acetylation and phosphorylation within its DNA binding domain and hinge region (24–26). PXR may also utilize these phosphatases and deacetylases to repress the HNF4α-mediated activation of the SULT1E1 gene, which presents an interesting possibility for future investigations to decipher the details of this repression mechanism.
The liver is an estrogen-targeted organ, although whether or not hepatic estrogen metabolism critically regulates liver function in an autocrine and or paracrine fashion remains to be further explored. Estrogen receptor α (ERα) KO mice developed hepatic insulin resistance by up-regulating lipogenic enzymes in the liver (27). Protein S is a major risk factor for developing thrombosis and the liver is the site of its production. ERα activation is known to repress the protein S gene in mouse liver (28). Estrogens regulate hepatic bile acid synthesis and a sustained activation of ERα by 17α-ethynylestradiol produces hepatotoxicity in mice (29). Utilizing Pxr−/− mice, we recently determined that PXR represses the hepatic Sult1e1 gene during fasting (data not shown). CAR, a closely related nuclear xenobiotic-activated receptor, was reported to activate the SULT1E1 gene in mouse liver (11). In human primary hepatocytes, on the other hand, drug activation of CAR resulted in repression of the SULT1E1 gene (30) (Supplementary Figure S7). Moreover, similar to PXR, CAR repressed the 5.3 kb SULT1E1 promoter and the expression of the endogenous SULT1E1 gene in Huh7 cells (Supplementary Figure S8). In addition to SULT1E1, PXR and CAR can also regulate the expression of enzymes that metabolize estrogens, such as CYP3A4, following activation by an overlapping group of xenobiotics and therapeutics. Thus, PXR and CAR may co-operate in regulating hepatic levels of active estrogens, thereby affecting the physiology and/or pathophysiology of human liver.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramutal Research Program of the National Institutes of Health and National Institute of Environmental Health Sciences: Z01ES1005-01. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the NIEHS sequence core for excellent assistance. We also thank Mr Rick Moore at NIEHS for technical assistance.
REFERENCES
- 1.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 2.Kodama S, Negishi M. Pregnane X receptor PXR activates the GADD45β gene, eliciting the p38 MAPK signal and cell migration. J. Biol. Chem. 2011;286:3570–3578. doi: 10.1074/jbc.M110.179812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodama S, Koike C, Negishi M, Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol. Cell. Biol. 2004;24:7931–7940. doi: 10.1128/MCB.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodama S, Moore R, Yamamoto Y, Negishi M. Human nuclear pregnane X receptor cross-talk with CREB to repress cAMP activation of the glucose-6-phosphatase gene. Biochem. J. 2007;407:373–381. doi: 10.1042/BJ20070481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1α. Functional implications in hepatic cholesterol and glucose metabolism. J. Biol. Chem. 2004;279:45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- 6.Song WC, Moore R, McLachlan JA, Negishi M. Molecular characterization of a testis-specific estrogen sulfotransferase and aberrant liver expression in obese and diabetogenic C57BL/KsJ-db/db mice. Endocrinology. 1995;136:2477–2484. doi: 10.1210/endo.136.6.7750469. [DOI] [PubMed] [Google Scholar]
- 7.Khor VK, Dhir R, Yin X, Ahima RS, Song WC. Estrogen sulfotransferase regulates body fat and glucose homeostasis in female mice. Am. J. Physiol. Endocrinol. Metab. 2010;299:657–664. doi: 10.1152/ajpendo.00707.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong MH, Jiang H, Liu P, Lawson JA, Brass LF, Song WC. Spontaneous fetal loss caused by placental thrombosis in estrogen sulfotransferase-deficient mice. Nat. Med. 2005;11:153–159. doi: 10.1038/nm1184. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki M, Ishida H, Shiotsu Y, Nakata T, Akinaga S, Takashima S, Utsumi T, Saeki T, Harada N. Expression level of enzymes related to in situ estrogen synthesis and clinicopathological parameters in breast cancer patients. J. Steroid Biochem. Mol. Biol. 2009;113:195–201. doi: 10.1016/j.jsbmb.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Utsumi T, Yoshimura N, Takeuchi S, Ando J, Maruta M, Maeda K, Harada N. Steroid sulfatase expression is an independent predictor of recurrence in human breast cancer. Cancer Res. 1999;59:377–381. [PubMed] [Google Scholar]
- 11.Qatanani M, Zhang J, Moore DD. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology. 2005;146:995–1002. doi: 10.1210/en.2004-1350. [DOI] [PubMed] [Google Scholar]
- 12.Gong H, Guo P, Zhai Y, Zhou J, Uppal H, Jarzynka MJ, Song WC, Cheng SY, Xie W. Estrogen deprivation and inhibition of breast cancer growth in vivo through activation of the orphan nuclear receptor liver X receptor. Mol. Endocrinol. 2007;21:1781–1790. doi: 10.1210/me.2007-0187. [DOI] [PubMed] [Google Scholar]
- 13.Gong H, Jarzynka MJ, Cole TJ, Lee JH, Wada T, Zhang B, Gao J, Song WC, DeFranco DB, Cheng SY, et al. Glucocorticoids antagonize estrogens by glucocorticoid receptor-mediated activation of estrogen sulfotransferase. Cancer Res. 2008;68:7386–7393. doi: 10.1158/0008-5472.CAN-08-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue K, Negishi M. Nuclear receptor CAR requires early growth response 1 to activate the human cytochrome P450 2B6 gene. J. Biol. Chem. 2008;283:10425–10432. doi: 10.1074/jbc.M800729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda A, Matsui K, Yamamoto Y, Pedersen LC, Sueyoshi T, Negishi M. Thr176 regulates the activity of the mouse nuclear receptor CAR and is conserved in the NR1I subfamily members PXR and VDR. Biochem. J. 2005;388:623–630. doi: 10.1042/BJ20041572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J. Biol. Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- 17.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J. Biol. Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 18.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell. Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue K, Borchers CH, Negishi M. Cohesin protein SMC1 represses the nuclear receptor CAR-mediated synergistic activation of a human P450 gene by xenobiotics. Biochem. J. 2006;398:125–133. doi: 10.1042/BJ20060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 21.Teferedegne B, Green MR, Guo Z, Boss JM. Mechanism of action of a distal NF-κB-dependent enhancer. Mol. Cell. Biol. 2006;26:5759–5770. doi: 10.1128/MCB.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura K, Moore R, Negishi M, Sueyoshi T. Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J. Biol. Chem. 2007;282:9768–9776. doi: 10.1074/jbc.M610072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 25.Guo H, Gao C, Mi Z, Wai PY, Kuo PC. Phosphorylation of Ser158 regulates inflammatory redox-dependent hepatocyte nuclear factor-4α transcriptional activity. Biochem. J. 2006;394:379–387. doi: 10.1042/BJ20051730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, Sladek FM. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol. Endocrinol. 2007;21:1297–1311. doi: 10.1210/me.2006-0300. [DOI] [PubMed] [Google Scholar]
- 27.Bryzgalova G, Gao H, Ahren B, Zierath JR, Galuska D, Steiler TL, Dahlman-Wright K, Nilsson S, Gustafsson JA, Efendic S, et al. Evidence that oestrogen receptor-α plays an important role in the regulation of glucose homeostasis in mice: insulin sensitivity in the liver. Diabetologia. 2006;49:588–597. doi: 10.1007/s00125-005-0105-3. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki A, Sanda N, Miyawaki Y, Fujimori Y, Yamada T, Takagi A, Murate T, Saito H, Kojima T. Down-regulation of PROS1 gene expression by 17β-estradiol via estrogen receptor α (ERα)-Sp1 interaction recruiting receptor-interacting protein 140 and the corepressor-HDAC3 complex. J. Biol. Chem. 2010;285:13444–13453. doi: 10.1074/jbc.M109.062430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto Y, Moore R, Hess HA, Guo GL, Gonzalez FJ, Korach KS, Maronpot RR, Negishi M. Estrogen receptor α mediates 17α-ethynylestradiol causing hepatotoxicity. J. Biol. Chem. 2006;281:16625–16631. doi: 10.1074/jbc.M602723200. [DOI] [PubMed] [Google Scholar]
- 30.Lambert CB, Spire C, Claude N, Guillouzo A. Dose- and time-dependent effects of phenobarbital on gene expression profiling in human hepatoma HepaRG cells. Toxicol. Appl. pharmacol. 2009;234:345–360. doi: 10.1016/j.taap.2008.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.