Abstract
In eukaryotes, exposure to stress conditions causes a shift from cap-dependent to cap-independent translation. In trypanosomatids, environmental switches are the driving force of a developmental program of gene expression, but it is yet unclear how their translation machinery copes with their constantly changing environment. Trypanosomatids have a unique cap structure (cap-4) and encode four highly diverged paralogs of the cap-binding protein, eIF4E; none were found to genetically complement a yeast mutant failing to express eIF4E. Here we show that in promastigotes, a typical cap-binding complex is anchored through LeishIF4E-4, which associates with components of the cap-binding pre-initiation complex. In axenic amastigotes, expression of LeishIF4E-4 decreases and the protein does not bind the cap, whereas LeishIF4E-1 maintains its expression level and associates with the cap structure and with translation initiation factors. However, LeishIF4E-1 does not interact with eIF4G-like proteins in both life stages, excluding its involvement in cap-dependent translation. Using pull-down assays and mass-spectrometry, we identified a novel, non-conserved 4E-Interacting Protein (Leish4E-IP), which binds to LeishIF4E-1 in promastigotes, but not in amastigotes. Yeast two-hybrid and NMR spectroscopy confirmed the specificity of this interaction. We propose that Leish4E-IP is a translation regulator that is involved in switching between cap-dependent and alternative translation pathways.
INTRODUCTION
Translation initiation of most mRNAs in higher eukaryotes proceeds through a cap-dependent mechanism, whereby the small ribosomal subunit associates with the cap-binding eukaryotic translation Initiation Factor 4F (eIF4F) complex at the 5′-end of mRNAs. eIF4F consists of a cap-binding protein, eIF4E; a large scaffold MIF4G-domain protein, eIF4G; and a DEAD-box RNA helicase, eIF4A, which promotes RNA secondary structure unfolding in the 5′-UTR for scanning by the complex until the first AUG codon is reached. In higher eukaryotes, eIF4G recruits eIF3, that is associated with the 40S ribosomal subunit. eIF4G also interacts with the Poly(A)-Binding Protein (PABP) at the 3′-end of the mRNA, allowing a transient circularization of the mRNA (1).
Assembly of the cap-binding complex can be globally regulated by the eIF4E-binding protein, 4E-BP, which is a conserved, small and mostly unstructured protein of about 10 kDa (2,3). 4E-BP and eIF4G both contain a Y(X4)LΦ motif, where X is any amino acid and Φ is a hydrophobic residue, that is responsible for binding to eIF4E (4,5). 4E-BP binds eIF4E when it is in a hypophosphrylated state (6), whereas phosphorylation of 4E-BP by the mTOR kinase reduces its affinity to eIF4E (7). Thus activation of the mTOR kinase pathway increases cap-dependent translation. While eIF4E, eIF4G, eIF4A and PABP homologs have been identified in the genome databases of most eukaryotes, so far 4E-BP remains unidentified in members of the Trypanosomatidae family as well as in Caenorhabditis elegans.
Leishmania are unicellular protozoans with a complex life cycle. They reside as flagellated promastigotes in the alimentary canal of sand-fly vectors. After being transmitted into the mammalian host through a blood-meal from the female vector, they are engulfed by macrophages and other cells of the immune system. The promastigotes then transform into obligatory intracellular and non-motile amastigotes (8). During their life cycle the parasites are exposed to highly variable and hostile environments. The temperature shift caused by entry into mammalian hosts induces a heat shock response that is an integral part of Leishmania life cycle. In higher eukaryotes, short term heat excess represses cap-dependent translation (9), and prolonged heat shock eventually causes cell death. In Leishmania, such conditions initially cause growth arrest (10), and then lead to a developmental switch that generates thermo-resistant organisms.
Leishmania parasites are ancient eukaryotes, known for their unique molecular features. Protein coding genes in these organisms are constitutively transcribed as part of the large polycistronic chromosomal units, that are further processed by trans-splicing and polyadenylation into mature monocistronic mRNAs (11,12). During trans-splicing, a conserved spliced leader RNA (SL RNA) is joined to all mRNAs (13), providing a unique cap structure (14). This cap is highly modified in the first four nucleotides (m7Gpppm36,6,2′Apm2′Apm2′Cpm23,2′U), as compared to the simple m7GTP cap used in higher eukaryotes, and is denoted cap-4 (14).
Since protein coding genes in Leishmania are not regulated by transcriptional activation, translation regulation is believed to play a key role in driving the differential pattern of gene expression throughout the life cycle, after transmission from the insect vector to the mammalian host. We formerly characterized four cytoplasmic eIF4E orthologs of Leishmania (LeishIF4E1-4), and showed that they are highly diverged from their counterparts in higher eukaryotes. Indeed, none of the LeishIF4E paralogs could complement a yeast mutant that fails to express its own eIF4E (15). We further identified the parasite eIF4G homolog, LeishIF4G-3, as the scaffold protein of the LeishIF4F cap-binding complex (16). LeishIF4G-3 is one of six proteins identified in the Leishmania genome that contain a MIF4G domain, but all show a low degree of homology to mammalian eIF4GI. The interaction between LeishIF4E-4 and LeishIF4G-3 is mediated by an eIF4E binding peptide within the LeishIF4G-3 (YPGFSLD), which resembles only in part the previously mentioned consensus eukaryotic motif (5,17,18). Both proteins are eluted from a m7GTP-sepharose column and comigrate on sucrose gradients in fractions that contain the pre-initiation complex of translation. Thus LeishIF4E-4 and LeishIF4G-3 could function as components of the eIF4F complex. After exposure to elevated temperatures the majority of LeishIF4E-4 shifts to the upper part of the gradient, whereas LeishIF4E-1 changes its migration profile on sucrose gradients and enters heavier fractions (16). This pattern of migration led us to examine in detail the changes in the translation machinery that take place during parasite stage differentiation.
In this work we characterize the LeishIF4E-1 and LeishIF4E-4-binding partners in the different life stages of Leishmania amazonensis. We show that the LeishIF4E-4 complex participates in the translation initiation process in promastigotes, but during heat shock and in amastigotes its expression decreases and it fails to bind both LeishIF4G-3 and the cap structure. Unlike LeishIF4E-4, the expression level of LeishIF4E-1 under these conditions remains unchanged and the protein continues to bind m7GTP. However, LeishIF4E-1 fails to interact with any MIF4G domain protein in both life stages. This suggests that, at least for some of the genes expressed in amastigotes, translation initiation may proceed in a cap-independent manner, a process that is also common under stress conditions in higher eukaryotes. We further report a novel 4E-Interacting Protein (Leish4E-IP) that associates with LeishIF4E-1 in promastigotes, but not in amastigotes. This new Leish4E-IP could therefore direct the stage-specific function of LeishIF4E-1.
MATERIALS AND METHODS
Organisms
Leishmania amazonensis and Leishmania major (Friedlin strain) promastigotes were cultured in Schneider's medium (pH 7.0) supplemented with 10% fetal calf serum (FCS), 4 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin at 25°C.
Host free differentiation and maintenance of L. amazonensis axenic amastigotes was performed in Schneider's medium (pH 5.5) supplemented with 20% fetal calf serum (FCS), 4 mM l-glutamine, 60 U/ml penicillin and 60 µg/ml streptomycin. The cells were transferred into the acidic medium and grown at 25°C for 6 days and then transferred to 33°C, as described previously (19,20).
Vectors and transfection
For pull-down experiments using L. amazonensis, a Streptavidin-binding Peptide (SBP) tagging vector pX-H-SBP-H was used. This vector contains an SBP-tag flanked by two intergenic regions derived from the Hsp83 genomic cluster (denoted ‘H’), that promote expression in both life forms. Foreign genes were cloned upstream to the SBP tag. For experiments with L. major a Tandem Affinity Purification (TAP) tagging vector pSNSAP1 (21) was used, placing the tag in the C-terminus of the target gene. See Supplementary ‘Materials and Methods’ section for plasmids construction details.
Leishmania amazonensis cells were transfected as previously described (22) with pX-H-LeishIF4E1-SBP-H, pH-LeishIF4E4-SBP-H and pH-LeishIF4EIP-SBP-H and stable cell lines were selected using 100 µg/ml G-418. Leishmania major cells were similarly transfected with pSNSAP1-LeishIF4E-1 or pSNSAP1-LeishIF4E-4, and stable cell lines were selected using 200 µg/ml G-418.
In vivo pull-down analysis of the LeishIF4F complex
For pull-down assays using transgenic L. amazonensis, cells (0.6–1 × 109) were harvested, washed, resuspended in Binding Buffer (BB) [35 mM HEPES, pH 7.5, 10 mM MgCl2, 100 mM KCl, 1 mM DTT, 2 mM iodoacetamide and a cocktail of protease inhibitors (Sigma)] and lysed by sonication. Supernatants were agitated with streptavidin-Sepharose beads (GE Healthcare) for 2 h at 4°C. The beads were washed with BB containing 0.1% NP-40 and proteins were eluted with BB containing 2 mM biotin. Leishmania major transgenic cells (0.6–1 × 109) were extracted and the tagged proteins were purified over streptavidin–sepharose, as described above. The final eluate was loaded on a m7GTP-Sepharose column, which was washed with BB containing 0.1% NP-40 and then with the same buffer containing 0.1 mM GTP. Proteins were eluted with BB containing 0.1% NP-40 containing 0.5 M NaCl. The endogenous cap-binding proteins and their associated partners were isolated from wild-type L. amazonensis cells. Lysates were loaded on a m7GTP-Sepharose column, washed and eluted as described above. In all cases the eluted proteins were precipitated by TCA, resolved by SDS–PAGE (10–15%) and subjected to western blot analysis with specific antibodies against the different LeishIF4F subunits.
Mass spectrometry analysis
Proteins that were pulled down with the tagged LeishIF4E-1 of LeishIF4E-4 from L. amazonensis cell lines (3–4 × 109 cells) were precipitated by TCA and further resolved on 10% SDS–PAGE. Gels were stained by Coomassie-blue and the lanes were split into three or four segments that were each subjected to LC-MS/MS analysis on the Orbitrap (Thermo) mass spectrometer. Proteins were identified by BLAST analysis against the L. major database using Sequest 3.31 software (Smaller Protein Center, Technion, Haifa). Control experiments were performed on non-transfected wild-type cells, and the identified proteins were eliminated from the complete list (this list is given in Supplementary Table S2).
Monitoring protein expression levels
Late log promastigotes grown at 25°C, promastigotes that were exposed to elevated temperatures (2 h at 33°C) and axenic amastigotes at least 8 days after differentiation, were compared. The cells were washed in PBS, resuspended in BB and lysed by addition of 1% Triton X-100. The cell debris was precipitated and the supernatant was collected for analysis. Protein concentrations were determined by the BCA kit (Thermo). Equal protein amounts (50 µg) were resolved on 10–15% SDS–PAGE and then subjected to western analysis with specific antibodies against the different LeishIF4F subunits. The differentiation state was monitored by antibodies against specific markers—GP46 (a gift from Ch. Jaffe, Hebrew University, Israel) and HSP100 (a gift from J. Clos, Bernhard Nocht Institute for Tropical Medicine, Germany). GP46 is a convenient promastigote specific marker for L. amazonensis, which is completely repressed in amastigotes of this species (20) and Hsp100 is an amastigote-specific protein, and is therefore a useful indicator of stage transformation (23). Band intensities were determined using ImageJ software.
Yeast two-hybrid assay
A yeast two-hybrid assay was performed using the commercial GAL4 Two-Hybrid Phagemid Vector Kit (Stratagene) following the manufacturer's instructions. The ORFs of LeishIF4E-1 through -4, LeishIF4E-4 W163A, LeishIF4A-1, LeishPABP-1 and LeishIF4G-3 were cloned into the GAL4-binding domain vector (pBD) using EcoRI and SalI sites (for a list of primers used, see Supplementary Table S1). In addition, the ORFs of LeishIF4G-1 through -6, Leish4E-BP, LeishIF4E-1, LeishIF4E-4 and LeishIF4E-4(Δ1-86) were cloned into the GAL4 activation domain vector (pAD) through BamHI and XbaI or EcoRI and SalI sites (for a list of primers used, see Supplementary Table S1). A point mutation in LeishIF4E-4 was generated using the Phusion enzyme (Finnzymes). All constructs were confirmed by DNA sequencing.
The yeast strain YRG-2 [Mata ura352 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::UASGAL1-TATA GAL1-HIS3 URA3::UASGAL4 17mers(x3) TATACYC1-lacZ] was co-transformed with specific pAD and pBD constructs. Yeast transformants were cultured in Liquid SD-2 (-Trp/-Leu) medium at 30°C overnight, diluted to a final concentration of OD600 = 0.15 and growth was continued to OD600 = 0.5. Yeast were spotted on SD-2 (-Trp/-Leu) and SD-3 (-Trp/-Leu/-His) containing or not 1 mM 3-amino-1,2,4-triazole (3-AT) (Sigma) plates. pAD-WT and pBD-WT or pGAL-WT and pLaminC plasmids were used, respectively, as positive or negative controls, following the manufacturer's instructions.
LeishIF4E-1 NMR titration experiments with LeishIF4E-IP
His-tagged recombinant LeishIF4E-1 was expressed in Escherichia coli using M9 media supplemented with 15N-ammonia as the only nitrogen source. Expression was carried out in 2H2O to obtain partially deuterated protein, which yields sharper resonances and results in better spectroscopic behavior. The protein was purified over a nickel-agarose column (Quiagen) in 50 mM Na2HPO4, 50 mM NaH2PO4, 500 mM NaCl, 2 mM DTT, pH 7.4 containing increasing concentrations of imidazole from the lysis buffer (10 mM) to the elution buffer (500 mM). The His-tag was then cleaved using the TEV protease, and the protein was further purified over a Superdex 75 16/60 gel filtration column (Amersham) using a buffer containing 50 mM Na2HPO4, 50 mM NaH2PO4, 100 mM NaCl and 2 mM DTT at pH 7.4. LeishIF4E-1 was concentrated to 100 µM and incubated with 5 mM m7GTP. Excess m7GTP was removed using a PD-10 column (GE Healthcare) and the buffer was exchanged to NMR buffer comprising 50 mM Na2HPO4, 50 mM NaH2PO4, 100 mM NaCl, 2 mM DTT and 8% D2O at pH 6.5. NMR spectra were recorded at 298 K on a Varian Inova 600-MHz spectrometer equipped with a cryoprobe. TROSY versions of HSQC spectra were recorded with 64 complex points in the indirect 15N dimension and 1024 complex points in the direct dimension. The concentration of the LeishIF4E-IP 15 residue peptide (VRTMYTREELLRIAT, synthesized by Peptide2.0 inc.) was gradually increased from 50 to 500 µM, giving a molar ratio of 0.5–5 relative to LeishIF4E-1.
RESULTS
The LeishIF4E-4 complex of promastigotes
We and others, have previously shown that LeishIF4E-4 interacts with LeishIF4G-3 (16,24). To gain insight into the role of LeishIF4E-4 in translation initiation we sought to identify its binding partners. We initially focused on the promastigote stage. For this purpose a Streptavidin-binding Peptide tag (SBP, 46 amino acids) was added at the C-terminus of LeishIF4E-4 and the fusion protein was expressed in transgenic L. amazonensis promastigotes. The tagged LeishIF4E-4 and its associated proteins were purified from transgenic parasites by affinity purification over Streptavidin–sepharose, and the eluted proteins were analyzed by mass spectrometry (LC–MS/MS). LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1 were part of the tagged complex (Table 1), which was confirmed by western blot analysis, using specific antibodies (Figure 1A). In addition to the LeishIF4F subunits, LeishIF4E-4 associated with additional translation initiation factors, including LeishPABP-1 and -2, LeishIF5, LeishIF5B, subunits of LeishIF3 and LeishIF2, as well as ribosomal proteins (Table 1 and Supplementary Table S2). Additional proteins known to be associated with the translation machinery were identified, including the Polypyrymidine Tract-Binding Protein 1 (PTB1), a pluripotent protein that has been implicated in translation (25); its Leishmania homolog was shown to interact with LeishIF4G-3 in a yeast two hybrid system (David M., unpublished data). The precipitated complex also contained two homologs of a yeast protein known to be involved in translation initiation, Ded1p (26), and several RNA helicases of yet unknown function. A large number of peptides were derived from the translation elongation machinery, LeishEF1, LeishEF2 and LeishEF-TU (Table 1). Multiple proteins associated with other cellular functions were also identified (Supplementary Table S2). These results strongly suggest that LeishIF4E-4 is part of the functional translation initiation complex in promastigotes. The pull down analysis also revealed a small number of peptides derived from LeishIF4E-3 and LeishIF4G-4. These two proteins interact with each other [(24) and Manor et al., in preparation], but they are unlikely to play a role in translation initiation, since LeishIF4E-3 does not bind cap-4 (15). LeishIF4E-3 is part of a large nuclease-resistant complex of approximately 80S, which is associated with stress granules in Trypanosoma brucei (27).
Table 1.
Annotated translation initiation factors associated either with LeishIF4E-1 or LeishIF4E-4
| GeneDB ID | Annotation | Promastigotes |
Axenic Amastigotes | |
|---|---|---|---|---|
| 4E1 | 4E4 | 4E1 | ||
| LmjF27.1620 | LeishIF4E-1 | 18,19 | 2 | 13,46 |
| LmjF30.0450 | LeishIF4E-4 | 17,20 | ||
| LmjF16.1600 | LeishIF4G-3 | 33,31 | ||
| LmjF01.0770-80 | LeishIF4A-1 | 6,14 | 8,17 | 11,9 |
| LmjF35.5040 | LeishPABP-1 | 9,9 | 19,23 | |
| LmjF35.4130 | LeishPABP-2 | 10,7 | 8,12 | 6,4 |
| LmjF36.6060 | LeishIF4G-4 | 4 | 6 | 3 |
| LmjF38.2500 | LeishIF4E-3 | 3,3 | ||
| LmjF35.3980 | Leish4E-IP | 4,5 | 7,9 | |
| LmjF03.0980 | LeishIF2-alpha | 6,9 | 8,10 | 8,4 |
| LmjF36.4930 | LeishIF2-beta | 2 | ||
| LmjF09.1070 | LeishIF2-gamma | 2,7 | 5,8 | 6,2 |
| LmjF17.1290 | LeishIF3b | 5,3 | 3 | 2 |
| LmjF36.6980 | LeishIF3c | 3,3 | 2 | 3,2 |
| LmjF28.2310 | LeishIF3e | 3 | ||
| LmjF36.3880 | LeishIF3i | 4 | 2 | |
| LmjF32.2180 | LeishIF3k | 2 | ||
| LmjF34.0350 | LeishIF5 | 3,6 | 5 | 4 |
| LmjF33.2740 | LeishIF5B | 13 | 2,15 | 5 |
| LmjF08.0550 | translation initiation factor-like protein | 8 | 5 | 4,2 |
| LmjF04.1170 | LeishPTB1 | 6 | 6 | 4,2 |
| LmjF34.0840 | LeishEF1-beta | 2 | ||
| LmjF09.0970 | LeishEF1-gamma | 5,5 | 4 | 5,2 |
| LmjF36.0180-90 | LeihsEF2 | 20,24 | 13,27 | 15,11 |
| LmjF18.0740 | LeishEF-TU | 4,6 | 5 | |
| LmjF32.0400 | LeishDed1p-1 | 11,19 | 12,23 | 12,12 |
| LmjF35.3100 | LeishDed1p-2 | 3,7 | 12 | 8,4 |
The table shows a list of translation factors that were pulled down by tagged LeishIF4E-1 and LeishIF4E-4 in promastigotes, and proteins that were pulled down by LeishIF4E-1 in axenic amastigotes. Proteins were subjected to mass spectrometry analysis. The number of identified peptides for each protein is indicated. The results represent data from two independent experiments. The complete list of proteins is provided in Supplementary Table S2.
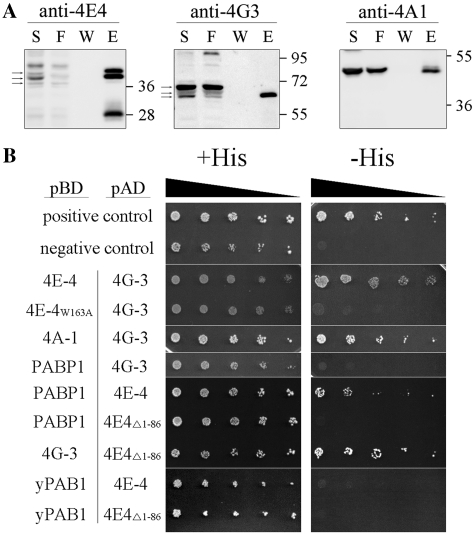
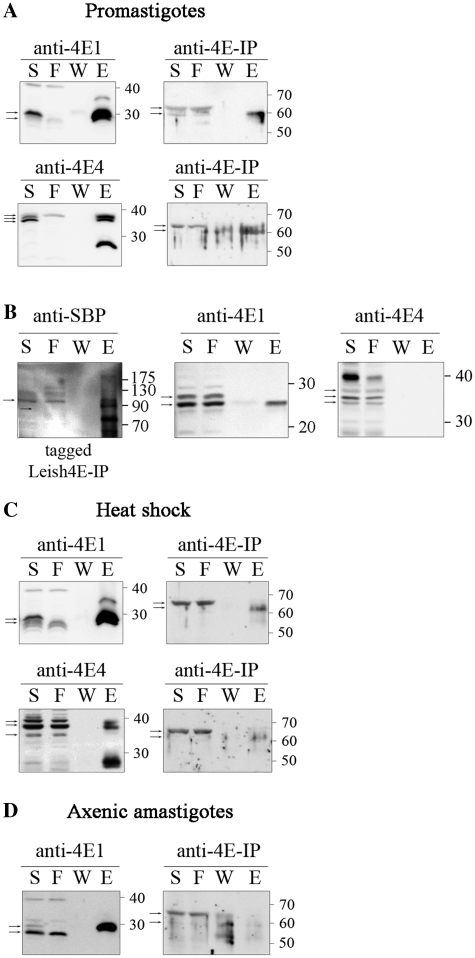
Figure 1.
Proteins associated with LeishIF4E-4 in promastigotes. (A) Pull-down experiments with the SBP-tagged LeishIF4E-4 from L. amazonensis promastigotes was done using affinity purification over streptavidin–Sepharose beads. Aliquots of the soluble extract (S, 1%), the flow-through (F, 1%), the final wash (W, 20%) and eluted proteins (E, 20%) were separated by SDS–PAGE (10–15%) and subjected to western blot analysis using specific antibodies against LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1. Densitometric analysis that was normalized to the total protein amounts showed that the elution fraction contained 16, 9 and 3.4% of the total LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1, respectively. The arrows indicate the specific reaction of the antibodies. (B) The yeast two-hybrid assay was performed by cotransfecting wild-type YRG-2 yeast strain with pBD fused to LeishIF4E-4, LeishIF4E-4 W163A mutant, LeishIF4A-1, LeishPABP-1, LeishIF4G-3 or yPAB1 and pAD-LeishIF4G-3, pAD-LeishIF4E-4 or pAD-LeishIF4E-4Δ1-86. Cells were cultured under restrictive (−His) and non-restrictive (+His) growth conditions, and spotted as described in ‘Materials and Methods’ section. Positive and negative controls were supplied by the manufacturer and expression of the tested proteins was verified by western blot analysis (Supplementary Figure S7).
We used a yeast two-hybrid assay to characterize direct associations between components of the LeishIF4E-4 complex. Residues in the LeishIF4G-3 that are required for the interaction with LeishIF4E-4 have been formerly characterized (16). Based on sequence analysis and alignment of conserved regions between LeishIF4E-4 and heterologous eIF4Es (17,28), Trp163 in LeishIF4E-4 was highlighted as a key residue responsible for the interaction with LeishIF4G-3 (Supplementary Figure S1). The yeast two-hybrid assay shows that the interaction between LeishIF4E-4 and LeishIF4G-3 is indeed disrupted by the W163A mutation in LeishIF4E-4. The assay also confirmed the interaction between LeishIF4A-1 and LeishIF4G-3 (Figure 1B), which is the functional ortholog of eIF4A in Leishmania (29).
The higher eukaryote scaffold protein eIF4G is also known to interact with PABP. Since LeishPABP-1 was pulled-down by the tagged LeishIF4E-4, we sought to understand how LeishPABP-1 is associated with the eIF4F complex in Leishmania. This was especially relevant knowing that LeishIF4G-3 has a short amino-terminus of only 50 amino acids upstream of the MIF4G domain, while the human eIF4GI has an extended amino terminus of 760 amino acids that interacts with PABP during translation initiation (30,31). Yeast two-hybrid experiments show that LeishIF4G-3 does not interact with LeishPABP-1, but that the latter protein is recruited to the cap-binding complex through LeishIF4E-4 (Figure 1B). Such an association has never been reported in higher eukaryotes. This interaction was mediated by the long, non-conserved N-terminus of LeishIF4E-4 [86 amino acid (Supplementary Figure S2)]. LeishIF4E-4 lacking this region (LeishIF4E-4Δ1−86) failed to interact with LeishPABP-1 in a yeast two hybrid assay, while the association between the truncated LeishIF4E-4 and LeishIF4G-3 was unaffected (Figure 1B). Furthermore, the interaction between LeishIF4E-4 and LeishPABP-1 was specific to Leishmania, and was not observed for the yeast PAB1. It is therefore most likely that the LeishIF4E-4 N-terminus is responsible for mRNA circularization during translation initiation, emphasizing yet another difference between the cap-binding complex of Leishmania and higher eukaryotes. These results vary from a former report claiming an interaction between LeishIF4G-3 and LeishPABP-1 (32). However, the reported interaction was obtained using radiolabeled proteins that were generated in an in vitro transcription-translation system, where RNA could serve as a mediator for this interaction.
Proteins that associate with LeishIF4E-1 in promastigotes
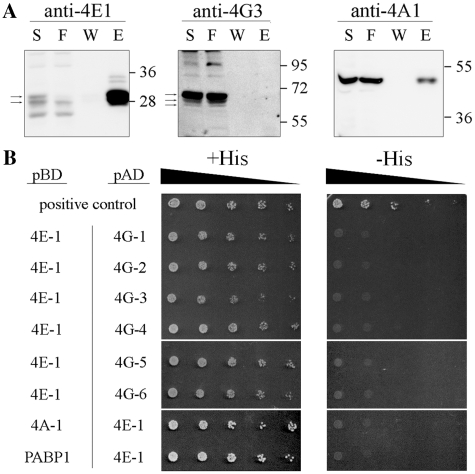
LeishIF4E-1 is another eIF4E homolog in Leishmania that was previously considered as a candidate to function in the translation initiation process (15,16). To investigate the role of LeishIF4E-1 in promastigotes the protein was SBP-tagged, affinity purified over Streptavidin–sepharose and the eluted proteins were analyzed by mass-spectrometry. LeishIF4E-1 pulled down several translation initiation factors (Table 1), including LeishIF4A-1 (verified by western blot analysis, Figure 2A), LeishPABP-1 and -2, subunits of eIF3 and eIF2 and other proteins that are related to translation. Proteins associated with LeishIF4E-1 were mostly similar to the ones associated with LeishIF4E-4 as described above. However, in this assay, neither LeishIF4G-3 nor any other eIF4G candidates were pulled-down (Table 1 and Figure 2A). This was verified by a yeast two-hybrid assay, in which all Leishmania MIF4G containing proteins, LeishIF4G-1 to -6, respectively (LmjF30.1150, LmjF15.1320, LmjF16.1600, LmjF36.6060, LmjF10.1080 and LmjF36.5160) failed to interact with LeishIF4E-1 (Figure 2B). Table 1 indicates a minor representation of LeishIF4G-4 peptides, but a direct interaction with LeishIF4E-1 was not reproduced by western blot analysis (Supplementary Figure S3). Furthermore, LeishIF4E-1 did not interact directly with LeishIF4A-1 or with LeishPABP-1, thus ruling out unexpected interactions within the LeishIF4E-1 complex.
Figure 2.
Proteins associated with LeishIF4E-1 in promastigotes. (A) Pull-down experiments with the SBP-tagged LeishIF4E-1 from L. amazonensis promastigotes was done using affinity purification and further analysis as described in Figure 1A. The antibodies used were directed against LeishIF4E-1, LeishIF4G-3 and LeishIF4A-1. Densitometric analysis showed that the elution fraction contained 24, 0 and 3% of the total LeishIF4E-1, LeishIF4G-3 and LeishIF4A-1, respectively. (B) The yeast two-hybrid assay was performed by co-transfecting wild-type YRG-2 yeast strains with pBD fused to LeishIF4A-1, LeishPABP-1 or LeishIF4E-1 and with pAD fused to LeishIF4G-1 through -6 or to LeishIF4E-1. The cells were cultured and spotted as described in ‘Materials and Methods’ section. Expression of the tested proteins was verified by western blot analysis (Supplementary Figure S7).
Characterization of translation initiation factors in amastigotes
We used an axenic amastigote system of L. amazonensis to understand if and how translation initiation is affected by parasite stage differentiation. As previously described, differentiation to amastigotes in certain Leishmania species can be induced under axenic conditions in cultured parasites following exposure to acidified growth media and elevated temperatures. These conditions mimic the environment that the parasites experience within the mammalian host, and trigger a gene expression profile that mimics the differentiation process (10,23,33,34).
Axenic amastigote-like cells resemble the intracellular parasites in their basic morphology, and can proliferate for prolonged periods. Here we show that L. amazonensis cells that were shifted from 25 to 33°C and from neutral to an acidic milieu (pH 5.5) rounded up after 24 h, and that their flagellum was adsorbed (Supplementary Figure S4). Expression of Hsp100, an amastigote marker (23), increased upon temperature elevation, whereas expression of GP46, a promastigote specific antigen of L. amazonensis (20), decreased under these conditions to a level that was almost undetectable in axenic amastigotes (Figure 3A). Growth of the amastigote-like cells was monitored for eleven days and compared to that of promastigotes (Supplementary Figure S4). The doubling time of promastigotes was 7–9 h while that of axenic amastigotes was ~10-fold longer, in line with former reports on proliferation rates during in vitro differentiation (23). In agreement with this observation, translation in axenic amastigotes was also ~10-fold lower than in promastigotes, as measured by metabolic labeling of the cells (data not shown). At different time periods after differentiation, axenic amastigotes could dedifferentiate into the promastigote stage (after 24 h of incubation at 26°C), supporting the functionality of the system and long-term viability of the amastigotes. In addition, there was no difference between the growth patterns of wild-type and transgenic axenic amastigotes (data not shown).
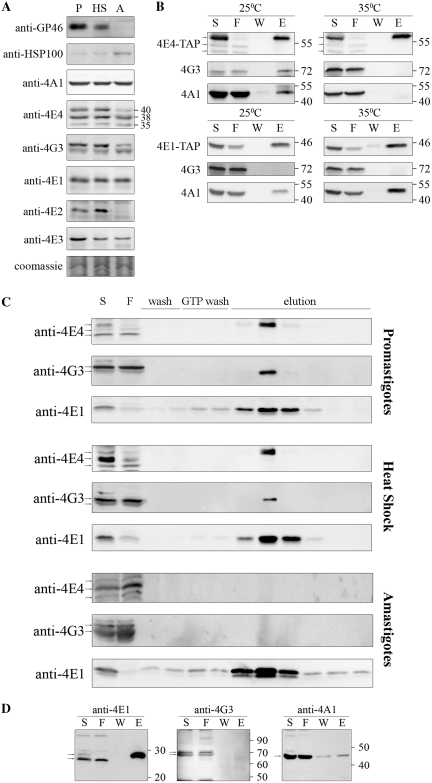
Figure 3.
Characterization of the LeishIF4F complex during stage differentiation. (A) Expression levels of LeishIF4F subunits in wild-type L. amazonensis, exposed or not to heat shock and differentiated in vitro. Cells were lysed and samples with 50 µg protein from the supernatants were resolved on SDS–PAGE (10–15%) and subjected to western blot analysis using specific antibodies against GP46, HSP100, LeishIF4E-1 to -4, LeishIF4G-3 and LeishIF4A-1. P, promastigotes from late log phase cultures; HS, late log promastigotes following a transient exposure to 33°C for 2 h; A, axenic amastigotes 9 days after differentiation. Coomassie staining of Tubulin was used to verify the protein loads. (B) Pull-down experiments with TAP tagged LeishIF4E-1 or LeishIF4E-4 from L. major promastigotes, exposed or not to heat shock (35°C, 2 h). Cell extracts were affinity purified over streptavidin-Sepharose beads and then over m7GTP-Sepharose column. The pulled-down proteins were analyzed as described in Figure 1A, using specific antibodies against LeishIF4E-1, LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1. (C) m7GTP pulldown assays with wild-type L. amazonensis promastigotes, promastigotes after 2 h at 33°C and axenic-amastigotes, 9 days after differentiation. The cells were lyzed and loaded on a m7GTP-Sepharose column. The eluted proteins were analyzed as described in Figure 1A, using specific antibodies against LeishIF4E-1, LeishIF4E-4 and LeishIF4G-3. (D) Pull-down experiments in axenic-amastigotes of L. amazonensis expressing SBP tagged LeishIF4E-1, 9 days after differentiation. Proteins were purified and analyzed as described in Figure 1A, using specific antibodies against LeishIF4E-1, LeishIF4G-3 and LeishIF4A-1.
Since the cap-dependent translation machinery is known to be temperature-sensitive in higher eukaryotes, its adaptation to axenic amastigotes was examined. To monitor possible changes, we focused on the LeishIF4F complex. We measured the expression level of its subunits along the differentiation process, and monitored their ability to assemble on the 5′ cap. The steady-state level of LeishIF4A-1 remained unchanged through short (2 h) and prolonged heat shock treatments, which simulate the process of differentiation into amastigote-like cells (Figure 3A). This is in agreement with other reports describing equivalent levels of expression for TbEIF4AI in procyclic and bloodstream forms of T. brucei (29). The short-term heat shock treatment did not lead to drastic changes in LeishIF4E-4 and LeishIF4G-3 expression. However, their expression in axenic amastigotes clearly decreased (Figure 3A). In agreement with our findings, proteomic studies performed in L. donovani show a slight reduction in the levels of LeishIF4E-4 and LeishIF4G-3 during early differentiation (2.5–10 h), while at later time points (15–144 h), expression of both proteins is reduced below the detection level (23). The LeishIF4E-4 and LeishIF4G-3 specific antibodies recognize more than one band in our assays; these could possibly represent post-translationally modified forms of the proteins. In both cases the band with the largest molecular weight was reduced by ~70% in differentiated cells, whereas the lower molecular weight bands did not show major changes (Figure 3A). Supplementary Figure S5 shows that the larger form of LeishIF4E-4 was exclusively responsible for the interaction with FLAG-tagged LeishIF4G-3 in promastigotes.
We tested the effect of increased temperatures on the association between LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1 in L. major cells expressing a TAP-tagged LeishIF4E-4. The tagged complex derived from cells grown at 26°C and after 2 h exposure to 35°C was affinity purified first over Streptavidin–sepharose and then over m7GTP-sepharose. Western blot analysis of the eluted proteins indicated that the interaction between subunits of the LeishIF4E-4 complex was weakened following heat shock (Figure 3B). This observation is supported by former polysome profiling experiments in heat-shocked cells, in which LeishIF4E-4 shifted to the top of the sucrose gradients, indicating that its association with the translation initiation complex was impaired (16). Similar results were obtained with L. amazonensis exposed to short term heat shock treatment. Pull-down experiments of tagged LeishIF4E-4, performed in axenic amastigotes of L. amazonensis, could not be completed since the protein was hardly detectable in these cells, which were maintained during prolonged periods at elevated temperatures. The use of a proteasome inhibitor (MG132) did not lead to elevated levels of the tagged protein (data not shown). To overcome this problem, the endogenous LeishIF4E-4 complex from wild-type axenic amastigotes was purified over a m7GTP column (Figure 3C). Unlike in promastigotes grown at 26°C, or after a short heat shock treatment (33°C, 2 h), LeishIF4E-4 from axenic amastigotes failed to bind to m7GTP-sepharose. Antibodies against this protein interact with three bands, which could represent products of post-translational modifications. The two smaller bands fail to bind the cap structure in all life forms, and only the largest band is eluted from the m7GTP column. It therefore appears that this specific form of LeishIF4E-4 is responsible for the cap-binding activity. As expected, LeishIF4G-3 was also absent from the eluate in amastigotes. We therefore conclude that LeishIF4E-4 loses its ability to bind the 5′ cap after long-term exposure to heat shock conditions.
Unlike LeishIF4E-4 and LeishIF4G-3, the steady-state level of LeishIF4E-1 remained unchanged before and after heat-shock treatment and upon differentiation to amastigote-like cells (Figure 3A). LeishIF4E-1 maintained its ability to bind m7GTP-sepharose in axenic amastigotes, indicating that this feature is insensitive to elevated temperatures, as observed for LeishIF4E-4 (Figure 3C). LeishIF4E-1 could also pull down LeishIF4A-1 during heat shock and in axenic amastigotes (Figure 3B and D). Mass-spectrometry analysis of proteins that were copurified with the SBP-tagged LeishIF4E-1 from axenic amastigotes excluded the interaction with any MIF4G-domain protein, despite the presence of a variety of translation initiation factors in the pulled-down experiments in both life forms (eIF2 and eIF3 subunits, Table 1). However, LeishPABP-1 was absent in the amastigote pull-down, unlike LeishPABP-2, which was detected in both life forms. It appears that LeishIF4E-1 continues to play an as-yet unidentified role in axenic amastigotes.
Characterization of a novel 4E-binding protein in Leishmania
In addition to the annotated proteins that were associated with LeishIF4E-1, an unidentified ‘hypothetical’ protein (LmjF35.3980) was present in the pull-down experiments in both life stages. A BLAST search yielded no homologs of this protein outside the Trypanosomatidae family. It was highly conserved between different Leishmania species, but showed only partial homology with its orthologs from T. brucei or T. cruzi (Supplementary Figure S5). A secondary structure prediction (provided by the Psi-Pred server) suggests that the protein is mostly unstructured (Supplementary Figure S5).
The role of LmjF35.3980 and its mode of association with the cap-binding proteins in Leishmania were examined by the yeast two-hybrid assay. LmjF35.3980 showed a clear association with LeishIF4E-1 and weaker interaction with LeishIF4E-4. We designated this novel protein as Leishmania 4E-Interacting Protein (Leish4E-IP). We detected no binding of this novel protein to LeishIF4E-2, LeishIF4E-3, LeishIF4A-1, LeishIF4G-3 or LeishPABP-1 (Figure 4A).
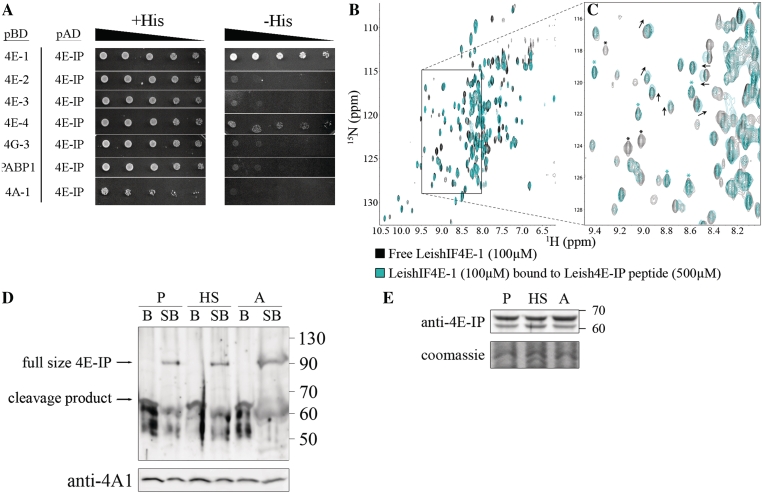
Figure 4.
A novel 4E-IP homolog in Leishmania interacts with LeishIF4E-1 in vitro. (A) Yeast two-hybrid assay was performed by co-transfecting wild-type YRG-2 yeast strain with pBD fused to LeishIF4E-1 through -4, LeishIF4G-3, LeishPABP-1 and LeishIF4A-1 and pAD-Leish4E-IP. The cells were cultured and spotted as described in ‘Materials and Methods’ section. Expression of the tested proteins was verified by western blotting (Supplementary Figure S7). (B) Overlays [15N, 1H]TROSY-HSQC spectra of 15N-labeled LeishIF4E-1 (100 µM) bound to the human cap (m7GTP, 5 mM) without (black) and with (cyan) a non-labeled LeishIF4E-IP peptide (500 μM, VRTMYTREELLRIAT). In this figure, each peak corresponds to a specific LeishIF4E-1 amino acid. A region displaying chemical shift differences between LeishIF4E-1 unbound (black) and bound (cyan) to LeishIF4E-IP is boxed, and an enlargement is shown in (C). Examples of peaks that disappeared upon LeishIF4E-IP peptide binding are indicated by a black asterisk, while some remaining unaffected are indicated by a cyan asterisk. (D) Leish4E-IP appears to be easily cleaved in all life forms. Wild-type L. amazonensis promastigotes grown at normal temperatures (P), promastigotes after heat shock treatment at 33°C for 2 h (HS), and axenic amastigotes, 9 days after differentiation (A) were lyzed, washed and only the supernatants were used (B lanes); or lyzed by sample buffer without any prior treatment (SB lanes). The lysates were separated by SDS–PAGE (10–15%) and subjected to western blot analysis using specific antibodies against Leish4E-IP. Western blot analysis with anti-LeishIF4A-1 was used as a loading control. (E) Leish4E-IP is constitutively expressed in different Leishmania life forms. Wild-type L. amazonensis cells were treated and analyzed as described in Figure 3A, using specific antibody against Leish4E-IP. P, late log phase promastigotes; HS, late log promastigotes after 2 h of heat shock; A, axenic amastigotes, 9 days after differentiation.
All proteins known to bind eIF4E interact through a conserved sequence motif of Y(X)4LΦ, where X is variable and Φ is hydrophobic (5,18). In mammalian cells, this motif adopts a helical structure that binds on the dorsal side of eIF4E (17). We also found a consensus Y(X)4LΦ eIF4E-binding peptide at the very N-terminus of Leish4E-IP. We used NMR titration to investigate whether a short Leish4E-IP peptide encompassing this motif (4-VRTMYTREELLRIAT-18) was sufficient to interact with LeishIF4E-1. Recombinant LeishIF4E-1 was expressed in E. coli and purified using a nickel-agarose resin. Since it was previously shown that LeishIF4E-1 binds m7GTP and cap-4 with similar affinity (15), but only m7GTP is commercially available, m7GTP was added to LeishIF4E-1 to stabilize the protein. A [15N,1H]TROSY-HSQC spectrum (35) was recorded using uniformly 15N-labeled LeishIF4E-1 (Figure 4B). The unlabeled Leish4E-IP peptide was then added in increasing concentrations that ranged between 50 and 500 µM, giving a molar ratio of 0.5–5 relative to LeishIF4E-1. This resulted in chemical shifts in a select subset of resonances (Figure 4C), while other peaks remained unaffected. Moreover, peak broadening also occurred at specific residues in LeishIF4E-1, to such an extent that peaks representing the bound form of the protein were no longer observed. This is another indication for the interaction between the two binding partners. Taken together, these results confirm that the Leish4E-IP peptide is sufficient to interact with LeishIF4E-1.
Leish4E-IP proved to be highly sensitive to proteolysis. This was concluded from western analysis of cell extracts that were subjected to different methodologies. Specific antibodies raised against the recombinant Leish4E-IP verified the presence of the full-length protein in whole cell extracts that were disrupted by SDS-sample buffer (83 kDa protein that migrates as ~90 kDa on SDS–PAGE). However, if the cells disrupted with 1% Triton X-100 were further fractionated by centrifugation, Leish4E-IP was cleaved to smaller polypeptides of 50–65 kDa, despite the presence of a various combinations of protease inhibitors or inhibition of the proteasome, in different buffer compositions (Figure 4D and data not shown). Furthermore, purified recombinant Leish4E-IP showed a similar pattern of cleavage (data not shown), suggesting that it could be due to the predicted unstructured nature of this protein.
Investigation of LeishIF4E-IP expression during differentiation showed that the steady state level of LeishIF4E-IP remained unchanged in both life stages (Figure 4E). The interaction profile of Leish4E-IP was examined by pull-down assays of SBP-tagged LeishIF4E-1 or LeishIF4E-4 in promastigotes followed by western blot analysis. In this life stage Leish4E-IP was bound to both LeishIF4E-1 and LeishIF4E-4 (Figure 5A). However, the binding to LeishIF4E-4 is most likely non-specific, since LeishIF4E-IP is present in high quantity in the wash fraction (bottom panel). This is supported by the reciprocal pull-down experiment performed with SBP tagged Leish4E-IP, which revealed an interaction only with LeishIF4E-1 but not with LeishIF4E-4 (Figure 5B).
Figure 5.
Leish4E-IP interacts with LeishIF4E-1 and LeishIF4E-4 in promastigotes. Pull-down analysis with SBP tagged LeishIF4E-1, LeishIF4E-4 or Leish4E-IP from L. amazonensis was done using affinity purification and analysis as described in Figure 1A. Aliquots of the soluble extract (S, 1%), the flow-through (F, 1%), the final wash (W, 40% for A, C, D and 20% for B) and eluted proteins (E, 40% for A, C, D and 20% for B) were separated by SDS–PAGE (10–15%) and subjected to western blot analysis using specific antibodies against LeishIF4E-1, LeishIF4E-4, Leish4E-IP and SBP. The pull-down analysis was performed in promastigotes (A and B), promastigotes after 2 h at 33°C (C) and in axenic amastigotes, 9 days after differentiation (D) Densitometric analysis of panel A showed that the elution fraction contained 10.4 and 14% of LeishIF4E-1 and Leish4E-IP, respectively (top) and 8 and 12% of LeishIF4E-4 and Leish4E-IP, respectively (bottom). The wash fraction of Leish4E-IP that was pulled down by LeishIF4E-4 contained 7% of total protein, suggesting that the eluted fraction contained some protein that came down non-specifically.
Evaluation of the Leish4E-IP-binding partners after 2 h of heat shock (33°C) indicated a decrease in its ability to interact with both LeishIF4E-1 and LeishIF4E-4 (Figure 5C). In fully differentiated axenic amastigotes, the interaction between Leish4E-IP and LeishIF4E-1 was hardly detectable, unlike in promastigotes (Figure 5D). As indicated before, such an analysis was not possible for the tagged LeishIF4E-4, as it was undetectable. These observations point to a promastigote specific interaction between LeishIF4E-1 and Leish4E-IP.
DISCUSSION
In this study we sought to identify and define changes in the translation machinery of Leishmania that are associated with stage differentiation and chose to focus on the cap-binding complex. Cap-binding proteins are central to translation initiation and its regulation; they function as general factors [eIF4E, (36)], or as gene-specific regulators that form a bridge between protein complexes on 3′-UTRs and the translation machinery (1,37). Many organisms express several isoforms of eIF4E, and in most cases their role has been deciphered only partially (38–40).
The Leishmania genome contains four paralogs of eIF4E, but none of them can complement a yeast strain deficient of this protein (15), indicating their evolutionary divergence (15,41). LeishIF4E-1 and LeishIF4E-4 were formerly implicated in translation initiation (15,16). This is consistent with a recent report that the T. brucei orthologs TbEIF4E1 and TbEIF4E4 can replace each other in RNAi assays (these are not yet available for Leishmania), since only their co-silencing in procyclic parasites is lethal and provokes arrest of translation (24). LeishIF4E-3 is unlikely to serve as a homolog of the conventional translation initiation factor, since it has a very low affinity for the trypanosomatid cap-4. Furthermore, unlike typical translation initiation factors, it migrates as a nuclease resistant ~80S particle over sucrose gradients (15,42). LeishIF4E-2 can interact mainly with cap-4 and not with m7GTP. However, it co-migrates with nuclease sensitive polysomal fractions (15) and therefore also deviates from the pattern observed for the consensus eIF4E from higher eukaryotes (42). Finally, both LeishIF4E-2 and LeishIF4E-3 are down-regulated in axenic amastigotes (Figure 3A), excluding their potential function during translation in this life stage. In this study, we therefore focused on the role of LeishIF4E-1 and LeishIF4E-4 in translation.
Interactions within the LeishIF4E-4 complex highlight this isoform as the general translation initiation factor in promastigotes. Affinity purification of the tagged LeishIF4E-4 and analysis of its binding partners by mass-spectrometry supports this conclusion. LeishIF4E-4 pulls down LeishIF4G-3 and LeishIF4A-1, both of which are part of LeishIF4F complex. The central role of LeishIF4E-4 in translation is further supported by the presence of LeishPABP-1 [reported to be involved in translation (32)], LeishIF2 and LeishIF3 subunits in its macromolecular complex. Direct interactions between LeishIF4E-4 and its typical binding partners were detected using a yeast two hybrid assay. LeishIF4G-3 was found to interact with LeishIF4E-4 and LeishIF4A-1, but failed to bind LeishPABP-1. In higher eukaryotes the amino-terminus of eIF4GI contains a PABP binding domain (31), which LeishIF4G-3 lacks. Using a deletion mutant of LeishIF4E-4, we showed that its extended amino-terminus is responsible for recruiting LeishPABP-1 to the initiation complex. Such interaction was never reported in other eukaryotes, further emphasizing the divergence of the translation machinery in trypanosomatids.
It was impossible to perform a pull-down analysis of the tagged LeishIF4E-4 in axenic amastigotes, since the tagged protein was not expressed in this life stage, despite the fact that intergenic regions of the Hsp83 gene cluster drove its expression, to ensure the stability and translatability of its transcript. Using an alternative experimental approach, we monitored the ability of the endogenous, wild-type LeishIF4E-4 and LeishIF4G-3 in axenic amastigotes, to copurify over m7GTP-sepharose. Indeed, LeishIF4E-4 from axenic-amastigotes was unable to bind the cap structure, and as a consequence, LeishIF4G-3 was also absent from the eluted fractions. The migration profile of wild-type LeishIF4E-4 in SDS–PAGE suggests that it is subject to post-translational modifications. In that perspective, reciprocal pull-down analysis shows that only the largest molecular weight species can bind the cap-structure (Figure 3C) and LeishIF4G-3 in promastigotes (Supplementary Figure S6). This form is mostly abolished in axenic amastigotes, in accordance with the decrease of the association between LeishIF4E-4 and LeishIF4G-3 at elevated temperatures (Figure 3B). Taken together, these data suggest that the function of LeishIF4E-4 in amastigotes is impaired, while LeishIF4E-1 is the only eIF4E isoform that is equally expressed in all life stages. LeishIF4E-1 also binds m7GTP and continues to associate with other initiation factors in the amastigote life stage. We therefore examined a possible role of LeishIF4E-1 in translation in both life stages.
Analysis of proteins that were pulled down by tagged LeishIF4E-1 in promastigotes and axenic amastigotes, gave a profile of translation factors, suggesting that this protein has a role in translation. In a previous study, LeishIF4E-1 increased its ability to form high molecular weight complexes in heat-shocked cells and entered heavier fractions in sucrose gradients after exposure of the parasites to elevated temperatures (16). However, LeishIF4G-3 was not present among the proteins that were pulled down by the tagged LeishIF4E-1, suggesting that the latter does not participate in the traditional cap-dependent translation. Although we previously detected an interaction between LeishIF4E-1 and LeishIF4G-3 using recombinant proteins (16), the current in vivo analysis using pull-down as well as yeast two hybrid experiments did not support this interaction. No other MIF4G domain protein was found to be associated with LeishIF4E-1, and the occasional presence of a few LeishIF4G-4 peptides in the mass spectrometry analysis was likely not due to a direct interaction between the two proteins (Figure 2B and Supplementary Figure S3). This result is in accordance with a recently published paper on the T. brucei eIF4E homologs (24).
In higher eukaryotes, disassembly of the eIF4F complex and the use of alternative pathways for translation initiation are implemented by the 4E-binding protein (4E-BP). However, no 4E-BP homologue could be identified in Leishmania or other trypanosome genome databases. Our pull down experiments highlighted a novel 4E-Interacting Protein (Leish4E-IP) that does not resemble the consensus 4E-BP from higher eukaryotes. The interaction between Leish4E-IP and LeishIF4E-1 was observed in reciprocal pull-down experiments, in a yeast two-hybrid assay, and in NMR titration experiments using a consensus 4E-BP found at the amino-terminus of Leish4E-IP. This peptide resembles the consensus YXXXLϕ motif found in the 4E-BP and eIF4G of higher eukaryotes, but varies from the partially conserved 4E-binding peptide characterized in LeishIF4G-3 (16). The difference between the two peptides can explain why the binding of Leish4E-IP to LeishIF4E-4 is weaker than to LeishIF4E-1. Prediction of the secondary structure of Leish4E-IP (83 kDa) suggests that the newly identified protein is mostly unstructured, except for a small region in its N-terminus. This feature is typical of 4E-BPs from higher eukaryotes (~10 kDa) (2). The Leish4E-IP homologs in T. brucei and T. cruzi are only partially conserved (~50% similarity), in agreement with the prediction that they are unstructured.
In higher eukaryotes, 4E-BP binds eIF4E only when it is unphosphorylated. Phosphorylation of 4E-BP by mTOR causes its disassembly from the cap-binding protein (6,7). In Leishmania, the prolonged exposure to elevated temperatures during axenic differentiation reduced the binding of Leish4E-IP to LeishIF4E-1, to a level that was hardly detectable. Preliminary results of changes in the phospho-proteome during differentiation show that Leish4E-IP becomes phosphorylated during stage differentiation (Zilberstein and Tsygankov, personal communication). In such a case, phosphorylation of Leish4E-IP in amastigotes could release the bound LeishIF4E-1 and promote its binding to cap-4. Taken together, these results suggest that Leish4E-IP has properties similar to the higher eukaryote 4E-BP, although its exact function in Leishmania remains to be determined.
In summary, we describe the developmental changes that occur in the translation machinery of different life stages of the Leishmania parasites. In promastigotes, translation is mostly, though not exclusively, cap-dependent, and is driven by LeishIF4E-4, which anchors an eIF4F-like complex. However, the LeishIF4E-4/LeishIF4G-3 interaction and the ability of LeishIF4E-4 to bind to the cap structure decrease at elevated temperatures as part of adaptation to the mammalian host. In parallel, alternative pathways such as cap-independent translation, or other non-traditional mechanisms for translation could be activated, in which LeishIF4E-1 is most likely involved. The nature of LeishIF4E-1 function is however still unclear. LeishIF4E-1 could be regulated in a stage-specific manner by Leish4E-IP, which has no counterparts in higher eukaryotes. We propose that in promastigotes Leish4E-IP binds LeishIF4E-1 and keeps it inactive. Upon differentiation to amastigotes, this binding is no longer detectable and LeishIF4E-1 is released. These findings are yet another reminder of the resourceful nature of these very ancient eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US-Israel Binational Foundation (BSF, grant number 2007287 to M.S. and G.W.); Israel Science Foundation (ISF, grant No 395/09 to M.S.); National Institutes of Health (NIH, grant CA068262 to G.W.); Funding for open access charge: Fonds de la Recherche en Santé au Québec (FRSQ) (to M.L.). Funding for open access charge: BSF, ISF, NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dan Zilberstein for providing data prior to their publication. The authors acknowledge Charles Jaffe and Joachim Clos for providing specific antibodies against GP46 and Hsp100, respectively.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher CM, McGuire AM, Gingras AC, Li H, Matsuo H, Sonenberg N, Wagner G. 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry. 1998;37:9–15. doi: 10.1021/bi972494r. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher CM, Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JC., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 7.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacks DL, Perkins PV. Identification of an infective stage of Leishmania promastigotes. Science. 1984;223:11417–11419. doi: 10.1126/science.6701528. [DOI] [PubMed] [Google Scholar]
- 9.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 10.Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol. Biochem. Parasitol. 2005;141:99–108. doi: 10.1016/j.molbiopara.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 12.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 13.Liang XH, Haritan A, Uliel S, Michaeli S. Trans- and cis-splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- 15.Yoffe Y, Zuberek J, Lerer A, Lewdorowicz M, Stepinski J, Altmann M, Darzynkiewicz E, Shapira M. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot. Cell. 2006;5:1969–1979. doi: 10.1128/EC.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoffe Y, Léger M, Zinoviev A, Zuberek J, Darzynkiewicz E, Wagner G, Shapira M. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res. 2009;37:3243–3253. doi: 10.1093/nar/gkp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 18.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cysne-Finkelstein L, Temporal RM, Alves FA, Leon LL. Leishmania amazonensis: long-term cultivation of axenic amastigotes is associated to metacyclogenesis of promastigotes. Exp. Parasitol. 1998;89:58–62. doi: 10.1006/expr.1998.4276. [DOI] [PubMed] [Google Scholar]
- 20.Hodgkinson VH, Soong L, Duboise SM, McMahon-Pratt D. Leishmania amazonensis: cultivation and characterization of axenic amastigote-like organisms. Exp. Parasitol. 1996;83:94–105. doi: 10.1006/expr.1996.0053. [DOI] [PubMed] [Google Scholar]
- 21.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laban A, Wirth DF. Transfection of Leishmania enriettii and expression of chloramphenicol acetyltransferase gene. Proc. Natl Acad. Sci. USA. 1989;86:9119–9123. doi: 10.1073/pnas.86.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2008;22:590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- 24.Freire ER, Dhalia R, Moura DM, da Costa Lima TD, Lima RP, Reis CR, Hughes K, Figueiredo RC, Standart N, Carrington M, et al. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol. Biochem. Parasitol. 2011;176:25–36. doi: 10.1016/j.molbiopara.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, Spriggs RV, Willis AE. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 29.Dhalia R, Marinsek N, Reis CR, Katz R, Muniz JR, Standart N, Carrington M, de Melo Neto OP. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res. 2006;34:2495–2507. doi: 10.1093/nar/gkl290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 31.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Costa Lima TD, Moura DM, Reis CR, Vasconcelos JR, Ellis L, Carrington M, Figueiredo RC, de Melo Neto OP. Functional characterization of three Leishmania poly(a) binding protein homologues with distinct binding properties to RNA and protein partners. Eukaryot. Cell. 2010;9:1484–1494. doi: 10.1128/EC.00148-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zilberstein D, Shapira M. The role of pH and temperature in the development of Leishmania parasites. Annu. Rev. Microbiol. 1994;48:449–470. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- 34.Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pervushin KV, Wider G, Riek R, Wuthrich K. The 3D NOESY-[(1)H,(15)N,(1)H]-ZQ-TROSY NMR experiment with diagonal peak suppression. Proc. Natl Acad. Sci. USA. 1999;96:9607–9612. doi: 10.1073/pnas.96.17.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi B, Cameron A, Jagus R. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004;271:2189–2203. doi: 10.1111/j.1432-1033.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- 37.Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T, Rhoads RE. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 2000;275:10590–10596. doi: 10.1074/jbc.275.14.10590. [DOI] [PubMed] [Google Scholar]
- 39.Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001;128:3899–3912. doi: 10.1242/dev.128.20.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 2005;25:100–113. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhalia R, Reis CR, Freire ER, Rocha PO, Katz R, Muniz JR, Standart N, de Melo Neto OP. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 2005;140:23–41. doi: 10.1016/j.molbiopara.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Hiremath LS, Hiremath ST, Rychlik W, Joshi S, Domier LL, Rhoads RE. In vitro synthesis, phosphorylation, and localization on 48 S initiation complexes of human protein synthesis initiation factor 4E. J. Biol. Chem. 1989;264:1132–1138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.