Abstract
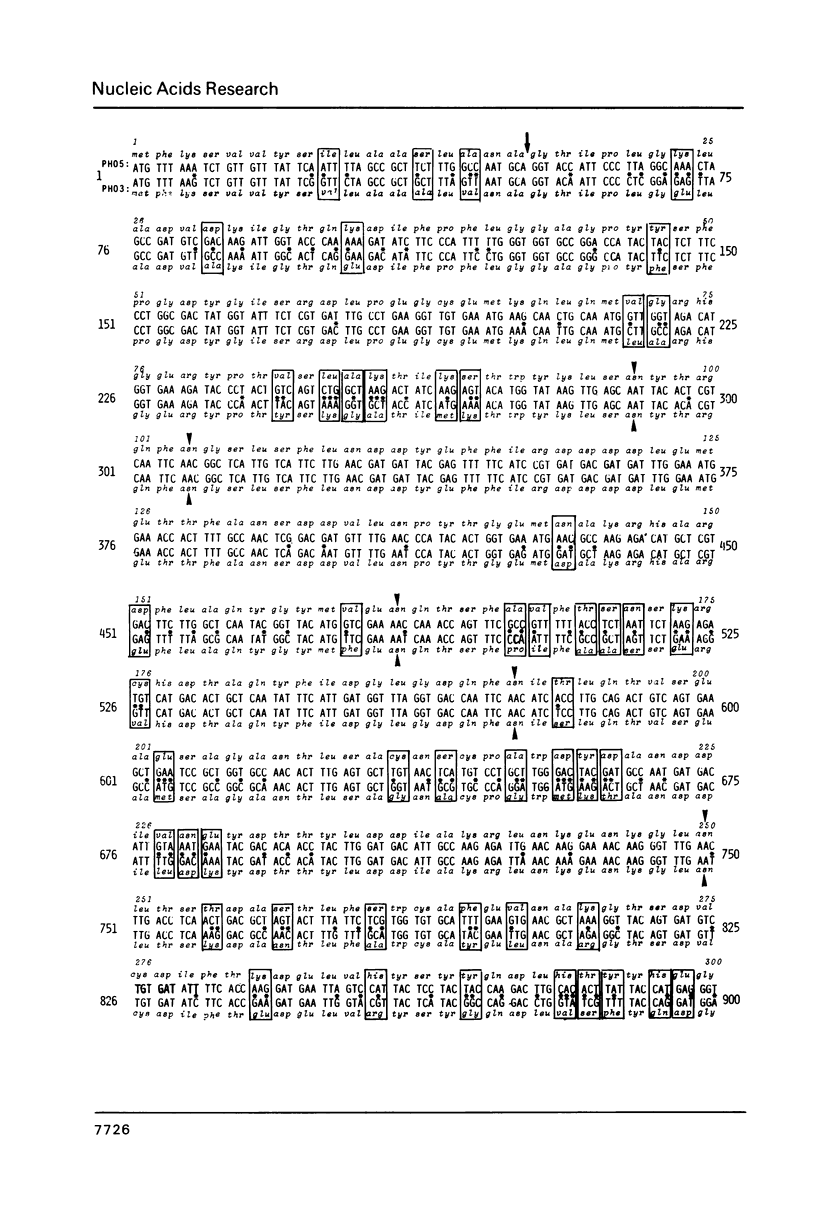
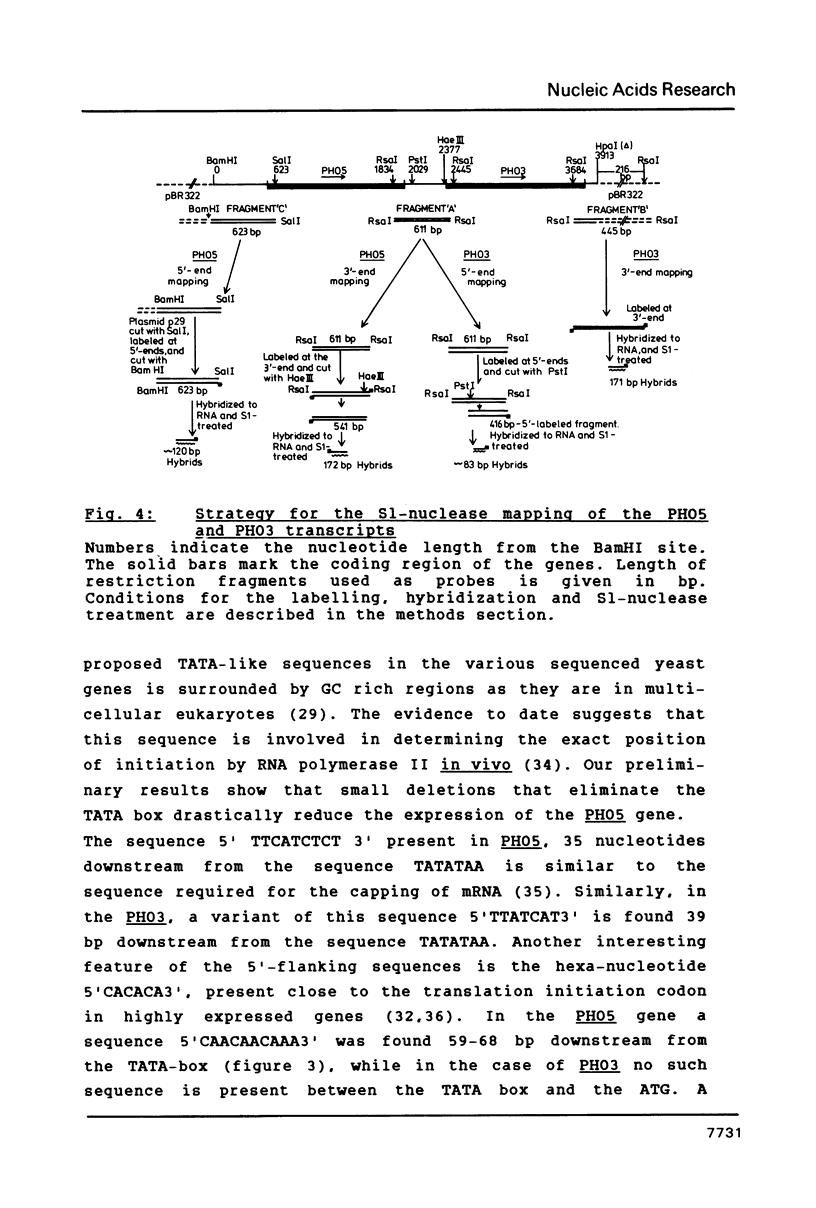
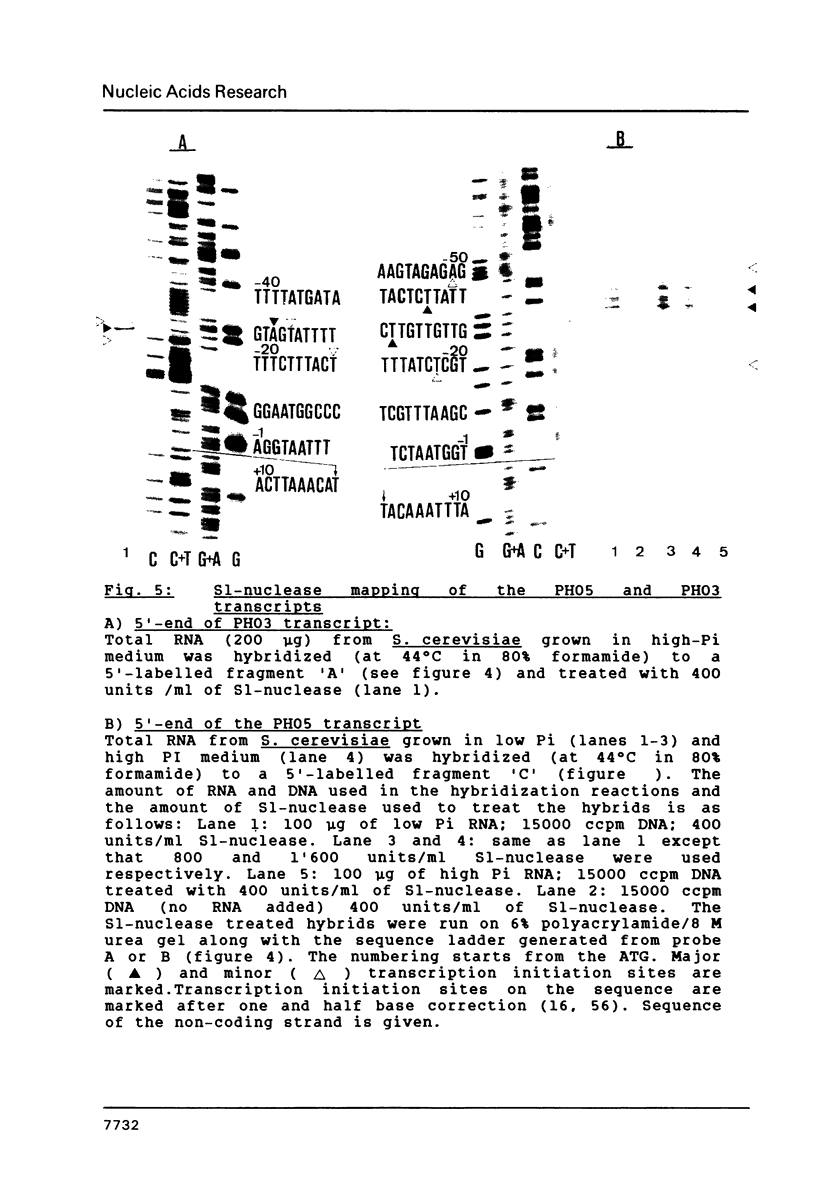
We have sequenced the genetically linked genes for repressible (PHO5) and and constitutive (PHO3) acid phosphatase from S. cerevisiae. Both genes are located on a 3.91 Kb BamHI and HpaI fragment, in the order (5') PHO5, PHO3 (3'). The mRNA transcripts have been analysed by S1-nuclease mapping. They show heterogenous initiation sites. Each of the PHO5 and PHO3 genes codes for 467 amino acids as deduced from the DNA sequence. The coding regions of the two genes show homology both at the nucleotide (82%) and the amino acid (87%) level. In the coding sequences, long stretches of homologous regions are flanked by small non-homologous regions. The nucleotide homology (65%) extends to some length into the 5' and 3' non-coding flanking sequences. Further upstream sequences are unrelated. The comparison of the NH2-terminal amino acid sequence deduced from the nucleotide sequence, with that of purified repressible acid phosphatase revealed the presence of a putative signal peptide.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen N., Thill G. P., Kramer R. A. RNA and homology mapping of two DNA fragments with repressible acid phosphatase genes from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):562–569. doi: 10.1128/mcb.3.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima K., Oshima T., Kubota I., Nakamura N., Mizunaga T., Toh-e A. The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res. 1983 Mar 25;11(6):1657–1672. doi: 10.1093/nar/11.6.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Gene conversion: some implications for immunoglobulin genes. Cell. 1981 Jun;24(3):592–594. doi: 10.1016/0092-8674(81)90082-9. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Boer P., Van Rijn H. J., Reinking A., Seryn-Parvé E. P. Biosynthesis of acid phosphatase of baker's yeast. Characterization of a protoplast-bound fraction containing precursors of the exo-enzyme. Biochim Biophys Acta. 1975 Feb 19;377(2):331–342. doi: 10.1016/0005-2744(75)90314-9. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Lemire J. M., Cannon L. E., Halvorson H. O. In vitro synthesis of repressible yeast acid phosphatase: identification of multiple mRNAs and products. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4504–4508. doi: 10.1073/pnas.77.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J. F., Stewart J. W., Sherman F. The cyc1-11 mutation in yeast reverts by recombination with a nonallelic gene: composite genes determining the iso-cytochromes c. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6334–6338. doi: 10.1073/pnas.78.10.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Perrin F., Seidel R. The actin gene in yeast Saccharomyces cerevisiae: 5' and 3' end mapping, flanking and putative regulatory sequences. Nucleic Acids Res. 1981 Dec 11;9(23):6339–6350. doi: 10.1093/nar/9.23.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. Structural comparison of two nontandemly repeated yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1980 Mar 25;255(6):2596–2605. [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. The primary structure of a glyceraldehyde-3-phosphate dehydrogenase gene from Saccharomyces cerevisiae. J Biol Chem. 1979 Oct 10;254(19):9839–9845. [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Knecht R., Seemüller U., Liersch M., Fritz H., Braun D. G., Chang J. Y. Sequence determination of eglin C using combined microtechniques of amino acid analysis, peptide isolation, and automatic Edman degradation. Anal Biochem. 1983 Apr 1;130(1):65–71. doi: 10.1016/0003-2697(83)90650-4. [DOI] [PubMed] [Google Scholar]
- Kramer R. A., Andersen N. Isolation of yeast genes with mRNA levels controlled by phosphate concentration. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6541–6545. doi: 10.1073/pnas.77.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal M., D'Eustachio P., Ruddle F. H., Arnheim N. Human nucleolus organizers on nonhomologous chromosomes can share the same ribosomal gene variants. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5744–5748. doi: 10.1073/pnas.78.9.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhaber S. A., Goossens M., Kan Y. W. Homology and concerted evolution at the alpha 1 and alpha 2 loci of human alpha-globin. Nature. 1981 Mar 5;290(5801):26–29. doi: 10.1038/290026a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyhack B., Bajwa W., Rudolph H., Hinnen A. Two yeast acid phosphatase structural genes are the result of a tandem duplication and show different degrees of homology in their promoter and coding sequences. EMBO J. 1982;1(6):675–680. doi: 10.1002/j.1460-2075.1982.tb01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunaga T., Noguchi T. The role of core-oligosaccharide in formation of an active acid phosphatase and its secretion by yeast protoplasts. J Biochem. 1982 Jan;91(1):191–200. doi: 10.1093/oxfordjournals.jbchem.a133676. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. A position effect in the control of transcription at yeast mating type loci. Nature. 1981 Jan 22;289(5795):244–250. doi: 10.1038/289244a0. [DOI] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981 Aug;25(2):461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Ollo R., Auffray C., Morchamps C., Rougeon F. Comparison of mouse immunoglobulin gamma 2a and gamma 2b chain genes suggests that exons can be exchanged between genes in a multigenic family. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2442–2446. doi: 10.1073/pnas.78.4.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Cheng C. C., Brownlee G. G. Sequence analysis of eukaryotic mRNA. Prog Nucleic Acid Res Mol Biol. 1976;19:123–134. doi: 10.1016/s0079-6603(08)60914-9. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmincke C. D., Herrmann K., Hausen P. Size of primary transcripts in Ehrlich ascites cells as measured by tetraphosphate determination. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1994–1998. doi: 10.1073/pnas.73.6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier P. H., Bothwell A. L., Mueller-Hill B., Baltimore D. Multiple differences between the nucleic acid sequences of the IgG2aa and IgG2ab alleles of the mouse. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4495–4499. doi: 10.1073/pnas.78.7.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber A. M., Schweingruber M. E. Purification and identification of inactive forms of repressible and constitutive acid phosphatase in yeast. Biochim Biophys Acta. 1982 Aug 6;717(2):203–209. doi: 10.1016/0304-4165(82)90170-2. [DOI] [PubMed] [Google Scholar]
- Sharma C. B., Lehle L., Tanner W. N-Glycosylation of yeast proteins. Characterization of the solubilized oligosaccharyl transferase. Eur J Biochem. 1981 May;116(1):101–108. doi: 10.1111/j.1432-1033.1981.tb05306.x. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Transcription of the his3 gene region in Saccharomyces cerevisiae. J Mol Biol. 1981 Nov 5;152(3):535–552. doi: 10.1016/0022-2836(81)90267-9. [DOI] [PubMed] [Google Scholar]
- Thill G. P., Kramer R. A., Turner K. J., Bostian K. A. Comparative analysis of the 5'-end regions of two repressible acid phosphatase genes in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):570–579. doi: 10.1128/mcb.3.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To-E A., Ueda Y., Kakimoto S. I., Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973 Feb;113(2):727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A., Inouye S., Oshima Y. Structure and function of the PHO82-pho4 locus controlling the synthesis of repressible acid phosphatase of Saccharomyces cerevisiae. J Bacteriol. 1981 Jan;145(1):221–232. doi: 10.1128/jb.145.1.221-232.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A., Kakimoto S. Genes coding for the structure of the acid phosphatases in Saccharomyces cerevisiae. Mol Gen Genet. 1975 Dec 30;143(1):65–70. doi: 10.1007/BF00269421. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Hereford L., Grunstein M. Histone H2B genes of yeast encode two different proteins. Cell. 1980 Dec;22(3):799–805. doi: 10.1016/0092-8674(80)90556-5. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]




