Abstract
Genome duplication in free-living cellular organisms is performed by DNA replicases that always include a DNA polymerase, a DNA sliding clamp and a clamp loader. What are the evolutionary solutions for DNA replicases associated with smaller genomes? Are there some general principles? To address these questions we analyzed DNA replicases of double-stranded (ds) DNA viruses. In the process we discovered highly divergent B-family DNA polymerases in phiKZ-like phages and remote sliding clamp homologs in Ascoviridae family and Ma-LMM01 phage. The analysis revealed a clear dependency between DNA replicase components and the viral genome size. As the genome size increases, viruses universally encode their own DNA polymerases and frequently have homologs of DNA sliding clamps, which sometimes are accompanied by clamp loader subunits. This pattern is highly non-random. The absence of sliding clamps in large viral genomes usually coincides with the presence of atypical polymerases. Meanwhile, sliding clamp homologs, not accompanied by clamp loaders, have an elevated positive electrostatic potential, characteristic of non-ring viral processivity factors that bind the DNA directly. Unexpectedly, we found that similar electrostatic properties are shared by the eukaryotic 9-1-1 clamp subunits, Hus1 and, to a lesser extent, Rad9, also suggesting the possibility of direct DNA binding.
INTRODUCTION
DNA replication is one of the most fundamental processes in all living entities. The replication of genomic DNA has to be not only accurate but also very efficient. To achieve this, free-living organisms from all three domains of life and some viruses use multicomponent protein machines termed DNA replicases. A DNA replicase consists of a DNA polymerase and accessory subunits including a DNA sliding clamp and a clamp loader (1). A sliding clamp is a ring-shaped polymerase processivity factor, which needs to be loaded onto the DNA by a clamp loading complex. Once loaded, a DNA sliding clamp encircles the DNA double helix serving as a mobile tether for the replicative DNA polymerase. The attachment to the DNA-loaded sliding clamp transforms the polymerase into an extremely processive enzyme that can synthesize thousands of nucleotides without falling off the DNA (2).
It is striking that despite the mechanistic uniformity of replication of the DNA double helix, the replicative DNA polymerases, central players in this process, are not universally conserved. Both sequence (3) and structure (4,5) analyses led to the conclusion that bacterial replicative polymerases on one hand and eukaryotic/archaeal polymerases on the other hand evolved independently from different ancestral proteins. The catalytic α-subunit of the bacterial replicative polymerase (polIIIα) belongs to the C-family of DNA polymerases (PolC). Eukaryotic and archaeal replicative polymerases belong to the unrelated B-family (PolB). In addition, a unique D-family polymerase was found to participate together with a B-family polymerase in DNA replication in euryarchaea (6–8). In dsDNA viruses the diversity of replicative polymerases is even larger. In addition to canonical B-family polymerases that initiate DNA synthesis from the 3′ terminus of the RNA primer (PolBr), some viruses encode protein-primed DNA B-family polymerases (PolBp) that use a hydroxyl group supplied by a protein (9). A-family DNA polymerases (PolA) that play only a limited/specialized role in DNA synthesis of cellular organisms also participate in viral genome replication (10). Although distinct, the A-family is distantly related to the B-family (11). Interestingly, while B- and A-family replicative DNA polymerases in dsDNA viruses are common, C-family polymerases have been detected only in a handful of bacteriophages (12).
In contrast to the disparity of replicative DNA polymerases, their processivity factors (DNA sliding clamps) are conserved in all cellular organisms and T4-like phages (13). The bacterial DNA sliding clamp (polIIIβ) is a homodimer, while eukaryotic and archaeal Proliferating Cell Nuclear Antigen (PCNA) is a homotrimer with few archaea having a heterotrimeric PCNA (14). The gp45 sliding clamp in T4-like phages, like eukaryotic and the majority of archaeal PCNAs, is a homotrimer. Eukaryotes also have an additional PCNA-like heterotrimeric DNA sliding clamp, the 9-1-1 complex, which specializes in DNA repair processes (15). Despite differences in oligomeric state (dimer or trimer) all these DNA sliding clamps represent structurally similar rings with pseudo 6-fold symmetry and a central hole large enough to fit the DNA double helix. Replicative DNA polymerases and other proteins usually interact with DNA sliding clamps through the hydrophobic pocket formed by the interdomain connector (16). In addition to the ring-shaped gp45 DNA sliding clamp in T4-like phages, the viral world has produced alternative recipes of how to increase the DNA replication processivity. For instance, processivity factors in herpesviruses are structurally similar and have the identical domain composition as PCNA or gp45, but they do not form rings. UL42 acts as a monomer representing one-third of a ring (17), while UL44 and BMRF1 form C-shaped dimers that correspond to two thirds of a ring (18,19). Another virus-specific example is the recruitment of a host protein, unrelated to DNA sliding clamps (Escherichia coli thioredoxin), to serve as the DNA polymerase processivity factor in the T7 phage (20).
Ring-type DNA sliding clamps need protein complexes known as clamp loaders for their loading onto DNA (1). All subunits of cellular clamp loaders belong to the AAA+ protein superfamily. Although the exact subunit composition may differ, the core of all known clamp loaders is a pentameric protein complex with at least one subunit being different from the remaining four. Archaeal and eukaryotic clamp loaders are quite similar. They are composed of one large and four small subunits. In eukaryotes all four small subunits are different, while in archaea they usually are identical or, in few cases, are represented by two types (21). The bacterial clamp loader consists of δ, δ′ and three copies of the γ/τ subunit. A clamp loader in T4-like phages is composed of four copies of gp44 and a single copy of gp62 protein.
In free-living cellular organisms the combination of a DNA polymerase and a DNA sliding clamp with its loader appears to be a universal solution to the replicase processivity problem (1). In contrast, many dsDNA viruses do not encode processivity factors, and some do not even have their own DNA polymerases, totally relying on DNA replication machinery of the host. Could it be that the size of a genome is an important factor determining the need for a processive DNA replicase? Perhaps there is an approximate genome size threshold, above which the processivity properties of a replicase become critical? dsDNA viruses are an excellent model group for addressing such fundamental questions as they represent a wide range of genome sizes (from ~5 up to ~1200 kb) and a large variety of genome replication strategies.
In this study, using data derived from the sequenced genomes of dsDNA viruses, we examined the presence and the type of viral DNA replicases in the context of their genome size. To this end we used sensitive homology detection methods to identify DNA polymerases, processivity factors and clamp loaders encoded in viral genomes. We detected a number of previously uncharacterized components of DNA replicases and explored their properties using a variety of computational methods. Our results establish that the presence and the type of DNA replicase components are linked with the viral genome size.
MATERIALS AND METHODS
Viral databases
Viral protein and genome data were downloaded from NCBI URLs ‘http://www.ncbi.nlm.nih.gov/protein/?term=dsDNA+viruses,+no+RNA+stage’ and ‘http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=35237’ respectively. Family Polydnaviridae was excluded from the analysis because these viruses have a distinct genome organization (split in small segments), and their genome acts only as a vector for transmission of parasitic wasp genes (22).
Genome filtering
To obtain a more representative genome set, highly similar genomes were removed. All genomes were compared to each other using LAST (v128) (23), and genomes with local sequence identity >70% were filtered out. Repetitive genomic regions were identified and ignored during the comparison.
Genome translations
All the genomes of dsDNA viruses were subjected to the six-frame translation using Virtual Ribosome (24) and the standard genetic code translation table. In addition to annotated open reading frames (ORF), all previously unassigned ORFs longer than 60 residues were retained for further analysis.
Sequence similarity searches and the identification of conserved domains
Standard sequence searches were performed using BLAST and PSI-BLAST (25) with default parameters in non-redundant (nr) databases installed locally and updated weekly. To identify conserved domains in viral protein and ORF sequences, each of them was searched against the CDD profile database (26) using RPS-BLAST (25) with default parameters. In addition, profile Hidden Markov Models (HMMs) were constructed for each viral sequence for searches against the library of profile HMMs of known protein structures (PDB). Profile HMMs were constructed using the buildali.pl script and HHmake algorithm from the HHsearch (v1.5.0) software suite (27). The profile HMM construction with buildali.pl included running three iterations of PSI-BLAST search against the nr90 (nr filtered to maximum 90% sequence identity) database using the E-value = 1e-03 inclusion threshold. HHsearch with default parameters was then used to search the pdb70 database of profile HMMs (ftp://toolkit.lmb.uni-muenchen.de/HHsearch/databases/) installed locally. In addition, locally generated profiles (profile HMMs) for individual DNA replicase components from multiple sequence alignments were appended to CDD and pdb70 databases. RPS-BLAST and HHsearch hits, with E < 0.1 and probability >50%, respectively, were extracted from the results and analyzed for the presence of DNA polymerases, DNA sliding clamps and clamp loaders. Unreliable hits to replicase components were further validated with additional approaches such as COMA server (28) or GeneSilico MetaServer (29).
Sequence clustering
DNA replicase components were clustered according to their pairwise similarity using CLANS (30). The similarity in CLANS is represented with P-values derived from BLAST or PSI-BLAST E-values. For clustering divergent proteins (all DNA polymerases and all DNA sliding clamps), their pairwise similarity was quantified using PSI-BLAST. For each sequence, CLANS was configured to run two iterations of PSI-BLAST using the E = 1e-03 inclusion threshold against the reference database (nr80) to generate a sequence profile. The last PSI-BLAST iteration with the obtained profile was performed against the database of sequences to be clustered. In our case this was the database of either viral DNA polymerases or sliding clamps. To partition the largest subset of B-family polymerases (the PolBrCore cluster) into distinct groups, CLANS was based on a direct BLAST all-against-all sequence comparison.
Multiple sequence alignments
Multiple sequence alignments were constructed with MAFFT (31) optimized for accuracy (parameter L-INS-i). If sequences had homologs with known structures PROMALS3D (32) with default parameters was used instead.
Homology modeling
Alignments between the sequence to be modeled (target) and a related structure (template) were constructed with PSI-BLAST-ISS (33), COMA server (28) or GeneSilico MetaServer (29). Uncertain alignment regions were modified manually, during an iterative modeling process (34). Protein 3D models were constructed from target-template alignments using Modeller 9v7 (35). Models were evaluated visually for significant flaws. In addition, the model quality was estimated using ProsaWeb (36) by comparing Prosa Z-scores of models with those of corresponding templates.
Analysis of electrostatic properties
Calculation of theoretical isoelectric points (pIs) for DNA sliding clamps and their homologs was performed using the ‘Isoelectric point’ program from the EMBOSS software package (37). Sequences of sliding clamps and their homologs were collected by performing PSI-BLAST searches against the nr70 database. Non-conserved N- and C-termini were removed from the sequences before the pI calculation. Surface electrostatic potential maps were computed with APBS (v1.2.1), which was accessed through the PyMol APBS Tools2 plug-in (http://www.pymolwiki.org/index.php/APBS). Prior to computation, all heteroatoms and water molecules from PDB files were removed. Both models and PDB structures were prepared for calculations using PDB2PQR (v1.5) (38) with the AMBER force field.
RESULTS
DNA replicase components and the genome size
We analyzed the available fully sequenced genomes of dsDNA viruses for the presence of DNA replicase components. In all, genomes of 808 viruses including 458 (57%) bacteriophages, 317 (39%) eukaryotic and 33 (4%) archaeal viruses were analyzed. Specifically, we looked for DNA polymerases, polymerase processivity factors (DNA sliding clamps) and clamp loader subunits. We detected DNA polymerases in about half of the analyzed viral genomes. In addition to either known or previously annotated enzymes, for the first time we identified highly divergent DNA polymerases in phiKZ-like bacteriophages. We found a significantly smaller fraction of genomes (<20%) coding for homologs of DNA sliding clamps that may serve as DNA polymerase processivity factors. We newly discovered remote homologs of cellular DNA sliding clamps in Microcystis phage Ma-LMM01 and the Ascoviridae family. DNA sliding clamps that form rings (PCNA, polIIIβ, gp45) need a multimeric clamp loader for their loading onto DNA. In line with this prerequisite, we detected clamp loader subunits only in genomes carrying genes of DNA sliding clamp homologs. Yet, surprisingly, not all PCNA or polIIIβ homologs are accompanied by clamp loader subunits.
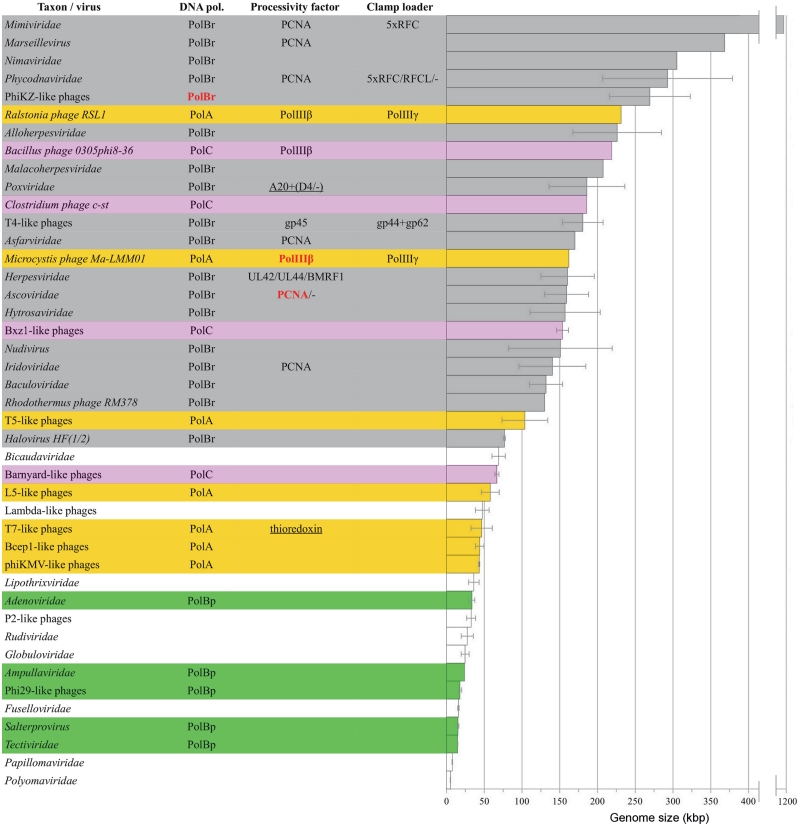
Overall, the results revealed a great variety of DNA replicase components and their combinations in dsDNA viruses (for the complete list see File 1 in Supplementary Data). The variety is much larger than it is in all three domains of cellular life combined and seemingly without any discernible pattern. However, we reasoned that if the increase in viral genome size requires improved processivity properties of a DNA replicase we should be able to detect this dependency even in the face of this overwhelming variety. Indeed, the arrangement of viral taxonomic groups according to their average genome size revealed a clear trend (Figure 1). Viruses having smallest genomes (<40 kb) either have a B-family protein-primed DNA polymerase or do not have a DNA polymerase at all. Viruses with larger genomes (40–140 kb) have their own DNA polymerases more often. These polymerases usually belong to A-, rarely to B- or C-families. Viruses having largest genomes (>140 kb) always encode DNA polymerases (most often B-family RNA-primed), frequently have processivity factors and sometimes clamp loader subunits.
Figure 1.
DNA replicase components in dsDNA viral genomes. Viral taxonomic groups are arranged by their average genome size. DNA pol., DNA polymerase type; PolA, A-family; PolBr, B-family DNA polymerase that uses RNA as a primer; PolBp, B-family DNA polymerase that uses protein as a primer; PolC, C-family. Coloring scheme: white, no polymerases found; green, PolBp; yellow, PolA; gray, PolBr; pink, PolC. Newly identified replicase components are labeled in bold red font. Processivity factors, non-homologous to the cellular ones, are underlined. Minus sign indicates that the processivity factor is missing in some viruses within the taxonomic group. Error bars indicate standard deviation from the mean genome size.
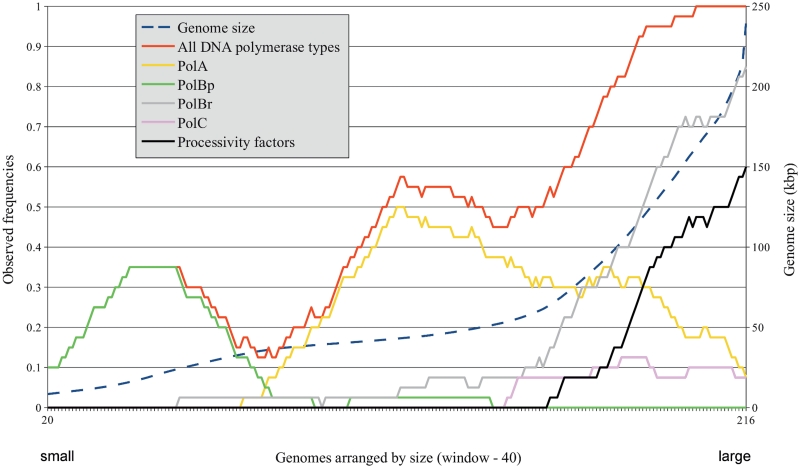
However, the representation of various viral taxonomic groups differs significantly. In addition, some taxons show quite large variation of the genome size. Therefore, we next asked whether or not the observed pattern of distribution of replicase components depends on the taxonomic classification of viruses. To address this question, we arranged individual genomes according to their size without dividing into taxonomic groups and plotted the observed frequency of a particular DNA replicase component against the moving average of the genome size (Figure 2). To reduce sample bias in this analysis, we performed pairwise genome comparisons and retained only 236 viral genomes that were <70% identical to each other. Again, the plot showed a clear relationship between DNA replicase components and the genome size, indicating that this is a general property and not the result of taxon-specific division.
Figure 2.
Dependence between the observed frequencies of viral DNA replicase components and the genome size of dsDNA viruses. X-axis—genomes arranged by their size (from smallest to largest); major y-axis (left)—observed frequencies of various DNA replicase components in viral genomes; minor y-axis (right)—genome size (kb). The genome size and the observed frequencies of DNA replicase components were averaged using the moving window of 40 genomes and a single-genome step. Broken blue line corresponds to the averaged genome size. Solid lines correspond to averaged observed frequencies of individual DNA replicase components: all DNA polymerase types, red; PolBp, green; PolA, yellow; PolBr, gray; PolC, pink; known and predicted processivity factors, black.
Having established a general dependency of the presence and the type of viral DNA replicase components on the genome size (Figures 1 and 2), we were nonetheless puzzled by the substantial number of seeming exceptions. While DNA polymerases are present in all taxonomic groups above the certain genome size, processivity factors and clamp loaders are not. If we assume that DNA replicase processivity properties become more important as the genome size increases, how to rationalize the absence of DNA sliding clamps and clamp loaders in some taxons with the large average genome size? To address this question, we performed a detailed analysis of sequence and structure properties of DNA polymerases, sliding clamp homologs and clamp loader subunits. Results of this analysis for each of the three components of DNA replicases are presented in separate sections below.
DNA polymerases
Major DNA polymerase groups
We identified DNA polymerases in 415 out of the 808 analyzed genomes of dsDNA viruses. The majority of DNA polymerases (255 genomes) belong to B-, less frequently (132) to A-, and very rarely (28) to the C-family. No polymerases of the archaeal D-family were detected. B-family polymerases are present in viruses that infect organisms from all three domains of life. In contrast, we found A- and C-family polymerases only in bacteriophage genomes. The greatest diversity by far is among B-family members, followed by the distantly related A-family (Figure 3). Most proteins belonging to the evolutionary unrelated C-family are fairly similar to each other.
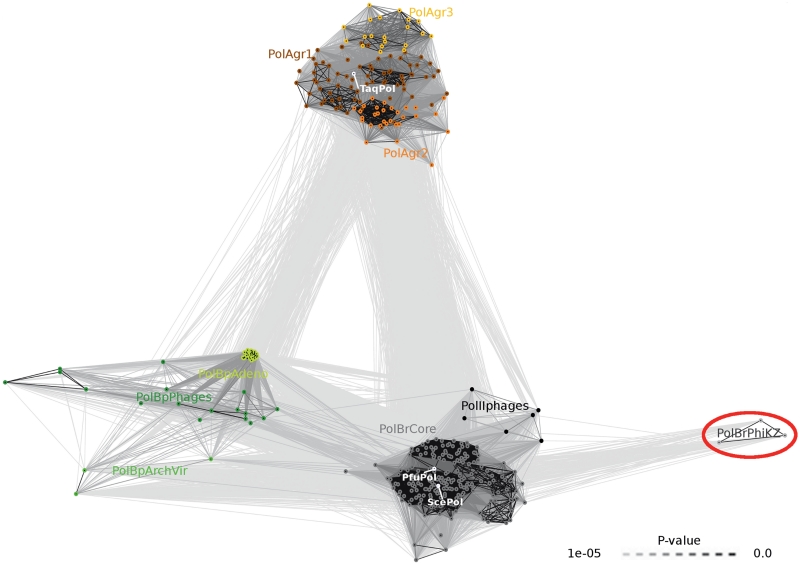
Figure 3.
DNA polymerases of A- and B-families clustered by the pairwise sequence similarity. Nodes represent individual sequences. Lines connect sequences with P ≤ 1e-05. Line shading corresponds to P-values according to the scale in the bottom-right corner (light and long lines connect distantly related sequences). A-family DNA polymerases are represented using shades of orange, PolBp—shades of green, PolBr—shades of gray; well-known cellular DNA polymerases are shown in white. Newly identified DNA polymerases are marked with the red ellipse. ArchVir, archaeal viruses; Adeno, Adenoviridae; gr, group; PhiKZ, phiKZ-like phages; Pfu, Pyrococcus furiosus; Sce, Saccharomyces cerevisiae; Taq, Thermus aquaticus.
Based on sequence similarity, PolB polymerases can be divided into three distinct clusters: one including protein-primed (PolBp), and two that include RNA-primed (PolBr) polymerases (Figure 3). PolBp DNA polymerases include mutually highly similar adenoviral polymerases (PolBpAdeno) and significantly more diverse subgroups from bacteriophages (PolBpPhages) and archaeal viruses (PolBArchVir). The largest of the two PolBr clusters contains the majority of viral RNA-primed DNA polymerases of B-family (PolBrCore group) and the small PolIIphages subgroup. The PolBrCore group, typified by polymerases from T4-like phages and Herpes Simplex virus 1, is closely related to eukaryotic and archaeal polymerases (e.g. yeast Polδ and the archaeal Pfu DNA polymerase). Members of the PolIIphages subgroup can be distinguished from the main PolBrCore group by the characteristic motif (‘NTDG’) in the polymerase active site and a higher similarity to E. coli PolII. The small second PolBr cluster consists of highly divergent PolBrPhiKZ polymerases identified in this study for the first time.
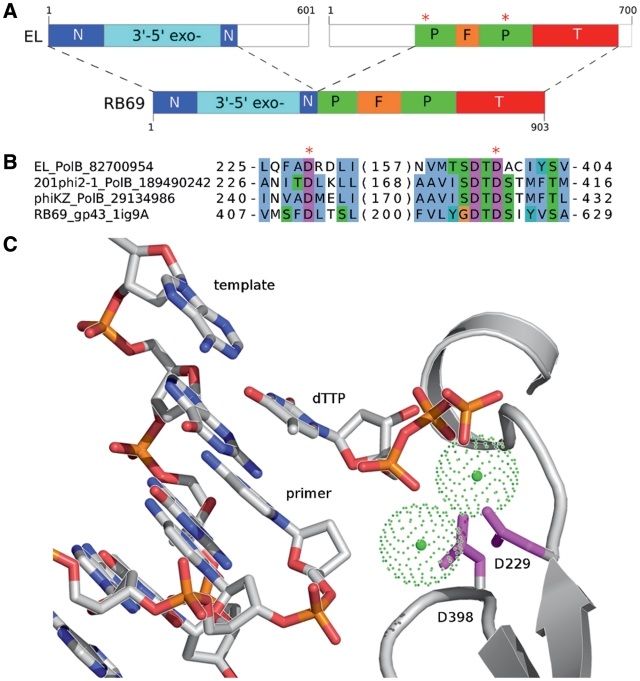
PhiKZ-like viruses have a genome that is almost twice as large as that of T4 phage (e.g. Pseudomonas phage 201phi2—317 kb, T4—169 kb), yet no DNA polymerases were found in their genome sequences during previous analyses (39–41). Since our initial data suggested that the absence of a polymerase gene in viral genomes of this size is highly unlikely, we performed a particularly thorough analysis of the genomes of PhiKZ-like phages. Not surprisingly, standard homology detection methods (BLAST, RPS-BLAST and PSI-BLAST) failed to detect statistically significant similarity between predicted proteins of these phages and any known polymerases. Only when we applied very sensitive homology search methods based on profile-profile comparison, we were able to identify putative polymerases. Thus, HHsearch (27) matched Pseudomonas phage EL hypothetical protein (gi: 82700954) and the RB69 (T4-like) phage DNA polymerase gp43 with high statistical significance (89% probability). COMA (42) for the same phage EL protein also identified a B-family DNA polymerase (from Thermococcus sp.) as the best match (E = 4e-07). The putative EL polymerase and its homologs in the other two phiKZ-like phages apparently include all the polymerase domains characteristic of gp43 except for the N-terminal region, which harbors the 3′–5′ exonuclease domain. Interestingly, the 3′–5′ exonuclease domain in these phages has been detected previously as a separate ORF (41). Thus, 3′–5′ exonuclease and polymerase activities in these phages appear to reside in two separate polypeptide chains (Figure 4). To further validate the polymerase assignment we analyzed the motifs, essential for the DNA polymerase function. Both sequence motifs harboring active site residues are conserved between RB69 gp43 and predicted polymerases in all three phiKZ-like phages (Figure 4B). In particular, as illustrated with a 3D model of the predicted EL polymerase active site, both aspartates (Figure 4C) involved in the coordination of metal ions are absolutely conserved. B-family polymerases often interact with corresponding DNA sliding clamps through a short C-terminal sequence motif. Predicted polymerases of phiKZ-like phages at the very C-terminus feature a consensus motif, which may be considered to represent a variant of the clamp-binding motif (16). The functional significance of this motif (‘TRLISDFY’, key hydrophobic positions are underlined) is not obvious, as aromatic residues in one of the three polymerases are substituted with hydrophilic ones. We also did not find homologs of sliding clamps in the genomes of the phiKZ-like group. However, there is a chance that corresponding proteins are encoded in the genomes, but their sequences might have diverged beyond recognition.
Figure 4.
Comparison of DNA polymerases from phiKZ-like phages and the RB69 phage. (A) Correspondence of structural domains in Pseudomonas phage EL 3′–5′ exonuclease (gi: 82700984) and DNA polymerase with those in the RB69 DNA polymerase. N, N-terminal; P, palm; F, fingers; T, thumb. Red stars indicate positions of the active site aspartates (D229 and D398). The correspondence was derived using COMA server. Unaligned regions are represented as the white boxes. (B) Alignment of the DNA polymerase active site motifs. For each sequence, the beginning and end positions are indicated. Numbers in parenthesis correspond to the number of residues omitted from the alignment. Sequence labels consist of the phage acronym, the protein name, and the gi number (PDB code in the case of RB69). (C) A 3D model of the Pseudomonas phage EL DNA polymerase active site complexed with the primed DNA and the incoming dTTP based on the ternary complex of the RB69 DNA polymerase and the DNA (PDB code: 1ig9). A fragment of the polymerase active site is shown in cartoon representation. Side chains of the active site aspartates coordinating two metal ions (green spheres) are shown as pink sticks.
A-family DNA polymerases could be subdivided into three groups. The most diverse group, PolAgr1, contains phages such as phiKMV, L5, N4, T5, SPO1, RSL1 and Ma-LMM01. Interestingly, the SPO1 DNA polymerase has the additional uracil-DNA glycosylase (UDG) domain at its N-terminus. It has been hypothesized that the UDG domain may serve as the intrinsic polymerase processivity factor (43). According to our analysis, the T5 DNA polymerase, which is highly processive (44), also has the UDG domain-like extension at the N-terminus. Taking into account that UDG (D4) in complex with A20 confers DNA polymerase processivity in eukaryotic vaccinia virus (45), the role of the UDG domain as the intrinsic polymerase processivity factor is quite likely. Groups 2 and 3 consist of T7-like and Bcep1-like viruses respectively.
Viral C-family DNA polymerases have domain organization similar to that of E. coli polIIIα (4). The conservation extends from the N-terminal PHP domain and includes the polymerase active site as well as the ‘fingers’ domain. However, the C-terminal region following the ‘fingers’ domain does not show significant similarity to the E.coli replicative polymerase suggesting that it may include different structural domains. Only the DNA polymerase from Bacillus phage 0305phi8-36 (gi: 154622917) appears to extend sequence conservation past the ‘fingers’ domain and into the OB-domain. In addition, this polymerase has a sequence motif (1131-EEDLL-1135) that aligns to the polIIIβ interaction motif in E. coli polIIIα (920-QADMF-924) suggesting that it may utilize a DNA sliding clamp to achieve the processivity. Incidentally, the Bacillus phage 0305phi8-36 has the largest genome of those found to carry a C-family polymerase, and the only one among them in which we found a polIIIβ homolog (gi: 154622720).
Distinct subgroups of RNA-primed B-family DNA polymerases
The application of a more stringent clustering procedure (using CLANS coupled with BLAST instead of PSI-BLAST) revealed a number of subgroups within the large PolBrCore cluster (Supplementary Figure S1). Since most PolBrCore polymerases are present in viruses with fairly large genomes, we analyzed polymerase sequences from poorly characterized subgroups to obtain hints as to the possible DNA replication processivity mechanisms. Polymerases of T4-like phages and herpesviruses that utilize DNA sliding clamps as processivity factors are known to possess characteristic clamp-binding motifs at their C-termini (16). Therefore, we looked for the presence of any clamp-binding motifs in all remaining subgroups. We readily identified a putative PCNA-interacting motif (the consensus sequence QxxIxxFF, where x is any amino acid) within the C-terminus of phycodnaviral DNA polymerases. In other subgroups we either did not find any clamp-binding motifs, the alignments of C-terminal regions were too variable or the number of sequences was too small to make a definite conclusion. In addition to clamp-binding motifs we looked for the presence of additional domains. It turned out that the members of three outlying subgroups (Malacoherpesviridae, Alloherpesviridae and Nimaviridae families; Supplementary Figure S1) feature additional sequence regions compared with typical PolBrCore representatives. Although we were unable to confidently assign any known functional/structural domains to these additional polymerase regions, their very presence suggests that these three viral families may have evolved alternative processivity mechanisms for the efficient replication of their large genomes.
Processivity factors
Diversity and taxonomic distribution
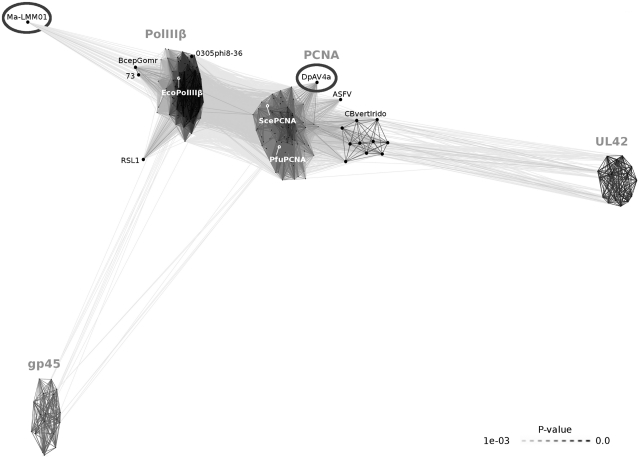
Similarly as in the case of DNA polymerases, we asked whether each of the analyzed viral genomes encodes a polymerase processivity factor. In particular, we looked for homologs of either cellular (PCNA and polIIIβ) or viral (gp45, UL42, UL44 and BMRF1) DNA sliding clamps. As a result, in addition to already characterized or annotated sliding clamps, we discovered two new putative processivity factors: a PCNA homolog in the family Ascoviridae and a polIIIβ homolog in the Ma-LMM01 phage. All sliding clamp homologs identified in viral genomes were pooled together with representatives of cellular sliding clamps (PCNA and polIIIβ) and clustered. The results shown in Figure 5 indicate that, just like DNA polymerases, viral DNA sliding clamp homologs are significantly more diverse than their cellular counterparts. Two major clusters correspond to PCNA and polIIIβ families. PolIIIβ homologs were found only in phages, while all PCNA homologs (except for PCNA from the archaeon Natrialba phage PhiCh1 and some baculoviruses) were found in eukaryote-infecting nucleo-cytoplasmic large DNA viruses (Figure 1). PCNA homologs from iridoviruses infecting cold-blooded vertebrates form a distinct subgroup in the PCNA cluster (Figure 5, CBvertIrido). In addition to two major clusters corresponding to PCNA and polIIIβ families, there are two compact outlying groups: gp45 and UL42. Gp45 includes DNA sliding clamps from T4-like phages, UL42 is found in Herpesviridae, with both groups having structurally characterized representatives (17,46). Three additional divergent families of viral sliding clamps (UL44, BMRF1 and G8R) are not included in Figure 5 as the clustering procedure was unable to link these families and any other clamps. However, it is known that herpesviral UL44 and BMRF1 are structurally similar to UL42 and other DNA sliding clamps (18,19). G8R is a remote PCNA homolog (47) found in vaccinia virus and other members of the Chordopoxvirinae subfamily, however, it does not act as a processivity factor in DNA replication (48).
Figure 5.
DNA sliding clamps and their homologs grouped by the pairwise sequence similarity. Sliding clamps of model cellular organisms are labeled in white. Newly identified sliding clamp homologs are marked with ellipses. Ma-LMM01, Microcystis phage Ma-LMM01; RSL1, Ralstonia phage RSL1; 73, Pseudomonas phage 73; BcepGomr, Burkholderia phage BcepGomr; 0305phi8-36, Bacillus phage 0305phi8-36; Eco, Escherichia coli; ASFV, African swine fever virus; DpAV4a, Diadromus pulchellus ascovirus 4a; CBvertIrido, cold-blooded vertebrate animal iridoviruses.
During the search for PCNA homologs we identified PCNA in ascovirus DpAV-4a as one of the unassigned ORFs (File 1 in Supplementary Data) after the six-frame translation of the genome. We also found highly divergent PCNA homologs in two other ascoviruses, HvAV-3e (gi: 134287330) and SfAV-1a (gi: 11932043 and 11932044). However, those sequences align poorly to cellular PCNAs and seem to be incomplete. In addition, the putative PCNA in SfAV-1a is split into two ORFs. These observations suggest that PCNA homologs in HvAV-3e and SfAV-1a ascoviruses are likely non-functional. In some viruses we detected not single, but several copies of PCNA. Phycodnaviridae family viruses Ehv-86 and PBCV-1 have two, Mimivirus has three PCNAs (Supplementary Table S1). However, one PCNA from PBCV-1 and Mimivirus (PBCV1_PCNA1 and MimiPCNA1, respectively) is more similar to PCNAs that are present as single copies in the phycodnavirus Ostreococcus virus OsV5 and CroV, a recently sequenced relative of Mimivirus (49). Therefore, it might be expected that PCNA1 sequences of PBCV-1 and Mimivirus represent orthologs essential for viral DNA replication. On the other hand, PBCV1_PCNA2 and Ehv86_PCNA2 are most similar to PCNAs from algae; therefore, it is likely that they have been acquired from the host. MimiPCNA2 and MimiPCNA3 show the highest similarity to MimiPCNA1 and most probably are the result of multiple gene and genome duplication events, inferred to have occurred during Mimivirus evolution (50).
We detected polIIIβ homologs in only twelve phages. Of the 12 polIIIβ homologs, 7 have a typical length and five are shorter, covering only the second and third domains of polIIIβ (Supplementary Figure S2). A full-length distant polIIIβ homolog in Ma-LMM01 phage was identified (the HHsearch probability of 96%) for the first time. The Ma-LMM01 polIIIβ is coded (locus tag: MaLMM01_gp176) near other DNA replication proteins (51), supporting its putative processivity factor function.
A number of the identified viral sliding clamp homologs may have been acquired through the horizontal gene transfer (patchy taxonomic distribution, high similarity to corresponding host proteins, the absence of a DNA polymerase in the viral genome). For example, only nine out of 53 baculoviruses have PCNA homologs, and seven of those show high similarity to PCNAs from mosquitoes and moths (Supplementary Figure S3). For one of baculoviruses, Autographa californica nucleopolyhedrovirus (AcMNPV), it has been shown that its own PCNA is not required for genome replication (52). As polIIIβ and PCNA homologs, likely acquired through horizontal gene transfer (Supplementary Table S2), are either known or can be assumed to be dispensable for DNA replication, we did not include them in the summary presented in Figures 1 and 2.
Unexpectedly, we did not find homologs of any known processivity factors in some viral families with the large average genome size. These include eukaryotic Nimaviridae, Alloherpesviridae, and Malacoherpesviridae families as well as phiKZ-like phages and Clostridium phage c-st (Figure 1). However, as discussed in the ‘Polymerases’ section, DNA polymerases of the three eukaryotic viral families are atypical B-family members with additional uncharacterized domains (Supplementary Figure S1). The Clostridium phage c-st DNA polymerase is one of the C-family polymerases having a divergent C-terminal region. These observations suggest that viruses from these families may use different mechanisms to ensure DNA replication processivity. In the case of PhiKZ-like phages, whether or not processivity factors are indeed absent from their genomes remains an open question.
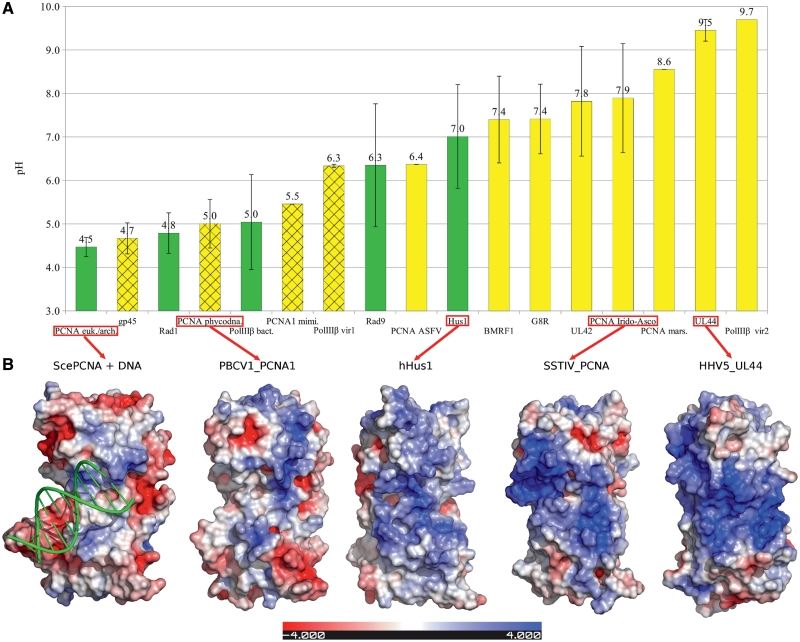
Electrostatic properties
DNA sliding clamp distribution in viral genomes (Figure 1) shows that Bacillus phage 0305phi8-36 and several families of eukaryotic viruses carrying correspondingly polIIIβ and PCNA genes in their genomes totally lack clamp loader subunits. Since a clamp loader is needed to open and load ring-shaped polIIIβ or PCNA onto DNA, this finding raised a question as to how these sliding clamps may function. One possibility is that these viruses use a clamp loader of the host. Another possibility is that these clamps do not form a closed ring and, similarly to UL42 or UL44, bind DNA directly without the need for a clamp loader. While the first possibility cannot be explored using computational approaches, the second one can.
One of the observed differences between non-ring sliding clamps (e.g. UL42, UL44) and the ring-forming ones (PCNA, polIIIβ) is that the former have an increased positive charge located on the DNA-binding face (53,54). To explore the electrostatic properties of all the identified viral sliding clamp homologs, we calculated their theoretical pIs. In addition, we constructed 3D models for representatives of viral PCNA homologs (Supplementary Table S3) and analyzed electrostatic properties of their surfaces. The obtained data was then compared to structurally and functionally characterized cellular and viral processivity factors (Figure 6 and Supplementary Table S4). It turned out that pIs of sliding clamp homologs show a striking correlation with the presence/absence of clamp loader subunits in corresponding viral families. Thus, Phycodnaviridae and Mimivirus PCNAs, predicted to be orthologous, have electrostatic properties similar to ring-shaped sliding clamps. In contrast, electrostatic properties of G8R and PCNAs of Asfarviridae (ASFV), Irido-Asco viruses and Marseillevirus are more similar to herpesviral non-ring processivity factors. Phycodnaviridae and Mimivirus have RFC homologs, while Asfarviridae, Irido-Asco viruses and Marseillevirus do not. A similar correlation is observed for sliding clamp homologs in bacteriophages. PolIIIβ homologs in phages Ma-LMM01 and RSL1 (Figure 6, PolIIIβ vir1) show much lower pI values than polIIIβ in Bacillus phage 0305phi8-36 (PolIIIβ vir2). Phages Ma-LMM01 and RSL1 do encode clamp loader subunits, while Bacillus phage 0305phi8-36 does not. Hence, based on the electrostatic properties, DNA sliding clamp homologs from Phycodnaviridae and Mimiviridae are expected to form rings, while PCNA homologs in the remaining families and polIIIβ from the Bacillus phage 0305phi8-36 are likely to bind the DNA directly, in a manner that does not require clamp loaders. According to pI values, PolIIIβ homologs of Ma-LMM01 and RSL1 phages are at the intermediate position between the characterized ring-forming and non-ring sliding clamps. However, the presence of clamp loader subunits (polIIIγ) in the corresponding genomes suggests that the closed-ring polIIIβ structure is more likely.
Figure 6.
Electrostatic properties of processivity factors and their homologs. (A) Average theoretical pIs of DNA sliding clamp subunits from cellular organisms (green bars) and viruses (yellow bars). Bars with the grid pattern correspond to viral sliding clamp homologs that are accompanied by clamp loader subunits in the genome. (B) Electrostatic potential maps of solvent accessible surface of five representatives (red color indicates negative, blue—positive potential; scale units—KbT/ec). All structures are shown in the same orientation as the ScePCNA complexed with DNA (PDB code: 3k4x). arch., Archaea; asco., —Ascoviridae; ASFV, African swine fever virus; euk., Eukarya; hHus1, Homo sapiens Hus1 (PDB code: 3g65), HHV5_UL44, Human Herpesvirus 5 UL44 (PDB code: 1t6l); irido., Iridoviridae; PCNA mars., Marseillevirus PCNA (gi:284504238); PCNA mim., Mimivirus PCNA (gi:55664866); PBCV1_PCNA1, Paramecium bursaria Chlorella Virus-1 PCNA1 (gi:9631761); Sce, S. cerevisiae; SSTIV_PCNA, Soft-shelled turtle iridovirus PCNA (gi:228861299); PolIIIβvir1, polIIIβ from Microcystis phage Ma-LMM01 and Ralstonia phage RSL1 (gi respectively: 117530347, 189233246); PolIIIβvir2, polIIIβ from Bacillus phage 0305phi8-36 (gi: 154622720).
To our surprise, we found that electrostatic properties of human checkpoint protein Hus1 and to a lesser degree of Rad9, but not of Rad1, are also similar to non-ring viral processivity factors (Figure 6). Previously, experiments have established that Rad9, Hus1 and Rad1 form a heterotrimeric PCNA-like complex (the 9-1-1 checkpoint complex), and that they do not self-multimerize (55). In addition, it has been shown that different individual subunits can interact in a pairwise manner (55). Our results combined with these experimental data suggest that Hus1 and perhaps Rad9 might also bind DNA directly as monomers or as components of heterodimeric subcomplexes. Unfortunately, there does not seem to be any available experimental data on DNA-binding properties of Hus1, Rad9 and Rad1.
Clamp loaders
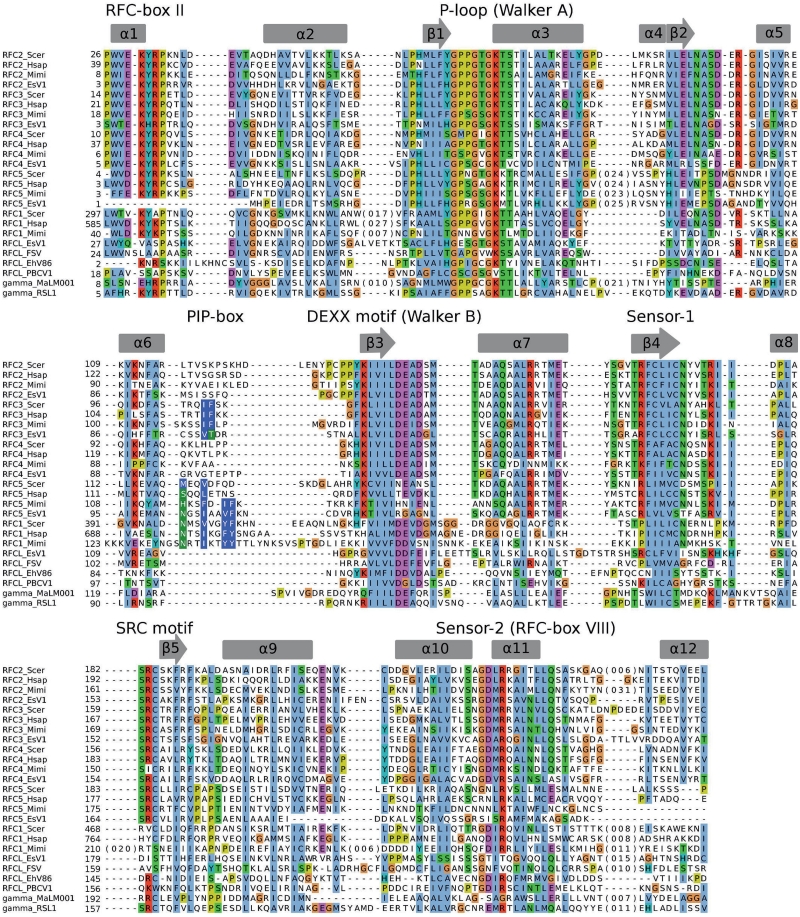
Compared to DNA polymerases and sliding clamps, homologs of clamp loader subunits are present in the fewest number of viral genomes. However, their genomic distribution appears to be highly non-random. We detected clamp loader subunits only in viruses with the largest genomes and only in those that also code for homologs of DNA sliding clamps. Moreover, as indicated above, the presence of clamp loader subunits correlates with electrostatic properties of DNA sliding clamps in the corresponding viral families. Hence, we found homologs of RFC subunits only in Mimivirus and Phycodnaviridae, the only two families that have PCNAs with electrostatic properties similar to those of ring-forming cellular PCNAs (Figures 1 and 6). Mimivirus and its relative CroV code all five RFC subunits. Members of Phycodnaviridae family have only the largest RFC subunit homolog, similar to the archaeal large RFC subunit (RFCL). The exceptions include EsV-1, which encodes all five RFC subunits, and two other viruses (Ostreococcus virus OsV5 and Ostreococcus tauri virus 1) that do not have any RFC subunit. Interestingly, the genomes of the latter two viruses are among the smallest in the family. Homologs of bacterial clamp loader subunits were identified in only two phages, RSL1 and Ma-LM001. In each case we found only a homolog of a single clamp loader subunit, polIIIγ. Both polIIIγ homologs have conserved P-loop, DEXX and SRC motifs (Figure 7) suggesting that they are active ATPases. Again, polIIIβ homologs in these two phages have significantly lower pIs than polIIIβ in Bacillus phage 0305phi8-36, lacking any clamp loader subunit (Figure 6). T4-like clamp loaders consisting of gp44 and gp62 subunits were identified only in T4-like phages.
Figure 7.
Alignment of eukaryotic and viral clamp loader subunits. Sequence alignment is based on multiple structure superposition of experimental X-ray structures and homology models obtained using MUSTANG (66). Secondary structure of the yeast RFC3 subunit (PDB code: 1sxj) is shown above the alignment. PBCV1, Paramecium bursaria Chlorella Virus-1; EsV1, Ectocarpus siliculosus virus 1; EhV86, Emiliania huxleyi virus 86; FSV, Feldmania species virus; Hsap, Homo sapiens; MaLM001, Microcystis phage Ma-LM001; Mimi, Mimivirus; RSL1, Ralstonia phage RSL1; Scer, Saccharomyces cerevisiae.
All five RFC subunits from Mimivirus and the phycodnavirus EsV-1 are similar to corresponding human and yeast proteins (Figure 7) and have motifs for both ATP binding (P-loop) and hydrolysis (DEXX-motif). However, there are few differences compared to eukaryotic RFC. Collectively, structural studies of yeast RFC–PCNA complex (56) and biochemical experiments (57,58) indicate that RFC1, RFC3 and RFC5 interact with the corresponding hydrophobic pockets of PCNA protomers. Human and yeast RFC1, RFC3 and RFC5 have progressively ‘weaker’ PCNA-interaction (PIP-box) motifs (Figure 7), correlating with the decreasing PCNA-binding strength (56–58). In Mimivirus RFC1 and RFC3 PIP-boxes follow the same trend, but the PIP-box in RFC5 is more like the one in RFC1. Interestingly, EsV-1 has the ‘strongest’ PCNA-interaction motif in RFC5 followed by RFC3, and no PIP-box in the RFC large subunit. Notably, a similar non-canonical distribution of the PIP-box ‘strength’ between RFC1, RFC3 and RFC5 is also observed in some eukaryotes (Supplementary Figures S4–S6). Other phycodnaviruses including FSV, EhV-86 and Chlorella viruses have only a homolog of the RFC large subunit, which, similarly to EsV-1 RFCL, has no apparent PIP-box (Figure 7). At least in Chlorella viruses RFCL appears to be the inactive ATPase because of non-canonical substitutions in P-loop and the DEXX motifs, which are essential for ATP-binding and hydrolysis in the AAA+ protein family (59).
DISCUSSION
Our results show that the presence and the nature of DNA replicases encoded in the genomes of dsDNA viruses is related to the genome size. This relationship can be defined as the tendency to encode polymerase processivity components in addition to the DNA polymerase more often as the genome size increases.
Viruses having genomes smaller than ~40 kb most often do not have their own DNA polymerases. However, if they do, it is usually a PolBp type DNA polymerase. Interestingly, this is seen in viruses infecting organisms from all domains of life. Coupled with the observation that PolBp polymerases disappear completely from larger viral genomes (Figures 1 and 2), this suggests that properties of protein-primed B-family DNA polymerases might be optimal for this genome size range.
As the genome size increases (~40–140 kb) A-family polymerases take over. However, it is not clear whether the dominance of A-family polymerases in this genome size range is significant. The reason is that we detected A-family polymerases only in bacteriophages, and this particular size range is overrepresented with bacteriophage genomes. Nonetheless, even if we ignore the polymerase type, the typical feature of genomes in this size range is the lack of DNA sliding clamp homologs. It has been shown that E. coli polymerase I (A-family) is stimulated by the polIIIβ clamp (60). Therefore, the absence of sliding clamp homologs cannot be explained by the inability of polA to utilize sliding clamp as a processivity factor. Moreover, in two phages (Ma-LMM01 and RSL1) with large genomes (>150 kb) we detected an A-family polymerase, a polIIIβ homolog and a clamp loader subunit suggesting that the polIIIβ homolog may function as a processivity factor together with polA. On the other hand, some bacteriophages have evolved the increased processivity of A-family polymerases without using DNA sliding clamps. One such solution is the recruitment of thioredoxin from the host as observed in T7-like phages (61). The UDG-like domain in DNA polymerases of SPO1-like and T5-like phages may well be another solution, which is yet to be addressed experimentally.
The genome size range of 140 kb and larger is represented by eukaryotic viruses and bacteriophages. They all have their own DNA polymerases, typically of B-family. Our discovery of evolutionary distant DNA polymerases in phiKZ-like phages has eliminated the only seeming exception to this rule. DNA replicases in this size range often include DNA polymerase processivity factors and sometimes clamp loaders. Initially, there does not seem to be any discernible pattern as to the presence or absence of sliding clamp homologs and clamp loaders (Figure 1). However, if we consider properties of DNA polymerases, homologs of sliding clamps and the presence of clamp loader subunits we get a fairly coherent picture.
Thus, we did not find any sliding clamp homologs in several groups of large dsDNA viruses. However, their DNA polymerases either have additional uncharacterized domains or non-homologous regions. It may be that these polymerases either possess an increased intrinsic processivity due to these additional/altered regions or use alternative processivity factors. On the other hand, the fact that we did not find any sliding clamp homolog in phiKZ-like phages is somewhat puzzling. Their polymerases, although evolutionary distinct, seem to possess a typical B-family architecture. In addition, two of the three polymerases at their C-termini feature a putative signature of a clamp-binding motif. It is quite possible that processivity factors are encoded in genomes of phiKZ-phages, but are too strongly diverged to be detected with current methods.
As it comes to the viral families that do have homologs of DNA sliding clamps, the intriguing finding was that a number of these families completely lack clamp loader subunits. However, the subsequent analysis of electrostatic properties of sliding clamp homologs was quite revealing. It showed that PCNA homologs from Irido-Asco, Asfar-viruses and Marseillevirus as well as a polIIIβ homolog from Bacillus phage 0305phi8-36, all have elevated pIs (Figure 6A). Models of several representatives showed that most of the increased positive charge is localized to the DNA-interacting face (Figure 6B). This property is typical for well-characterized herpesviral processivity factors. They do not form rings; instead they bind DNA directly as monomers (UL42) or dimers (UL44). This suggests a similar direct DNA-binding mode for the sliding clamp homologs with the elevated pI and without clamp loaders in corresponding genomes. In this regard it is interesting to point out that we observed a similarly increased positive charge on the DNA interaction side of two components of the human 9-1-1 complex, Hus1 and, to a lesser degree, Rad9. In contrast, Rad1, the third component of the complex, has electrostatic properties similar to those of cellular PCNAs. This observation suggests that Hus1 and perhaps Rad9 could also bind DNA as monomers or as components of heterodimeric subassemblies and serve either as recruitment platforms or processivity factors for other proteins. Interestingly, there is genetic data supporting possible additional and different roles for Hus1, Rad9 and Rad1. Experimental data on telomere maintenance in Schizosaccharomyces pombe revealed that Rad1 mutants had telomere-shortening defects, whereas Hus1 and Rad9 mutants had normal telomere lengths (62). More recently, it was shown that for carrying its telomere maintenance function, Rad1 requires the presence of either Hus1 or Rad9 (63). An interesting possibility is that the different electrostatic properties of Rad9, Hus1 and Rad1 revealed in this study may be responsible for the observed differences in mutant phenotypes.
Findings concerning viral clamp loaders are perhaps most puzzling compared to other replicase components. Only three eukaryotic viruses have a complete set of five RFC subunits corresponding to the eukaryotic clamp loader, RFC. As expected for functional RFC, all three viruses have characteristic P-loop and DEXX motifs in RFC1-4 subunits and also feature PCNA-interacting (PIP-box) motifs in RFC1, RFC3 and RFC5. The analysis of their PIP-boxes led to an interesting observation that the distribution of ‘strength’ of the PCNA-binding motifs across the three RFC subunits can be different in comparison to human or yeast RFC (Figure 7). In other words, it appears that in the course of evolution the ‘strength’ of PCNA-binding motifs in RFC1, RFC3 and RFC5 may evolve differently. This idea is also supported by the observation that, in contrast to human and yeast, RFC5 sequences in some other eukaryotes (Supplementary Figures S4–S6) feature a canonical PCNA-binding motif, while RFC1 has a strongly reduced one. Several members of Phycodnaviridae family have only a single homolog of the RFC large subunit. From studies with human and yeast RFC it is known that the RFC large subunit determines the specificity for the clamp (1). For example, RFC1 determines specificity for PCNA, while Rad17—for the 9-1-1 complex. Thus, it may be that the viral homolog of the large RFC subunit recruits four small RFC subunits of the host to form a pentameric complex specific for binding and loading viral PCNA. However, these RFC large subunits seem to completely lack PCNA-binding motifs and some have non-canonical ATPase motifs. It has been shown that the mutation in the ATP-binding motif of the large RFC subunit in yeast does not affect PCNA loading (64). Therefore, the ATPase activity may also be dispensable in viral RFC large subunits. It is not clear, though, how to reconcile the absence of a PCNA-binding motif with the expected specificity for the viral PCNA. Two large phages, Ma-LMM01 and RSL1, that have a bacterial clamp loading subunit homolog, polIIIγ, additionally have an A-family DNA polymerase and a homolog of polIIIβ sliding clamp. In these two cases it is also not clear what is the composition of the functional replicase. Does the viral polIIIγ recruit host clamp loader subunit(s) to produce a functional clamp loader specific for the viral polIIIβ? Or perhaps the composition of these clamp loaders is analogous to the T4 clamp loader, which is made of four copies of gp44 (polIIIγ homolog) and a single taxon-specific subunit gp62 (no detectable homologs outside the T4-like group)? To address these questions, computational methods can hardly substitute laboratory experiments.
Overall, our observed connection between the virus genome size and DNA replicase components might help in predicting the expected type and completeness of replicase components for newly sequenced viral genomes. In addition, our observations for DNA replicases in dsDNA viruses perhaps may have a more general significance. For example, symbiotic bacteria belonging to genus Hodgkinia and Carsonella have presently the smallest known cellular genomes of 144 and 160 kb size, respectively (65). It turns out that neither has a DNA sliding clamp or a clamp loader. However, somewhat larger genomes of symbionts Sulcia cicada (277 kb), Buchnera Cc (416 kb) and Nanoarchaeum equitans (491 kb) already have the complete set of DNA replicase components. With more large viral and small cellular genomes available, it will be interesting to see how universal the observed relationship is.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute (55005627); Ministry of Education and Science of Lithuania. Funding for open access charge: European Community's Seventh Framework Programme (FP7-REGPOT-2009-1 project 245721 ‘MoBiLi’).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Giedrius Sasnauskas, Edita Sužiedėlienė and Ana Venclovienė for comments and suggestions.
REFERENCES
- 1.Indiani C, O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell. Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc. Natl Acad. Sci. USA. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 6.Cann IK, Komori K, Toh H, Kanai S, Ishino Y. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl Acad. Sci. USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishino Y, Komori K, Cann IK, Koga Y. A novel DNA polymerase family found in Archaea. J. Bacteriol. 1998;180:2232–2236. doi: 10.1128/jb.180.8.2232-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berquist BR, DasSarma P, DasSarma S. Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1. BMC Genet. 2007;8:31. doi: 10.1186/1471-2156-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamtekar S, Berman AJ, Wang J, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 11.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 12.Koonin EV. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct. 2006;1:39. doi: 10.1186/1745-6150-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruck I, O'Donnell M. The ring-type polymerase sliding clamp family. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-1-reviews3001. REVIEWS3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams GJ, Johnson K, Rudolf J, McMahon SA, Carter L, Oke M, Liu H, Taylor GL, White MF, Naismith JH. Structure of the heterotrimeric PCNA from Sulfolobus solfataricus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:944–948. doi: 10.1107/S1744309106034075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl Acad. Sci. USA. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuccola HJ, Filman DJ, Coen DM, Hogle JM. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
- 18.Appleton BA, Loregian A, Filman DJ, Coen DM, Hogle JM. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell. 2004;15:233–244. doi: 10.1016/j.molcel.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Murayama K, Nakayama S, Kato-Murayama M, Akasaka R, Ohbayashi N, Kamewari-Hayami Y, Terada T, Shirouzu M, Tsurumi T, Yokoyama S. Crystal structure of epstein-barr virus DNA polymerase processivity factor BMRF1. J. Biol. Chem. 2009;284:35896–35905. doi: 10.1074/jbc.M109.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh S, Hamdan SM, Cook TE, Richardson CC. Interactions of Escherichia coli thioredoxin, the processivity factor, with bacteriophage T7 DNA polymerase and helicase. J. Biol. Chem. 2008;283:32077–32084. doi: 10.1074/jbc.M805062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YH, Lin Y, Yoshinaga A, Chhotani B, Lorenzini JL, Crofts AA, Mei S, Mackie RI, Ishino Y, Cann IK. Molecular analyses of a three-subunit euryarchaeal clamp loader complex from Methanosarcina acetivorans. J. Bacteriol. 2009;191:6539–6549. doi: 10.1128/JB.00414-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federici BA, Bigot Y. Evolution of immunosuppressive organelles from DNA viruses in insects. In: Pontarotti P, editor. Evolutionary Biology - Concepts, Molecular and Morphological Evolution. Springer Berlin Heidelberg; 2010. pp. 229–248. [Google Scholar]
- 23.Frith MC, Wan R, Horton P. Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res. 2010;38:e100. doi: 10.1093/nar/gkq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wernersson R. Virtual ribosome–a comprehensive DNA translation tool with support for integration of sequence feature annotation. Nucleic Acids Res. 2006;34:W385–W388. doi: 10.1093/nar/gkl252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 28.Margelevičius M, Laganeckas M, Venclovas Č. COMA server for protein distant homology search. Bioinformatics. 2010;26:1905–1906. doi: 10.1093/bioinformatics/btq306. [DOI] [PubMed] [Google Scholar]
- 29.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 31.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margelevičius M, Venclovas Č. PSI-BLAST-ISS: an intermediate sequence search tool for estimation of the position-specific alignment reliability. BMC Bioinformatics. 2005;6:185. doi: 10.1186/1471-2105-6-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venclovas Č, Margelevičius M. The use of automatic tools and human expertise in template-based modeling of CASP8 target proteins. Protein Struct. Funct. Bioinformatics. 2009;77:81–88. doi: 10.1002/prot.22515. [DOI] [PubMed] [Google Scholar]
- 35.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 38.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R, Robben J, Mesyanzhinov V, Krylov VN, Volckaert G. Genome comparison of Pseudomonas aeruginosa large phages. J. Mol. Biol. 2005;354:536–545. doi: 10.1016/j.jmb.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 40.Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, Krylov VN, Volckaert G. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J. Mol. Biol. 2002;317:1–19. doi: 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- 41.Thomas JA, Rolando MR, Carroll CA, Shen PS, Belnap DM, Weintraub ST, Serwer P, Hardies SC. Characterization of Pseudomonas chlororaphis myovirus 201varphi2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology. 2008;376:330–338. doi: 10.1016/j.virol.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margelevičius M, Venclovas Č. Detection of distant evolutionary relationships between protein families using theory of sequence profile-profile comparison. BMC Bioinformatics. 2010;11:89. doi: 10.1186/1471-2105-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006;30:321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 44.Andraos N, Tabor S, Richardson CC. The highly processive DNA polymerase of bacteriophage T5. Role of the unique N and C termini. J. Biol. Chem. 2004;279:50609–50618. doi: 10.1074/jbc.M408428200. [DOI] [PubMed] [Google Scholar]
- 45.Druck Shudofsky AM, Silverman JE, Chattopadhyay D, Ricciardi RP. Vaccinia virus D4 mutants defective in processive DNA synthesis retain binding to A20 and DNA. J. Virol. 2010;84:12325–12335. doi: 10.1128/JVI.01435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 47.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyle K, Traktman P. Poxviruses. In: Raney KD, Gotte M, Cameron CE, editors. Viral Genome Replication. Springer US; 2009. pp. 225–247. [Google Scholar]
- 49.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl Acad. Sci. USA. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suhre K. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J. Virol. 2005;79:14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida T, Nagasaki K, Takashima Y, Shirai Y, Tomaru Y, Takao Y, Sakamoto S, Hiroishi S, Ogata H. Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J. Bacteriol. 2008;190:1762–1772. doi: 10.1128/JB.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kool M, Ahrens CH, Goldbach RW, Rohrmann GF, Vlak JM. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl Acad. Sci. USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komazin-Meredith G, Santos WL, Filman DJ, Hogle JM, Verdine GL, Coen DM. The Positively charged surface of herpes simplex virus UL42 mediates DNA binding. J. Biol. Chem. 2008;283:6154–6161. doi: 10.1074/jbc.M708691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loregian A, Sinigalia E, Mercorelli B, Palu G, Coen DM. Binding parameters and thermodynamics of the interaction of the human cytomegalovirus DNA polymerase accessory protein, UL44, with DNA: implications for the processivity mechanism. Nucleic Acids Res. 2007;35:4779–4791. doi: 10.1093/nar/gkm506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J. Biol. Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 56.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 57.Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, Hingorani MM, O'Donnell M. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J. Biol. Chem. 2003;278:50744–50753. doi: 10.1074/jbc.M309206200. [DOI] [PubMed] [Google Scholar]
- 58.Yao NY, Johnson A, Bowman GD, Kuriyan J, O'Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J. Biol. Chem. 2006;281:17528–17539. doi: 10.1074/jbc.M601273200. [DOI] [PubMed] [Google Scholar]
- 59.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 60.Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl Acad. Sci. USA. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bedford E, Tabor S, Richardson CC. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc. Natl Acad. Sci. USA. 1997;94:479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dahlen M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol. Biol. Cell. 1998;9:611–621. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khair L, Chang YT, Subramanian L, Russell P, Nakamura TM. Roles of the checkpoint sensor clamp Rad9-Rad1-Hus1 (911)-complex and the clamp loaders Rad17-RFC and Ctf18-RFC in Schizosaccharomyces pombe telomere maintenance. Cell Cycle. 2010;9:2237–2248. doi: 10.4161/cc.9.11.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt SL, Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J. Biol. Chem. 2001;276:34784–34791. doi: 10.1074/jbc.M011633200. [DOI] [PubMed] [Google Scholar]
- 65.McCutcheon JP. The bacterial essence of tiny symbiont genomes. Curr. Opin. Microbiol. 2010;13:73–78. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.