Abstract
Emmprin is a multifunctional glycoprotein expressed by cancer cells and stromal cells in the tumor microenvironment. Through both direct effects within tumor cells and promotion of tumor-stroma interactions, emmprin induces tumor cell invasiveness and regional angiogenesis. The Kaposi’s sarcoma-associated herpesvirus (KSHV) is a common etiology of cancers arising in the setting of immune suppression, including Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL). However, whether emmprin expression and function are regulated by KSHV or other oncogenic viruses in the tumor microenvironment to promote viral cancer pathogenesis remains unknown. Fibroblasts and endothelial cells support latent KSHV infection and represent cellular components of KS lesions. Therefore, we utilized primary human fibroblasts and endothelial cells to determine whether KSHV itself regulates emmprin expression, and whether KSHV-emmprin interactions mediate cell invasiveness. We found that KSHV promotes fibroblast and endothelial cell invasiveness following de novo infection through the upregulation of emmprin, and that this effect is mediated by the KSHV-encoded latency-associated nuclear antigen (LANA). We further validated these findings through our observations that emmprin promotes invasiveness, as well as colony formation, by PEL cells derived from human tumors. Collectively, these data implicate KSHV activation of emmprin as an important mechanism for cancer progression and support the potential utility of targeting emmprin as a novel therapeutic approach for KSHV-associated tumors.
Keywords: KSHV, CD147, Kaposi’s sarcoma, lymphoma
Introduction
The membrane-associated protein emmprin (Extracellular Matrix MetalloPRoteinase INducer; CD147; basigin) induces matrix metalloproteinase (MMP) synthesis by both fibroblasts and tumor cells, thereby promoting tumor cell invasiveness and angiogenesis in the local environment (1). However, it remains unknown whether emmprin plays a role in viral cancer pathogenesis, or whether viruses themselves directly regulate emmprin expression. The Kaposi’s sarcoma-associated herpesvirus (KSHV) is an important etiologic agent for cancers preferentially arising in the setting of immune suppression, including primary effusion lymphoma (PEL) (2) and Kaposi’s sarcoma (KS) (3). Following de novo infection of target cells, KSHV-encoded proteins induce the expression and secretion of multiple factors, including MMPs, with established importance for tumor cell invasiveness and angiogenesis (4,5). The KSHV-encoded latency-associated nuclear antigen (LANA) tethers viral episomes to host cell chromatin and acts as a regulator of transcription for various cellular and viral genes (6–8), but a role for LANA in perpetuating cell invasiveness has not been defined. Since fibroblasts and endothelial cells support latent KSHV infection and represent cellular components of KS lesions (9,10), we used primary human fibroblasts and endothelial cells, as well as PEL cells derived from human tumors, to determine whether KSHV, and possibly LANA, regulates emmprin expression and associated cell invasiveness.
Materials and Methods
Cell culture and infection assays
Two KSHV-infected, patient-derived PEL cell lines, BCP-1 and BCBL-1 cells, were kindly provided by Dr. Dirk Dittmer (University of North Carolina, Chapel Hill), and both RT-PCR and immunofluorescence assays were used to verify the uniform presence of KSHV episomes within these cells through the identification LANA expression as described elsewhere (11). PEL cells were maintained in RPMI 1640 media (Gibco) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (pH 7.5), 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine, 0.05 mM β-mercaptoethanol, and 0.02% (wt/vol) sodium bicarbonate. Human foreskin fibroblasts (HFF) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% FBS, 100 U/mL penicillin and 100 µg/mL streptomycin. Human umbilical vein endothelial cells (HUVEC) were grown in DMEM/F-12 50/50 medium (Cellgro) supplemented with 5% FBS and 0.001 mg/mL Puromycin (Sigma). To obtain KSHV for infection experiments, BCBL-1 cells were incubated with 0.6 mM valproic acid for 6 days, and the concentration of infectious viral particles within concentrated culture supernatants determined prior to infection experiments as described previously (12). Using an MOI = 10, immunofluorescence assays (IFA) revealed that approximately 80–90% of HFF and HUVEC exhibited positive LANA staining 12–24 h after viral incubation (Fig. S1).
Immunofluorescence assays
Briefly, 1×104 per well of either HFF or HUVEC were seeded in eight-well chamber slides (Nunc) and incubated with purified virions at MOI=10 in the presence of 8 µg/mL polybrene (Sigma-Aldrich) for 2 h at 37°C. Following overnight culture, cells were incubated in 1:1 methanol-acetone at 20°C for fixation and permeabilization, then with a blocking reagent (10% normal goat serum, 3% bovine serum albumin, and 1% glycine) for an additional 30’. Cells were then incubated for 1 h at 25°C with 1:1000 dilution of a rat anti-LANA monoclonal antibody (ABI) followed by 1:100 dilution of a goat anti-rat secondary antibody conjugated to Texas Red (Invitrogen). For nuclear localization, cells were subsequently counterstained with 0.5 µg/mL 4’,6-diamidino-2-phenylindole (DAPI, Sigma) in 180 mM Tris-HCl (pH 7.5). Slides were washed once in 180 mM Tris-HCl for 15 min and prepared for visualization using a Leica TCPS SP2 AOBS confocal microscope.
PCR
Total RNA was isolated using the RNeasy Mini kit according to the manufacturer’s instructions (QIAGEN) to identify LANA transcripts. cDNA was synthesized from equal total RNA using SuperScript III First-Strand Synthesis SuperMix Kit (Invitrogen) according to the manufacturer’s procedures. The primers designed for target genes were the following: LANA sense 5’ TCCCTCTACACTAAACCCAATA 3’; LANA antisense 5’ TTGCTAATCTCGTTGTCCC 3’; β-actin sense 5’ GGAAATCGTGCGTGACATT 3’; β-actin antisense 5’ GACTCGTCATACTCCTGCTTG 3’. The PCR was performed in a DNA thermal cycler (Gene Amp PCR System 9700, Applied Biosystems) under conditions of 94°C for 5 min, 35 cycles of 94°C for 30 sec, 54°C for 30 sec, and 72°C for 60 sec.
Transfection assays
HFF and HUVEC were transfected with control pcDNA3.1 or pcDNA3.1-LANA vectors in 12-well plates for 48 h using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency was determined through co-transfection of a lacZ reporter construct and quantification of β-galactosidase activity using a commercially available enzyme assay according to the manufacturer’s instructions (Promega). For RNA silencing, HFF, HUVEC and PEL cells were transfected for 48 h with either emmprin- or control non-target-siRNAs (ON-TARGET plus SMART pool, Dharmacon) using a commercially available transfection reagent (Dharmacon) according to the manufacturer’s instructions. 3 independent transfections were performed for each experiment, and all samples were analyzed in triplicate for each transfection.
Immunoblotting
Total cell lysates (20 µg) were resolved by 10% SDS–PAGE, transferred to nitrocellulose membranes, and immunoblotted with antibodies for emmprin (BD Pharmingen), MMP 1, 2, 7 and 9 (Santa Cruz), and β-Actin (Sigma) as a loading control. Immunoreactive bands were developed using a commercial enhanced chemiluminescence reaction (Perkin-Elmer), and visualized by autoradiography.
Transwell invasion/migration assays
Matrigel Invasion Chambers (Becton Dickinson) were hydrated for 4 h at 37°C with culture medium. Following hydration, media in the bottom of the well was replaced with fresh media, then 5 × 104 HFF or HUVEC were plated in the top of the chamber. After 24 h, cells were fixed with 4% formaldehyde for 15 min at room temperature and chambers rinsed in PBS prior to staining with 0.2% crystal violet for 10 min. After washing the chambers, cells at the top of the membrane were removed and cells at the bottom of the membrane counted using a phase contrast microscope. For PEL cell migration assays, procedures were performed as described above except that 5 × 106 cells were plated in the top of the chamber, and 10 ng/mL hVEGF (Lonza) was added to the lower well as a chemoattractant as previously described (5). Relative invasion/migration was determined for cells in experimental groups as follows: relative invasion = # invading cells in experimental group / # invading cells in control groups.
Colony formation assays
Colony formation assays were performed as previously described (13). Briefly, 96-well plates were seeded first with 2 × 104 HFF per well, then subsequently with either BCP-1 or BCBL-1 cells ranging from 1 to 100 cells per well (8 replicates per dilution). After 14 d, colony formation was tabulated as the percentage of wells containing individual colonies comprised of >1,000 cells.
Statistical Analysis
Significance for differences between experimental and control groups was determined using the two-tailed Student's t-test (Excel 8.0).
Results and Discussion
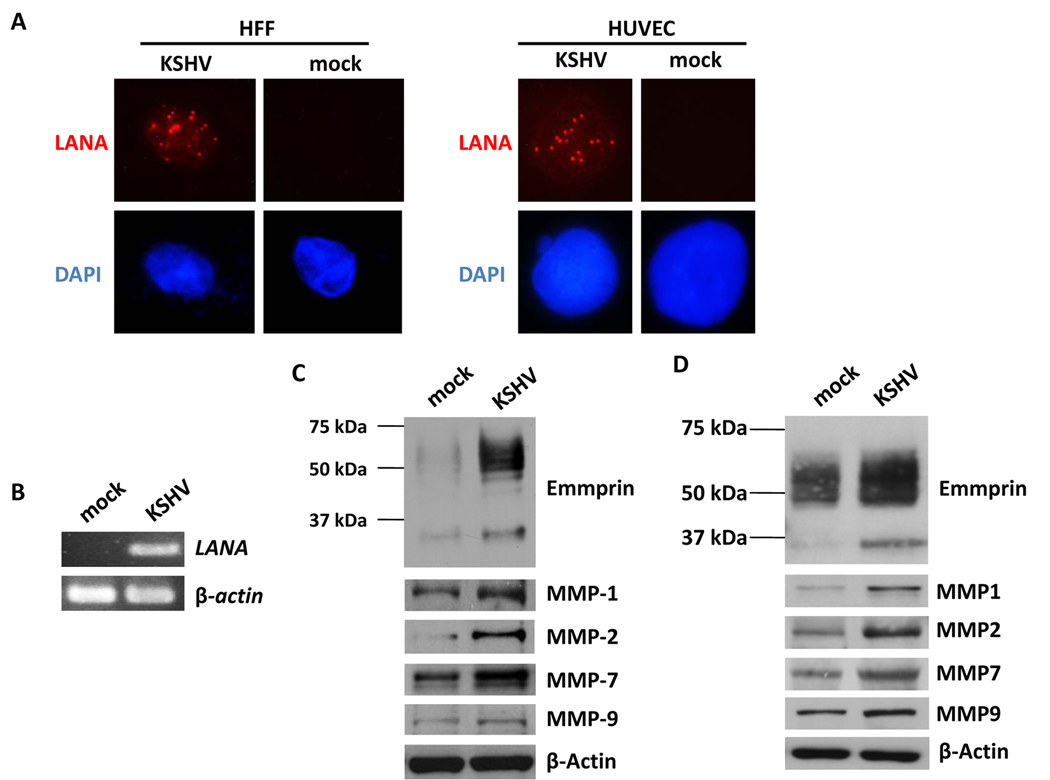
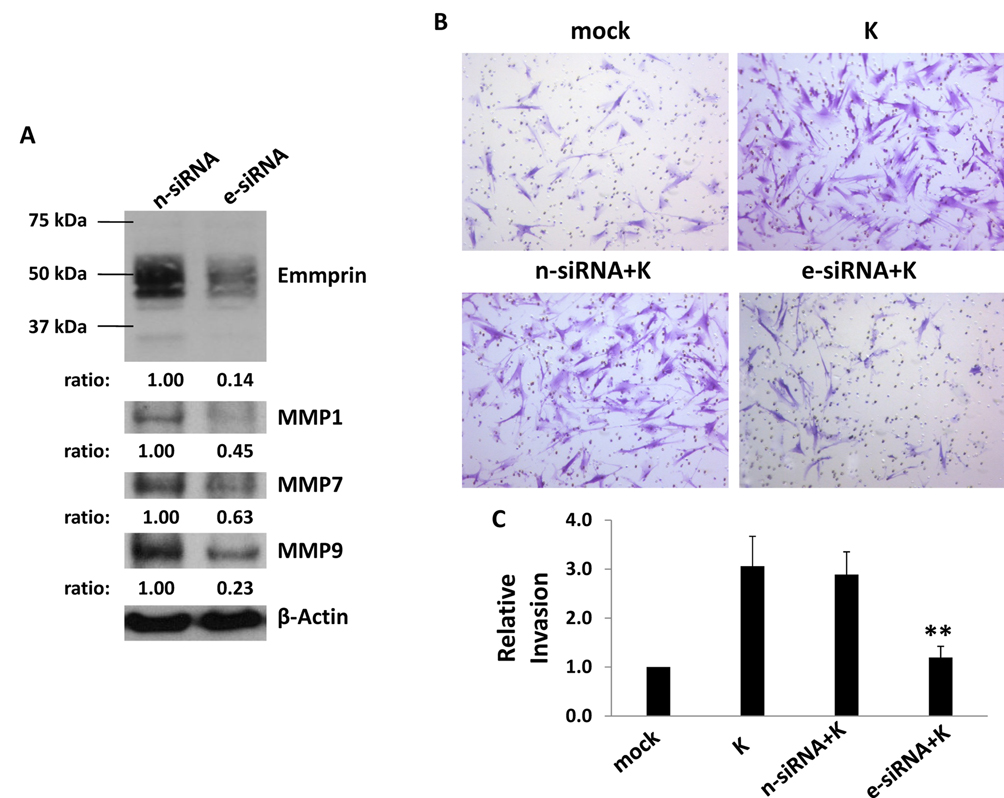
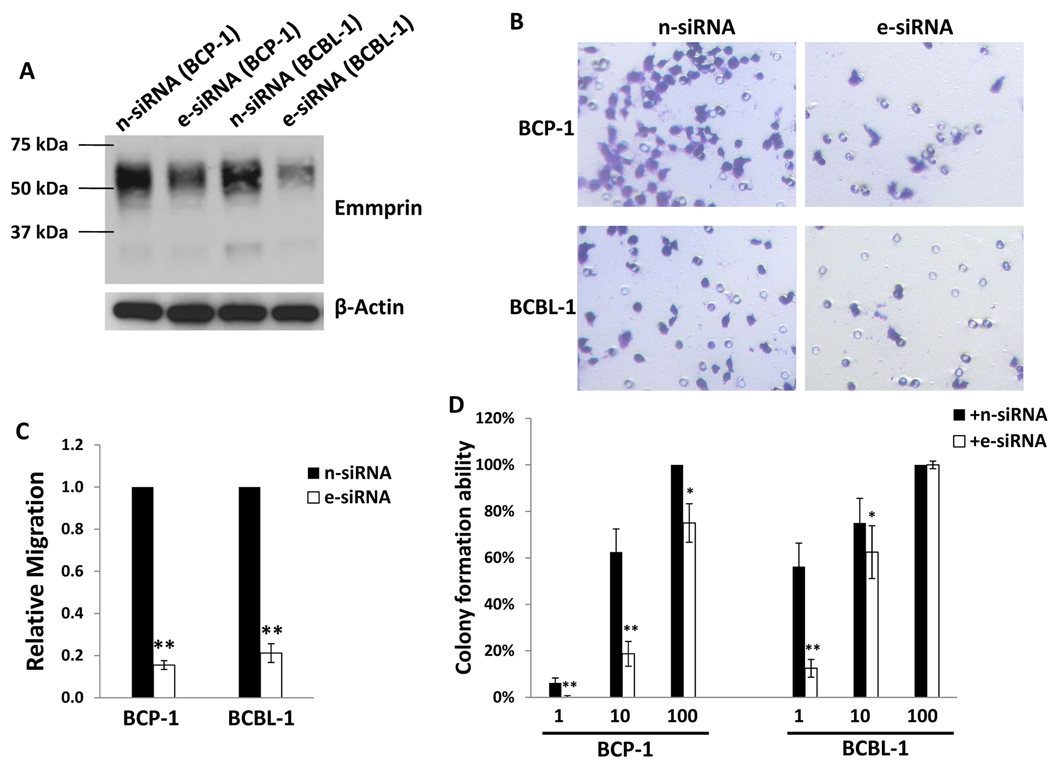
We first verified establishment of KSHV infection within HFF and HUVEC using LANA expression as a surrogate marker for infection (Fig. 1A, B; Fig. S1). Subsequent immunoblot analyses revealed increased expression of emmprin, as well as MMPs associated with emmprin activation, in both of these cell types (Fig. 1C, D). Of note, both high MW (~65 kDa) and low MW (~35 kDa) emmprin glycoforms were elevated in KSHV-infected cells, but particularly the mature high MW glycoform associated with the biological activity of emmprin and induction of MMP secretion (14). After establishing an efficient RNA interference protocol for reducing expression of emmprin in HFF and HUVEC, we confirmed that reduction of emmprin expression caused a corresponding reduction in MMP expression using HFF (Fig. S2). Subsequent transwell invasion assays confirmed that KSHV infection significantly increased invasiveness for HFF and HUVEC, and that emmprin-siRNA significantly reduced this effect (Fig. 2A–D). To validate these findings using human KSHV-associated tumor cells, we established an RNA interference protocol for reducing emmprin expression in PEL cell lines (Fig. 3A). Using modified transwell migration and colony formation assays validated previously for virus-infected lymphoma cells (5,13), we found that emmprin-siRNA significantly reduced the capacity for both migration (Fig. 3B,C) and colony formation (Fig. 3D) for two independent KSHV-infected PEL cell lines.
Figure 1. KSHV induces expression of emmprin and MMPs following de novo infection.
(A) HFF and HUVEC were incubated with either media (mock) or purified KSHV for 2 h, and following a subsequent 24 h incubation, IFA were performed as in Methods for identification of LANA expression indicated by typical intranuclear, punctate staining (red dots). Cells were counterstained with DAPI to identify nuclei (blue). (B) LANA transcripts were identified by RT-PCR (data shown for HFF). (C,D) Immunoblots were performed using HFF (C) and HUVEC (D) 24 h after viral incubation to identify expression of high MW (~65 kDa) and low MW (~35 kDa) glycoforms of emmprin, representative MMPs associated with emmprin activation, and β-Actin as an internal control.
Figure 2. Targeting emmprin suppresses KSHV promotion of cell invasiveness.
Cells were transfected with either control non-target (n-siRNA) or emmprin-specific siRNA (e-siRNA), and after 48 h, HFF (A) or HUVEC (C) were incubated with media (mock) or KSHV (K) for 2 h. Subsequent transwell invasion assays were performed 24 h later, and invasiveness for HFF (B) and HUVEC (D) in each group was determined relative to control cells as in Methods. Error bars represent the S.E.M. for three independent experiments. ** = p<0.01 for comparison of invasion for e-siRNA-transfected/KSHV-infected cells versus n-siRNA-transfected/KSHV-infected control cells.
Figure 3. Targeting emmprin reduces migration and colony formation for primary effusion lymphoma (PEL) cells.
(A) BCP-1 and BCBL-1 cells were transfected with either control (n-siRNA) or emmprin-specific siRNA (e-siRNA) for 48 h, and emmprin expression subsequently identified by western blot. (B,C) Invasiveness of emmprin-siRNA-transfected PEL cells was assessed relative to control transfectants using a modified transwell invasion assay and calculations detailed in Methods. (D) PEL cells were transfected as above, and wells seeded with either BCP-1 or BCBL-1 cells ranging from 1 to 100 cells per well. Colony formation assays were performed as in Methods. Error bars represent the S.E.M. for three independent experiments. *=p<0.05, ** = p<0.01 for comparison of colony formation for e-siRNA-transfected cells versus n-siRNA-transfected control cells.
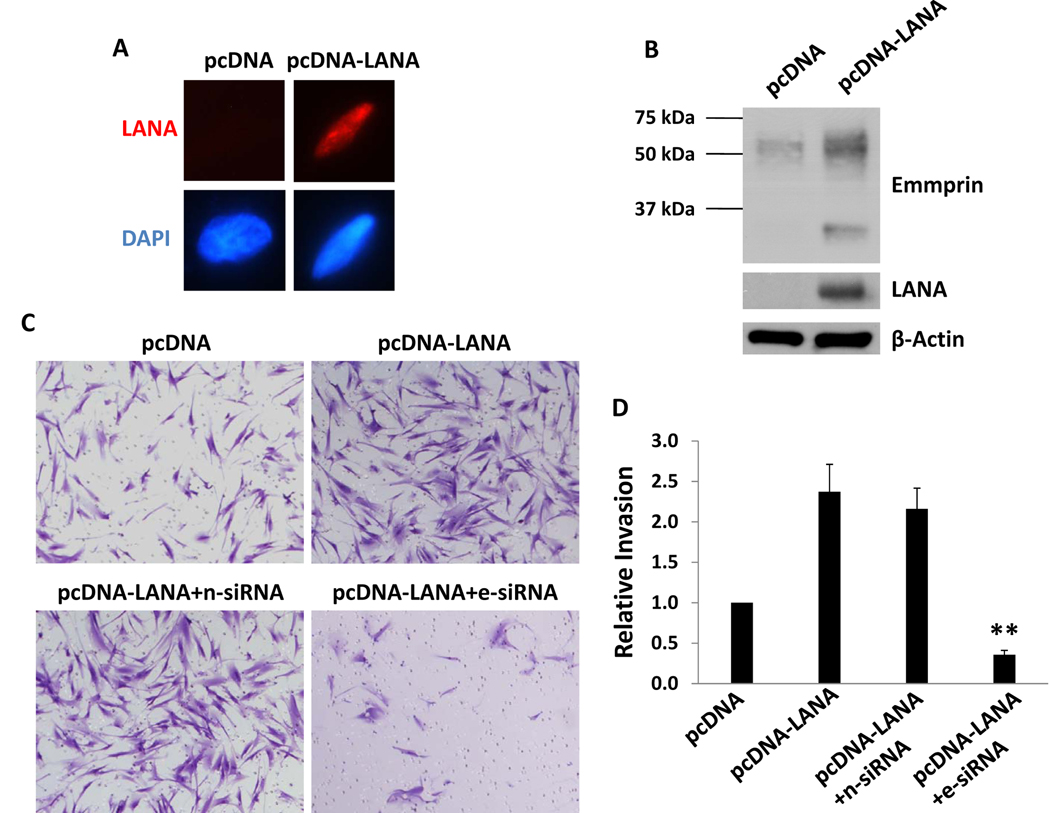
Since others have shown that de novo infection of primary human cells by KSHV results in the predominant expression of latent viral genes including LANA (7,8), we hypothesized that LANA induces emmprin expression. After establishing transient transfection assays conferring intranuclear expression of LANA within HFF (Fig. 4A), we found that ectopic LANA expression significantly increased emmprin expression in these cells, especially the high MW glycoform (Fig. 4B). Subsequent assays revealed that LANA expression significantly increased HFF invasiveness, and that emmprin-siRNA suppressed this effect (Fig. 4C, D).
Figure 4. LANA induces emmprin expression and cell invasiveness.
(A,B) HFF were transfected with either control (pcDNA) or LANA-encoding (pcDNA-LANA) vectors for 48 h, and LANA or emmprin expression subsequently confirmed by IFA (A) and/or western blot (B). (C,D) HFF were co-transfected with pcDNA-LANA and either control non-target (n-siRNA) or emmprin-specific siRNA (e-siRNA), and invasion assessed as previously described. Error bars represent the S.E.M. for three independent experiments. ** = p<0.01 for comparison of invasiveness for e-siRNA/pcDNA-LANA co-transfected cells versus n-siRNA/pcDNA-LANA co-transfected control cells.
These data provide the first evidence that upregulation of emmprin by KSHV, and more specifically KSHV-encoded LANA, plays an important role in promoting KSHV-associated cancer pathogenesis. Existing data support a role for emmprin as a co-factor for infection or pathogenesis related to the human immunodeficiency virus (16), coronaviruses (17) and Hepatitis C (18), but to our knowledge, data supporting the direct regulation of emmprin by oncogenic viruses has not yet been reported. In addition, LANA has not been previously implicated in the induction of cell motility and invasiveness by KSHV. Cooperative interactions between emmprin and the hyaluronan receptor CD44 on the cell surface (19) initiate activation of downstream signal transduction events relevant to cell mobility and proliferation and which coincidentally are initiated following de novo KSHV infection (1, 9). Future studies should, therefore, confirm whether targeting emmprin directly or through competitive inhibition of hyaluronan-CD44 interactions as previously described (20) would selectively disrupt cancer pathogenesis associated with KSHV.
Supplementary Material
HFF (A,B) and HUVEC (C,D) were incubated with either media alone or purified KSHV for 2 h, and following subsequent 24 h incubation, IFA were performed as in Methods for identification of LANA expression indicated by typical intranuclear, punctate staining (red dots). Cells were counterstained with DAPI to identify nuclei (blue). To verify the percentage of cells expressing LANA, at least 100 cells were counted from each group for three independent experiments with representative images shown.
HFF (A) and HUVEC (B) were transfected with either control non-target (n-siRNA) or emmprin-specific siRNA (e-siRNA), and after 48 h, cells were incubated with purified KSHV for 2 h. 24 h after viral incubation, expression of emmprin and representative MMPs was determined by western blot.
Acknowledgements
We would like to acknowledge Dr. Dirk Dittmer (University of North Carolina, Chapel Hill) for providing PEL cells, Dr. Rolf Renne (Shands Cancer Center, University of Florida, Gainesville, FL) for providing LANA overexpression constructs, Dr. Yusuf Hannun (Medical University of South Carolina, Charleston, SC) for providing LacZ constructs, and Dr. Narayan Bhat (Medical University of South Carolina, Charleston, SC) for providing HFF. This work was supported by grants from the National Institutes of Health (K08-1CA103858 to CP), the South Carolina COBRE for Oral Health (P20-RR-017696; CP subproject investigator), and the MUSC Hollings Cancer Center (core grant P30-CA-138313).
References
- 1.Yan L, Zucker S, Toole BP. Roles of the multifunctional glycoprotein, emmprin (basigin CD147), in tumour progression. Thromb Haemost. 2005;93:199–204. doi: 10.1160/TH04-08-0536. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.Qian LW, Xie J, Ye F, Gao SJ. Kaposi's sarcoma-associated herpesvirus infection promotes invasion of primary human umbilical vein endothelial cells by inducing matrix metalloproteinases. J Virol. 2007;81:7001–7010. doi: 10.1128/JVI.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai YH, Wu MF, Wu YH, et al. The M type K15 protein of Kaposi's sarcoma-associated herpesvirus regulates microRNA expression via its SH2-binding motif to induce cell migration and invasion. J Virol. 2009;83:622–632. doi: 10.1128/JVI.00869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupin N, Fisher C, Kellam P, et al. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci U S A. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, et al. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 8.Renne R, Barry C, Dittmer D, Compitello N, Brown PO, Ganem D. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akula SM, Naranatt PP, Walia NS, Wang FZ, Fegley B, Chandran B. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) infection of human fibroblast cells occurs through endocytosis. J Virol. 2003;77:7978–7990. doi: 10.1128/JVI.77.14.7978-7990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Offermann MK. Kaposi's sarcoma and HHV-8. Trends Microbiol. 1996;4:419. doi: 10.1016/0966-842x(96)84953-5. [DOI] [PubMed] [Google Scholar]
- 11.Parsons CH, Adang LA, Overdevest J, et al. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J Clin Invest. 2006;116:1963–1973. doi: 10.1172/JCI27249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Z, Freitas E, Sullivan R, et al. KSHV-encoded microRNAs upregulate xCT to facilitate infection and the survival of KSHV-infected cells during oxidative stress. PLoS Pathog. 2010;6:e1000742. doi: 10.1371/journal.ppat.1000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mack AA, Sugden B. EBV is necessary for proliferation of dually infected primary effusion lymphoma cells. Cancer Res. 2008;68:6963–6968. doi: 10.1158/0008-5472.CAN-08-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang W, Chang SB, Hemler ME. Links between CD147 function, glycosylation, and caveolin-1. Mol Biol Cell. 2004;15:4043–4050. doi: 10.1091/mbc.E04-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogden SR, Noto JM, Allen SS, et al. Matrix metalloproteinase-7 and premalignant host responses in Helicobacter pylori-infected mice. Cancer Res. 2010;70:30–35. doi: 10.1158/0008-5472.CAN-09-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushkarsky T, Zybarth G, Dubrovsky L, et al. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc Natl Acad Sci U S A. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Mi L, Xu J, et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shackel NA, McGuinness PH, Abbott CA, Gorrell MD, McCaughan GW. Insights into the pathobiology of hepatitis C virus-associated cirrhosis: analysis of intrahepatic differential gene expression. Am J Pathol. 2002;160:641–654. doi: 10.1016/S0002-9440(10)64884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toole BP, Slomiany MG. Hyaluronan, CD44 and Emmprin: partners in cancer cell chemoresistance. Drug Resist Updat. 2008;11:110–121. doi: 10.1016/j.drup.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misra S, Ghatak S, Zoltan-Jones A, Toole BP. Regulation of multidrug resistance in cancer cells by hyaluronan. J Biol Chem. 2003;278:25285–25288. doi: 10.1074/jbc.C300173200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HFF (A,B) and HUVEC (C,D) were incubated with either media alone or purified KSHV for 2 h, and following subsequent 24 h incubation, IFA were performed as in Methods for identification of LANA expression indicated by typical intranuclear, punctate staining (red dots). Cells were counterstained with DAPI to identify nuclei (blue). To verify the percentage of cells expressing LANA, at least 100 cells were counted from each group for three independent experiments with representative images shown.
HFF (A) and HUVEC (B) were transfected with either control non-target (n-siRNA) or emmprin-specific siRNA (e-siRNA), and after 48 h, cells were incubated with purified KSHV for 2 h. 24 h after viral incubation, expression of emmprin and representative MMPs was determined by western blot.