Abstract
Mammalian behavior and physiology undergo daily rhythms that are coordinated by an endogenous circadian timing system. This system has a hierarchical structure, in that a master pacemaker, residing in the suprachiasmatic nucleus of the ventral hypothalamus, synchronizes peripheral oscillators in virtually all body cells. While the basic molecular mechanisms generating the daily rhythms are similar in aIl cells, most clock out-puts are cell-specific. This conclusion is based on genomewide transcriptome profiling studies in several tissues that have revealed hundreds of rhythmically expressed genes. Cyclic gene expression in the various organs governs overt rhythms in behavior and physiology, encompassing sleep-wake cycles, metabolism, xenobiotic detoxification, and cellularproliferation. As a consequence, chronic perturbation of this temporal organization may lead to increased morbidity and reduced lifespan.
Keywords: circadian clock, suprachiasmatic nucleus, peripheral oscillator, PAR bZip transcription factor, albumin d-element-binding protein, hepatic leukemia factor, thyrotroph embryonic factor, xenobiotic detoxification
Abstract
La conducta y la fisiología de los mamíferos dependen de ritmos diarios que están coordinados por un sistema circadiano endógeno. Este sistema tiene una estructura jerárquica, en la cual un marcapasos maestro, localizado en el núcleo supra-quiasmático del hipotálamo ventral, tiene la potencialidad de sincronizar osciladores periféricos en casi todas las células del cuerpo. Estudios acerca del perfil del transcriptoma de todo el genoma en diversos tejidos han demostrado que cientos de genes se expresan de una manera rítmica. La expresión cíclica de genes en diversos órganos controla determinados ritmos del comportamiento y de la fisiología, incluyendo el ciclo sueño vigilia, el metabolismo, la detoxificación xenobibtica y la proliferación celular. Como consecuencia, la alteración crónica de esta organización temporal puede llevar a un aumento de la morbilidad y a una reducción de la expectativa de vida. Sin embargo, aun se requiere de mucho trabajo de laboratorio para poder establecer relaciones inequívocas entre la desorganización de la fisiología circadiana y la aparición de la enfermedad.
Abstract
La physiologie et le comportement des mammifères suivent des rythmes quotidiens coordonnés par un système circadien de synchronisation endogène. Ce système est hiérarchisé en ce sens que son pacemaker principal, situé dans le noyau suprachiasmatique de l'hypothalamus ventral, synchronise des oscillateurs périphériques dans presque toutes les cellules de l'organisme. Alors que les mécanismes moléculaires qui sont à la base des rythmes endogènes sont semblables dans toutes les cellules, les conséquences du fonctionnement des horloges biologiques diffèrent quant à elles selon les cellules. Des études du transcriptome du génome entier dans plusieurs tissus ont montré des centaines de gènes exprimés de façon rythmique. L'expression cyclique de gènes dans différents organes gouverne des rythmes mesurables en physiologie et en comportements, comme les cycles veille-sommeil, le métabolisme, la détoxication xénobiotique et la prolifération cellulaire. Une perturbation chronique de cette organisation temporelle peut donc augmenter la morbidité et réduire la durée de vie. D'autres travaux de laboratoire sont néanmoins nécessaires pour confirmer les relations de cause à effet entre des modifications de la physiologie circadienne et la survenue de la maladie.
Biologlcal clocks are devices that can measure time In the absence of environmental timing cues, such as changes In light Intensity, temperature, or humidity.1 The discovery of circadian clocks dates back to 1729, when the French astronomer Jean Jacques Ortous de Malran observed that mimosa plants continued to open and close their leaves in a daily manner when kept in the absence of sunlight.2 Obviously, other environmental oscillations such as daily temperature fluctuations could have driven the cyclic leaf openings in de Mairan's experiment, thereby challenging his conclusion about the existence of a mimosa clock. However, in 1832 the Swiss physician and botanist Augustin Pyrame de Candolle demonstrated that in constant light mimosa plants opened and closed their leaves with a cycle of 22 hours rather than 24 hours.3 This observation provided irrefutable evidence that the leaf movement rhythm was not merely driven by cyclic environmental cues depending on the earth's rotation, but by a self-sustained biological clock. Incidentally, “circadian” is derived from the Latin words “circa diem” and indicates that circadian clocks can measure days only approximately. Hence, the phase of circadian oscillators must be corrected daily to stay in resonance with geophysical time. The photoperiod (ie, daily variations in light intensity) is the primary Zeitgeber for the synchronization of circadian clocks.1, 4-6
Since the discovery of endogenous timekeepers in plants, such devices have been found in virtually all investigated light-sensitive organisms, encompassing cyanobacteria, fungi, protozoans, algae, insects, and mammals. Model systems in which the molecular makeup of circadian oscillators is being dissected in detail have been established for several species across the phyla. Thus, during the past two decades, impressive progress in the understanding of circadian clockworks has been made in the cyanobacterium Synechococcus elongatus,7 the filamentous fungus Neurospora crassa 8 the green plant Arabidopsis thaliana,9 the dipterian insect Drosophila melanogaster,10 and the mouse Mus musculus.11, 12 In these organisms many essential clock genes have been identifled, and their biochemical and genetic interactions studied. Originally, negative feedback loops in clock gene expression have been thought to underlie the rhythm generation in all of these species.1 However, breathtaking work on cyanobacterial oscillators has recently challenged this paradigm. In this photosynthetic micro-organism, the transcription of virtually all genes undergoes robust daily oscillations, and these depend on an operon encompassing the three clock genes kaiA, kaiB, and kaiC. 13 Kondo and coworkers have now shown that circadian oscillations in KaiC phosphorylation and dephosphorylation persist in the absence of transcription and translation,14 and that this phosphorylation clock can be reconstituted in the test tube with just the three clock proteins KaiA, KaiB, and Kai C, and adenosine triphosphate (ATP).15, 16 In this cell-free assay, self-sustained and temperature-compensated cycles of KaiC phosphorylation can be observed for nearly a week. The clock components identified in cyanobacteria, fungi, plants, and animals do not exhibit obvious similarities, suggesting that circadian clocks may have evolved independently in different phyla.17 Nevertheless, the clockwork circuitry of insects and vertebrates share most clock components and must therefore have a common origin. Owing to the powerful genetic tools available in the fruit fly Drosophila melanogaster, many important concepts of animal circadian oscillators have first been elaborated in this insect. These include the first unambiguous demonstration of single genes affecting circadian behavior in a Mendelian manner18 and of a negative feedback loop in gene exprèssion driving circadian oscillations.19 In the late 1990s, comparative genomics has unveiled several mammalian orthologs of essential Drosophila clock genes, and genetic loss of function studies in mice confirmed essential roles of these mammalian orthologs in clock function.11 In this review article, the focus will be on the molecular and cellular makeup of the mammalian circadian timing system, on the mechanisms involved in its phase entrainment, and on emerging pathways through which It can Influence clrcadlan physiology. The way that chronic perturbatlons of circadlan rhythms might Increase morbidity in humans, and why knowledge of circadian physiology may be clinically relevant, will also be discussed. The Box lists functional definitions of some of the terms used.
Box. Functional definition of some terms utilized for the description of molecular mechanisms involved in the control of gene expression.
| Transcription factors are proteins that control the activity of genes. These proteins recognize specific DNA sequences located within promoter sequences (= regulatory sequences located immediately in front of the gene) and/or enhancer sequences (= regulatory sequences that can be located upstream, within, or downstream of the gene). Activators are transcription factors that increase transcriptions when bound to promoter or enhancer sequences, while repressors are transcription factors that decrease transcription when occupying such sequences. Frequently, transcription factors bind to specific DNA elements as pairs. When such pairs are composed of two identical proteins, they are called homodimers, when they consist of two different proteins, they are called heterodiiners. For example, the two activators CLOCK and BMALI bind as heterodirners to socalled E-Box motifs (with the sequence CACGTG), located within promoter and enhancer sequences of the genes mPerl and mPerJ (murine Period gene 1 and murine Period gene 2). |
| The repression of gene transcription is also called “silencing Epigenetic silencing” is a stable form of silencing that involves covalent modifications of chromatin constituents, either as methyl groups on cytosine bases of the DNA or as various posttranslational modifications on certain amino acids within histories- Histories are proteins that package DNA into protein-DNA complexes called chromatin within cell nuclei. |
| Orthologs are genes that show a high DNA sequence similarity between different organisms - say humans, mice, and flies - and that perform equivalent function in these organisms. |
Molecular clockwork circuitry in mammals
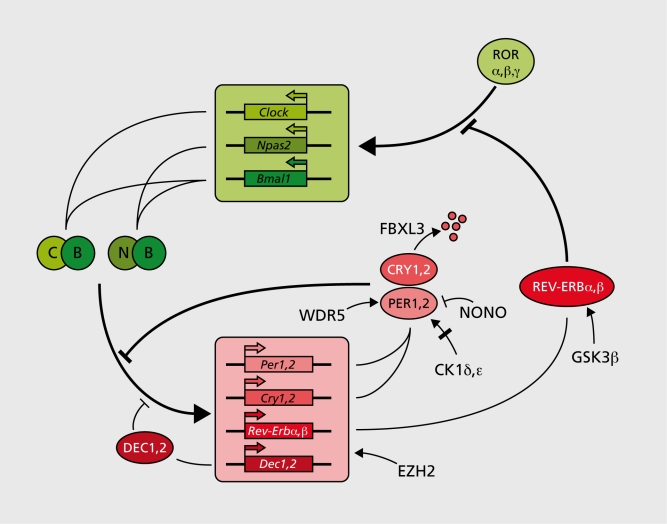
Although circadian physiology and behavior in mammals have been studied for many decades,20 the first circadian genes (Clock, Per1, and Per2) were discovered only 10 years ago. Since then, many genes required for normal clock function have been added to the list. The approaches used in these endeavors are outlined in Table I.21-45 In analogy with early work on the Drosophila circadian oscillator these genes have been assembled into an ever more complex clockwork circuitry (Figure 1). The four transcriptional repressor-encoding genes Cry1, Cry2, Per1, and Per2 are the centerpieces of this molecular oscillator.5 Transcription of these genes is activated via the binding of BMALlCLOCK or BMAL1-NPAS2 heterodimers to Ebox motifs of Cry and Per promoter and enhancer regions. As a consequence, Cry and Per messenger ribonucleic acid (mRNA) and protein levels rise, and once they have reached critical concentrations, CRY and PER proteins form heterotypic complexes. PER-CRY complexes directly interact with BMAL1-CLOCK or BMAL1-NPAS2 heterodimers and thereby attenuate the transactivation potential of these transcription factors.5, 28 BMAL1-CLOCK/NPAS2 heterodimers bind their target E-box sequences In a clrcadlan cycle with an opposite phase to that of CRY-PER accumulation.22 This Is compatible with a scenario In which PER-CRY complexes Impede the binding of BMAL1-CLOCK/NPAS2 heterodimers to their cognate deoxyribonucleic adlc (DNA) sequences. A secondary mechanism, Involving the orange-domain basic helix-loop-helix proteins DEC1 and DEC2 may reinforce the clrcadlan E-box binding of BMAL1-CLOCK/NPAS2 heterodimers.47 DEC1 and DEC2, both transcriptional repressors, can establish direct proteinproteln Interactions with BMAL1 and thereby sequester this essential clock component Into an Inactive complex. In addition, DEC proteins can compete with BMAL1-CLOCK heterodimers for E-box binding, and hence diminish E-box-dependent activation of BMAL1-CLOCK target gene expression. Although In mammals the function of DEC1 and DEC2 In circadian rhythm generation has not yet been firmly established by genetic loss-of -function experiments, this has recently been accomplished for the Drosophila ortholog clockwork orange (CWO).48-50 Post-translational mechanisms modulating the stability and/or activity of PER and CRY proteins also play pivotal roles In circadian clock function. For example, phosphorylation of PER1 and PER2 by casein kinase 1 (CK1δ or CKlε) can modulate the period of circadian gene expression,26 and REV-ERBα has been shown to be stabilized when phosphorylated by glycogen synthase kinase 3p (GSK3β).45 In addition to phosphorylation, sumoylatlon and acetylation appear to participate in the functional finetuning of clock components. Sumoylation of BMAL1 proceeds In a clrcadlan fashion and correlates with the temporal transactivatlon efficiency of CLOCK-BMAL1 heterodimers.51 Moreover, CLOCK contains a histone acetyl transferase (HAT) activity that Is required for normal clrcadlan rhythm generation.52
Figure 1. Feedback loop model for mammalian circadian oscillator. The transcription of Per and Cry genes is activated by heterodimers between BMAL1 (B) and either of the two related proteins CLOCK (C) or NPAS2 (N). The polycomb protein EZH2 interacts with these heterodimers and thereby facilitates their action. The accumulation and activity of PER and CRY proteins are also influenced by phosphorylation by protein kinases (CK1δ, ε), by ubiquitination via a complex containing the F-box protein FBXL3 (specific for CRYs), by the histone methyl transferase binding protein WDR5, and by MONO, an RNAand DNA binding protein. DEC1 and DEC2 (D1, 2) compete with BMAL1-CLOCK/NPAS2 heterodimers for E-box binding and thereby reduce E-box-mediated transactivation. A accessory feedback loop, employing the nuclear orphan receptors RORα, RORβ, and RORγ (RORα,β, γ) as activators, and REV-ERBα and REV-ERβ (REV-ERBα, β) as repressors, regulates the circadian transcription of Bmal1 See text for further explanations.
Table I. Isolation of mammalian circadian clock genes and mutant phenotypes. Mammalian circadian clock genes have been identified and isolated using various approaches. Their protein products function in the following transcriptional and post-translational mechanisms: CRY1, 2, PER1, 2, DEC1, 2, REV-ERBα, β, γ act as transcriptional repressors. CLOCK, NPA52, BMAL1, and RORα , β, γ are transcriptional activators. CK, δ, ε, and GSK3β are protein kinases. FBXL3 is a substrate recognition protein of an ubiquitin ligase complex. EZH2 is a member of the polycomb protein family, which probably keeps chromatin regions in a transcription-poised state. NONO is an RNA-DNA-binding protein which attenuates the action of PER proteins through yet unidentified mechanisms. CIPC has no recognizable functional peptide motif. period length; nd, not determined; DNA, deoxyribonucleic acid; RNA, ribonucleic acid.
| Approach | Gene | Phenotype (wheel running) | References |
| Genomics | Period 1 (Per1) | KO: τ shorter to arrhythmic | 21-23 |
| Period 2 (Per2) | KO: τ shorter to arrhythmic | 21, 23 | |
| Per1/Per2 | KO arrhythmic | 21, 23 | |
| Cryptochrome 1 (Cry1) | KO: τ shorter | 24, 25 | |
| Cryptochrome 2 (Cry2) | KO: τ longer | 24, 25 | |
| Cry1/Cry2 | KO: arrhythmic | 24, 25 | |
| Casein kinase 1δ (Ck1δ | nd | 26 | |
| Casein kinase 1ε (Ck1ε) | (see Tau mutation, below) | 26 | |
| Npas2 | KO: mild circadian phenotypes | 27 | |
| Npas2/Clock | KO: arrhythmic | 28 | |
| Genetics | |||
| ENU screen | Clock | Antimorph: τ longer to arrhythmic | 29 |
| Overtime (FbxI3Ovtm) | Hypomorph or null: τ longer | 30-32 | |
| Spontaneous mutation | Tau (Ck1ε, hamster) | Hypomorph: τ shorter | 33 |
| Yeast-two-hybrid assay | Bmal1 | KO arrhythmic | 34, 35 |
| Cipc | nd (but depletion in fibroblasts shortens τ) | ||
| Biochemistry | |||
| Protein-DNA interactions | Rev-erbα | KO: τ shorter | 36, 37 |
| Rev-erbβ | |||
| Rorα | nd (also identified in a screen for as Bmal1 activators) | 38, 39 | |
| Rorβ | KO: τ longer | 40, 41 | |
| Rorγ | nd (also identified in a screen for as Email activators) | 39, 42 | |
| Protein-protein | Nono | nd (but required for clock function in fibroblasts) | 43 |
| Interactions with Mper1 | |||
| CLOCK-BMAL1 | Ezh2 | nd (but required for clock function in fibroblasts) | 44 |
| REV-ERB | Gsk3β | nd (but required for normal clock function in fibroblasts) | 45 |
Circadian oscillators are likely to be influenced by the cells' metabolic state, and the temporal coordination of metabolism may actually be a major purpose of circadian clocks. In keeping with this Idea, McKnight and coworkers have shown that, at least In cell-free assays, the binding of CLOCK-BMALf and NPAS2-BMAL1 heterodimers to E-box motifs Is strongly Influenced by the ratio of reduced to oxidized nicotinamide adenosine dinucleotide (NAD) cofactors.53 In turn, this ratio is determined by the cell's metabolic condition, In particular by the reduction of pyruvate to lactate. Intricate molecular Interactions have also been proposed between heme metabolism and the clockwork circuitry. NPAS2 is a heme-blnding protein,54, 55 and binding of carbon monoxide (CO) to heme-bound Iron strongly reduces the affinity of NPAS2 for DNA.54 Lee and colleagues proposed that In liver NPAS2 regulates the clrcadlan expression of aminolevulinic acid synthase (ALAS1) In a feedback loop directly coupled to heme anabollsm and catabolism. In their model the expression of ALAS1, the rate-limiting enzyme In the synthesis of heme, is stimulated by a NPAS2-heme-PER2 ternary complex.56 The resulting accumulation of excess heme then Induces the expression of heme oxygenase, the rate-limiting enzyme In heme catabolism. Heme oxygenase breaks heme down to carbon monoxide (CO) and blliverdln, and the released CO Inhibits the transactivatlon potential of NPAS2 by binding to its heme cofactor. In turn, this leads to a downregulatlon of Alasl transcription. In liver, this accessory, metabolic feedback loop of NPAS2 activity may work In parallel or In synergy with the more classical feedback loop exemplified In Figure 1.
A hierarchical network of ceIlular clocks
The suprachiasmatic nucleus
In the late 1970s, lesion studies In laboratory rodents Indicated that the suprachlasmatic nuclei (SCN), two small groups of neurons located In the ventral hypothalamus above the optic chiasma, play an Important role In circadian behavior.57 In fact, surgical bilateral SCN ablation resulted In Immediate arrhythmlcity In rats.58 Nevertheless, these experiments did not unequivocally discriminate between a pacemaker and a relay function of the SCN. The breakthrough was accomplished by transplantation experiments by Ralph and coworkers, using wild-type and Tau mutant hamsters that free-ran with a period length of 24 and 20 hours, respectively, when kept in constant darkness.59 Fetal SCN tissue grafted Into the third ventricle of SCN-lesioned animals rescued clrcadlan rhythms In locomotor activity, and the period length was determined by the donor tissue. These results clearly Identified the SCN as the central clrcadlan pacemakers In mammals, several years before the first mammalian clock genes were Identified. Subsequently, organ and cell culture experiments indicated that circadlan rhythm generation is a cell-autonomous property of SCN neurons. However, although dissociated SCN neurons displayed robust rhythms In electrical firing frequency, the Intercellular variability In period lengths was enormous.60, 61 Hence, In Intact animals cellular SCN oscillators must be coupled by Intercellular communication. Oscillator coupling Is not only important for the synchronization of Individual neurons, but also renders SCN neurons much more resilient to genetic perturbations. Kay and colleagues have recently shown that mPERldeficient SCN neurons lose their rhythm In clock gene expression when cultured as Individual cells, but exhibit robust dally cycles In gene expression when kept In organotypic slice cultures.62 Cellular crosstalk probably Involves both neuronal and paracrine signaling. The Importance of the latter has been revealed by gene knockout experiments. For example, mice deficient for the vasoactive intestinal peptide (VIP) and pituitary adenylate cyclase activating peptide (PACAP)receptor VPAC2 are nearly arrhythmic, In spite of ongoing rhythms in Individual cells.63, 64
Since the SCN can measure time only approximately, It must be resynchronized dally. This synchronization Is accomplished by the photoperiod via conventional rod and cone photoreceptors In the outer retinal layer and, In addition, a very small fraction of melanopsin containing ganglion cells In the Inner retina.65 Mice devoid of rods and cones are visually blind, but owing to melanopsin containing ganglion cells they can still synchronlze their circadian clocks. Only when the melanopsin gene Is disrupted In these mice are they free-running with their intrinsic period length, when kept In dally light-dark cycles. Photic cues perceived In the retina are transmitted to the SCN via the retlna-hypothalamic tract. Synaptic release of glutamate and PACAP leads to an Influx of Ca++. This triggers the activation of a variety of protein kinases In postsynaptic SCN neurons, which In turn elicits the activation of immediate early transcription factors, such as cyclic adenosine monophosphate (cAMP) response element binding protein (CREB). CREB then binds to cAMP response elements (CRE) In the promoter and enhancer regions of Per1 and Per2 genes and thereby provokes an increase of the repressors PER1 and PER2.66, 67 Light-Induced Per expression during dawn and dusk advance and delay, respectively, the phase of clrcadlan PER accumulation, and this keeps circadian rhythms tuned to the photoperiod.68
Under certain circumstances behavioral rhythms do not require an Intact SCN.
When laboratory rodents like mice, rats, or hamsters, are offered food during a restricted time period during the day, they entrain to the imposed feeding schedule and anticipate feeding, as manifested by wheel running bouts several hours before getting access to meals. After food Is offered ad libitum again, the animals still display anticipatory behavior for a few days, indicating that the foodentrained oscillator (FEO) can free-run during a limited time span. Despite considerable efforts, the FEO has not yet been associated unequivocally with an anatomical region In the brain or elsewhere. 69
The “methamphetamine-sensltive circadian oscillator” (MASCO) is perhaps even more mysterious than the FEO. In 1987 Honma and colleagues noticed that the administration of methamphetamine in drinking water rescued behavioral rhythmlcity In SCN-lesioned rats.70 More recently, this was also demonstrated for mice.71, 72 Of note, methamphetamine also restores rhythmic locomotor activity In Clock À19 mutant mice.73 Strikingly, chronic methamphetamine treatment of rats engenders a splitting of locomotor activity from other circadian outputs. For example, rPerl expression In SCN neurons and plasma melatonin rhythms are not affected by methamphetamine in rats that are kept under 12-hour light-dark cycles, but the period length of locomotor activity Is considerably lengthened In these animals.74 Moreover, rPer1, rPer2, and rBmal1 expression was found to be completely phaseInverted In the caudate-putamen and the parietal cortex of methamphetamlne-treated rats. These unexpected findings are open to speculation, but I find the following scenario worth considering: In untreated animals with an intact SCN, the MASCO-containlng brain region may be a relay center In the processing of SCN outputs to dally rest-activIty cycles. SCN lesion may lead to a desynchronizatlon of cellular oscillators In this relay station, manifesting Itself In the loss of clrcadlan rhythmiclty Methamphetamine may enhance crosstalk between MASCO-containing cells, perhaps by facilitating intercellular oscillator coupling via signaling through dopamine or nicotinic receptors.75, 76 Of note, dopamine has been shown to activate mltogen-activated protein kinase (MAPK) and cAMP CREB,77 both known to be Involved In the phase resetting of cellular oscillators. Once phase coherence Is reached, the MASCO may now drive locomotor activity cycles independently of the SCN.
Peripheral oscillators
The examination of clrcadlan clock gene expression In the late 1990s revealed an unexpected result. Several groups observed that virtually all examined peripheral tissues transcribe Cry, Per, Bmal1, and Rev-erb α genes In a cyclic fashion.78, 79 More importantly, robust rhythms In clock gene expression was able to be demonstrated In serum-shocked fibroblasts and tissue expiants.78, 80 Furthermore, real-time recording of fluorescence or bioluminescence revealed that Individual cultured fibroblasts harbor self-sustained and cell-autonomous oscillators similar to those operative In SCN neurons.81 Caused by differences in period length, peripheral cell oscillators rapidly desynchronlze in culture or In organs of SCN-lesioned animals.82 Elegant experiments by BIttman and colleagues suggest that the SCN must probably synchronize each individual hepatocyte every day in order to maintain phase coherence in the liver.83
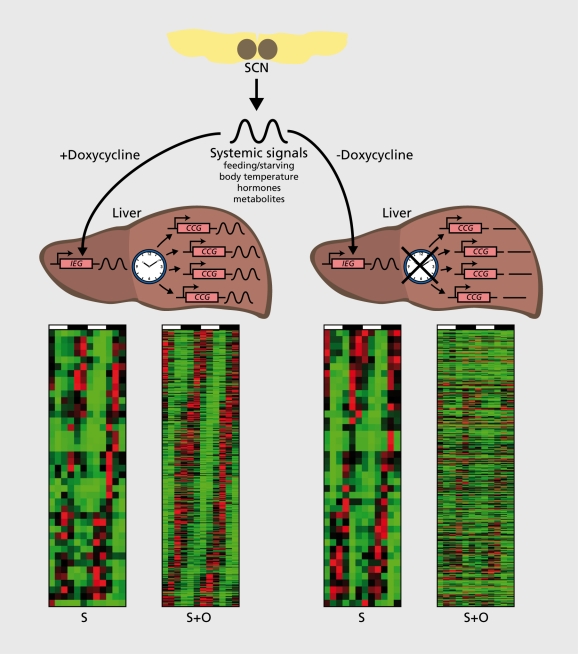
Daily feeding-fasting cycles appear to be the dominant Zeitgebers for several organs, Including liver, kidney, pancreas, and heart muscle.84-86 In addition, glucocorticoid hormones, whose plasma concentrations oscillate with a strong daily amplitude in laboratory rodents and humans, and probably many other systemic timing cues, contribute to the phase entralnment of peripheral clocks.87-90 One approach towards the identification of such signals in liver was recently reported by Kornmann and coworkers.91 The rationale of this strategy, Illustrated In Figures 1 and 3, makes the following assumption: The SCN drives the rhythmic activity and/or abundance of systemic signals that, In turn, modulate the diurnal activity of Immediate early genes. In a mouse strain with conditionally active hepatocyte oscillators (Figure 2), systemlcally driven genes are rhythmically expressed Irrespective of whether the liver clocks are running or arrested. Under these premises, such genes could be Identified using genome-wide transcriptome profiling (Figure 3). Indeed the mRNA encoding mPER2, an essential clock component, was amongst the 30 systemlcally regulated clrcadlan transcripts, suggesting that mPer1 expression can be regulated by both systemic signals and local oscillators. Interestingly, the temporal expression of mPer2 was in phase with that of several heat shock protein genes and In antiphase with that of genes specifying F-box (recognition components of ubiqultin llgase complexes) and cold-Induced RNA binding proteins. Based on these findings, it Is tempting to speculate that the regulation of Immediate early gene expression by body temperature rhythms may be Involved In the synchronization of hepatocyte clocks. However, since heat shock transcription factor 1 (HSF1), the purported regulator of Hsp and, perhaps, mPer1 expression, can also be activated by feeding and reactive oxygen species (ROS), this pathway may also be implicated in the phase entrainment of peripheral docks by feeding-fasting rhythms.
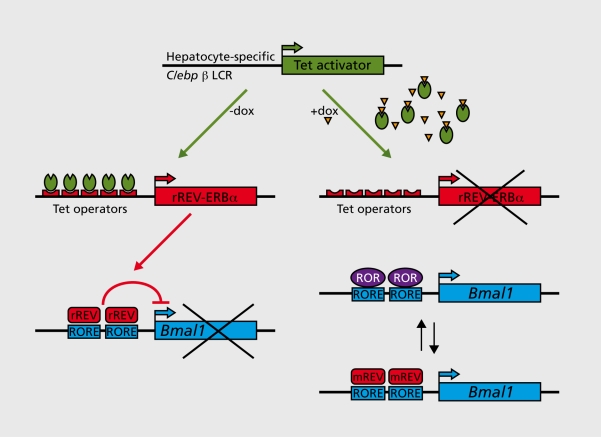
Figure 2. A mouse with conditionally active hepatocyte clocks. A transgenic mouse was generated in which the function of hepatocyte circadian oscillators requires the tetracycline derivative doxycycline (Dox). Hepatocyte-specific, Dox-dependent overexpression of REV-ERBα (rREV= rat REV-ERBα) was achieved by placing a rat REV-ERBa (rREV-ERBα) cDNA transgene under the control of seven tetracycline operators. In the liver of transgenic mice expressing the Tet activator from the hepatocyte-specific Clebpβ locus control region (LCR), rREV-ERBα transcription is constitutively high in the absence of the tetracycline analog Dox. This leads to a constitutive repression of Bmal1 transcription and thereby to an attenuation of circadian oscillator function, since BMAL1 is required for circadian rhythm generation. The addition of Dox to the food or drinking water abolishes the binding of Tet activators to their operators of the rREV-ERBα transgene promoter, and thereby provokes the reactivation of Bmail transcription. Under these conditions, the circadian regulation of Bmal1 transcription is accomplished by a rhythmic exchange of ROR activators and endogenous REV-ERB repressors (mREV= murine REV-ERBα), as in wild-type mice (see Figure 1). Hence, circadian hepatocyte oscillators are operative in the presence of Dox.
Figure 3. Systemically and oscillator-driven circadian liver genes. Circadian liver transcripts were identified in the transgenic mice presented in Figure 2, using genome-wide Affymetrix microarray hybridization with liver RNAs harvested at 4-hour intervals over 2 days from doxycycline-treated and untreated animals (see ref 84). This technique enables the simultaneous quantification of mRNA levels for virtually all of the 15 000 genes that are active in the liver. In doxycycline-treated mice, core clock and clock-controlled genes (CCGs) as well as systemically controlled genes, are expressed in a circadian manner. In mice not receiving doxycycline in the food, only systemically controlled genes are rhythmically expressed. The heat maps below the livers are phase maps of cyclically accumulating transcripts (red for high expression, green for low expression). S stands for mRNAs whose rhythmic transcription is controlled by systemic cues, and S+O stands for transcripts whose rhythmic transcription can be controlled by either systemic cues or hepatocyte oscillators. Comparison of the S+O heat maps of untreated and doxycycline-treated mice reveals that the circadian expression of most genes requires functional hepatocyte clocks. RNA, ribonucleic acid; mRNA, messenger ribonucleic acid.
Interestingly, most forebrain structures apart from the SCN and the pineal gland display relatively shallow oscillations in the expression of core dock and dock-controlled genes. For example Dbp mRNA accumulation fluctuates with an approximately 100-fold amplitude In liver, but only with an approximately 3-fold amplitude In most brain regions.92, 93 This low amplitude could reflect either an inefficient synchronization of brain cell clocks, or an Intrinsic difference between neuronal and non-neuronal cell clocks. I favor the first interpretation, given the similarity In the molecular makeup of oscillators In all examined cell types. Conceivably, the chemical timing cues involved In the synchronization of peripheral oscillators - and all brain cell clocks except those operative In SCN neurons must be considered as peripheral clockstraverse the blood-brain barrier Inefficiently. As a consequence, only a subpopulation of brain cells may be phase-entrained by these cues, and the compound rhythms determined for brain cell populations would thus have a low amplitude. The reduced amplitude of brain circadian oscillations may be physiologically meaningful. In fact, many enzymes participating In neurotransmitter homeostasis, such as glutamate decarboxylase, aromatic amino acid decarboxylase, branched chain amino acid 2-oxoglutarate aminotransferase, lamda-amlnobutyric acid (GABA) transaminase, glycine cleavage enzyme, L-serine racemase, and histidine decarboxylase, require the vitamin B6 derivative pyridoxal phosphate (PLP) as a coenzyme.94 The expression of pyridoxal kinase, the enzyme phosphorylating pyridoxal to PLP, Is Influenced by the three strongly circadian PAR bZIp transcription factors DBP, HLF, and TEE Indeed, a large fraction of PAR bZip triple knockout mice succumb to spontaneous and sound induced epileptic seizures, supposedly due to the Impaired expression of pyridoxal kinase. In the liver of wild-type animals, pyridoxal kinase mRNA and PLP levels oscillate about 2.5-fold and 1.5-fold, respectively, during the day.92 Even this moderate fluctuation may be hazardous In the brain.
Molecular analysis of circadian outputs: metabolism and detoxification
Genome-wide transcrlptome profiling studies have uncovered large repertoires of genes undergoing circadian expression cycles In a variety of organs. Depending on the tissue and the stringency of the algorithms used in the data-mining of DNA mlcroarray data, the fraction of rhythmically expressed genes varies between 2% and 10 %.91, 95-101 The majority of cyclically accumulating transcripts encode polypeptides with tissue-specific functions, supporting the notion that different organs must fulfil different temporally controlled tasks. Liver Is clearly the tissue whose circadian transcrlptome has been examined most thoroughly. As expected, many cyclically expressed transcripts encode key enzymes of major metabolic pathways involved In food processing and energy homeostasis, including fatty acid and carbohydrate metabolism, cholesterol utilization, and bile acid synthesis, and xenobiotic detoxification. Similar to what has been concluded for the ultradlan metabolic cycle of yeast,102-104 there Is some obvious logic in the circadian organization of metabolism. It seems sensible to anticipate the expression of enzymes and regulators of xenobiotlc detoxification before feeding, which Inevitably Is associated with the absorption of toxins (eg, plant alkaloids, coumarin, etc). Similarly, It Is safer to produce and secrete bile acids into the Intestine only when they are needed for the emulslficatlon of absorbed lipids, than throughout the day. Bile acids act as detergents, and the chronic exposure of the Intestinal wall to these aggressive substances may have adverse effects on epithelial cells. Accordingly, the expression of cholesterol 7a hydroxylase (CYP7A), the rate-limiting enzyme In the conversion of cholesterol to bile acids, Is under tight circadian control.105, 106 Sugar metabolism may also be optimized by circadian regulation. After carbohydrate-rich meals, glucose Is polymerized into glycogen, which in liver serves as a rapidly available “fuel” store to mobilize glucose for brain and blood cells during the postabsorptive phase. Obviously, It would be counterproductive if glycogen synthase and glycogen phosphorylase were simultaneously active throughout the day, and glycogen synthase and glycogen phosphorylase are therefore expressed In an anticycllc manner.107
As highlighted by these examples, biological clocks can coordinate metabolism through three principles: (i) anticipation of metabolic pathways to optimize food processing; (ii) limitation of metabolic processes with adverse side effects to time periods when they are needed; and (iii) sequestration of chemically Incompatible reactions to different time windows. During the past 10 years, detailed molecular regulatory pathways have been unraveled through which this temporal coordination can be achieved. Obviously, circadian clocks do not function In Isolation, but work In tight cooperation with Inducible regulatory processes. For example, llgand-dependent nuclear receptors, such as FXR, LXR, PXR, and CAR,108-110 their coregulators SHP, SRC-1, DRIP205, CBP, and PGC-1,108, 111 the sterol-sensing transcription factor SREBP, and Its régulators SCAP and INSIG1/2112-114 cooperate In an intimate fashion with circadian clock components In the temporal control of cholesterol/bile acid metabolism.
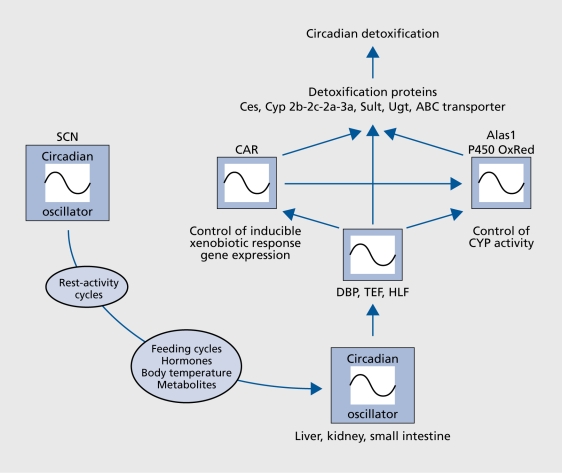
Xenobiotic detoxification, the Inactivatlon and elimination of food toxins and drugs, has been known to be circadian for decades.114 The daytime-dependent differences in drug sensitivity can be remarkable. For example, In mice the dose at which 50% of the animals die after the administration of the anticancer drug 5-fluorouracil is twice higher at ZT05 as compared with ZT17.116 Moreover, the probability of succumbing to a single constant dose of tumor necrosis factor alpha Injected at regular Intervals during the day oscillates approximately 10-fold.117 All In all, day time dependent toxicity has been established for over 30 anticancer therapeutics In laboratory rodents.117 Owing to the availability of mutant mouse models for various core clock and clock-controlled genes, some genetic circuits linking circadian oscillators to xenoblotic detoxification could be deciphered. One such pathway, Involving DBP, HLF, and TEF, the three members of the PAR bZIp transcription factor, is Illustrated In Figure 4. In liver, kidney, and small Intestine, the accumulation of all three of these proteins follows a robust circadian rhythm that Is controlled both on the transcriptional and post-translational level.93, 119-121 DBP, TEF, and HLF must execute partially overlapping functions, since disruption of only one or two of the genes encoding these transcription factors does not result In strong phenotype changes under laboratory conditions.92, 93, 122, 123 However, mice deficient In all three PAR bZlp proteins age at an accelerated rate and die prematurely. Genome-wide transcrlptome profiling revealed that these transcription factors govern the circadian accumulation and/or activity of circadian regulators and enzymes Involved In xenobiotic detoxification pathways (Figure 4). As a consequence, PAR bZip-deficient mice are exquisitely sensitive to xenobiotic compounds such as barbiturates and anticancer drugs.123
Figure 4. A clock output pathway regulating circadian xenobiotic detoxification. The SCN master pacemaker synchronizes circadian oscillators in peripheral organs, such as liver, kidney and small intestine. The molecular signaling pathways involved in this process are still poorly understood, but they are related to feeding fasting cycles, body temperature rhythms, cyclic hormone secretion (eg, glucocorticoids), and possibly the rhythmic accumulation of metabolites. The molecular clocks in liver, kidney and small intestine govern the circadian expression of the PAR bZip transcription factors DBP, HLF, and TEF, which in turn modulate the rhythmic expression of regulators and enzymes involved in detoxification (see ref 122 for details). SCN, suprachiasmatic nuclei.
As reported by Antoch and colleagues, mice homozygous for a Bmal1 null allele or a Clock dominant-negative mutant allele also display Impaired resistance against xenobiotic drugs such as cyclophosphamide.124 These authors concluded that daytime dependent responses of the drug targets (eg, the hematopoietic system), rather than circadian drug metabolism, was the rate-limiting parameter in circadian sensitivity to cyclophosphamide. Clearly, more experiments with additional drugs will be required to examine the entire spectrum of mechanisms involved in the circadian sensitivity to xenobiotics. Whatever their outcome will be, such studies will hopefully contribute to the awareness that the time of day should be taken into consideration when designing regimens for therapeutic treatments.
Carcadian rhythm disruption in humans and other animals
Given the wide spectrum of physiological processes influenced by the circadian clock, it is not surprising that disruption of circadian timing can manifest itself in a plethora of physical and mental symptoms and morbidities. Sleep disturbances, caused by jet lag, have probably been experienced by all transatlantic travelers. Jet lag reflects the limited phase-shifting capacity of the suprachiasmatic nucleus.125 Sudden 1-hour phase delays and advances, such as the ones caused by switching from summer time to winter time and vice versa, should not disrupt the circadian cycle, since these phase changes are well within the synchronization capacity of the clock. However, several days are required to adapt the circadian pacemaker to abrupt and large daytime changes caused by transatlantic flights. Jet lag not only affects sleep-wake cycles, but also peripheral organs, such as the gastrointestinal tract, liver, pancréas, and the kidney.126 As a consequence, heavy meals absorbed at “inadequate” daytimes after a transatlantic flight may cause indigestion. Moreover, during the jet lag period “poorly timed” urine production by the kidney may increase the frequency of urination during night hours. Adaptation is achieved faster after westbound journeys than after eastbound journeys, presumably since the SCN has a greater capacity for phase delays than phase advances.125 This was documented in a rather objective manner by examining the performance of top-class German athletes after transatlantic flights to Atlanta (westbound) or Osaka (eastbound). Jet lag-associated drops in performance disappeared after 5 days in Atlanta, but only after 7 days in Osaka.127
While occasional episodes of jet lag have probably no consequences on morbidity, chronic jet lag suffered by nurses and flight attendants on rotation shift work during extended time periods has been reported to significantly increase breast cancer risk.128 Moreover, mice subjected to light-dark regimens causing chronic jet lag show a sharp increase in morbidity and mortality.129 If animals kept under such conditions receive tumor grafts, the tumors proliferate more rapidly than in control mice.130 The molecular mechanisms linking circadian rhythms to tumor biology remain to be elucidated, but several observations hint towards the implication of Per genes. Thus, a large fraction of mPerl mutant mice die of cancer, most frequently of spontaneous lymphomas.131, 132 Perhaps relevant to the increased breast cancer incidence in women with chronically disrupted circadian rhythms, Chen and coworkers reported that 56 out of 59 tumor samples from Taiwanese woman displayed strongly deregulated PER1, PER2, and PER3 gene expression.133 In these tumors, epigenetic silencing through DNA methylation, rather than mutations was responsible for the reduced levels of PER proteins.
Perturbation of circadian clock function can also cause psychiatric ailments, SAD (seasonal affective disorder) being probably the most common among them. SAD manifests itself by a profound depression and is particularly frequent in Northern countries during winter time.134 In many cases it can be cured simply by the administration of strong artificial light during early morning hours.135, 136 The successful treatment of SAD with light suggests that this mood disorder is caused by an impairment of circadian clock synchronization, either because of insufficient luminosity or deregulated melatonin secretion during wintertime.134
In addition to the serious physical and psychic illnesses mentioned above, there are more innocuous manifestations of aberrant circadian clock functions. Human subjects have individual preferences for their activity phase and, accordingly, can be classified into “chronotypes.”137 Due to socioeconomic constraints many chronotypes can only adopt their favorite lifestyle during weekends and vacations.138 “Morning larks” choose to get up early in the morning and go to bed relatively early at night, while “night owls” prefer to stay in bed longer and to remain active during a good part of the night. The most extreme forms of these behaviors are known as advanced sleep phase syndrome (ASPS) and delayed sleep phase syndrome (DSPS), respectively.139 In one form of familial advanced sleep phase syndrome (FASPS) a mutation in the hPER2 gene was identified as the culprit.140 The mutant hPER2 protein carries a glycine residue instead of a serine residue at position 662. This mutation prevents a phosphorylation, normally occurring on S662, which triggers further phosphorylation by casein kinases 1S/ε (CK18 and CK1ε) at nearby serine residues C-terminal to S662. In the absence of these phosphorylations, mPER2 accumulates to lower than normal levels, resulting in a shortening of the period length and, as a consequence, in a daily phase advance. These molecular events could be successfully reproduced in transgenic mice141 and cultured fibroblasts141 expressing transgenes specifying S662G mutant proteins. The successful dissection of molecular mechanisms responsible for FASPS in animal and even cellular model systems exemplifies the power of reductionist approaches in tackling seemingly complex behavioral traits.
Conclusions
Although the first circadian clock was discovered almost 280 years ago, the mechanisms involved in biological timekeeping remained a mystery for the following two and a half centuries. Owing to the development of powerful genetic, genomic, and molecular tools during the past few decades, clock genes were able to be identified, isolated, and studied in several model systems. These technical advances converted circadian rhythm research from a purely phenomenological to a molecular and mechanistic discipline. In one organism, cyanobacteria, a temperature-compensated clock ticking for over a week could be reconstituted with purified recombinant proteins in the test tube. The chemical and biophysical analysis of protein-protein interactions and kinetics of enzyme activities in this simple assay system will probably allow us to understand how biological time-keeping can work at atomic resolution. While additional decades may be required to reach a similar state of sophistication in the analysis of mammalian clockwork function, the progress made in this field has been nevertheless extraordinary. During the past 10 years, an impressive repertoire of molecular cogwheels has been established, and we are beginning to understand how these cogwheels are intertwined. The discovery of cell-autonomous and self-sustained molecular oscillators in virtually every body cell led to a paradigm change of how the clockwork circuitry governs overt rhythms in behavior and physiology. It now appears that the mammalian timing system resembles an extensive and hierarchically structured web of cellular oscillators, whose phases must be coordinated at the single cell level by the master pacemaker in the SCN. We are also beginning to understand how molecular clocks in individual peripheral cells cooperate with cell typespecific and inducible mechanisms to optimize metabolism and physiology. Despite these advances, an important and scientifically challenging issue remains to be addressed. Although evolution-based arguments leave little doubt as to the importance of a well-functioning circadian clock for survival under natural conditions, it has been difficult to show its contribution to fitness of mammalian organisms in the laboratory. The association of increased morbidity to clock gene mutations does not address this issue in a satisfactory fashion, since such genes may execute important functions unrelated to circadian rhythm generation (for example control of ossification by clock genes143, 144). In cyanobacteria (Synechococcus elongatus)145, 146 and a green plant (Arabidopsis thaliana)147 the benefit of circadian timing was demonstrated by an ingenious and convincing strategy. In both species, a clock resonating with imposed light-dark cycles has been shown to increase performance and fitness. Since, depending on the imposed environmental conditions, the same clock gene mutation can be beneficial or deleterious in such experiments, the observed phenotypes must thus be caused by a rhythm-related property of the gene mutation under study. Eventually this approach should succeed in mammals as well, given the availability of mutant mice and hamsters with aberrant period length.
Selected abbreviations and acronyms
- ASPS
advanced sleep phase syndrome
- cAMP
cyclic adenosine monophosphate
- CRE
cAMP response elements
- CREB
cAMP response element binding protein
- DNA
deoxyribonucleic acid
- DSPS
delayed sleep phase syndrome
- FASPS
familial advanced sleep phase syndrome
- FEO
food-entrained oscillator
- MAPK
Mitogen-activatedprotein kinase
- MASCO
methamphetamine-sensitive circadian oscillator
- mRNA
messenger ribonucleic acid
- RNA
ribonucleic acid
- SAD
seasonal affective disorder
- SCN
suprachiasmatic nuclei
I thank my colleagues Hans Reinke, Charna Dibner, and Gad Asher for comments on the manuscript and Nicolas Roggi for the artwork. Research in my laboratory is supported by the Swiss National Science Foundation (through an individual research grant and the National Center of Competence in Research program grant Frontiers in Genetics), the State of Geneva, the Louis Jeantet Foundation of Medicine, the Bonizzi-Theler Stiftung, and the 6th European Framework Project EUCLOCK.
REFERENCES
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.De Mairan JJD. Observation botanique. Histoire de l'Académie Royale des Sciences. 1729:35–36. [Google Scholar]
- 3.de Candolle AP. Physiologie végétale; ou Exposition des forces et des fonctions vitales des végétaux: pour servir de suite à l'organographie végétale, et d'introduction à la botanique géographique et agricole. Paris, France: Bechet; 1832 [Google Scholar]
- 4.Gehring W., Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol. 2003;57 (suppl 1):S286–S289. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM., Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 6.Wijnen H., Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 7.Woelfle MA., Johnson CH. No promoter left behind: global circadian gene expression in cyanobacteria. J Biol Rhythms. 2006;21:419–431. doi: 10.1177/0748730406294418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzen C., Liu Y. The Neurospora crassa circadian clock. Adv Genetics. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- 9.Gardner MJ., Hubbard KE., Hotta CT, Dodd AN., Webb AA. How plants tell the time. Biochem J. 2006;397:15–24. doi: 10.1042/BJ20060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu W., Hardin PE. Circadian oscillators of Drosophila and mammals. J CellSci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 11.Ko CH., Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 (special issue 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 12.Stratmann M., Schibler U. Properties, entrapment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki H., Kondo T. Circadian timing mechanism in the prokaryotic clock system of cyanobacteria. J Biol Rhythms. 2004;19:436–444. doi: 10.1177/0748730404269060. [DOI] [PubMed] [Google Scholar]
- 14.Tomita J., Nakajima M., Kondo T., Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science (New York, NY). 20054;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama H., Nishiwaki T., Nakajima M., Iwasaki H., Oyama T., Kondo T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima M., Imai K., Ito H., et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science (New York, NY). 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 17.Harmer SL., Panda S., Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Devel Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 18.Konopka RJ., Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardin PE., Hall JC., Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 20.Richter CP. A behavioristic study of the activity of the rat. Comp Psychol Monogr. 1922;1:1–55. [Google Scholar]
- 21.Bae K., Jin X., Maywood ES., Hastings MH., Reppert SM., Weaver DR. Differential functions of mPerl, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 22.Cermakian N., Monaco L., Pando MP., Dierich A., Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Periodl gene. EMBOJ. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng B., Albrecht U., Kaasïk K., et al. Nonredundant roles of the mPerl and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 24.van der Horst GT., Muijtjens M., Kobayashi K., et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 25.Vitaterna MH., Selby CP., Todo T., et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallego M., Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 27.Reick M., Garcia JA., Dudley C., McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science (New York, NY). 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 28.DeBruyne JP., Weaver DR., Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King DP., Zhao Y., Sangoram AM., et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busino L., Bassermann F., Maïolïca A., et al. SCFFbxB controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science (New York, NY). 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 31.Godinho SI., Maywood ES., Shaw L., et al. The after-hours mutant reveals a role for FbxI3 in determining mammalian circadian period. Science (New York, NY). 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 32.Siepka SM., Yoo SH., Park J., et al. Circadian mutant Overtime reveals Fbox protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1123. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowrey PL., Shimomura K., Antoch MP., et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science (New York, NY). 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gekakis N., Staknis D., Nguyen HB., et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science (New York, NY). 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 35.Zhao WN, Malinin N., Yang FC., et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat Cell Biol. 2007;9:268–275. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 36.Preitner N., Damiola F., Luis Lopez M., et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 37.Ueda HR., Chen W., Adachî A., et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 38.Akashi M., Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock BrnaH. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 39.Sato TK., Panda S., Miraglia LJ., et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 40.Andre E., Conquet F., Steïnmayr M., Stratton SC., Porciatti V., Becker-Andre M. Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBOJ. 1998;17:3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masana Ml., Sumaya IC., Becker-Andre M., Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. Am J Physiol. 2007;292:R2357–R2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 42.Jetten AM., Kurebayashi S., Ueda E. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol. 2001;69:205–247. doi: 10.1016/s0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- 43.Brown SA., Ripperger J., Kadener S., et al. PERIOD1 -associated proteins modulate the negative limb of the mammalian circadian oscillator. Science (New York, NY). 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 44.Etchegaray JP., Yang X., DeBruyne JP., et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 45.Yin L., Wang J., Klein PS., Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science (New York, NY). 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 46.Ripperger JA., Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 47.Honma S., Kawamoto T., Takagï Y., et al. Ded and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 48.Kadener S., Stoleru D., McDonald M., Nawathean P., Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim C., Chung BY., Pitman JL., McGill JJ., Pradhan S., Lee J., et al. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumoto A., Ukaï-Tadenuma M., Yamada RG., et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo JJ., Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1 . Science (New York, NY). 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 52.Doi M., Hirayama J., Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransf erase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 53.Rutter J., Reick M., Wu LC., McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science (New York, NY). 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 54.Dioum EM., Rutter J., Tuckerman JR., Gonzalez G., Gilles-Gonzalez MA., McKnight SL. NPAS2: a gas-responsive transcription factor. Science (New York, NY). 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 55.Mukaiyama Y., Uchida T., Sato E., et al. Spectroscopic and DNA-binding characterization of the isolated heme-bound basic helix-loop-helix-PAS-A domain of neuronal PAS protein 2 (NPAS2), a transcription activator protein associated with circadian rhythms. FEBSJ. 2006;273:2528–2539. doi: 10.1111/j.1742-4658.2006.05259.x. [DOI] [PubMed] [Google Scholar]
- 56.Kaasik K., Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature, 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 57.Kawamura H., Ibuka N. The search for circadian rhythm pacemakers in the light of lesion experiments. Chronobiologia. 1978;5:69–88. [PubMed] [Google Scholar]
- 58.Ibuka N., Kawamura H. Loss of circadian rhythm in sleep-wakefulness cycle in the rat by suprachiasmatic nucleus lesions. Brain Res. 1975;96:76–81. doi: 10.1016/0006-8993(75)90574-0. [DOI] [PubMed] [Google Scholar]
- 59.Ralph MR., Foster RG., Davis FC., Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science (New York, NY). 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 60.Liu C., Weaver DR., Strogatz SH., Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 61.Welsh DK., Logothetïs DE., Meister M., Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 62.Liu AC., Welsh DK., Ko CH., et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harmar AJ., Marston HM., Shen S., et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 64.Maywood ES., Reddy AB., Wong GK., et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 65.Panda S. Multiple photopigments entrain the Mammalian circadian oscillator. Neuron. 2007;53:619–621. doi: 10.1016/j.neuron.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 66.Gillette MU., Mitchell JW. Signaling in the suprachiasmatic nucleus: selectively responsive and integrative. Cell Tissue Res. 2002;309:99–107. doi: 10.1007/s00441-002-0576-1. [DOI] [PubMed] [Google Scholar]
- 67.Jakubcakova V., Oster H., Tamanini F., et al. Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron. 2007;54:831–843. doi: 10.1016/j.neuron.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 68.Albrecht U., Zheng B., Larkin D., Sun ZS., Lee CC. MPerl and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 69.Landry GJ., Yamakawa GR., Mistlberger RE. Robust food anticipatory circadian rhythms in rats with complete ablation of the thalamic paraventricular nucleus. Brain Res. 2007;1141:108–118. doi: 10.1016/j.brainres.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 70.Honma K., Honma S., Hiroshige T. Activity rhythms in the circadian domain appear in suprachiasmatic nuclei lesioned rats given methamphetamine. Physiology Behav. 1987;40:767–774. doi: 10.1016/0031-9384(87)90281-2. [DOI] [PubMed] [Google Scholar]
- 71.lijima M., Nïkaido T., Akiyama M., Morïya T., Shibata S. Methamphetamine-induced, suprachiasmatic nucleus-independent circadian rhythms of activity and mPer gene expression in the striatum of the mouse. EurJNeurosci. 2002;16:921–929. doi: 10.1046/j.1460-9568.2002.02140.x. [DOI] [PubMed] [Google Scholar]
- 72.Tataroglu O., Davidson AJ., Benvenuto LJ., Menaker M. The methamphetamine-sensitive circadian oscillator (MASCO) in mice. J Biol Rhythms. 2006;21:185–194. doi: 10.1177/0748730406287529. [DOI] [PubMed] [Google Scholar]
- 73.Masubuchi S., Honma S., Abe H., Nakamura W., Honma K. Circadian activity rhythm in methamphetamine-treated Clock mutant mice. EurJNeurosci. 2001;14:1177–1180. doi: 10.1046/j.0953-816x.2001.01749.x. [DOI] [PubMed] [Google Scholar]
- 74.Masubuchi S., Honma S., Abe H., et al. Clock genes outside the suprachiasmatic nucleus involved in manifestation of locomotor activity rhythm in rats. EurJNeurosci. 2000;12:4206–4214. [PubMed] [Google Scholar]
- 75.Garcia-Rates S., Camarasa J., Escubedo E., Pubill D. Methamphetamine and 3,4-methylenedioxymethamphetamine interact with central nicotinic receptors and induce their up-regulation. Toxicol Appl Pharmacol. 2007Inpress doi: 10.1016/j.taap.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 76.Kokoshka JM., Vaughan RA., Hanson GR., Fleckenstein AE. Nature of methamphetamine-induced rapid and reversible changes in dopamine transporters. Eur J Pharmacol. 1998;361:269–275. doi: 10.1016/s0014-2999(98)00741-9. [DOI] [PubMed] [Google Scholar]
- 77.Yan Z., Feng J., Fienberg AA., Greengard P. D dopamine receptors induce mitogen-activated protein kinase and cAMP response element-binding protein phosphorylation in neurons. Proc Natl Acad Sci U S A. 1999;96:11607–11612. doi: 10.1073/pnas.96.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Balsalobre A., Damïola F., Schïbler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 79.Zylka MJ., Shearman LP., Weaver DR., Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 80.Yamazaki S., Numano R., Abe M., et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science (New York, NY). 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 81.Yagita K., Tamanini F., van Der Horst GT., Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science (New York, NY). 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 82.Yoo SH., Yamazaki S., Lowrey PL., et al. PERlOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 20043;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo H., Brewer JM., Lehman MN., Bittman EL. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker. J Neurosci. 2006;26:6406–6412. doi: 10.1523/JNEUROSCI.4676-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Damïola F., Le Minh N., Preïtner N., Kornmann B., Fleury-OIela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hara R., Wan K., Wakamatsu H., Aida R., et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 86.Stokkan KA., Yamazaki S., Tei H., Sakaki Y., Menaker M. Entrainment of the circadian clock in the liver by feeding. Science (New York, NY). 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 87.Balsalobre A., Brown SA., Marcaccï L., et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science (New York, NY). 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 88.Balsalobre A., Marcaccï L., Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 89.Le Minh N., Damïola F., Tronche F., Schutz G., Schïbler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBOJ. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reddy AB., Maywood ES., Karp NA., King VM., Inoue Y., Gonzalez FJ., et al. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007;45:1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- 91.Kornmann B., Schaad O., Bujard H., Takahashi JS., Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gachon F., Fonjallaz P., Damïola F., et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lopez-Molina L., Conquet F., Dubois-Dauphin M., Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBOJ. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clayton PT. B6-responsive disorders: a model of vitamin dependency. J Inher Metabol Dis. 2006;29:317–326. doi: 10.1007/s10545-005-0243-2. [DOI] [PubMed] [Google Scholar]
- 95.Akhtar RA., Reddy AB., Maywood ES., et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 96.Duffield GE., Best JD., Meurers BH., Bittner A., Loros JJ., Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of Mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 97.Kornmann B., Preïtner N., Rïfat D., Fleury-OIela F., Schibler U. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res. 2001;29:E51–1. doi: 10.1093/nar/29.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McCarthy JJ., Andrews JL., McDearmon EL., et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007In press doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Panda S., Antoch MP., Miller BH., Su AI., Schook AB., Straume M., et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 100.Storch KF., Lipan O., Leykin I., Viswanathan N., Davis FC., Wong WH., et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 101.Walker JR., Hogenesch JB. RNA profiling in circadian biology. Methods Enzymoi. 2005;393:366–376. doi: 10.1016/S0076-6879(05)93016-4. [DOI] [PubMed] [Google Scholar]
- 102.Chen Z., Odstrcil EA., Tu BP., McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science (New York, NY). 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 103.Klevecz RR., Bolen J., Forrest G., Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci U S A. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tu BP., Kudlïckï A., Rowicka M., McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science (New York, NY). 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 105.Chiang JY., Miller WF., Lin GM. Regulation of cholesterol 7 alpha-hydroxylase in the liver. Purification of cholesterol 7 alpha-hydroxylase and the immunochemical evidence for the induction of cholesterol 7 alpha-hydroxylase by cholestyramine and circadian rhythm. J Biol Chem. 1990;265:3889–3897. [PubMed] [Google Scholar]
- 106.Noshiro M., Nishïmoto M., Okuda K. Rat liver cholesterol 7 alpha-hydroxylase. Pretranslational regulation for circadian rhythm. J Biol Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- 107.lshikawa K., Shimazu T. Daily rhythms of glycogen synthetase and phosphorylase activities in rat liver: influence of food and light. Life Sci. 1976;19:1873–1878. doi: 10.1016/0024-3205(76)90119-3. [DOI] [PubMed] [Google Scholar]
- 108.Eloranta JJ., KuIIak-Ublick GA. Coordinate transcriptional regulation of bile acid homeostasis and drug metabolism. Arch Biochem Biophys. 2005;433:397–412. doi: 10.1016/j.abb.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 109.Handschin C., Meyer UA. Regulatory network of lipid-sensing nuclear receptors: roles for CAR, PXR LXR, and FXR. Arch Biochem Biophys. 2005;433:387–396. doi: 10.1016/j.abb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 110.Moore DD., Kato S., Xïe W., et al. International Union of Pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor alpha, farnesoid X receptor β, liver X receptor alpha, liver X receptor β, and vitamin D receptor. Pharmacol Rev. 2006;58:742–759. doi: 10.1124/pr.58.4.6. [DOI] [PubMed] [Google Scholar]
- 111.Pineda Torra I., Freedman LP., Garabedian MJ. Identification of DRIP205 as a coactivator for the Farnesoid X receptor. J Biol Chem. 2004;279:36184–36191. doi: 10.1074/jbc.M405126200. [DOI] [PubMed] [Google Scholar]
- 112.Brown MS., Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A. 1998;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McPherson R., Gauthier A. Molecular regulation of SREBP function: the Insig-SCAP connection and isoform-specific modulation of lipid synthesis. Biochemistry Cell Biol. 2004;82:201–211. doi: 10.1139/o03-090. [DOI] [PubMed] [Google Scholar]
- 114.Wang X., Sato R., Brown MS., Hua X., Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 115.Radzialowski FM., Bousquet WF. Daily rhythmic variation in hepatic drug metabolism in the rat and mouse. J Pharmacol Exp Ther. 1968;163:229–238. [PubMed] [Google Scholar]
- 116.Burns ER., Beland SS. Effect of biological time on the determination of the LD50 of 5-fIuorouradl in mice. Pharmacology. 1984;28:296–300. doi: 10.1159/000137977. [DOI] [PubMed] [Google Scholar]
- 117.Hrushesky WJ., Langevin T., Kim YJ., Wood PA. Circadian dynamics of tumor necrosis factor alpha (cachectin) lethality. J Exp Med. 1994;180:1059–1065. doi: 10.1084/jem.180.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Levi F. Chronotherapy of cancer: biological basis and clinical application. Pathologie-Biologie. 1994;42:338–341. [PubMed] [Google Scholar]
- 119.Falvey E., Marcacci L., Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- 120.Fonjallaz P., Ossipow V., Wanner G., Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 121.Wuarin J., Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- 122.Franken P., Lopez-Molina L., Marcacci L., Schibler U., Tafti M. The transcription factor DBP affects circadian sleep consolidation and rhythmic EEG activity. J Neurosci. 2000;20:617–625. doi: 10.1523/JNEUROSCI.20-02-00617.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gachon F., Olela FF., Schaad O., Descombes P., Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 124.Gorbacheva VY., Kondratov RV., Zhang R., Cherukurï S., Gudkov AV., Takahashï JS., et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Khalsa SB., Jewett ME., Cajochen C., Czeisler CA. A phase response curve to single bright light pulses in human subjects. J Physiol. 2003;549(Pt3):945–952. doi: 10.1113/jphysiol.2003.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Waterhouse J., Reilly T., Atkinson G., Edwards B. Jet lag: trends and coping strategies. Lancet. 2007;369:1117–1129. doi: 10.1016/S0140-6736(07)60529-7. [DOI] [PubMed] [Google Scholar]
- 127.Lemmer B., Kern RI., Nold G., Lohrer H. Jet lag in athletes after eastward and westward time-zone transition. Chronobiol Int. 2002;19:743–764. doi: 10.1081/cbi-120005391. [DOI] [PubMed] [Google Scholar]
- 128.Moser M., Schaumberger K., Schernhammer E., Stevens RG. Cancer and rhythm. Cancer Causes Control. 2006;17:483–487. doi: 10.1007/s10552-006-0012-z. [DOI] [PubMed] [Google Scholar]
- 129.Davidson AJ., Sellix MT., Daniel J., Yamazaki S., Menaker M., Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Filipski E., Delaunay F., King VM., et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 2004;64:7879–7885. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 131.Fu L., Pelicano H., Liu J., Huang P., Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 132.Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- 133.Chen ST., Choo KB., Hou MF., Yeh KT., Kuo SJ., Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 134.McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wirz-Justice A. Light therapy for depression: present status, problems, and perspectives. Psychopathology. 1986;19(suppl2):136–141. doi: 10.1159/000285145. [DOI] [PubMed] [Google Scholar]
- 136.Wïrz-Justïce A., Graw P. [Phototherapy]. Therapeutische Umschau. 2000;57:71–75. doi: 10.1024/0040-5930.57.2.71. [DOI] [PubMed] [Google Scholar]
- 137.Roenneberg T., Wïrz-Justïce A., Merrow M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. 2003;18:80–90. doi: 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- 138.Wittmann M., Dinich J., Merrow M., Roenneberg T. Social jetlag: misalignment of biological and social time. Chronobiol Int. 2006;23:497–509. doi: 10.1080/07420520500545979. [DOI] [PubMed] [Google Scholar]
- 139.Ebisawa T. Circadian rhythms in the CNS and peripheral clock disorders: human sleep disorders and clock genes. J Pharmacol Sci. 2007;103:150–154. doi: 10.1254/jphs.fmj06003x5. [DOI] [PubMed] [Google Scholar]
- 140.Toh KL., Jones CR., He Y., Eïde EJ., Hïnz WA., Vïrshup DM., et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science (New York, NY). 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 141.Xu Y., Toh KL., Jones CR., Shin JY., Fu YH., Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 20072;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vanselow K., Vanselow JT., Westermark PO., et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bunger MK., Walisser JA., Sullivan R., et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 144.Fu L., Patel MS., Bradley A., Wagner EF., Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 145.Ouyang Y., Andersson CR., Kondo T., Golden SS., Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci U S A. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Woelfle MA., Ouyang Y., Phanvijhitsiri K., Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 147.Dodd AN., Salathïa N., Hall A., et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science (New York, NY). 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]