Abstract
U12 snRNA is analogous to U2 snRNA of the U2-dependent spliceosome and is essential for the splicing of U12-dependent introns in metazoan cells. The essential region of U12 snRNA, which base pairs to the branch site of minor class introns is well characterized. However, other regions which are outside of the branch site base pairing region are not yet characterized and the requirement of these structures in U12-dependent splicing is not clear. U12 snRNA is predicted to form an intricate secondary structure containing several stem–loops and single-stranded regions. Using a previously characterized branch site genetic suppression assay, we generated second-site mutations in the suppressor U12 snRNA to investigate the in vivo requirement of structural elements in U12-dependent splicing. Our results show that stem–loop IIa is essential and required for in vivo splicing. Interestingly, an evolutionarily conserved stem–loop IIb is dispensable for splicing. We also show that stem–loop III, which binds to a p65 RNA binding protein of the U11-U12 di.snRNP complex, is essential for in vivo splicing. The data validate the existence of proposed stem–loops of U12 snRNA and provide experimental support for individual secondary structures.
INTRODUCTION
In metazoans, precursor messenger RNA (pre-mRNA) splicing requires numerous trans-acting protein factors and several small nuclear RNAs (snRNAs). Removal of U2-type introns requires the U1, U2, U4, U5 and U6 snRNAs as well as hundreds of associated splicing factors assembled into a large dynamic complex known as spliceosome (1,2). Similarly, the splicing of minor or U12-dependent introns requires an analogous set of five snRNAs, namely U11, U12, U4atac, U5 and U6atac (3–6). A large number of RNA motifs and RNA–RNA interactions appear to be conserved between major and minor spliceosomal snRNAs and these motifs are shown to be essential for splicing in their respective spliceosomes.
The U2 and U12 snRNAs appear to play a central role in major and minor spliceosomes, respectively. U2 snRNA binds the branch site sequences by canonical base pairing interactions, which are necessary for U2-dependent intron splicing (7). Similarly, U12 snRNA binds to the branch site of a U12-dependent intron by RNA-RNA base pairing essential for splicing (3,8). Besides binding to the branch site sequence, U2 snRNA also base pairs with U6 snRNA to form an intermolecular helix required for U2-dependent splicing (9–12). In an analogous manner, the region of U12 snRNA containing first 13 nucleotides (light grey shaded area in SLI, Figure 1) is predicted to form helix I by base pairing with U6atac snRNA (6). The existence of this structure is inferred by cross-linking experiments and appears to be essential for U12-dependent splicing (6,13,14). In the process of the assembly of U12-dependent spliceosome, U11 snRNA base pairs with the 5′ splice site and U12 snRNA base pairs with the branch site (3–5,15). These interactions are analogous to U1 and U2 snRNA interactions with their corresponding splice sites in the major spliceosome. In the major spliceosome formation, U1 and U2 snRNAs base pair with the 5′ splice site and branch site as mono small nuclear ribonucleoproteins (snRNPs), respectively (16–20). In contrast, U12 snRNA is not only found as single snRNP, it also occurs as 18S U11/U12 di.snRNP (21). The formation of the minor spliceosomal complex requires a preformed U11/U12 di.snRNP complex (22). Simultaneous recognition of the 5′ splice site and branch site by U11/U12 snRNAs as di.snRNP complex suggests a molecular mechanism which is unique to the recognition of U12-dependent introns.
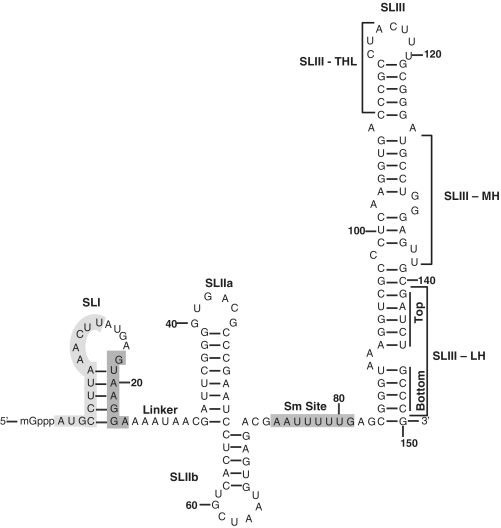
Figure 1.
Secondary structure model of human U12 snRNA. The structure is based on the earlier predicted models (21,30,31,56). SLI, Stem–Loop I; SLIIa, Stem–Loop IIa; SLIIb, Stem–Loop IIb; SLIII, Stem–Loop III; SLIII-THL, Stem–Loop III-Terminal Helix and Loop; SLIII-MH, Stem–Loop III-Middle Helix; SLIII-LH, Stem–Loop III-Lower Helix. The light shaded grey region (nucleotides 1–13, SLI) is predicted to bind U6atac snRNA and the dark shaded region (nucleotides 18–24, SLI) binds to branch site of a U12-dependent intron. Predicted Sm site is also illustrated.
U12-dependent introns and their spliceosomal components including snRNAs and protein factors are conserved between plants and humans suggesting an early evolution of these introns (23–25). Initially, the U12-dependent introns were identified in metazoans including plants, cnidarians, insects and vertebrates including humans (26). U12-dependent snRNAs and traces of U12-dependent proteins are lacking in many species including budding yeast Saccharomyces cerevisiae and nematode Caenorhabditis elegans. Due to the availability of genome sequence data of various organisms and improved search algorithms, the more recent bioinformatics studies have identified U12-dependent introns and spliceosomal snRNAs in distant fungal and worm lineages (23). These studies have identified U12-dependent snRNAs in a number of diverged species including Acanthamoeba, Rhizopus, Phytophthora, Physarum and Trichinella (27,28). Comparative sequence analysis of U12 snRNA between various similar and diverged organisms revealed conserved and variable stretches of the RNA molecule (27). Except a few variable regions, the sequence and the predicted structure appear to be similar among U12 snRNAs of these species (23,29).
U12 snRNA was predicted to contain five stem–loops in the original proposed secondary structure (21). However, the more recent U12 snRNA predicted structure contains four stem- loops (Figure 1). The 3′ half of U12 snRNA is predicted to adopt an alternative structure, which leads to the loss of stem–loops IV and V (in the original structure) and gain in the length of stem–loop III (30). U12 snRNA has been predicted to contain stem–loop I (SLI), stem–loop IIa (SLIIa), a single-stranded region as linker between SLI and SLIIa, stem–loop IIb (SLIIb), Sm protein binding region and stem–loop III (SLIII) (Figure 1). Here, we describe SLIII in three distinct regions containing SLIII–terminal helix and loop (Figure 1, SLIII–THL), SLIII–middle helix (SLIII–MH) and SLIII–lower helix (SLIII–LH top and bottom). Based on key studies, it is apparent that at least two structural regions of U12 snRNA are essential for U12-dependent splicing. The 3′-stem of SLI containing nucleotides 18–24 (GUAAGGA) encompasses essential region which base pairs with the branch site region of a U12-dependent intron (31). The 5′-stem of SLI containing nucleotides 1–13 is predicted to base pair with U6atac snRNA to form helix I, which appears to be essential for the splicing of U12-dependent introns (6,13,24). Additionally, the apical stem–loop end of SLIII–THL (Figure 1) binds to p65 RNA binding protein, a unique U12-dependent spliceosomal protein of U11 and U12 di.snRNP complex (30,32).
The role and requirement of other structural elements of U12 snRNA, beyond the branch site interaction region, in U12-dependent splicing is not clear. In this study, we explored the validity of the proposed stem–loop structures and their requirement for in vivo U12-dependent splicing using a previously described branch site mutation suppression assay.
MATERIALS AND METHODS
Construction of U12 expression plasmids
The starting U12 snRNA expression plasmid has been described previously (8). This construct contains mutations GA23/24CU in the branch site interacting region to restore complementary base pairing to UC84/85AG branch site region of P120 U12-dependent intron F. For this study, second site mutations were introduced in U12 snRNA in the background of GA23/24CU. 5′ phosphorylated mutagenic oligonucleotides were used for site directed mutagenesis using the Change-IT mutagenesis kit (USB Corporation). The sequences of mutant snRNAs were confirmed by DNA sequencing.
Analysis of in vivo splicing
The P120 minigene plasmid, described previously, contains exons 5–8 and introns E, F and G of the human nucleolar protein P120 gene (24,33–35). The branch site of the U12-dependent intron F contains the UC84/85AG mutation, which abolishes U12-dependent splicing of this intron in vivo. The U12 snRNA GA23/24CU suppressor mutation was described previously (3,8). The details of the suppressor assay are illustrated in Supplementary Figure S1. Second site mutation containing U12 suppressor plasmids were co-transfected with UC84/85AG branch site P120 mutant into cultured CHO cells as described previously (3,8). For these experiments, 0.5 µg of P120 plasmid and 2 µg of each of second site mutation carrying U12 snRNA expression plasmids were added to CHO cells in 6-well plates. Where one or more U12 snRNA plasmids were omitted, a corresponding amount of pUC19 plasmid DNA was substituted. Total RNA was isolated from cells 36 h after transfection, reverse-transcribed and PCR-amplified as described (33,34,36). A total of 500 ng of DNase-treated RNA was reverse transcribed and PCR amplified using the Thermostable rTth Reverse Transcriptase RNA PCR kit (Applied Biosystems) and the following primers: forward: GGCCCGGGAAGCTGCTGCTGGGATC; reverse: CTTCTAAGAACTCCACCAGCTCAGA. One microliter of this amplified product was then subjected to nested PCR under the following conditions: 94°C for 3 min, 25 cycles of 94°C for 1 min and 60°C for 1 min followed by a final extension at 68°C for 5 min. The primers used for the nested PCR were as follows: forward: TTGTGCTGCCCCCTGCTGGGGAGATG and reverse: TCAGACAGAGGGAAGAGGTCCATGAG. The nested PCR products were analyzed by agarose gel electrophoresis. A reverse transcriptase minus control was kept in all experiments to monitor DNA template contamination. The DNA bands were visualized using ethidium bromide and scanned on Typhoon Image Scanner (GE Healthcare). Intensity of bands was quantified using the ImageJ software. For each lane, the band intensity of each product (unspliced, US; U12 spliced, U12S; U12-3′ cryptic spliced, U12-3′CS) was expressed as percentage of the total product. Each U12 snRNA suppressor was transfected a minimum of three times in two stocks of cells. Independent transfections and analyses gave substantially similar results.
RESULTS
Consensus secondary structure of U12 snRNA revealed conserved RNA domains
A number of organisms contain U12-dependent spliceosomal introns and corresponding snRNAs. Among these snRNAs, U12 snRNA has relatively evolutionarily conserved primary sequence and predicted structure. Variations are found in the primary sequence, which do not seem to affect the predicted secondary structure of U12 snRNA (28,29). To analyze primary sequence conservation and predict a consensus secondary structure, we performed multiple sequence analysis of U12 snRNA from 17 species spanning vertebrate, plant, insect, fungus and worm. The resultant consensus secondary structure is shown in Supplementary Figure S2. As evident from the figure, U12 snRNA primary sequence and secondary structure appear to be highly conserved in all the species, suggesting their important regulatory roles. As expected, the first 30 nucleotides are almost identical as this region spans the part of U12 snRNA (SLI), which forms putative helix I intermolecular base pairing with U6atac snRNA and also contains the nucleotides which base pair with the highly conserved branch site region of a U12-dependent intron. The SLI region appears to be identical in terms of sequence, size and structure among all the species. The nucleotide length differences between various U12 snRNAs were mainly adjusted in SLIIa and in the 3′-end of the U12 snRNA (Supplementary Figure S2). Interestingly, most nucleotide variations within helices were compensated by complementary nucleotide changes in the corresponding arm of the helix (Supplementary Figure S2).
Mutational analysis of SLI nucleotides 14–17
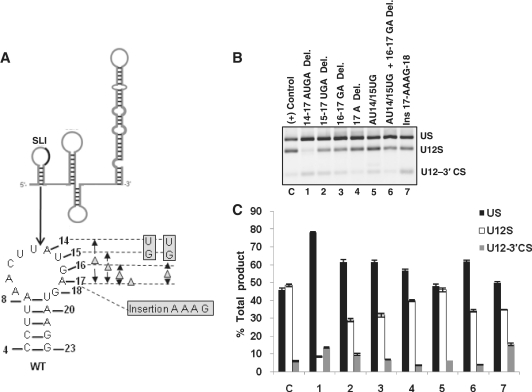
Both human U2 and U12 snRNAs contain SLI, which is comprised of nucleotides necessary for U2- and U12-dependent splicing, respectively. The SLI of U12 snRNA appears to have more thermodynamically stable secondary structure as compared to human U2 SLI. In U12 SLI, nucleotides 1–13 are predicted to form helix I with U6atac snRNA (light grey shaded nucleotides in the 5′-stem of SLI in Figure 1). The 3′-stem of SLI interacts with the branch site of a U12-dependent intron (Figure 1, dark grey shaded nucleotides from 18 to 24). These two regions are separated by four nucleotides (14–17) which appear to provide a ‘bridge’ between the two functionally important regions. These four nucleotides of U12 snRNA appear to juxtapose with AGAGAA box in human U6atac snRNA as it base pairs with the 5′ splice site and also forms the predicted intermolecular U6atac:U12 helix I structure (6,24). Evolutionarily conserved AGAGAA nucleotides in U6 snRNA have been shown to play a critical role in U2-dependent splicing (37,38). The requirement and the role of four nucleotides A14U15G16A17 between helix I and branch site binding region in in vivo splicing and in the functionality of U12 snRNA remains unclear. To test this, we made seven mutants in this region of U12 snRNA (Figure 2A). These were (the numbering also corresponds to lanes in Figure 2B and bars in Figure 2C): (1) 14–17 AUGA deletion, (2) 15–17 UGA deletion, (3) 16–17 GA deletion, (4) 17 A deletion, (5) AU14/15UG, (6) AU14/15UG + 16–17 GA deletion and (7) an insertion of four nucleotides AAAG between nucleotides A17 and G18 to increase the distance to 8 nucleotides. When these mutant snRNAs were tested for their in vivo suppression activity, a varied degree of splicing activity was observed. The 14–17 AUGA deletion mutant was the most defective for wild type (WT) splicing and produced mostly unspliced transcripts (lane 1 in Figure 2B, comparison of U12S bars of 1 and C in Figure 2C reveals that U12S product in 1 is approximately reduced by 5-fold as compared to control—C). The triple, double and single deletion mutants restored the suppression function of U12 snRNA as evident by the increasing amount of WT U12-dependent splicing (lanes 2–4 in Figure 2B and bars 2–4 in Figure 2C). Substitution mutant of U12 snRNA AU14/15UG had no effect on splicing from the WT splice sites and it produced similar splicing phenotype as that seen with positive control (compare lanes C and 5 in Figure 2B, also compare bars C and 5 in Figure 2C). Substitution mutant when combined with 16–17 deletion showed a partial suppression activity (lane 6 in Figure 2B, bar 6 in Figure 2C) similar to that seen with 16–17 deletion mutant (compare lanes 3 and 6 in Figure 2B and bars 3 and 6 in Figure 2C). Interestingly, an insertion of four random nucleotides between nucleotides 17 and 18 of U12 snRNA did not affect the suppression function of this mutant drastically (lane 7 in Figure 2B, bar 7 in Figure 2C). We conclude that the mutational analysis of the loop region of SLI of U12 snRNA shows that the loop nucleotides AUGA may be required for in vivo splicing activity. Our results suggest that A14U15G16A17 may be an important ‘spacer’ element when U12 snRNA is bound to U6atac snRNA to provide a bridging component of U12: pre-mRNA and U12:U6atac RNA:RNA interactions in U12-dependent splicing.
Figure 2.
(A) Line diagram of U12 snRNA predicted secondary structure. Solid black line within SLI region illustrates the region that was subjected to mutagenesis. The predicted structure/sequence of SLI and locations of the mutations in the loop region (nucleotides 14–17) are shown. These and all other mutations of U12 snRNA discussed in this study were made in the first site mutation GA23/24CU background. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated SLI mutants of U12 snRNA. Lane C (in this figure and all subsequent figures): a positive P120 branch site mutation suppression control using the mutant constructs shown in lane 4 of Supplementary Figure S1D. CHO cells were transfected with the UC84/85AG P120 intron F mutant and second site U12 mutants (indicated in each lane). Splicing phenotypes were obtained by RT–PCR. (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B). In this and all subsequent figures, error bars represent ± SE of three experiments. (Abbreviations in this figure and all subsequent figures: Del., deletion; Ins., insertion; US, unspliced; U12S, U12 spliced; U12-3′CS, U12-3′ cryptic spliced. A new abbreviation is explained wherever necessary).
Mutational analysis of single-stranded linker region
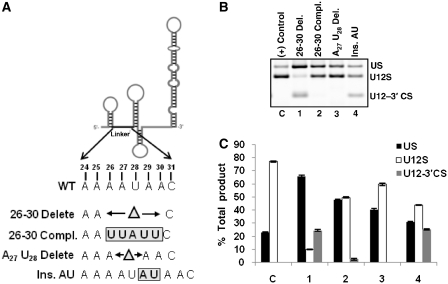
Next, we tested the requirement of a predicted single-stranded linker region between SLI and SLIIa, which consists of eight nucleotides (24–31). The role of this region in splicing is not clear. Whether this region has RNA or protein interacting partners is also unknown. We made four mutants (deletion, substitution and insertion) in the linker region of U12 snRNA as illustrated in Figure 3A. When these mutant snRNAs were tested for their requirement in in vivo splicing, only 26–30 deletion mutant was found to be most defective for splicing (lane 1 in Figure 3B, bar 1 in Figure 3C). The WT-spliced product (U12S) in lane 1 was reduced by 8-fold as compared to the U12S product in positive control lane (compare U12S bars of C and 1 in Figure 3C). U12 snRNA mutants with complementary nucleotide sequence alteration and two nucleotides deletion were active for their suppressor activity (lanes 2 and 3 in Figure 3B, bars 2 and 3 in Figure 3C). Interestingly, addition of two nucleotides reduced the WT splicing to approximately half of that seen with positive control (lanes 4 and C in Figure 3B, bars 4 and C in Figure 3C). The lack of suppressor activity of 26–30 deletion mutant supported the role of the linker region in U12-dependent in vivo splicing. This notion is also supported by the increase of the linker region length by two nucleotides, which compromised the splicing of the U12-dependent intron from WT splice sites (Figure 3B, lane 4). We conclude that the insertion, deletion and substitution mutants of the single-stranded linker region show its requirement for splicing activity in vivo and suggest that a distance constraint between SLI and SLIIa structures is important for splicing.
Figure 3.
(A) Schematic of U12 snRNA showing the location of the linker region. Series of mutations made in the linker region are illustrated. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated linker region mutants of U12 snRNA. (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B). (Compl., complementary).
Mutational analysis of SLIIa
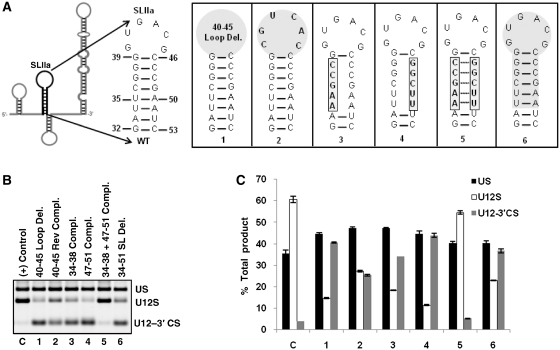
To test the requirement of SLIIa in splicing, we made six mutants (Figure 4A). In the first mutant, we deleted the terminal loop and the second mutant contained reverse complementary sequence of the loop (Figure 4A-1, 2). Complete deletion of the loop reduced the WT splicing activity to 1/4th of that seen with the positive control (compare lanes C and 1 in Figure 4B, bars C and 1 in Figure 4C). Second mutant with reverse complementary loop sequence reduced WT splicing activity to approximately half of that observed with the positive control (compare lanes C and 2 in Figure 4B, bars C and 2 in Figure 4C). Loss of suppressing activity of these two U12 snRNA mutants revealed the requirement of SLIIa terminal loop in U12-dependent in vivo splicing. Next, we tested the existence of helix and the requirement of proposed base pairing of SLIIa. For this, we constructed three more mutants spanning the 5′ and 3′ arms of the stem. For the first mutant of the helix, we mutated the 5′ stem sequence (nucleotides 34-38) to its complementary sequence (Figure 4A-3). Similarly, we constructed another mutant in which we mutated the 3′ stem sequence (nucleotides 47–51) to its complementary sequence (Figure 4A-4). We considered that if the predicted helix existed and was required for in vivo U12-dependent splicing, the levels of WT U12-dependent splicing would be reduced/abolished in the presence of these mutants. Indeed, when tested for their requirement for in vivo splicing, both mutants were defective for splicing (lanes 3 and 4 in Figure 4B, bars 3 and 4 in Figure 4C). In a subsequent mutant, we combined the complementary mutant sequences of both 5′ and 3′ arms of the helix of SLIIa to restore the proposed base pairing in compensatory base pairing manner (Figure 4A-5). When tested for in vivo splicing, this U12 suppressor mutant was able to restore splicing from WT splice junctions (lane 5 in Figure 4B, also compare U12S bars of C and 5 in Figure 4C), suggesting the existence of proposed base pairing and the requirement of the stem component of SLIIa for splicing. Finally, we tested the requirement of the whole SLIIa for in vivo splicing. For this, we deleted the region between nucleotides 34 and 51 (Figure 4A-6). As expected, this mutant was mostly inactive for WT splicing—the WT spliced product was reduced by ~3-fold as compared to that seen with the positive control (compare lanes C and 6 in Figure 4B and bars C and 6 in Figure 4C). This set of U12 suppressor mutants, thus confirmed the existence of SLIIa, the proposed base pairing and the requirement of the stem-loop for in vivo U12-dependent splicing.
Figure 4.
(A) Schematic of U12 snRNA showing the sequence and predicted secondary structure of WT SLIIa (nucleotides 32–53). Mutations made in the SLIIa region of U12 snRNA are illustrated. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated SLIIa mutants of U12 snRNA. Lane numbers 1 through 6 correspond to mutants 1 through 6 shown in (A). (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B). (Rev Compl., reverse complementary; Compl., complementary).
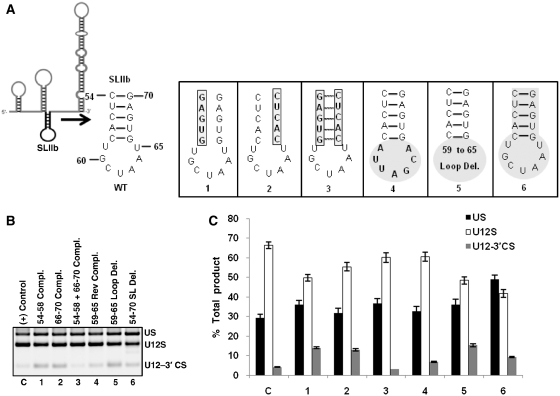
Mutational analysis of SLIIb
Next, we tested the predicted structure and the requirement of U12 SLIIb in the splicing of U12-dependent introns. SLIIb spans nucleotides 54–70 and contains 5 bp helix and 7 nt loop (Figure 5A). Based on multiple sequence comparative analysis, primary sequence and structure of SLIIb appear to be highly conserved from plants to humans (Supplementary Figure S2) (24). U12 snRNA SLIIb equivalent structure is also conserved in yeast, human and other U2 snRNA homologs (39,40). To test the existence of putative SLIIb, the predicted base pairing interactions constituting the helix and the functional requirement of SLIIb in splicing, we generated six mutants illustrated in Figure 5A. The first and second complementary sequence mutants were largely active for U12-dependent splicing as evident by the WT splicing levels similar to that of the positive control (compare lanes C, 1 and 2 in Figure 5B and the corresponding bars in Figure 5C). Splicing was restored to positive control levels when mutations in both arms of the stem restored the helix by compensatory base pairing interactions (compare lanes C and 3 in Figure 5B, bars C and 3 in Figure 5C). These results support the existence of SLIIb helix and potential base pairing interactions. Next, we tested the requirement of predicted loop region from nucleotides 59–65. The results of transfections (lanes 4 and 5 in Figure 5B, bars 4 and 5 in Figure 5C) show little loss of WT U12-dependent splicing suggesting redundancy of the loop region in splicing. Based on these five mutants of SLIIb, it became evident that drastic changes in the sequence and structure of SLIIb have modest or no effect on U12-dependent in vivo splicing. Hence, we made a sixth mutant of U12 snRNA in which we deleted the nucleotides 54–70 that constituted the entire predicted SLIIb (Figure 5A-6). Splicing phenotype obtained by the transfection of this mutant is shown in lane 6 of Figure 5B. Interestingly, SLIIb deletion mutant of U12 snRNA was functional for splicing, though slightly less efficient than the positive control (compare U12S bars of C and 6 in Figure 5C). Taken together, the mutational analyses of SLIIb suggest the existence of the stem-loop, yet a dispensable nature of this region for in vivo U12-dependent splicing.
Figure 5.
(A) Schematic of U12 snRNA showing the location of SLIIb. The sequence and predicted secondary structure of WT SLIIb (nucleotides 54–70) and the six SLIIb mutants are illustrated. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated SLIIb mutants of U12 snRNA. Lane numbers 1 through 6 correspond to mutants 1 through 6 shown in (A). (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B). (Compl., complementary; Rev Compl., reverse complementary).
Mutational analysis of SLIII
Next, we investigated if SLIII is essential for in vivo splicing of U12-dependent introns. For convenience, we divided SLIII into three distinct regions containing SLIII-THL, SLIII-MH and SLIII-LH (Figure 1). The apical loop and part of stem (nucleotides 113–121) in SLIII-THL region interact with p65 RNA binding protein (30). However, the existence and requirement of other helices of SLIII region have not been tested. To test this, we constructed a series of mutants of SLIII. Location of mutations and results are shown in Figures 6–8.
Figure 6.
(A) Schematic of U12 snRNA showing the sequence and predicted secondary structure of WT SLIII-THL. Mutations (1–6) made in the SLIII-THL region of U12 snRNA are illustrated. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated SLIII-THL mutants of U12 snRNA. Lane numbers 1 through 6 correspond to mutants 1 through 6 shown in (A). (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B). (Compl., complementary; Rev Compl., reverse complementary).
Figure 7.
(A) Schematic of U12 snRNA showing the sequence and predicted secondary structure of WT SLIII-MH. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated SLIII-MH mutants of U12 snRNA. (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B).
Figure 8.
(A) Schematic of U12 snRNA showing the sequence and predicted secondary structure of WT SLIII-LH. Mutations made in the SLIII-LH region of U12 snRNA are shown. (B) Splicing phenotypes of P120 branch site UC84/85AG mutant co-expressed with the indicated SLIII-LH mutants of U12 snRNA. Lane numbers 1 through 7 correspond to mutants 1 through 7 shown in (A). (C) Quantitative analysis of spliced/unspliced phenotypes from the gel image shown in (B). (Compl., complementary).
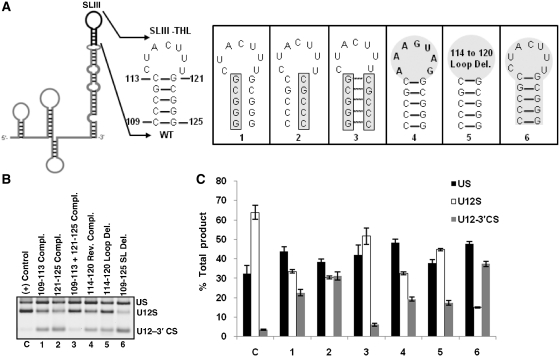
Mutational analysis of SLIII–THL
We tested the requirement of SLIII–THL structure in in vivo U12-dependent splicing by using a set of six mutants illustrated in Figure 6A. Mutants 1 and 2, which disrupted the base pairing in the helix, reduced WT splicing to approximately half of that observed with the positive control (compare lanes C, 1 and 2 in Figure 6B, bars C, 1 and 2 in Figure 6C). However, the combination mutant 3, which restored the helix, showed WT splicing activity comparable to that seen in positive control (compare lanes C and 3 in Figure 6B, bars C and 3 in Figure 6C). These data strongly support the existence of the helix region of SLIII–THL and also indicate the importance of this region for U12-dependent in vivo splicing. Next, we investigated the requirement of the loop element from nucleotides 114–120. For this, we constructed two mutants. In the first loop mutant, sequence CUACUUU was reverse complemented to AAAGUAG (Figure 6A-4), and in the second mutant, nucleotides 114–120 were deleted (Figure 6A-5). As seen in lanes 4 and 5 (Figure 6B), the loop mutants were active albeit the splicing efficiency was reduced to ~50%. Interestingly, the loop deletion mutant (Figure 6B, lane 5) appeared to have slightly better in vivo splicing activity as compared to the reverse complement loop mutant, which reduced WT splicing levels to approximately half of those seen in positive control (Figure 6C, bars C, 4 and 5). Finally, we examined the effect of the whole stem–loop deletion (nucleotides 109–125, Figure 6A-6) on splicing activity. As expected, this U12 mutant was largely inactive for WT splicing (lane 6 in Figure 6B, bar 6 in Figure 6C). The results of these mutants are not surprising as SLIII–THL has been shown to interact with the carboxyl terminal RRM domain of p65 RNA binding protein (30) and thus, play an essential role in splicing.
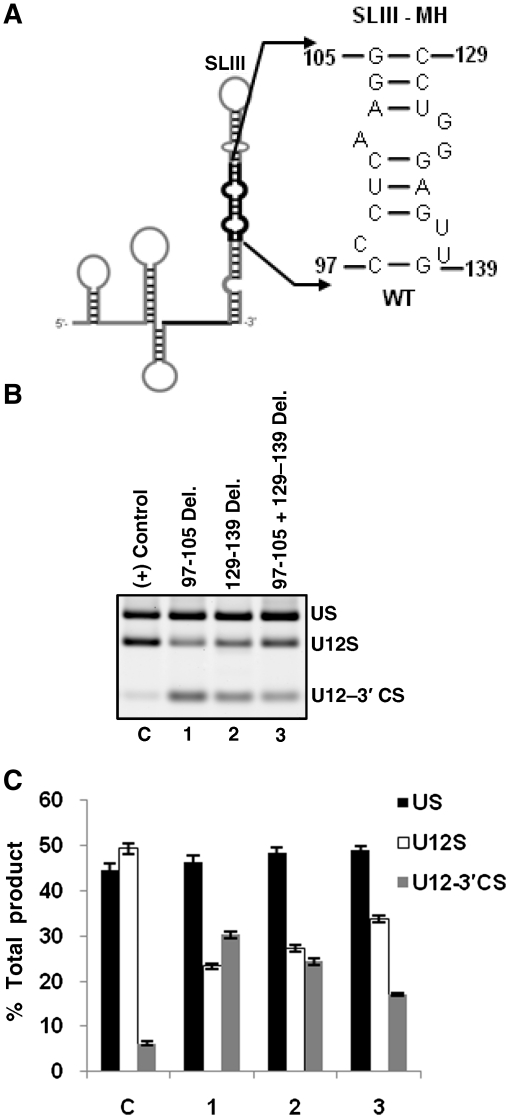
Mutational analysis of SLIII–MH
The middle helix of SLIII is predicted to be formed by nucleotides 97–105 and 129–139 in the 5′- and the 3′-arms of the helix, respectively (Figure 7A). Whether this region of SLIII has a functional role in U12-dependent splicing is currently unknown. To determine proposed secondary structure and the requirement of this region in splicing, we made three mutants: (1) 97–105 deletion, (2) 129-139 deletion and (3) combination of mutants 1 and 2 in which the whole SLIII-MH region (nucleotides 97–105 and nucleotides 129–139) was deleted (Figure 7A). In the first two mutants, we observed ~50% reduction in WT splicing but the combined mutant designed to shorten the length of SLIII improved the efficiency of splicing as compared to individual deletion mutants (lanes 1–3 in Figure 7B, bars 1–3 in Figure 7C). These data suggest the functional importance of the overall structure of SLIII–MH region in U12-dependent splicing.
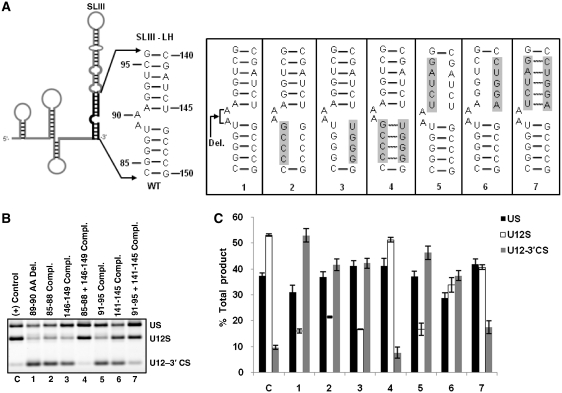
Mutational analysis of SLIII–LH
Finally, we tested the requirement of lower helix of SLIII for splicing. To this end, we constructed seven mutants in the region from nucleotides 84–96 and 140–150 (Figure 8A). Within this region two helices are predicted to be formed. One helix is proposed to form between nucleotides 84–88 and 146–150 and another helix between nucleotides 91–96 and 140–145 (Figure 8A). The first mutant generated in SLIII-LH region contained a deletion of a predicted A89A90 asymmetric bulge (Figure 8A-1). This mutant was largely inactive for in vivo U12-dependent splicing (lane 1 in Figure 8B, compare U12S bar of C and 1 in Figure 8C). Next three mutants were designed to test the predicted base pairing between nucleotides 85–88 and 146–149 at the bottom of the helix (Figure 8A-2, 3 and 4). The 8A-2 and 8A-3 mutants, which disrupted the base pairing, reduced WT splicing by ~3-fold as compared to that seen in positive control (lanes 2 and 3 in Figure 8B, also compare U12S bars of C, 2 and 3 in Figure 8C). In contrast, the 8A-4 mutant, which restored the base pairing, showed splicing activity similar to that of positive control (lanes C and 4 in Figure 8B, bars C and 4 in Figure 8C) thus providing evidence for the existence and functional importance of the bottom helix of SLIII–LH region. Similarly, we tested the existence and requirement of the base paired helix in the upper part of SLIII–LH region. For this, we constructed three mutants (Figure 8A-5, 6 and 7). The 91–95 complementary sequence mutant reduced WT splicing to ~1/3 rd compared to positive control (lanes C and 5 in Figure 8B, bars C and 5 in Figure 8C); its counterpart, the 141–145 complementary sequence mutant reduced WT splicing by approximately 40% (lane 6 in Figure 8B, bar 6 in Figure 8C). However, the U12 mutant containing the combination of these compensatory mutations restored WT splicing to almost positive control levels (lanes C and 7 in Figure 8B, bars C and 7 in Figure 8C). Combined together, the results of the mutational analysis of SLIII–LH region of U12 snRNA demonstrate the existence of the predicted base pairing in this region.
DISCUSSION
In this study, we used a previously characterized U12 snRNA branch site mutation suppressor and U12-dependent intron to study the requirement of specific structures and sequences of U12 snRNA in splicing. We focused mostly on combination mutants to test the existence of base pairing; insertion and deletion mutants to test the distance constraints between structural elements and finally, some deletion mutants to test if the entire element was dispensable for pre-mRNA splicing. Our results suggest that most of these RNA elements are important for the function of U12 snRNA.
Requirement of A14U15G16A17 of SLI suggests functionally important distance constraints between U12 branch site binding region and intermolecular helix I
The definitive role of the 5′-end of U12 snRNA from nucleotides 1 to 13 is not clear. This region, predicted to base pair with U6atac snRNA to form helix I, appears to be important for the function of U12 snRNA (14). On the other hand, the importance of U12 snRNA nucleotides 18–24 is unequivocally established. This region is indispensible for splicing because it base pairs with branch site of a U12-dependent intron (8,41). Within the 5′-end of U12 snRNA, we tested the function of a 4 nt region containing A14U15G16A17 (Figure 1). These nucleotides appear to form a bridge or linker which separates putative helix I from branch site binding region. We made seven mutants to learn if the length of the region or the identity of nucleotides is important for the function of U12 snRNA. Only one U12 suppressor mutant containing deletion of all four nucleotides was inactive for WT splicing (lane 1 in Figure 2B, bar 1 in Figure 2C). Interestingly, the AUGA deletion mutant was not only defective for WT splicing, the 3′ cryptic splicing was largely abolished as well (Figure 2B, lane 1). It is plausible that the interaction of this mutant to the branch site generated a stable, yet defective splicing complex by competing out the endogenous WT U12 snRNA and blocking the formation of splicing competent complex. Other mutants containing single, double or triple deletions or substitutions or insertions were functional for in vivo splicing (lanes 2–7 in Figure 2B, bars 2–7 in Figure 2C). Taken together, these data show that the nucleotide distance between intermolecular helix I forming region and branch site binding region may be an important feature necessary for the function of U12 snRNA in U12-dependent in vivo splicing.
Single-stranded linker region is important for U12 snRNA in vivo function
The single-stranded linker region between SLI and SLIIa (Figure 1) appears to provide a distance constraint with respect to the number of nucleotides that separate these two stem–loops. This is evident from our data—a complete deletion mutant (26–30 delete; Figure 3B, lane 1) and two nucleotide insertion mutant (Ins. AU; Figure 3B, lane 4) of linker region were deleterious for WT splicing while complementary sequence mutant (26–30 compl; Figure 3B, lane 2) was active for WT splicing. The sequence of linker region appears to be highly conserved among all U12 snRNAs that we have analyzed to predict consensus secondary structure in Supplementary Figure S2 and in earlier U12 snRNA predicted structures (28). However, there are variations observed within the sequence of this region in at least four other evolutionary divergent species. Homo sapiens U12 snRNA nucleotides 27A/31C within linker region have diverged to 27U/31A in T. spiralis and 27C/31A in A. thaliana, D. melanogaster and S. moellendorffii (28). We have not tested the functional significance of these changes in our U12 suppressor background; however, it is reasonable to believe that these changes may not be extremely deleterious for U12 snRNA function.
Conserved SLIIa is essential for in vivo U12 snRNA activity
We tested six mutants to study the requirement of SLIIa of U12 snRNA (Figure 4). These mutants were designed to study the requirement of loop region (nucleotides 40–45) and to test the existence of the predicted helix formed by nucleotides 34–38 and 47–51. SLIIa loop deletion mutant was largely inactive for U12-dependent splicing (lane 1 in Figure 4B, bar 1 in Figure 4C). Interestingly, the reverse complementary loop sequence mutant was less deleterious for splicing as WT splicing was restored to nearly half to that of control splicing (lane 2 in Figure 4B, bar 2 in Figure 4C). The loss of suppressor activity of SLIIa loop U12 mutants suggests the important role of the element in U12-dependent splicing. In yeast, SLIIa plays an important role in the recruitment of U2 snRNP to form the pre-spliceosome. SLIIa along with stem IIc appears to act as a structural conformation toggling switch, which is responsible for the recruitment of U2 snRNP (42,43). The evidence that SLIIa of U2 snRNA has a structural element, which is presumably important for splicing is found in the structural study of SLIIa loop (44). This study demonstrated the existence of a U-turn of consensus UNRN (N, any ribonucleotide; R, purine) sequence element in the loop of SLIIa. In U12 SLIIa loop also, a putative UNRN element appears to exist from U41, G42, A43 and C44 (Figure 4A-WT) and the deletion of this structure abolished the U12-dependent splicing (lane 1 in Figure 4B, bar 1 in Figure 4C). Interestingly, the reverse complement sequence of the loop also contained a UNRN element at U42, C43, A44, C45 (Figure 4A-2) which might explain the better but incomplete restoration of WT splicing efficiency of this mutant (lane 2 in Figure 4B, bar 2 in Figure 4C) as compared to loop deletion mutant. In summation, our data show that SLIIa is required for U12-dependent splicing. This is consistent with the observation that the U2 snRNA structural element, which is functionally equivalent to U12 SLIIa, is essential for splicing in human, yeast and amphibian (45–48).
The function of SLIIa region in U12-dependent splicing has not been investigated before. Although the SLIIa structure appears to be similar, the primary sequence of this region is not highly conserved evolutionarily among U12 snRNA sequences that we have analyzed in this study (Supplementary Figure S2) and also demonstrated previously by others (28). The loop region of SLIIa appears to be highly variable as A. thaliana, S. moellendorffii, P. infestans, D. melanogaster and S. mansoni have larger predicted loops (28) suggesting that variation in length of SLIIa can be adjusted in the loop region without compromising the functional aspects of the snRNA. Similarly, the primary sequence of the predicted helix region is not conserved; however, these non-conserved residues can retain the predicted helix by compensatory base pairing (Supplementary Figure S2) suggesting that the requirement of the structure is essential for U12 snRNA function. This is not unprecedented as both prokaryotic and eukaryotic ribosomal RNAs also appear to have variable loop regions of functional importance (49). Interestingly, insects appear to have larger predicted SLIIa regions (28) suggesting that this feature could very well be a derived feature of insect U12 snRNAs and may not be functionally important in mammals.
The phylogenetically conserved SLIIb is dispensable for in vivo U12-dependent splicing
The mutational analysis of SLIIb of U12 snRNA revealed that the primary sequence changes and potential structural modifications due to sequence changes in SLIIb have little or no effect on splicing (Figure 5). Our data suggest the redundant nature of SLIIb. It is surprising that the primary sequence and predicted secondary structure of SLIIb is highly conserved among U12 snRNAs (Supplementary Figure S2). It is likely that dispensable U12 snRNA sequences do not have any essential role in splicing; however, the conservation of primary sequence and predicted structure of SLIIb implies an important function. What could be the function of this region? It is plausible that the region might contribute to some other function of U12 snRNA that cannot be scored in our in vivo splicing assay. Also, since our assay system only tested for predictable RNA:RNA interactions in transient transfections using RT–PCR methods, the effect of U12 snRNA mutants on the formation of intermediate spliceosomal complexes could not be studied.
Many conserved yet functionally dispensable RNA structural elements are found in nature. Most noticeable are the stem–loops in yeast and human U2 snRNA. In yeast U2 snRNA, conserved loops B and B′ and loop C, which are similar to U12 SLIIb and SLIII are shown to be non-essential for in vivo and in vitro splicing (45). Similarly, in human U2 snRNA, bottom half of stem I is dispensable for splicing. In fact, it was suggested that some part of this structure may be deleterious for splicing (48). It is interesting to note that phylogenetically conserved regions of ribosomal RNAs are also found to be dispensable for translation (49,50). Similarly, deletion of large portions of peripheral RNA structures of budding yeast RNase MRP had no significant effect on substrate recognition and hence, the survival of the organism (51). Within spliceosomal context, conserved loop I of U5 snRNA is dispensable for both catalytic steps of pre-mRNA splicing (52). This study demonstrated that the substitution of conserved nucleotides comprising internal loop 2 or deletion of internal loop 1 had no significant effect on the ability of reconstituted U5 snRNPs to complement splicing (52). These functionally dispensable RNA structures may be required but not necessary for the adjustment of secondary RNA folding pathway and perhaps, for stabilizing some tertiary interactions. These structures may not perform any function by themselves but their presence may be required for promoting the folding and function of neighboring structures. Therefore, the whole molecule is presumably, under evolutionary pressure to retain these structures in the context of benefit to the entire process, in this case, splicing.
In vivo evidence for a single SLIII secondary structure
Of the two earlier predicted possible structures, our data support the later U12 snRNA structure which has an alternative, single and long SLIII. Compensatory combination and matching deletion mutations designed to restore upper (Figure 6A-3 and Figure 6B, lane 3), middle (Figure 7B, lane 3) and lower (Figure 8A-4, 7 and Figure 8B, lanes 4, 7) helices of SLIII region of U12 snRNA restored in vivo splicing. In summary, results of these mutations strongly suggested the existence of base paired helices at least in the upper and the lower end of the stem-loop. Interestingly, our data show that the asymmetrical A89A90 bulge at the bottom of SLIII is an important requirement for U12-dependent splicing. The deletion of both nucleotides abolished the function of suppressor U12 snRNA in splicing (Figure 8A-1 and Figure 8B, lane 1) suggesting the importance of either two adenosines in this position or the asymmetric bulge in splicing. Our in vivo data supporting single and long SLIII structure is consistent with the structure proposed by Benecke et al. (30).
The human 18S U11/U12 snRNP contains a set of seven proteins that are not found in U2 snRNP suggesting that the recognition of 5′ splice and branch sites of U12-dependent introns is distinct from U2-dependent introns (32,53). RNA binding p65 protein, a member of these proteins, forms a bridge between U11 and U12 snRNA by binding to another protein of the complex, p59 (30). p65 appears to bind to apical stem–loop region of U12 SLIII and is presumably, required for spliceosomal functions. In addition, the U11-48K protein recognizes the 5′ splice site of a U12-dependent intron and bridges with U11-59K protein (54). These studies convincingly elucidate the role of unique U11/U12 complex proteins in organizing the U11 and U12 snRNAs to their respective splice sites for proper formation of spliceosomal complexes and productive U12-dependent splicing. The role of other proteins including 35K, 31K, 25K, 20K is still not clear. It is tempting to speculate that some of these proteins might be binding to other structural elements of U11 and/or U12 snRNAs to form multiple RNA: protein and protein: protein interactions, which might be essential for splicing. Many of the U12 snRNA structural elements including SLIIa, linker region and lower helices of SLIII, especially A89A90 asymmetrical bulge may be essential for binding these proteins. These complex RNA structures along with the dispensable one may provide a core molecular scaffold required for catalytically active spliceosome. However, further experiments are required to test this hypothesis.
We have also tested the requirement of U12 snRNA Sm binding site by nucleotide substitution (GCCCCCA) mutation. As expected, this mutant lacked suppressor activity (data not shown). The Sm binding site is highly conserved among all snRNAs including that of major and minor spliceosomes. It is well documented that Sm site binds to several Sm proteins which are required for maturation and function of snRNAs (55) and studies have established the role and importance of Sm binding site in major and minor spliceosomal snRNAs (33,55).
The primary sequence homology between human U2 and U12 snRNAs is very limited. However, at secondary structure level, these snRNAs are organized into similar structural domains, in particular, intramolecular SLI, SLIIa and SLIIb secondary structures. U12 snRNA is central to U12-dependent splicing as it binds to U6atac snRNA to form helix I and recognizes the branch site of a U12-dependent intron. U12 snRNA branch site interacting sequence and its requirement in U12-dependent splicing is well documented (3,8). However, the requirement of other regions of U12 snRNA in U12-dependent splicing has not been studied. This study provides a comprehensive analysis of human U12 snRNA-predicted secondary structure outside the branch site region and the putative intermolecular helix I forming region. Several mutants were designed to test the predicted base pairing interactions of proposed helices. In addition, mutations were designed to test the requirement of loops as well as distance constraints between the predicted stem–loops. Interestingly, most mutants that were engineered to restore helices with compensatory base pairing were functional, suggesting the existence of predicted intramolecular helices. In summation, our data establish the functional requirements of SLIIa, SLIIb, SLIII and single stranded linker regions of U12 snRNA in U12-dependent in vivo splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation award (MCB-0842606 to G.C.S.). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Rick Padgett for U12 snRNA GA23/24CU expression plasmid and P120 minigene constructs.
REFERENCES
- 1.Guthrie C, Patterson B. Spliceosomal snRNAs. Annu. Rev. Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987;325:673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- 3.Hall SL, Padgett RA. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 4.Kolossova I, Padgett RA. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA. 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 5.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 6.Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 7.Zhuang Y, Weiner AM. A compensatory base change in human U2 SnRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- 8.Brock JE, Dietrich RC, Padgett RA. Mutational analysis of the U12-dependent branch site consensus sequence. RNA. 2008;14:2430–2439. doi: 10.1261/rna.1189008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- 10.Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 11.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 12.Wu JA, Manley JL. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991;352:818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- 13.Yu YT, Steitz JA. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc. Natl Acad. Sci. USA. 1997;94:6030–6035. doi: 10.1073/pnas.94.12.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frilander MJ, Steitz JA. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell. 2001;7:217–226. doi: 10.1016/s1097-2765(01)00169-1. [DOI] [PubMed] [Google Scholar]
- 15.Incorvaia R, Padgett RA. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA. 1998;4:709–718. doi: 10.1017/s1355838298980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black DL, Chabot B, Steitz JA. U2 as well as U1 small nuclear ribonucleoproteins are involved in pre-messenger RNA splicing. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 17.Heinrichs V, Bach M, Luhrmann R. U1-specific protein-C is required for efficient complex-formation of U1 SnRNP with a 5′ splice site. Mol. Biol. Rep. 1990;14:165. doi: 10.1007/BF00360459. [DOI] [PubMed] [Google Scholar]
- 18.Sumpter V, Kahrs A, Fischer U, Kornstadt U, Luhrmann R. Invitro Reconstitution of U1 and U2 SnRNPs from isolated proteins and SnRNA. Mol. Biol. Rep. 1992;16:229–240. doi: 10.1007/BF00419662. [DOI] [PubMed] [Google Scholar]
- 19.Will CL, Rumpler S, Gunnewiek JK, vanVenrooij WJ, Luhrmann R. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 1996;24:4614–4623. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Manley JL. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 21.Wassarman KM, Steitz JA. The low-abundance U11 and U12 small nuclear ribonucleoproteins (SnRNPs) interact to form a two-snRNP complex. Mol. Cell. Biol. 1992;12:1276–1285. doi: 10.1128/mcb.12.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell AG, Charette JM, Spencer DF, Gray MW. An early evolutionary origin for the minor spliceosome. Nature. 2006;443:863–866. doi: 10.1038/nature05228. [DOI] [PubMed] [Google Scholar]
- 24.Shukla GC, Padgett RA. Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA. 1999;5:525–538. doi: 10.1017/s1355838299982213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson CG, Brown JW. U12-dependent intron splicing in plants. Curr. Top. Microbiol. Immunol. 2008;326:61–82. doi: 10.1007/978-3-540-76776-3_4. [DOI] [PubMed] [Google Scholar]
- 26.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 27.Bartschat S, Samuelsson T. U12 type introns were lost at multiple occasions during evolution. BMC Genomics. 2010;11:106. doi: 10.1186/1471-2164-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez MD, Rosenblad MA, Samuelsson T. Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res. 2008;36:3001–3010. doi: 10.1093/nar/gkn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marz M, Kirsten T, Stadler PF. Evolution of spliceosomal snRNA genes in metazoan animals. J. Mol. Evol. 2008;67:594–607. doi: 10.1007/s00239-008-9149-6. [DOI] [PubMed] [Google Scholar]
- 30.Benecke H, Luhrmann R, Will CL. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J. 2005;24:3057–3069. doi: 10.1038/sj.emboj.7600765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montzka KA, Steitz JA. Additional low-abundance human small nuclear ribonucleoproteins - U11, U12, Etc. Proc. Natl Acad. Sci. USA. 1988;85:8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Luhrmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10:929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla GC, Cole AJ, Dietrich RC, Padgett RA. Domains of human U4atac snRNA required for U12-dependent splicing in vivo. Nucleic Acids Res. 2002;30:4650–4657. doi: 10.1093/nar/gkf609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla GC, Padgett RA. The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA. 2001;7:94–105. doi: 10.1017/s1355838201000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dietrich RC, Padgett RA, Shukla GC. The conserved 3′ end domain of U6atac snRNA can direct U6 snRNA to the minor spliceosome. RNA. 2009;15:1198–1207. doi: 10.1261/rna.1505709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla GC, Padgett RA. U4 small nuclear RNA can function in both the major and minor spliceosomes. Proc. Natl Acad. Sci. USA. 2004;101:93–98. doi: 10.1073/pnas.0304919101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun JS, Manley JL. The human U6 snRNA intramolecular helix: structural constraints and lack of sequence specificity. RNA. 1997;3:514–526. [PMC free article] [PubMed] [Google Scholar]
- 38.Valadkhan S, Manley JL. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McPheeters DS, Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992;71:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- 40.Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth M, Luhrmann R. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol. Cell. Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall SL, Padgett RA. Conserved sequences in a class of rare eukaryotic nuclear introns with nonconsensus splice sites. J. Mol. Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 42.Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stallings SC, Moore PB. The structure of an essential splicing element: stem loop IIa from yeast U2 snRNA. Structure. 1997;5:1173–1185. doi: 10.1016/s0969-2126(97)00268-2. [DOI] [PubMed] [Google Scholar]
- 45.Ares M, Igel AH. Mutations define essential and nonessential U2 RNA structures. Mol. Biol. Rep. 1990;14:131–132. doi: 10.1007/BF00360444. [DOI] [PubMed] [Google Scholar]
- 46.Hamm J, Dathan NA, Mattaj IW. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989;59:159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- 47.Vankan P, Hamm J, Dathan NA, Mattaj IW. Structure-function in Xenopus snRNPs. Mol. Biol. Rep. 1990;14:209–210. doi: 10.1007/BF00360477. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Manley JL. Multiple functional domains of human U2 small nuclear RNA: strengthening conserved stem I can block splicing. Mol. Cell Biol. 1992;12:5464–5473. doi: 10.1128/mcb.12.12.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 50.Nierhaus KH, Beyer D, Dabrowski M, Schafer MA, Spahn CM, Wadzack J, Bittner JU, Burkhardt N, Diedrich G, Junemann R, et al. The elongating ribosome: structural and functional aspects. Biochem. Cell Biol. 1995;73:1011–1021. doi: 10.1139/o95-108. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Zaman S, Langdon Y, Zengel JM, Lindahl L. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Res. 2004;32:3703–3711. doi: 10.1093/nar/gkh689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segault V, Will CL, Polycarpou-Schwarz M, Mattaj IW, Branlant C, Luhrmann R. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Luhrmann R, Query CC. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turunen JJ, Will CL, Grote M, Luhrmann R, Frilander MJ. The U11-48K protein contacts the 5′ splice site of U12-type introns and the U11-59K protein. Mol. Cell. Biol. 2008;28:3548–3560. doi: 10.1128/MCB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 56.Tarn WY, Yario TA, Steitz JA. U12 SnRNA in vertebrates - evolutionary conservation of 5′-sequences implicated in splicing of pre-messenger-RNAs containing a minor class of introns. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.