Abstract
The inhibitory switch (IS) domain of p21-activated kinase 1 (PAK1) stabilizes full-length PAK1 in an inactive conformation by binding to the PAK1 kinase domain. Competitive binding of small GTPases to the IS domain disrupts the autoinhibitory interactions and exposes the IS domain binding site on the surface of the kinase domain. To build an affinity reagent that selectively binds the activated state of PAK1, we used molecular modeling to re-engineer the isolated IS domain so that it was soluble and stable, did not bind to GTPases and bound more tightly to the PAK1 kinase domain. Three design strategies were tested: in the first and second case, extension and redesign of the N-terminus were used to expand the hydrophobic core of the domain and in the third case the termini were redesigned to be adjacent in space so that that the domain could be stabilized by insertion into a loop in a host cyan fluorescent protein (CFP). The best-performing design, called CFP-PAcKer, was based on the third strategy and bound the kinase domain of PAK1 with an affinity of 400 nM. CFP-PAcKer binds more tightly to a full-length variant of PAK1 that is stabilized in the ‘open’ state (Kd = 3.3 µM) than to full-length PAK1 in the ‘closed’ state (undetectable affinity), and binding can be monitored with fluorescence by placing an environmentally sensitive fluorescence dye on CFP-PAcKer adjacent to the binding site.
Keywords: Computational protein design, Rosetta, merocyanine dye, p21-activated kinase
INTRODUCTION
p21-activated kinase 1 (PAK1) is a serine/threonine kinase that regulates a diverse set of cellular activities including cytoskeletal remodeling, growth-factor and steroid-receptor signaling, transcription and mitosis.1 The activity of PAK1 is tightly regulated and the altered expression and activity of PAK1 has been implicated in several human cancers.2 Inactive PAK1 forms an autoinhibited homodimer that is stabilized by interactions between its inhibitory switch (IS) domain and the kinase domain. In the autoinhibited state, residues immediately C-terminal to the IS domain, called the KI segment, occupy the cleft between the N lobe and C lobe of the kinase domain and block the active site3 (Supplementary Fig. S1) (3). PAK1 can be activated by a variety of signaling molecules including the small GTPases Cdc42 and Rac1. In the GTP-bound state, Cdc42 and Rac1 bind to the p21-binding domain of PAK1 (PBD; also known as the Cdc42/Rac-interactive binding (CRIB) domain), which disrupts the autoinhibitory interaction between the IS domain and the kinase domain. The PBD (residues 70–113) and the IS domain (residues 81–136) structurally overlap, and therefore, binding of the GTPase to the PBD is mutually exclusive with binding between the IS domain and the kinase domain. Following GTPase binding, autophosphorylation further stabilizes the activated state.4
Because PAK1 activity is under tight spatio-temporal control, we are interested in creating a biosensor that can detect the localization and kinetics of PAK1 activation in living cells. Recently, a PAK1 biosensor (called Pakabi) was created by fusing yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) to the termini of PAK1.5 Upon activation and disruption of the interaction between the IS and kinase domains of PAK1 the distance between the PAK1 termini increases and the FRET signal between the fluorescent proteins decreases. Pakabi has been used to visualize PAK1 activation during cellular spreading and during the formation of cellular protrusions. One limitation of this type of sensor is that it is not detecting endogenous PAK1, but rather over -expressed protein. Additionally, to provide a larger FRET signal in the inactive state the first 64 residues of PAK1 were removed when creating Pakabi. As a result, Pakabi is missing binding sites that localize PAK1 to tyrosine kinase receptors via the adaptors Nck6 and Grb2.7 Localization of PAK1 to tyrosine kinase receptors has been shown to be important for actin remodeling, cell motility and the extension of membrane lamellae.
An alternative type of biosensor is one that detects the activation state of endogenous protein. One strategy for accomplishing this goal is to make use of proteins known as affinity reagents, that only bind to the protein of interest when it is in an activated state. Hahn and co-workers used this strategy to create a biosensor for activated Cdc42.8 An environmentally sensitive dye was covalently attached to a domain from the Cdc42 effector protein WASP that only binds activated Cdc42. The labeled protein shows a strong increase in fluorescence when binding endogenous, activated Cdc42, and has been used to study the spatio-temporal dynamics of Cdc42 activation in living cells. Here, we rationally design a modified fragment of the PAK1 autoinhibitory domain that can be used similarly to create a biosensor for activation of endogenous PAK1. In many cases biosensors cannot be created because there exist no known domains with appropriate binding characteristics; achieving this through the design of novel binding domains or improvement of naturally occurring binders would be important in extending the applicability of biosensors.
Upon PAK1 activation, the interaction between the IS domain and the kinase domain of PAK1 is broken, exposing the IS binding surface on the kinase domain. In previous work, we showed that a protein designed to interact with the IS binding site on the PAK1 kinase domain was sensitive to whether PAK1 was in the ‘open’ or ‘closed’ state.9 However, the designed protein only bound the ‘open’ state with an affinity of 100 µM, too weak to be used as the basis for a biosensor. Here, we examine if variants of the IS domain can be used as affinity reagents that differentiate active and inactive PAK. The wild type IS domain is unfolded in isolation and only has modest affinity for PAK1. To improve the stability of the IS domain and its affinity for PAK1, we used the molecular modeling software Rosetta to identify N- and C-terminal extensions that stabilize the domain and allow for favorable insertion in a host protein. The best performing redesign binds the PAK1 kinase domain with an affinity of 400 nM and has no measurable affinity for full-length PAK1 when it is in the ‘closed’ state. We also show that binding can be detected by placing an environmentally sensitive dye adjacent to the binding interface.
Results
The wild type IS domain is not a good affinity reagent
We first examined if the wild type IS domain could be used as an affinity reagent that is specific for activated PAK1. We expressed and purified residues 83–137 of PAK1 (WT_IS), which in the crystal structure of autoinhibited PAK1 interact with the C lobe of PAK1 kinase domain and form a small folded domain with three helices and a β-strand (Fig. 1). We did not include residues 138–149 of PAK1, which bind in the active site of the kinase. The circular dichroism spectrum of the purified IS domain showed that the domain does not fold in isolation (Fig. 2). Not surprisingly, the domain was prone to aggregation and proteolysis during expression and purification. In isothermal titration calorimetry experiments, the IS domain bound to the kinase domain of PAK1 with an affinity of only 4 µM, while biosensor-target interactions typically have a dissociation constant of less than 1 µM.
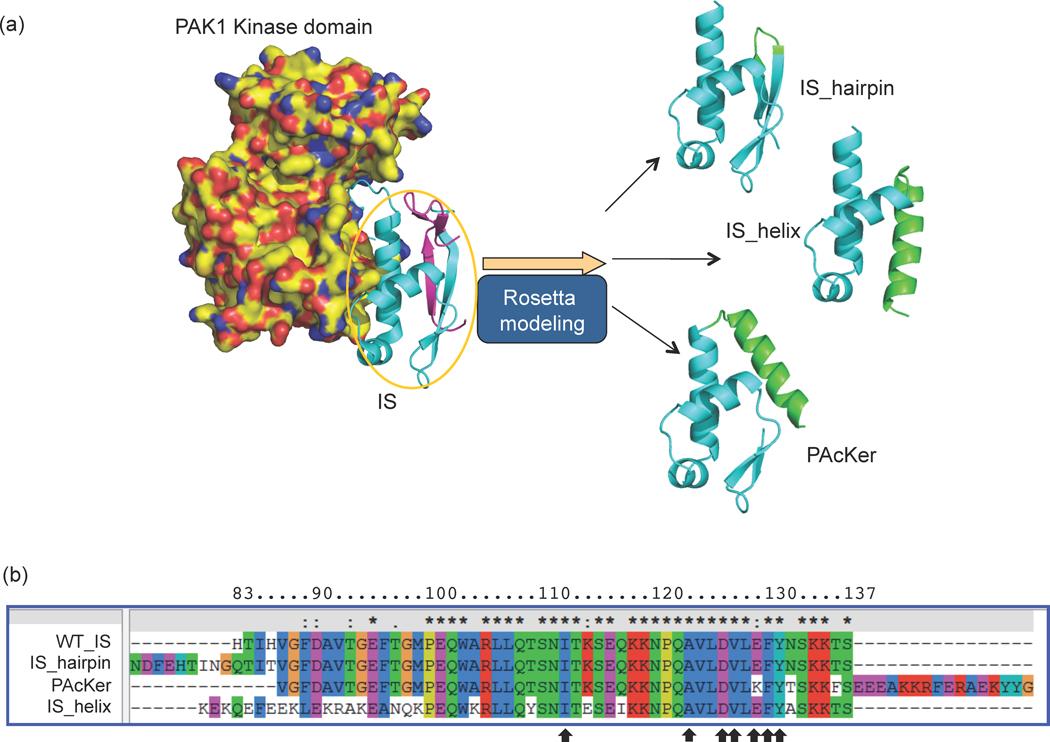
Fig. 1. Structural and sequence comparison of the designs.
All the designs were based on the IS domain (IS, cyan), which interacts with the C-lobe of the PAK1 kinase domain (yellow). In the crystal structure PAK1 forms a homodimer in an autoinhibited conformation with a β-strand from one IS domain interacting with the partner IS domain (magenta). The rest of the partner is not displayed for clarity. (a) Three alternative designs were tested, ‘IS_hairpin’, IS_helix’ and ‘PAcKer’. The new structural elements added to each variant are shown in green. The cyan regions were left largely intact except for some point mutations that allowed better packing with the new structural elements. (b) Multiple sequence alignment of the variants shows the new structural elements were at N-terminus or C-terminus while the residues involved in binding to the kinase domain were left unmutated (black arrows). The residue numbering is based on the primary sequence of full-length PAK1.
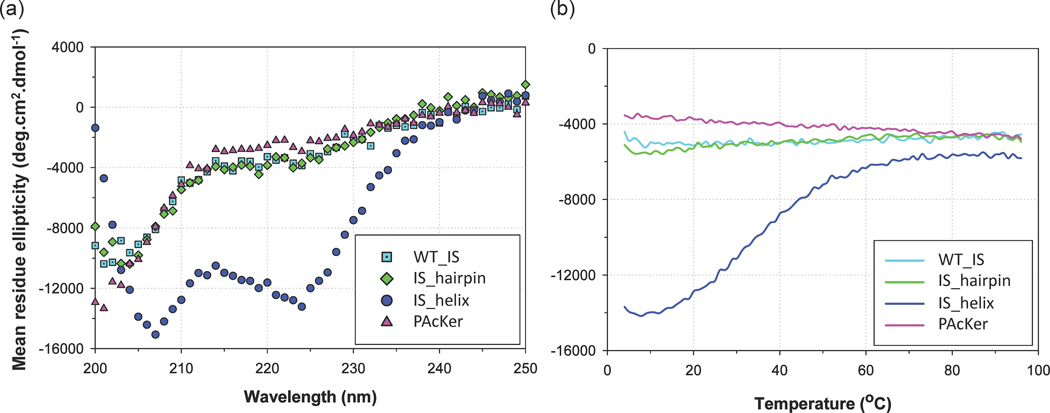
Fig. 2. Structural characterization of computational models and IS domain using circular dichroism.
(a) IS_helix shows strong helical content. Wild type IS domain (WT_IS), IS_hairpin and PAcKer are all unfolded. (b) IS_helix shows a modest cooperative thermal denaturation at 222 nm.
Design strategy
We tested three design strategies for stabilizing the IS domain (Fig. 1a). In all the designs, the goal was to keep the kinase-interacting residues intact while redesigning the surrounding residues to improve stability and destroy binding with Cdc42 and Rac1 (Fig. 1b).
The first strategy we tested was inspired by the crystal structure of PAK1 in its autoinhibited conformation (PDB Code: 1F3M). In this structure PAK1 forms a homodimer in which the IS domain from one chain binds to the kinase from the other chain. Additionally, the two IS domains interact with each other, residues 83–86 from the first IS domain form a β-strand that pairs with residues 82–85 from the second IS domain to form a small 2-stranded anti-parallel β-sheet. One face of the β-sheet packs against the rest of the IS domain to form a small hydrophobic core. To allow for a similar interaction in a monomeric IS domain we extended the N-terminus of the domain, starting from His 83, with a β-hairpin followed by a β-strand with the same sequence as residues 82–85 of PAK1 (Fig. 1a). The new β-strand is intended to mimic the residues from the second IS domain in the PAK1 crystal structure. This design was named IS_hairpin. To prevent competitive binding with the GTPases Cdc42 and Rac1, His-83 and His-86 (PDB Code: 1E0A10) and Rac (PDB Code: 2QME) were mutated to Gln and Thr respectively.
In a second approach, a 20-residue helix was designed off the N-terminus of trunc-WT_IS-1 (residues 100–137). The helix was designed to pack against the other helices of trunc-WT_IS-1 and expand the hydrophobic core of the domain. The hydrophobic effect is the driving force for protein folding and increasing buried hydrophobic surface area has been used previously to stabilize proteins.11 Our approach is novel in that we accomplish this by adding additional residues to the protein, rather than mutating pre-existing residues in the protein. This model was called IS_helix.
In a third approach, a 16-residue helical stretch was created at the C-terminus of trunc-WT_IS-2 (residues 87–137). The goal of this approach was to bring the N-terminus and C-terminus of the domain close to each other so that it can be inserted into a loop in the cyan fluorescent protein (CFP). The domain was inserted between residues Glu-172 and Asp-173 of CFP. A small GS linker was added between the N-terminal of designed domain and Glu-172 of CFP. Similarly, another GS linker was added between the C-terminal of designed domain and Asp-173 of CFP. There are two potential benefits of this approach. First, the host protein (CFP) may provide further stability to the IS domain by holding the N- and C-termini adjacent to each other. Second, biosensors often require a stable nonresponsive fluorophore attached to the responsive portion of the biosensor to use as a baseline signal for ratiometric imaging.12 As with the IS_helix design, the new helix in this case was packed against the rest of the domain and used to expand the hydrophobic core of the domain. Residues involved in Cdc42 and Rac interactions were also mutated in this design. This design was called PAcKer or CFP-PAcKer when inserted into CFP.
Design protocol
We used a computational protocol within the Rosetta molecular modeling program13; 14; 15 that iterates between structure and sequence optimization to design the new secondary structural elements described here.16 In brief, the protocol uses three- and nine-residue fragments drawn from the protein database to build loops or extensions of appropriate size. The fragments were picked based on target secondary structure propensity. During optimization of the helical extensions (IS_helix and PAcKer) or the designed hairpin (IS_hairpin) the new residues were allowed to adopt alternative conformations while the backbone of the rest of the domain was held fixed. Residues from the core of the domain that packed against the new helical extensions were allowed to adopt alternative side chain conformations and identities. Distance-based constraints were used to induce the new helical extensions to pack against the rest of the domain.
Expression and solubility of the designs
IS domain, IS_hairpin and PAcKer were expressed with an MBP (maltose binding protein) tag while IS_helix was expressed with a GST (glutathione S-transferase) tag in E. coli. PAcKer inserted into CFP (called CFP-PAcKer) was expressed with a 6×His tag. Expression levels were highest for IS_helix. Once the proteins were cleaved from GST or MBP, and concentrated, IS_helix showed no signs of aggregation up to a concentration of 1 mM, while wild-type IS and IS_hairpin had lower solubility (< 200 µM) and PAcKer showed an intermediate level of solubility.
Structural characterization of the designs
Far-UV circular dichroism (CD) scans showed that IS, IS_hairpin and PAcKer were primarily unfolded while IS_helix had strong helical content (minima at 208 nm and 222 nm, Fig. 2a). Circular dichroism was also used to monitor the thermal denaturation of IS_helix. The midpoint for unfolding was ~35 °C and the curve showed modest cooperativity as would be expected for a small protein. To further test the ability of IS_helix to form a stable folded structure, we performed a far-UV CD scan in the presence of trimethylamine N-oxide (TMAO), an osmolyte which has been shown to stabilize proteins.17 We saw a considerable increase in helical content at 0.5 M and 1.0 M of the osmolyte (Supplementary Fig. S2a), and in the presence of 2 M TMAO, the midpoint for unfolding was 55 °C (Supplementary Fig. S2b).
Binding affinity and suitability as an affinity reagent
The binding affinities of IS and the engineered proteins for the PAK1 kinase domain (a model for the active conformation of PAK1), PAK1 full length (a model for autoinhibited PAK1) and PAK1 full length with the mutations V127E and S144E (a model for full-length PAK1 in the ‘open’ state) were measured with isothermal titration calorimetry (ITC). Val-127 stabilizes the interaction between the IS domain and the kinase domain, and mutation to glutamate destabilizes this autoinhibitory interaction in full-length PAK1. We speculated that the mutation S144E will mimic the phosphorylation of Ser-144 that has been known to significantly contribute to PAK1 activation.4 IS_helix, IS_hairpin and wild-type IS (WT_IS) bound to the PAK1 kinase domain with affinities of 1.6 µM, 2.6 µM and 4 µM respectively. (Due to low ITC signal to noise ratio observed for kinase domain titrated into IS_helix, binding was reconfirmed using fluorescence polarization technique and was found to be 15 µM). It was surprising that IS_helix and WT_IS domain bound with such similar affinities, as the IS_helix was observed to be pre-folded while the WT_IS folds upon binding the kinase. One interpretation of these results is that IS_helix is not pre-ordered correctly and the structure must rearrange – at some cost to binding energy – to interact with the kinase domain. In this scenario, the reordering energy for IS_helix may be equivalent to the loss in conformational entropy that the IS domain undergoes upon binding.
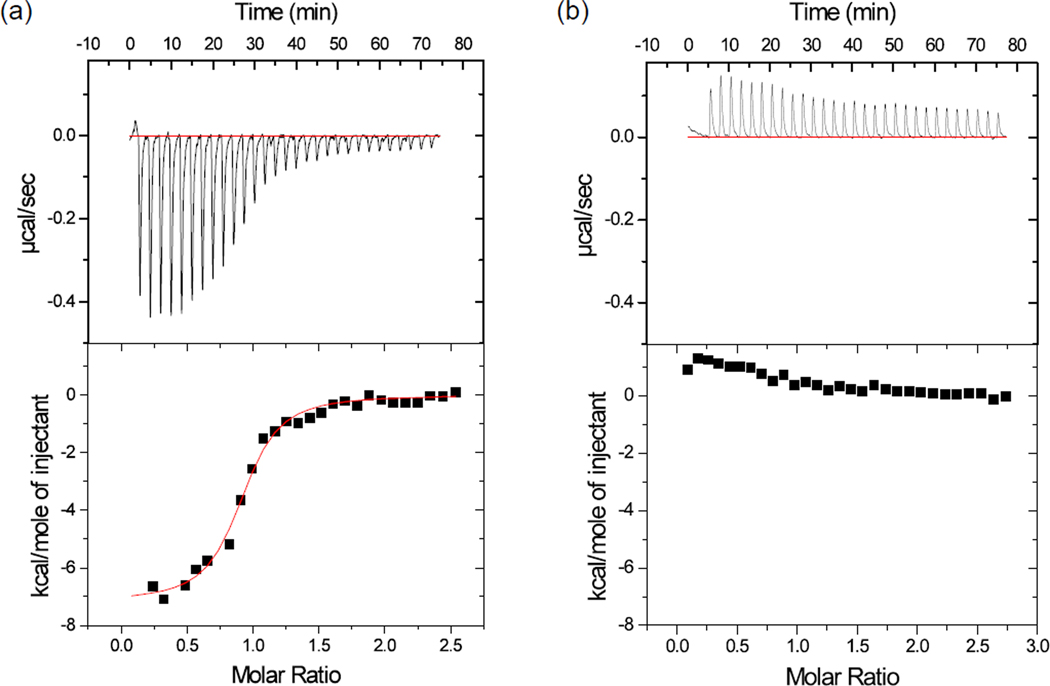
CFP-PAcKer bound to the PAK1 kinase domain with an affinity of 400 nM, 10-fold tighter than the IS domain (Fig. 3a). It did not bind to ‘closed’ full-length PAK1 (Fig. 3b), and the binding affinity for the open variant, PAK1V127E/S144E, was 3.3 µM (Supplementary Fig. S2). In order to test if CFP-PAcKer was binding to the kinase domain using the intended interaction surface, the valine on PAcKer that corresponded to residue 127 in full-length PAK1 was mutated to a glutamate. As expected, no binding was detected between CFP-PAcKerV127E and the kinase domain. Similarly, mutating Leu-470 on the PAK1 kinase domain to glutamate abolished binding between the two proteins (Supplementary Fig. S4). Leu-470 is at the center of the interface formed between the IS domain and kinase domain as seen in the crystal structure (PDB Code: 1F3M). These results indicate that CFP-PAcKer binds to the target surface area on PAK1, and that binding depends on the conformational state of PAK1.
Fig. 3. Isothermal titration calorimetry assays to determine the binding affinities of CFP-PAcKer against active and auto-inhibited conformations of PAK1.
(a) 208 µM of PAK1 kinase domain (model for active conformation) was titrated against 18 µM CFP-PAcKer. An equilibrium dissociation constant of 400 nM was observed with a molar ratio ~1. (b) 262 µM PAK1 full length (model for ‘close’ auto-inhibited form) was titrated against 21 µM CFP-PAcKer. The titration failed to show any appreciable heat released or absorbed. The data sets were drawn on similar scale for comparison.
To create CFP-PAcKer a helical extension was added to the C-terminus of the IS domain to bring the N- and C-terminus in closer proximity. To test if this new structural element was contributing to favorable binding with PAK1 we tested an alternative design in which trunc-WT_IS-2 domain (the starting sequence for PAcKer design) was inserted into CFP, but a glycine-serine linker (GSGSGS to residue 87 and GSGS to residue 137) was used to insert between Glu-172 and Asp-173 of CFP. This alternative design bound to the PAK1 kinase domain weaker than CFP-PAcKer, 1.65 µM compared to 400 nM (Supplementary Fig. S8).
PAK1 kinase domain induces fluorescence enhancement of a biosensor based on CFP-PAcKer (mero-CFP-Packer)
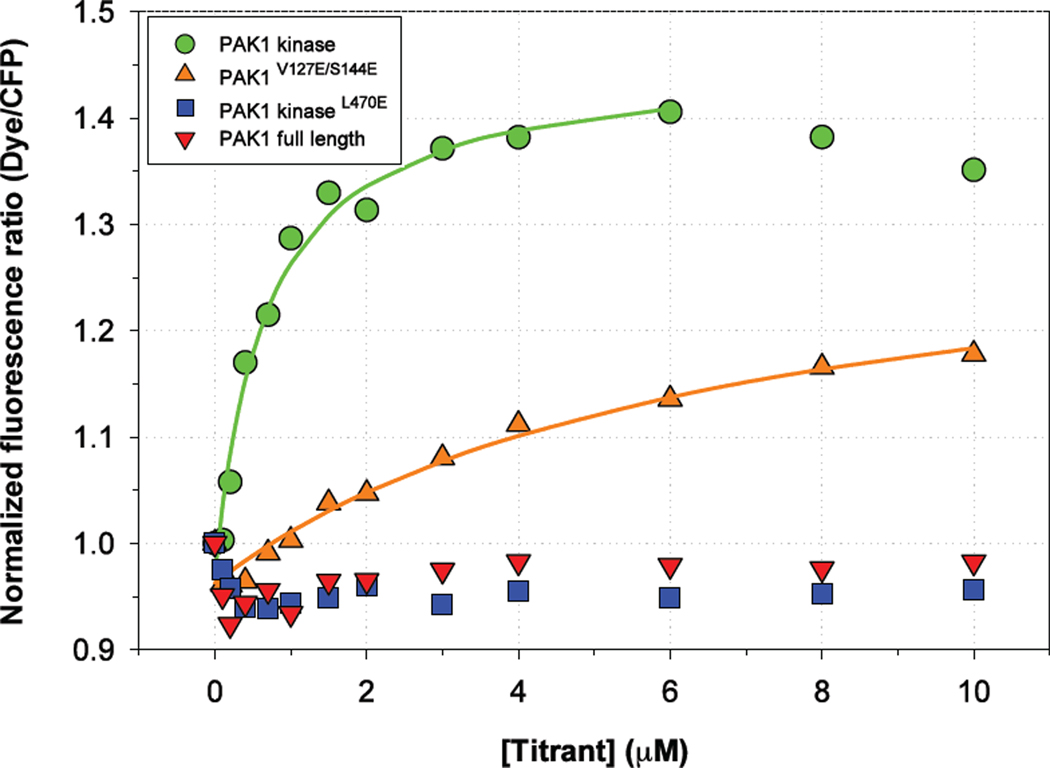
A lysine in CFP-PAcKer that corresponds to Lys-134 in the PAK1 sequence was mutated to a cysteine and covalently modified with an environment-sensitive merocyanine dye designed to respond to protein interaction.18 This position is adjacent to the predicted interface between CFP-PAcKer and PAK1. We therefore predicted that PAK binding would create a change in the local environment of the dye and lead to a change in fluorescence. We monitored both dye fluorescence (excitation wavelength 593 nm and emission wavelength 620 nm) and CFP fluorescence (excitation at 433 nm and emission at 474 nm) while titrating the biosensor with the kinase domain. The normalized ratio ((Dye/CFP)Titrant/(Dye/CFP)Buffer) fit to a ‘one-site binding’ curve gave a binding affinity of 600 nM, consistent with the equilibrium dissociation constant observed in ITC (Fig. 4). There was also a change in normalized fluorescence ratio when titrating with PAK1V127E/S144E (full-length PAK1 stabilized in the ‘open’ state). There was no change in normalized fluorescence ratio when mero-CFP-PAcKer was titrated with full-length PAK1 in the autoinhibited state or PAK1 kinaseL470E mutant. Additionally, when dye was conjugated to CFP-PAcKerV127E, there was no change in ratiometric fluorescence when titrating with the PAK1 kinase (Supplementary Fig. S5).
Fig. 4. Biosensor derived from CFP-PAcKer by conjugation with a solvent-sensitive merocyanine dye undergoes fluorescence increase in the presence of PAK1 kinase domain.
A net gain of ~40% in the normalized fluorescence ratio was observed in the case of PAK1 kinase domain when it was titrated in 200 nM of biosensor. PAK1 kinaseL470E and PAK1 full-length failed to show any appreciable change in fluorescence ratio. PAK1V127E/S144E (full-length ‘open’ PAK1) showed an intermediate level of change in fluorescence ratio.
Specificity of the CFP-PAcKer derived biosensor
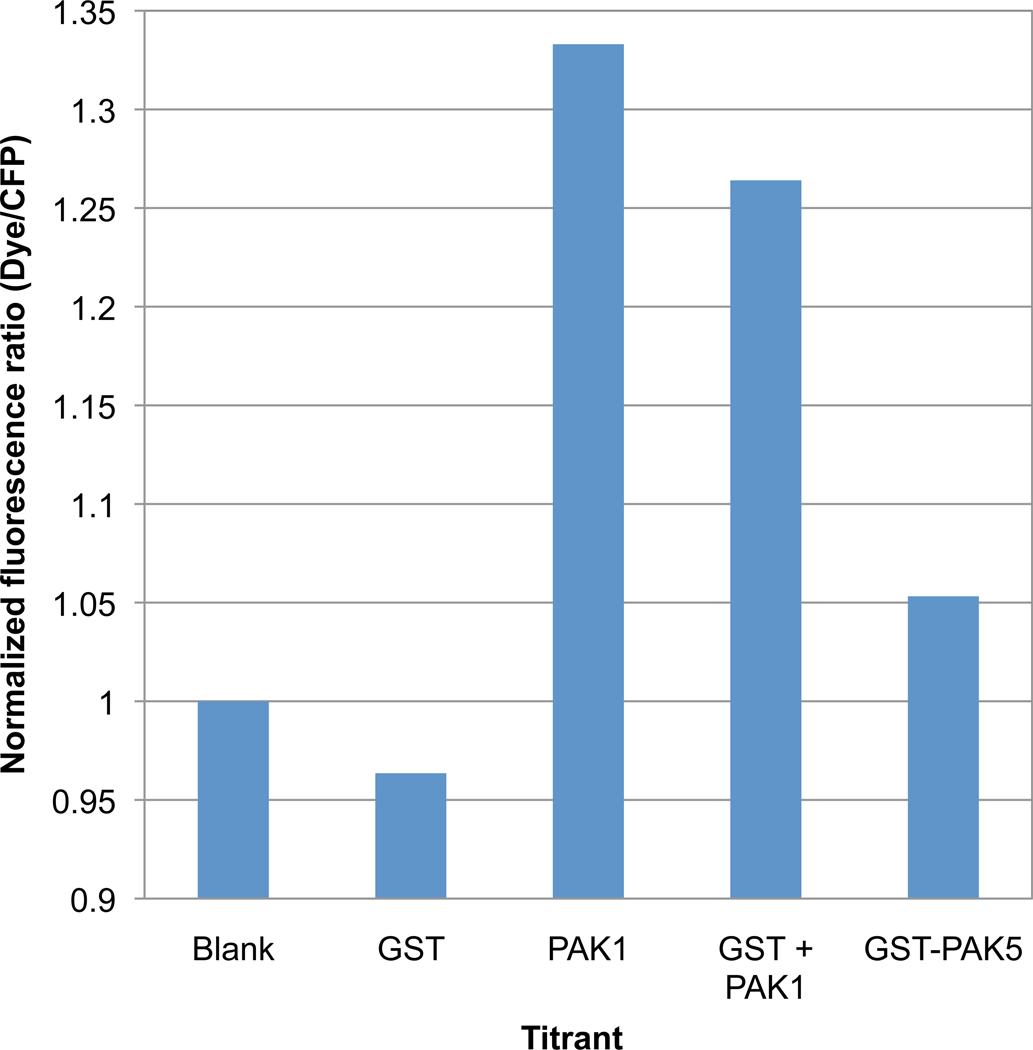
Mero-CFP-PAcKer is expected to interact with all group 1 PAKs (PAK 1–3) because the target binding surface on the kinase domain is identical in all three cases. Group 2 PAKs (PAK 4–6) are more divergent, and therefore, may bind more weakly to CFP-PAcKer. We tested the PAK5 kinase domain, expressed as a GST (Glutathione S-transferase) fusion, for binding to CFP-PAcKer. Isothermal titration calorimetry did not show any appreciable heat absorbed or released when the proteins were mixed (Supplementary Fig. S6) and CFP-PAcKer conjugated to merocyanine fluorescent dye showed only small changes in ratiometric fluorescence in the presence of 1 µM of GST-PAK5 kinase domain (Fig. 5). However, at higher concentrations of GST-PAK5 (> 8 µM) the change in normalized fluorescence was larger than with PAK1 (Supplementary Fig. S7). These results indicate that mero-CFP-PAcKer binds weaker to PAK5 than PAK1, but the change in fluorescence is greater when binding to PAK5. In addition to sequence divergence in the binding site, several of the residues surrounding the binding site vary in PAK1 and PAK5 and are expected to create different local environments for the merocyanine dye.
Fig. 5. CFP-PAcKer derived biosensor has a lower response with the PAK5 kinase domain.
PAK1 kinase domain and GST-PAK5 kinase domain at 1 µM was titrated into biosensor. For a good comparison, GST alone at 1 µM or equimolar PAK1 kinase domain and GST (1 µM each) were also titrated into assay mixture containing the biosensor. ‘Blank’ refers to 200 nM of the biosensor alone in assay buffer.
Discussion
We redesigned the IS domain of PAK1 for improved function as an affinity reagent. The focus of the design process was to stabilize tertiary structure formed by the IS domain when it binds the kinase domain of PAK1. In isolation, the IS domain is natively unfolded. The potential advantages of this approach are several: a well-folded domain should be more resistant to proteases, solubility should be improved by burying hydrophobic amino acids in the core of the domain, and pre-ordering the polypeptide in a conformation that is competent for binding should reduce losses in conformational entropy upon binding and increase binding affinity. The designed protein IS_hairpin did not fold in isolation and did not show a strong improvement in solubility. In contrast, IS_helix formed helical structure and was considerably more soluble. However, the redesign did not have enhanced affinity for PAK1, presumably because the structure was not pre-ordered in a conformation appropriate for binding.
Our most successful design, CFP-PAcKer, included a C-terminal extension modeled to bring the N- and C-termini of the IS domain adjacent to each other, which allowed for insertion into a loop in CFP. Inserting the domain into CFP led to increased affinity for PAK1. The rigid structure of the fluorescent protein likely holds the N- and C-termini of the IS domain adjacent to each other and stabilizes the domain in a binding competent conformation. CFP has high solubility and likely contributed to the enhanced solubility of the fused protein. CFP fluorescence can be used in ratiometric imaging to normalize fluorescence signals generated by merocyanine dyes attached to the IS domain. Instead of designing a helical extension to bring the N- and C-termini of the IS domain close in space, we also examined the use of linkers made from glycine and serine. This design did not show enhanced affinity for PAK1, perhaps because the flexible linkers prevented the preordering of the domain in a binding competent conformation.
The mero-CFP-PAcKer biosensor has the potential to be a powerful tool for biological studies, shedding light on PAK dynamics in living cells. With a practical affinity reagent in hand, next steps will include examination of different dyes and different dye attachment positions on the biosensor. Preliminary studies in cells showed localized fluorescence increases potentially attributable to PAK activity (data not shown), leading us to move forward with full validation demonstrating selectivity and robust response in vivo. This study demonstrates that a naturally occurring affinity reagent not suitable for biosensor studies can be optimized through rational engineering and modeling, expanding the reach and practicality of live cell biosensor designs.
Materials and methods
Modeling using Rosetta
To design IS_hairpin, IS_helix and PAcKer, we used Rosetta’s backbone sampling and design protocols.16; 19 Using the Robetta server,20 a customized library of fragments was created from the PDB for each three- and nine-residue window in a fasta sequence. A pseudo-sequence for the region to be modeled was used to bias the fragments to a desired secondary structure. For IS_hairpin, the query fasta sequence consisted of residues 79–84 (SDFEHT) from IS domain chain B (PDB Code: 1F3M), INGTTI for the canonical hairpin, and residues 86–137 from IS domain chain A (Supplementary Fig. S1a). For IS_helix the query fasta sequence was a poly-alanine sequence, A16 with (GS)2 sequence for linker followed by residues 100–137 of the IS domain (Chain A of PDB 1F3M). The query fasta sequence for PAcKer was created using sequence 87–137 of the IS domain (Chain A of PDB 1F3M) followed by GA13SG sequence. The fragments corresponding to the region to be modeled were then assembled using Monte Carlo simulated annealing technique. Fragment insertions were accompanied by a “wobble” operation where the backbone torsion angles (φ, ψ) were perturbed. Monte Carlo minimization protocol13; 21 was used to iterate between sequence optimization and structure prediction to optimize all-atom energy function in Rosetta.13 In brief the function is a linear weighted sum of 12-6 Lennard-Jones potential, the Lazaridis-Karplus implicit solvation model,22 an orientation dependent hydrogen-bonding potential,23 backbone-dependent rotamer probabilities,24 a knowledge-based electrostatic energy term,25 amino acid probabilities conditioned on φ and ψ space26 and reference energies that approximate the unfolded state energy of an amino acid.27
Plasmid constructs, gene synthesis and mutagenesis
Kinase dead mutants (K298R) of full-length PAK1 and PAK1V127E/S144E were cloned in pQE-80L (Qiagen) vector. Kinase domain (K298R mutant), residues 250–545 from PAK1 was also cloned in pQE-80L vector with an extra sequence (MRGSHHHHHHGSDYDIPTTENLYFQC) in N-terminus. The PAK1 kinaseL470E mutant was made by overlap extension using PCR.28 Kinase domain of PAK5, residues 425–718 was cloned in pGEX-4T-1 vector with an extra sequence (MHHHHHHSSGVDLGTENLYFQSM) in N-terminus and expressed as a GST (Glutathione S-transferase) fusion protein.
WT_IS domain (residue 83–137 in full-length PAK1 sequence), IS_hairpin and PAcKer were expressed as an MBP (maltose binding protein) fusion with a TEV protease cleavage site and cloned in pQE-80L vector so that 6×His-tag remained at N-terminus after the expression. IS_helix was cloned in pGEX-4T-1 vector so that the protein was expressed as GST fusion with a Thrombin cleavage site. CFP-PAcKer was cloned in pQE-80L vector with an N-terminus 6×His-tag.
Expression and protein purification
All proteins were expressed in BL21(DE3) pLysS cells. The cells were grown to an OD600 of 0.6–0.8 and then induced with 0.3 mM IPTG and further grown at 25 °C for 6 h, except for GST-PAK5 kinase which was expressed overnight at 20 °C. Cells were then disrupted using sonication and the resulting lysates were cleared by two rounds of centrifugation (18,000 × g) of 20 mins each. The supernatants with PAK1 variants and CFP-PAcKer were purified using a prepacked Ni-NTA column (HisTrap, HP, GE Healthcare) followed by an anion exchange step (Source 15Q beads, GE Healthcare). WT_IS domain, IS_hairpin and PAcKer were purified using a Ni-NTA column followed by overnight proteolysis using TEV protease and then again loading the product on Ni-NTA column. The second Ni-NTA affinity step removed the 6×His-tagged MBP and left the design variants (all with an extra GS sequence at N-terminus) as a flow through. The flow through was concentrated and loaded on a gel filtration column (Superdex 75, GE Healthcare). All the design variants eluted at similar elution volume (size corresponding to monomer) and the appropriate fractions were combined and concentrated (Amicon Ultra, Millipore). Protein concentration was estimated using theoretical molar extinction coefficients and absorbance at 280 nm. CFP-PAcKer concentration was estimated using CFP absorbance at 433 nm with an extinction coefficient of 44000 M−1 cm−1.
GST-tagged proteins, IS_helix and PAK5 kinase domain were purified using a prepacked column (GSTrap, HP, GE Healthcare). For IS_helix, the GST tag was cleaved off using Thrombin protease (Sigma-Aldrich). Post cleavage, the sample was loaded on an anion exchange column (HiTrap Q, GE Healthcare). IS_helix was collected as flow-through. GST-PAK5 kinase was further purified using an anion exchange step (Source 15Q beads, GE Healthcare).
Circular dichroism (CD)
CD data were collected on a Jasco J-815 CD spectrometer. Far-UV CD scans (250-200 nm) were performed with a protein concentration of 20 µM in a 1 mm cuvette. The temperature was maintained at 20 °C using a Peltier device. Variable temperature scans were carried out between 4–96 °C while measuring the ellipticity at 222 nm. All ellipticity data were corrected with a buffer blank and then converted to mean residue ellipticity. Osmolyte TMAO was dissolved in the same buffer and added to the protein. Ellipticity was subtracted from a buffer blank containing the same amount of TMAO.
Isothermal titration calorimetry (ITC)
The binding affinity of the designed proteins was measured using a VP-ITC isothermal titration calorimeter (MicroCal, GE Healthcare). The designs were placed in the cell at concentrations between 14 and 21 µM (IS_helix at a concentration of 50 µM). PAK1 variants were used as titrants at 12–15 fold higher concentrations. The proteins were exhaustively dialysed in 20 mM Na-Phosphate buffer, pH 7.4, 25 mM NaCl and 5 mM 2-mercaptoethanol (BME). For each experiment, 29 titrations of 10 µl were made. The data were analyzed using Origin50 software and fit using a model for ‘one-site binding’.
Fluorescence polarization assay
Due to a very low signal observed in ITC for IS_helix binding experiment, the binding affinity of IS_helix for PAK1 kinase domain was also measured using fluorescence polarization.29 Ser-36 in IS_helix sequence (corresponding to Ser-115 in full-length PAK1 sequence and Chain A of PDB 1F3M) was mutated to cysteine for conjugation of the thiol-reactive fluorescent probe Bodipy(507/545)-iodoacetamide (Molecular Probes). Fluorescence polarization assays were carried out on a Jobin Yvon Horiba Spex FluoroLog-3 instrument (Jobin Yvon Inc) performed in L-format with the excitation wavelength set at 508 nm and emission wavelength set at 545 nm. Bodipy conjugated IS_helix (in 50 mM Tris-Cl, pH 7.5, 5 mM BME) at a final concentration of 5 µM and volume 180 µl was titrated with PAK1 kinase domain (in 50 mM Tris-Cl, pH 7.5, 50 mM NaCl, 5 mM BME).
Conjugation of fluorescent dye to CFP-PAcKer
To enable detection of CFP-PAcKer binding to PAK1, a solvent sensitive merocyanine dye (18) was conjugated to CFP-PAcKer (mero-CFP-PAcKer). Freshly reduced (using 1 mM tris(2-carboxyethyl)phosphine or TCEP) CFP-PAcKer with L134C (residue numbering based on primary sequence of full-length PAK1 and Chain A of PDB 1F3M) mutation was buffer exchanged to 50 mM Na-Phosphate (pH 7.5) using a PD10 desalting column (Amersham Biosciences). 3–5 equivalents of merocyanine dye dissolved in DMSO (dimethyl sulfoxide) was added to 1 ml aliquot of protein (typical concentration 40–100 µM) in an eppendorf tube and immediately mixed by inverting the tube multiple times. The tube was then covered with aluminium foil and left under gentle mixing for 2 h at room temperature. The conjugation reaction was then terminated using an excess of BME (5 µl stock in 1 ml reaction mix). Unreacted dye was separated from the conjugated protein using a PD10 column equilibrated and then eluted with 20 mM Na-Phosphate (pH 7.4), 25 mM NaCl, 5 mM BME. Dye-labeled protein concentration was estimated using CFP absorbance at 433 nm (ε = 44000 M−1 cm−1). Dye concentration was estimated using dye absorbance at 593 nm (ε = 120000 M−1 cm−1). The conjugation efficiency (Dye concentration/CFP concentration) was ~100%.
Fluorimetric measurement of mero-CFP-PAcKer interaction with PAK1
Dye-labeled CFP-PAcKer (200 nM mero-CFP-PAcKer, 20 mM Na-Phosphate, pH 7.4, 25 mM NaCl, 5 mM BME) was titrated with PAK1 variants, 0–10 µM. The reaction mix for each titration point was made in a separate eppendorf tube and incubated for 10 mins at room temperature. Readings were taken on a Jobin Yvon Horiba Spex FluoroLog-3 instrument (Jobin Yvon Inc) with a 3 mm cuvette. The sample was excited at 433 nm (for CFP) and 593 nm (for dye) and the emission signal was measured at 474 nm (CFP signal) and 620 nm (merocyanine dye signal) respectively. The readings were taken from lower titrant concentration to higher without an intermediate cuvette washing step. Data accumulation took 30 minutes on an average from the start of experiment (0 titrant reading) to end of experiment (10 µM titrant reading). A normalized fluorescence ratio ((Dye/CFP)Titrant/(Dye/CFP)Buffer) was calculated and plotted against titrant concentration. The data was fit using SigmaPlot and Michaelis-Menten equation for ‘one-site binding’.
Experiments were also conducted with the GST-PAK5 kinase domain in a similar format.
Supplementary Material
Acknowledgements
We thank Ashutosh Tripathy from UNC Chapel Hill Macromolecular Interactions Facility for assistance with isothermal titration calorimetry. We also thank Evan Trudeau in Department of Pharmacology at UNC Chapel Hill for helping with plasmid constructs. The work was supported by NIH grants GM082288, GM057464 and GM073960.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 2.Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, Harrison SC. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 4.Chong C, Tan L, Lim L, Manser E. The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J Biol Chem. 2001;276:17347–17353. doi: 10.1074/jbc.M009316200. [DOI] [PubMed] [Google Scholar]
- 5.Parrini MC, Camonis J, Matsuda M, de Gunzburg J. Dissecting activation of the PAK1 kinase at protrusions in living cells. J Biol Chem. 2009;284:24133–24143. doi: 10.1074/jbc.M109.015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buday L, Wunderlich L, Tamas P. The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal. 2002;14:723–731. doi: 10.1016/s0898-6568(02)00027-x. [DOI] [PubMed] [Google Scholar]
- 7.Puto LA, Pestonjamasp K, King CC, Bokoch GM. p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem. 2003;278:9388–9393. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
- 8.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 9.Jha RK, Leaver-Fay A, Yin S, Wu Y, Butterfoss GL, Szyperski T, Dokholyan NV, Kuhlman B. Computational design of a PAK1 binding protein. J Mol Biol. 2010;400:257–270. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morreale A, Venkatesan M, Mott HR, Owen D, Nietlispach D, Lowe PN, Laue ED. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat Struct Biol. 2000;7:384–388. doi: 10.1038/75158. [DOI] [PubMed] [Google Scholar]
- 11.Kuhlman B, Baker D. Exploring folding free energy landscapes using computational protein design. Curr Opin Struct Biol. 2004;14:89–95. doi: 10.1016/j.sbi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Bright GR, Fisher GW, Rogowska J, Taylor DL. Fluorescence ratio imaging microscopy. Methods Cell Biol. 1989;30:157–192. doi: 10.1016/s0091-679x(08)60979-6. [DOI] [PubMed] [Google Scholar]
- 13.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 14.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu X, Wang H, Ke H, Kuhlman B. High-resolution design of a protein loop. Proc Natl Acad Sci U S A. 2007;104:17668–17673. doi: 10.1073/pnas.0707977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baskakov I, Bolen DW. Forcing thermodynamically unfolded proteins to fold. J Biol Chem. 1998;273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]
- 18.Toutchkine A, Kraynov V, Hahn K. Solvent-sensitive dyes to report protein conformational changes in living cells. J Am Chem Soc. 2003;125:4132–4145. doi: 10.1021/ja0290882. [DOI] [PubMed] [Google Scholar]
- 19.Rohl CA, Strauss CE, Chivian D, Baker D. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55:656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 20.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 22.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 23.Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J Mol Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 24.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 26.Bowers PM, Strauss CE, Baker D. De novo protein structure determination using sparse NMR data. J Biomol NMR. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- 27.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.