Abstract
Ligand-induced translocation of the G-protein-coupled receptor, neurokinin 3 (NK3-R), to the nucleus of hypothalamic neurons was reported using antibodies (ABs) raised against the C-terminal region of NK3-R. The current work was undertaken to substantiate the ability of NK3-R to enter the nucleus and identify which portion of the NK3-R molecule enters the nucleus. ABs directed at epitopes in the N-terminal and second extracellular loop of the rat NK3-R molecule were used to evaluate western blots of whole tissue homogenates and nuclear fractions from multiple brain areas. Specificity of the protein bands recognized by these ABs was demonstrated using CHO cells transfected with rat or human NK3-R. Both ABs prominently recognized a diffuse protein band of ~56–65kD (56kD=predicted size) and distinct ~70kD and 95kD proteins in homogenates of multiple brain areas. The ~95kD protein recognized by the extracellular loop AB was enriched in nuclear fractions. Recognition of these proteins by ABs directed at different regions of the NK3-R supports their identification as NK3-R. The size differences reflect variable glycosylation and possibly linkage to different cytosolic and nuclear proteins. Recognition of protein bands by both ABs in nuclear fractions is consistent with the full-length NK3-R entering the nucleus. Hypotension increased the density of the ~95kD band in nuclear fractions from the supraoptic nucleus indicating activity-induced nuclear translocation. Since NK3-R is widely distributed in the CNS, the presence of NK3-R in nuclei from multiple brain regions suggests that it may broadly influence CNS gene expression in a ligand-dependent manner.
Keywords: neurokinin 3 nuclear translocation, supraoptic nucleus, substantia nigra, caudate, ventral tegmentum
G-protein coupled receptors (GPCR) are cell membrane proteins that are characterized by 7 membrane spanning domains, extracellular domains that bind ligands, and intracellular domains that bind G-proteins allowing regulation of intracellular function by inducing a variety of signaling cascades. Classically, the G-protein subunits activate membrane-linked enzymes, such as adenyl cyclase or phospholipase C (PLC), which in turn initiate signaling cascades resulting in increases in intracellular calcium and activation of calcium-dependent and independent kinases (e.g. protein kinase C and A respectively; PKC and PKA). These kinases in turn may phosphorylate transcription factors to alter gene expression. Thus, these multi-step signaling cascades allow extracellular ligands for GPCRs to regulate gene expression. However, evidence is accumulating that GPCRs may be able to directly influence gene expression as a result of ligand-induced nuclear translocation of the GPCR itself.
We previously observed that immunoreactivity (IR) for the neurokinin 3 receptor (NK3-R), a GPCR activated by neurokinin B (NKB), was present in the nucleoplasm of neurons in the supraoptic (SON) and paraventricular nuclei (PVN) following activation of the neurons by hypotension or hyperosmolality (Howe et al., 2004; Haley and Flynn, 2006). A similar increase in nucleoplasm NK3-R-ir was observed following SON or ICV injections of the NK3-R agonist senktide (Howe et al., 2004; Haley and Flynn, 2006) suggesting that this reflected ligand-induced translocation of the receptor from the membrane to the nucleus. Immuno-electron microscopy confirmed that the NK3-R-IR was localized in chromatin-rich regions of the nucleus (Jensen et al., 2008). The Howe, Haley, and Jensen studies utilized two different antibodies (ABs), both directed at the COOH-terminal region of the NK3-R. The NK3-R identity of the IR observed with one of these ABs was supported by studies demonstrating that the nuclear NK3-R-IR was reduced by injection of NK3-R specific siRNA prior to osmotic stimulation, and that an NK3-AB targeting a region in the second extracellular loop (see Methods) immunoprecipitated the same protein recognized by the COOH-terminal AB (Jensen et al., 2008; Jensen et al., 2010).
The presence of NK3-R-IR in the nucleoplasm raises an important question: How do NK3-Rs get into the nucleus? The mechanism of import of macromolecules into the nucleoplasm is known to involve binding of chaperone proteins to nuclear localization sequences (NLS) with subsequent facilitation of the movement of these multiprotein complexes through the nuclear pore (Jans and Hubner, 1996). A putative NLS has been identified in the 8th helix region that connects the 7th transmembrane domain to the C-terminal tail of NK-3 (Lee et al., 2004). We also identified a second putative classical bipartite NLS in the C-terminal tail (aa 428–446 with the sequence RKKRATPRDPSFNGCSRR) (Howe et al., 2004). Binding of importin molecules to the NLS allows cytoplasmic molecules to enter the nucleus by passing through the nuclear pore. Jensen et al (2010) provide evidence that this is the mechanism by which NK3-R enters the nucleus: NK3-R associates with importin β–1, and immunoneutralization of importin β-1 reduces nuclear localization of NK3-R in an immortalized rat hypothalamic cell line (CLU209). However, this does not fully address the question of how a receptor embedded in the plasma membrane can translocate to the nucleus as we are suggesting for the NK3-R. Does it escape the membrane or does only a portion of the molecule enter the nucleus? Both possibilities have been suggested for growth factor receptors (Wells and Marti, 2002). In the case of other GPCRs that have been localized in the nucleus, size information indicates that nuclear AP1-R is full length (Lee et al., 2004). This is consistent with the NLS in AP1-R being located in the 3rd intracellular loop (Lee et al., 2004) rather than the C-terminal region as is the case for NK3-R. Evidence also suggests that nuclear PTH1R is full length, because the antibody used to demonstrate PTH1R-ir in the nucleus was directed to the N-terminus while the putative NLS sequence is located in the C-terminal tail (Tazawa et al., 2003). However, the nuclear localization of these receptors may not be initiated by exogenous ligand, because in both cases, ligands for the receptors are made in the same cell as the receptor and are also found in the nucleus. Thus, it is possible that they were not membrane-embedded prior to entering the nucleus which has been shown for the FGF receptor (Jans and Hassan, 1998). Recent evidence supporting nuclear trafficking of full-length OTR has been reported in cell lines (Kinsey et al., 2007). An alternate possibility, that only a portion of the protein enters the nucleus, remains feasible, because the existing evidence for nuclear localization of the NK3-R in vivo was obtained with ABs directed against peptide sequences in the C-terminal region that partially overlap with the NLS (Howe et al., 2004; Haley and Flynn, 2006; Jensen et al., 2008; Jensen et al., 2010). Therefore, it is possible that the C-terminal domain of the NK3-R is cleaved following ligand-induced internalization and enters the nucleus. To examine this possibility we raised ABs to other portions of NK3-R (N-terminal and 2nd extracellular loop). In this report, we present evidence that both ABs recognize NK3-R in nuclear fractions from multiple brain regions. This supports the interpretation that the full-length molecule enters the nucleus. It also suggests that NK3-R translocation is involved in diverse and important CNS functions.
Experimental Procedures
1) Animals
Male Sprague-Dawley rats [CRL:CD(SD)Br; Charles Rivers Laboratories, Wilmington, MA], female New Zealand White rabbits, and male, Ovis Aries sheep were used as donors for brain tissue (rats) or antibody generation (rabbits and sheep). All animal protocols used were performed in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Colorado School of Medicine or the University of Wyoming.
2) Antibody generation
The N-terminal peptide (aa 6–18 of rat NK3-R) was synthesized by Global Peptide (Ft. Collins, CO). A carboxy-terminal cysteine was added for linkage to keyhole limpet hemocyanin (KLH) for immunization and to permit coupling to a column for affinity purification. N- and C-termini were also modified by acetylation and amidation respectively. Two female New Zealand White rabbits were immunized with the N-terminal peptide. Following conjugation to KLH, 100μg of protein in PBS was mixed 1:1 with Freund’s complete adjuvant, and each rabbit received 1.0 ml of the mixture SQ in 3–4 sites on the upper back. Booster injections of the same dose of conjugated peptide mixed 1:1 with Freund’s incomplete adjuvant were given at monthly intervals. Blood was collected from the ear vein 2 weeks after immunization and the serum affinity purified using a peptide-coupled column (SulfoLink; Pierce Biotechnology, Rockford, IL) following the manufacture’s instructions. These affinity purified Abs were used for immunoblotting. Both rabbits generated ABs with comparable protein selectivity and affinity. All results shown here utilize AB from rabbit #2 (K2).
Sheep (n=3) were immunized in the University of Wyoming large animal facility with a peptide sequence (aa 180–192 ) on the second extracellular loop of rat NK3-R. The peptide sequence was synthesized by Genscript (Piscataway, NJ) and purified at the MacroMolecular Core Facility at the University of Wyoming. NK3-R peptide and KLH were incubated together for 1h and then passed through a desalting column. All injection volumes were 0.5 ml. Sheep were gently restrained during the brief immunization injections. For the first immunization, 50 μg NK3R-KLH conjugate (in 500 μl saline) was mixed with 500 μl Complete Freund’s Adjuvant and administered into 3 sites: right and left haunches (IM) and nape of neck (subcutaneous). One month later, sheep were administered a second immunization [100 μg NK3R peptide-KLH/sheep in Incomplete Freund’s Adjuvant (IFA)] and immunized monthly (150 μg NK3R peptide-KLH/sheep in IFA) for the next three months. Blood was collected from the jugular vein (100 ml) 7, 14 and 21 days after the final immunization. Whole serum was affinity purified using a peptide-coupled column (SulfoLink; Pierce Biotechnology, Rockford, IL) to yield a final concentration of 10 μg protein/ml. The AB used in these studies was obtained from animal #711.
3) NK3-R transfected CHO cells
CHO-K1 cells stably transfected with human NK3-R (hNK3-R) or rat NK3-R (rNK3-R) were used to demonstrate specificity of the NK3-R ABs. They were a gift from J.E. Krause (Neurogen, Branford, CT). Non-transfected CHO cells were obtained from American Tissue Type Culture Collection (Manassas, VA). The three cell lines were maintained at 37°C under 5%CO2 in Ham’s F-12 medium supplemented with 25mM HEPES, 20% fetal bovine serum, and 500 μg/ml G418 for the transfected cells and 5gm/L NaHCO3, 2 mM L-Glutamine, 10% fetal bovine serum, 200 U/ml penicillin, and 100 μg/ml streptomycin for the non-transfected cells. Cells were grown in T-75 flasks until confluent and then either subcultured, homogenized for Western blot analysis, frozen for stock, or replated into 35mm culture dishes for calcium imaging.
To confirm NK3R expression in the transfected CHO cell line, reverse transcription PCR (RT-PCR) was performed using mRNA from the CHO cell flasks (n=2). Total RNA was extracted from the flask of cells using Trizol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s protocols. Briefly, the media was removed from the cells and 2 ml of Trizol was added to the flask. The cells were vortexed for 5 min to lyse the cells. Trizol cell lysate (1 ml) was moved to a 2 ml Eppendorf tube and 0.2 ml of chloroform was added. The sample was mixed and left at room temperature for 5 min. The sample was then centrifuged 12,000 × g for 15 min. The aqueous phase was moved to a fresh tube and 0.5 ml of isopropyl alcohol was added. The sample was incubated at room temperature for 10 min and then centrifuged 12,000 × g for 10 min. The supernatant was removed and the RNA pellet was washed in 70% ethanol and then centrifuged 12,000 × g for 5 min. The supernatant was removed and the pellet was air dried to remove any ethanol left in the tube. The RNA pellet was then dissolved in 50 μl of RNAse free dH2O. The RNA was assayed for concentration using the Nanodrop spectrophotometer (Thermo Scientific Nanodrop Products, Wilmington, DE).
RT-PCR was performed using the extracted RNA with the Quantitect reverse transcription kit (Qiagen, Valencia, CA). RNA (1 μg) was incubated with genomic DNA wipeout buffer for 2 min at 42° C in a Minicycler (Bio-Rad, Hercules, CA) to remove genomic DNA contamination. The RT-PCR was then run following kit protocols to make the cDNA. Validated primer sets targeting the NK3R (177 bp amplicon), NK1R (164 bp amplicon), NK2R (91 bp amplicon), Neurokinin B (85 bp amplicon) and beta actin (131 bp amplicon) were purchased from SA Biosciences (Frederick, MD). PCR was run using 0.5 μg of the cDNA with the Core Taq polymerase kit (Qiagen, Valencia, CA) and the SA Biosciences primer sets. PCR product was electrophoresed at 110 V for 45 minutes on a 1% agarose gel. DNA bands were detected with ethidium bromide staining. Gels were imaged using the Gel Doc XRS imaging system with Quantity one software (Bio-Rad, Hercules, CA).
Functional expression of the NK3-Rs in the hNK3-R and rNK3-R cell lines was demonstrated by measurement of increases in intracellular [Ca++] ([Ca++]i) in response to senktide, because NK3-R is coupled to the Gq/ll G-protein and therefore, upon activation, induces Ca++ release from intracellular stores. Cells were incubated with 20 μM Fura-2AM for 1 hr at room temperature (Song et al., 2006). Fluorescence emitted at 510 nm was measured in response to 340 and 380 nm excitation. The ratio of 340:380 induced emission is proportional to the [Ca++]i (Grynkiewicz et al., 1985).
4) Cell/Tissue homogenates for WB analysis
CHO cells were scraped from the tissue culture flask and homogenized in cold protease-inhibitor buffer [tetrasodium pyrophosphate (20 mM), sodium phosphate (20 mM), MgCl (1 mM), EDTA (0.5 mM), sucrose (300 mM), benzamidine (0.8 mM), iodoacetamide (1.0 mM), levpeptin (1.1 uM), pepstatin A (0.7 uM), PMSF (0.23 mM), aprotinin (76.8 nM), pH 7.4] using a dounce homogenizer. Following centrifugation of the homogenate at 3,300 g for 15 min, the pellet was discarded and supernatant was collected and centrifuged at 50,000 g for 1 hour in a J2–21 Beckman centrifuge. The resulting pellet (membrane fraction) was resuspended in homogenizing buffer, mixed 1:1 with Laemmli buffer containng 5% β-mercaptoethanol, and heated at 90°C for 5min prior to loading on the gel.
Brain regions were obtained from decapitated, adult, male Sprague-Dawley rats as follows. SON was microdissected from the hypothalamus using the optic chiasm as a landmark. A rectangular block of tissue immediately rostral to optic chiasm approximately 1mm wide × 1mm deep × 3mm long was removed from each side of the brain using irridectomy scissors. The brain was then placed ventral surface up in a brain mold and 1.5–2.0 mm thick sections were prepared using landmarks on the ventral brain surface. The sections were then viewed under a dissecting microscope and tissue punches from the caudate nucleus (Caud), lateral septum (LS), medial preoptic area (MPOA), bed nucleus of stria terminalis (BNST), hypothalamic paraventricular nucleus (PVN), amygdala (Amyg), substantia nigra (SN), and ventral tegmental area (VTA) were prepared using landmarks from (Paxinos and Watson, 1982).
Whole tissue homogenates were prepared by directly homogenizing the microdissected SON and tissue punches in 1% STE (1%SDS,10 mM Tris-HCl, 1mM EDTA).
Nuclear extracts were prepared by homogenizing the tissue in a Dounce homogenizer containing Pierce Halt PIC and 1.0 ml Buffer #1 (0.32 M Sucrose, 3 mM CaCl2, 2 mM MgCl2, 0.1 mM EDTA, 0.1% Triton-X, 10 mM Tris-HCl, and 1 mM DTT). The homogenates were subjected to centrifugation at 1000×g for 15 min at 4C, the pellet was then re-suspended with 1.0 ml of Buffer #1/Pierce Halt PIC and centrifuged again as above. The pellet was treated with 750 μl buffer #1, 750 μl buffer #2 (1.8 M sucrose, 5 mM MgCl2, 0.1 mM EDTA, 10 mM Tris-HCl, and 0.5 mM DTT) and 10 μl Pierce Halt. Next the homogenate was overlaid on 1 ml of buffer #2 in an ultra-centrifuge tube and centrifuged at 19,000 rpm for 1h at 4°C using a Beckman Optima TL Ultracentrifuge with a swinging bucket rotor. After removing the supernatant, the pellet was re-suspended in 750 μl buffer #1 and repelleted at 1000×g for 15 min twice to ensure that the pellet was free of contamination. The resulting nuclear fraction was resuspended in 1% STE.
The samples were then mixed with an equal volume of Laemmli buffer with beta mercaptoethanol and heated at 90°C for 5 min. Blots were probed with an AB against calcium calmodulin protein kinase 1 (CamK), a cytoplasmic protein, to assess purity of nuclear cell fractions and as the loading control for whole tissue homogenates. The CamK AB was from Affinity BioReagents (0.5μg/ml). RNA polymerase (Pol II), a nuclear protein was used as the loading control for the nuclear fractions. The Pol II AB was from Santa Cruz (H-224; 1:500 dilution). Protein concentrations of the extracts were determined using Biosciences CB-X Protein Assay Kits according to manufacture’s instructions. N-glycosylation of NK3-R was assessed using PGNaseF (Sigma-Aldrich). Samples were mixed in an equal volume of Laemmli buffer then heated at 95°C for 5 minutes and incubated with PGNaseF (5U) and Triton-X-100 (2.5 ul) for 3 hrs at 37°C. Additional denaturation experiments were performed by heating samples in Laemmli buffer with 0.1 mM dithiothreitol (DTT) at 100°C for 30–60min.
5) Western blot
Proteins in the whole tissue, membrane fractions, and nuclear extracts were separated using 7.5% SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and incubated with NK3-AB or NK3-AB preblocked with peptide in 5% nonfat milk-PBS overnight at room temperature. NK3-ABs were used at 1:200 for K2, the N-terminal AB, and 0.02–2.0 μg/ml for 711, the 2nd extracellular loop. For peptide preblocking, AB was incubated with peptide for 5hrs on ice (1:1 peptide:AB for K2; 20:1 for 711). The nitrocellulose blot was then washed 3X for 10 min. in 0.1% Tween followed by 3X PBS rinses and incubated with donkey or goat anti-rabbit or anti-sheep IGG conjugated to peroxidase (1:5000) for 2 hrs at room temperature. Reactive protein bands are detected using the Pierce Supersignal West Femto Kit. AB controls include use of the non-transfected CHO cells and blocking of the primary NK3-R AB with the appropriate peptide antigen (see Results). When a single blot was reacted with multiple ABs, complete stripping of the preceding AB was confirmed prior to application of the next AB.
6) Western blot analysis
Western blots were imaged using a Kodak Digital Science Image Station 440CF. Multiple exposures were obtained of each blot and densitometrically analyzed using Image J, the freely available NIH image analysis software (http://rsbweb.nih.gov/ij/download.html). Comparisons of protein expression in control and hydralzine treated rats were performed by determining the relative intensity of the NK3-R protein bands (area under the densitometry curve) and normalizing to either the cytoplasmic protein, CamK, or the nuclear protein, Pol II, as protein loading controls. After normalizing to CamK or Pol II, density of NK3-R reactive bands was compared to that in whole tissue SON to determine the relative expression level in different brain regions. To compare nuclear content of NK3-R protein between control and hydralazine-treated rats, total nuclear protein extracted from pooled bilateral SON dissections from individual rats were run on one gel and band density differences analyzed by Student’s T-test.
7) Co-immunoprecipitation was performed on whole tissue homogenates of rat cortex as described previously (Jensen et al., 2010). The sheep 2nd extracellular loop AB (711) was covalently conjugated to MagnaBind Carboxyl Derivatized Beads (Thermo Scientific, Rockford, IL) according to the manufacturers directions using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl as the cross-linker. Following incubation of the tissue homogenate with the beads, protein was eluted from the AB (Jensen et al., 2010) and analyzed by western blot using the rabbit N-terminal NK3-R AB (K2). Eluate from beads incubated in the absence of sample where negative for protein based on Coomassie staining. Thus, the antibody did not elute from the beads.
Results
NK3-R-AB specificity demonstrated with NK3-R transfected CHO cells
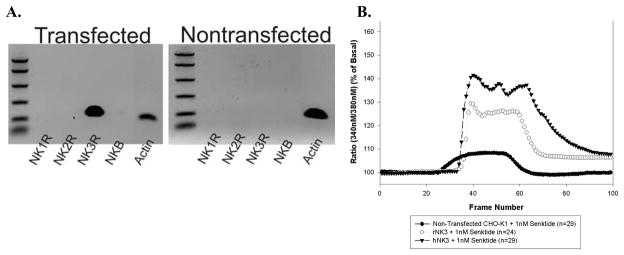
Expression of NK3-R in the transfected CHO lines was demonstrated using reverse transcriptase polymerase chain reaction (RT-PCR) and calcium imaging. As shown in Fig 1A, the NK3-R primers detected mRNA only in the cells transfected with rNK3-R. Calcium imaging studies with senktide (1 nM), an agonist for NK3-Rs confirmed the presence of functional NK3-R in the transfected lines (Fig 1B). Senktide induced a 130% and 140% increase in the 340:380 ratio in rNK3-R and hNK3-R CHO cells respectively that was blocked by the NK3-R antagonist, SB235375 [(Hay et al., 2002); gift from Glaxo Smith Kline Beecham (not shown)] and was significantly greater than observed in the non-transfected CHO cells in response to senktide (108% increase of 340:380 ratio).
Fig 1.
A. RNA expression profiles on the non-transfected and rat-NK3-transfected CHO cell lines. NK3-R RNA was only present in the rNK3-transfected cells. The CHO cells do not express the NK1R, the NK2R, or neurokinin B (NKB), the other members of the tachykinin receptor family. B. Changes in intracellular [Ca++] in non-transfected CHO cells or CHO cells transfected with the rat (rNK3) or human (hNK3) NK3-Rs in response to 1nM senktide. [Ca++]i was monitored with Fura-2 AM and the ratio of fluorescence emitted in response to excitation at 340 and 380 nM is presented. Senktide reached the cells at slightly different times in the 3 preparations (Frame 26 for non-transfected cells; Frame 32 for hNK3 cells; Frame 34 for rNK3 cells). Data represent the mean ratio of 340nm/380nm responses in 24–29 cells analyzed/culture.
The N-terminal AB (K2) labeled a diffuse protein band with a molecular weight ranging from ~65–85kD in membrane fractions from homogenates of SON and CHO cells transfected with the rat NK3-R (rNK3), but not in non-transfected CHO cells or CHO cells transfected with the human NK3-R (hNK3; Fig 2A). Differences in this peptide region between rat and human NK3 probably account for the rNK3 specificity to the AB (Shigemoto et al., 1990; Buell et al., 1992). Variable glycosylation probably accounts for both the diffuseness of the bands and the slight size difference between NK3-R in CHO cells transfected with rNK3 and SON (see below). As a further test of the specificity, the immunoreactivity was eliminated by preblocking the AB with the peptide sequence (Fig 2A, right side).
Fig 2.
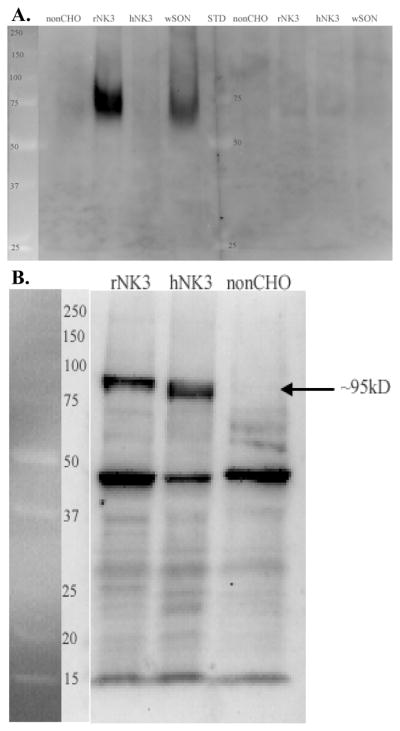
Western blot of SON and CHO cell homogenates reacted with the N-terminal AB (K2) or the 2nd extracellular loop AB (711). Size standards are shown and labeled (kD) on the left side of each blot. A. The N-terminal AB recognized broad protein bands ranging in size from ~65–75kD in SON and approximately 67–85kD in rat NK3-R transected CHO cells (rNK3). No bands were present in the non-transfected (non-CHO) and human NK3-R transfected CHO cells and both bands were absent when the AB was preblocked with the peptide used to generate the K2 AB (right side of figure). B. The 2nd extracellular loop AB labeled a prominent and distinct protein band ~90–95 kD in both rat and human transfected CHO cells (rNK3 and hNK3) but not in the non-transfected CHO cells. The AB also recognized a protein of ~47kD that is also prominent in the non-transfected CHO cells and therefore, not NK3-R.
The 2nd extracellular loop AB (711) labeled a distinct protein band with a molecular weight of ~90–95kD in membrane fractions from homogenates of CHO cells transfected with both rNK3 and hNK3, but this band was absent in the non-transfected CHO cells (Fig 2B). It also recognized a protein band of ~48kD, but this was not NK3-R specific as it was present in non-transfected CHO cells as well as the rNK3 and hNK3 CHO lines.
NK3-R expression in SON
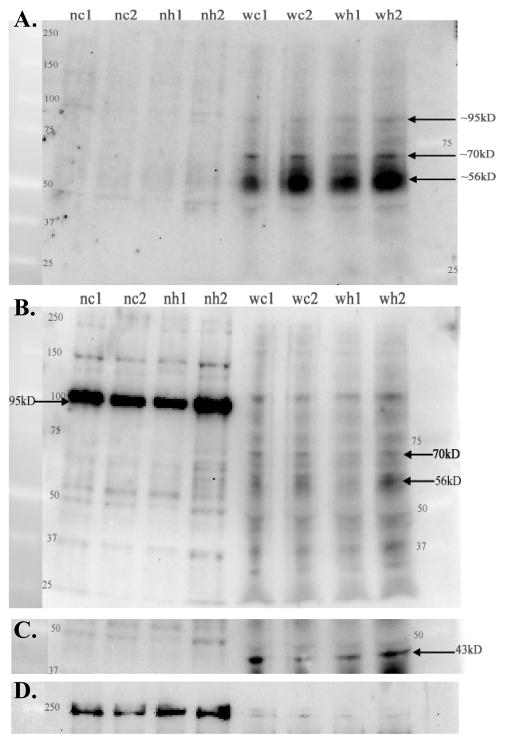
Proteins in whole tissue homogenates and nuclear fractions of microdissected SON from control and hydralazine treated rats were fractionated using SDS-PAGE and transferred to nitrocellulose (Figure 3). The resulting western blot was sequentially reacted with the N-terminal AB (K2), the 2nd extracellular loop AB (711), CamK AB, and Pol II AB with intervening stripping (Fig 3A–D). Complete stripping of the previous primary AB was verified prior to addition of the next AB. The N-terminal AB predominantly labeled a diffuse protein band of ~56–65kD in the whole tissue homogenate while the 2nd extracellular loop AB predominantly labeled a larger protein band in the nuclear fraction. Peptide preblocking of the ABs eliminated or markedly reduced the density of these bands (Fig 4A and B). The breadth of the diffuse (56–65kD) protein band recognized by the K2 AB was reduced to ~56–60kD by treatment with PGNaseF to remove N-linked glycosylation products (Fig 4C). This supports the interpretation that the diffuse (56–65kD) protein band is NK3-R (predicted size is 56kD) with variable glycosylation modifications. The protein identified predominantly in the nuclear fraction by the 2nd extracellular loop AB is larger (~95–100kD) possibly reflecting NK3R that is tightly associated with an importer or nuclear protein (Jensen et al., 2010). This interpretation is supported by the disappearance of this band following denaturation with 0.1mM DTT at 100°C for 40–60 min (Fig 4D). The failure of the same denaturating conditions to alter the ~70kD protein suggests that it represents native NK3-R folded in a configuration that slows its electrophoretic mobility in the gel (Fig 4D). Both ABs also reciprocally recognized the other protein although with much reduced affinity and therefore, fainter labeling (Fig 3A, B), and both ABs label a distinct ~70kD band that is present in whole tissue homogenates but not in nuclear fractions (Fig 3A, B). Co-immunoprecipitation demonstrated that the N-terminal and 2nd-extracellular loop ABs are immunoreactive for the the same proteins. As shown in Fig 4E, the 2nd extracellular loop AB (711) covalently linked to magnetic beads immunoprecipitated the N-terminal AB reactive ~95kD and 56kD proteins from a whole tissue homogenate. These data further support the interpretation gleaned from the CHO cell specificity data (Fig 1), that both ABs recognize NK3-R protein albeit with differing affinities depending on molecular modifications or binding partners. The absence of CamK-IR (43kD) in the nuclear fractions demonstrates that these fractions are free of cytoplasmic contaminants (Fig 3C). Pol II, the nuclear protein marker, was detectable in the whole tissue homogenate, but was greatly enriched (at least 36-fold based on densitometry) in the nuclear fractions (Fig 3D). Similarly, the ~95kD NK3-ir protein was greatly enriched in the nuclear fraction, but direct comparisons of the degree of enrichment of the ~95kD NK3 protein and Pol II in the nuclear fraction is not possible without knowing the affinity of the each AB for its ligand.
Figure 3.
Representative western blot of whole tissue homogenates (w) and nuclear fractions (n) of microdissected SON from control (c) and hydralazine (h) treated rats. 25 μg of protein was loaded per lane, separated using 7.5% SDS-PAGE, blotted to nitrocellulose and then sequentially reacted with K2, 711, CamK, and Pol II ABs with intervening stripping. A. The N-terminal AB (K2) labeled a prominent and diffuse protein band (~56–65kD), a distinct ~70kD protein, and faintly labeled a ~95kD protein in the whole tissue homogenate from both control and hydralazine treated rats. The 56–65kD and 95kD band are also faintly evident in nuclear fractions (also see Figs 6B and 7B of other brain regions where these bands are more distinct). B. The 2nd extracellular loop AB (711) also faintly labels the 56–65kD, 70kD, and 95–100kD bands in whole tissue homogenates, but the latter band shows a pronounced enrichment in nuclear fractions with this AB used at 2μg/ml. The 70kD band is absent in nuclear fractions with both ABs. The apparent slight difference in size of the ~95kD band in the nuclear and whole tissue preparations probably reflects the imperfection of the gel (e.g. the lanes ran slightly faster in the middle of the gel) and differences in the composition of the samples loaded on the gel (e.g. nuclear fraction vs straight homogenate). C. and D. The blot was then probed with ABs against CamK (C.) and Pol II (D.), the cytoplasmic and nuclear markers respectively, used to normalize protein loading for the whole tissue homogenate and nuclear fraction respectively. The absence of detectable CamK (43kD predicted size) in the nuclear fractions demonstrates low or absent cytoplasmic contamination. D. Pol II (240kD predicted size) was used as a nuclear protein marker. As expected, it is faintly present in the whole tissue preparation, and enriched (~36 fold) in the nuclear fraction.
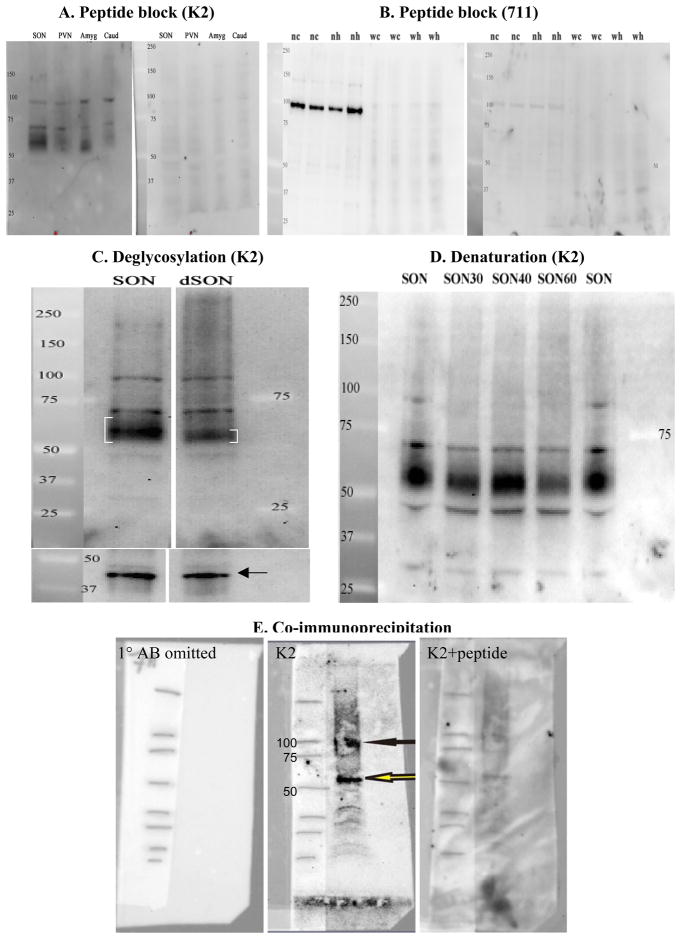
Figure 4.
Peptide blocking controls and effect of deglycosylation and denaturation. A. SDS-PAGE of duplicate lanes containing 25 ug of whole tissue homogenates of SON, PVN, amygdala (amyd), and caudate (Caud) were probed separately with the N-terminal (K2) antibody with (right half) or without (left half) peptide preblocking of the AB. Labeling of the 56–65kD, 70kD, and 95kD bands are evident in all lanes probed with the non-blocked AB, but not with the peptide-blocked AB. B. A blot containing 25μg of microdissected whole tissue homogenates (w) or nuclear fractions (n) of SON from control (c) or hydralazine-treated (h) rats was first probed with non-blocked 2nd exracellular loop (711 AB; 2 μg/ml; left side). Following stripping, the blot was reprobed with peptide-blocked AB (right side). Density of the prominent 95kD band evident in nuclear fractions is markedly reduced on the blot probed with peptide pre-absorbed AB. C. Comparison of NK3-R immunoreactivity in whole tissue homogenates of SON before (SON lane) and after incubation with PGNaseF to remove N-linked oligosaccharides (dSON lane). Note the ‘tightening’ (denoted by smaller brackets) of the diffuse 56–65kD seen in untreated SON (SON lane) following deglycosylation with PGNaseF (dSON lane). Following stripping, the blot was reprobed for the housekeeping protein, CamK, and demonstrated similar protein loading in the two lanes. These lanes were run on the same gel. A lane with lower protein loading was removed for clarity. D. Denaturing of the NK3-R positive ~95kD protein complex with DTT. The ~95kD band recognized by the K2 AB disappears from whole (w) tissue homogenates of SON during incubation with DTT at 100°C for 30, 40 or 60 minutes (SON30, SON40, SON60). In contrast, the ~70kD band withstands these denaturing conditions suggesting that it represents a stable configuration of native NK3-R. E. Co-immunoprecipitation of whole tissue homogenate from rat cortex. SDS-PAGE electrophoresis of eluate from magnetic beads covalently conjugated with 7ll AB (Jensen et al., 2010). Left panel: blot with primary AB omitted. The blot was reacted with 2ndary AB only. Middle panel: blot reacted with K2 AB. Right panel: blot reacted with peptide absorbed K2 AB. Solid black arrow indicates the higher molecular weight band that runs at ~95kD and the open arrow the native NK3 at 56kD. The absence of the 70kD band is likely due to ‘unfolding’ of this form during the co-IP protocol. These bands were almost eliminated by preincubating the K2 AB with its peptide (left panel). The high background is due to the necessity to increase the concentration of the secondary AB (1:2000) to visualize the K2 reactivity. Cortex was used in order to obtain sufficient protein for the co-IP.
Since hypotension was originally shown to increase NK3-R-IR in the nucleus (Howe et al., 2004), the effect of hydralazine-induced hypotension on NK3-R content was evaluated in whole tissue preparations and nuclear fractions of SON. Density of the 56–65kD, 70kD, and 95kD bands in Fig 3 as well as on additional blots (not shown) that contained whole tissue homogenates or nuclear fractions of SON and PVN from control and hydralazine-treated rats was determined and normalized to cytoplasmic or nuclear loading control proteins (CamK and Pol II respectively). Hydralazine-induced hypotension did not alter the density of the 56–65kD band detected with the N-terminal AB and normalized to CamK in whole tissue homogenates, but as shown in Fig 5, it did enhance the density of the ~95kD band detected by the 2nd extracellular loop AB in nuclear fractions of SON collected from animals exposed to 60 minutes of hydralazine-induced hypotension compared to SON nuclear fractions from control (normotensive) rats (Fig 5; t=3.78, p<0.01) when the AB concentration was reduced 100-fold to 0.02 μg/ml. Reducing the AB concentration was necessary to detect this increase, because the higher concentration of AB that was used in Fig 3B resulted in a maximal signal in nuclear fractions from normotensive control animals that obscured the hydralazine induced increase.
Figure 5.
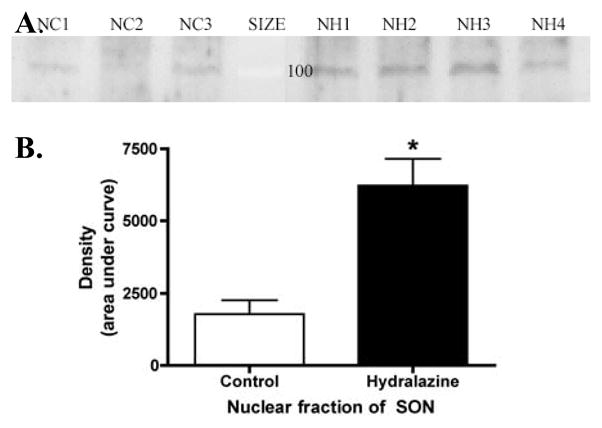
Effect of hydralazine treatment on nuclear content of NK3-R. A. Western blot of the nuclear (N) fraction of SON microdissected from individual control (C) or hydralazine (H) treated rats. All of the protein obtained from each rat was loaded in a single lane without correcting for protein content, because the individual microdissections contained a variable amount of the NK3-lacking tissue that surrounds SON. The blot was probed with a 100-fold lower concentration of the 2nd extracellular loop AB (711; 0.02 μg/ml) than was used in Fig 3B in order to detect increased protein content in the hydralazine-treated animals. Lanes NC1–3 were loaded with the total SON nuclear fraction obtained from control rats 1–3. Lanes NH1–4 were loaded with SON nuclear fraction from hydralazine treated rats. B. Densitometric analysis of the 95kD band demonstrated a significant increase in protein content following 60 minutes of hydralazine-induced hypotension (*p<0.05).
NK3-R expression in other brain regions
NK3-Rs are widely distributed in the brain (Stoessl and Hill, 1990; Stoessl, 1994; Ding et al., 1996; Shughrue et al., 1996; Yip and Chahl, 2000; Harrison et al., 2004), and intraventricular injection of senktide has been shown to induce fos expression in many of these areas (Smith and Flynn, 2000). As shown in Figures 6 and 7, we confirmed the expression of NK3-R in whole tissue and nuclear fractions obtained from punches of caudate/putamen, lateral septum, BNST, amygdala, medial preoptic area, PVN, substantia nigra, and ventral tegmental area using both the N-terminal and 2nd extracellular loop ABs.
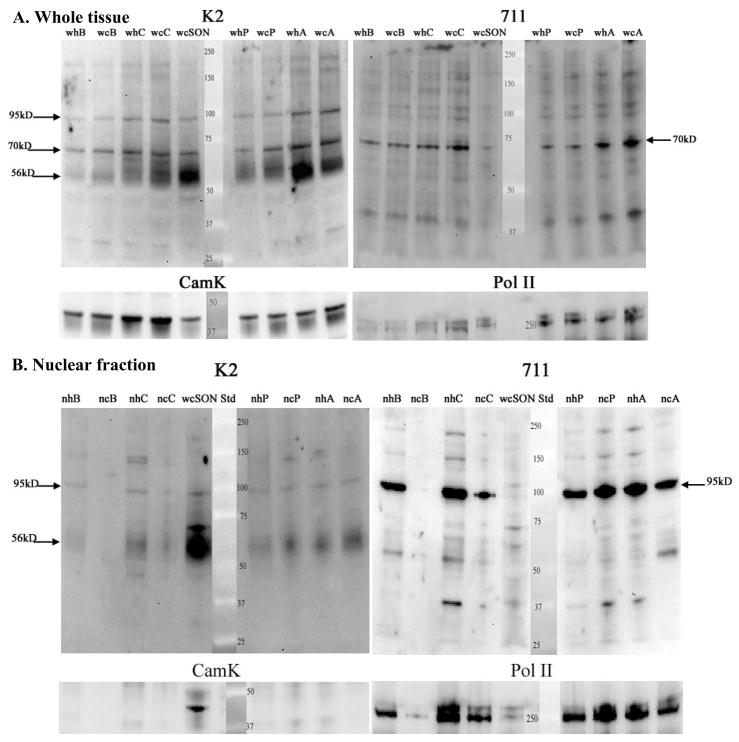
Figure 6.
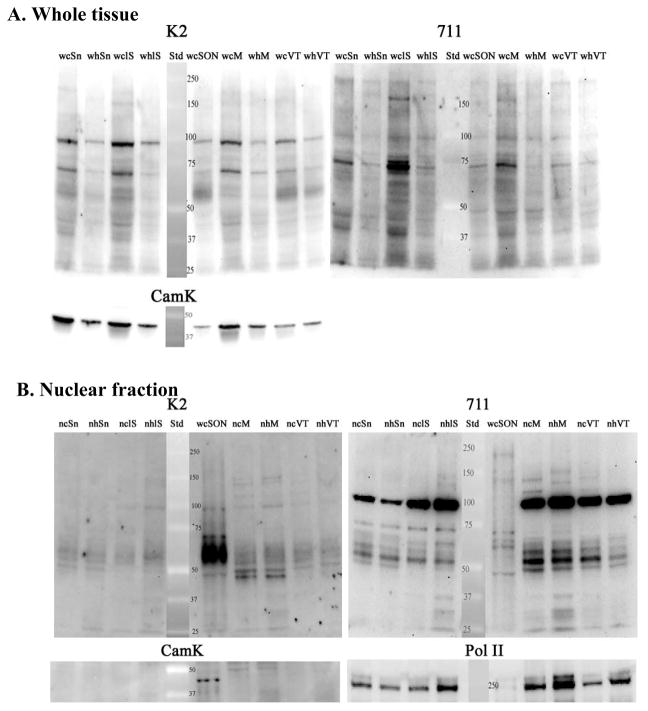
Comparison of whole tissue homogenates and nuclear fractions from tissue punches of BNST (B), Caudate (C), SON (S), PVN (P), amygdala (A). For the nuclear fraction, tissue punches were pooled from 3–4 animals. Gels were run, blotted and probed with ABs as in Figure 3 (e.g. sequential reaction with K2, 711, CamK, and Pol II ABs). A. Whole tissue homogenates probed with the N-terminal (K2) and 2nd extracellular loop (711) ABs. With the K2 AB, the 3 bands described in Fig 3 (56–65kD, 70kD, and 95kD) are evident in most brain regions, but regional differences in abundance are evident (see Fig 8). The 70kD band is prominently recognized by the 711 AB in whole tissue homogenates from most brain regions, but the affinity of this AB for the 56–65 kD band is much lower than the K2 AB. B. Nuclear fractions from the same regions as in Part A. Whole tissue homogenate of SON is included for comparison (wS; lane 4 in Fig 6B). The K2 AB detects a prominent 56–65kD band in nuclear fractions from BNST, Caudate, PVN, and Amygdala. This is not due to cytoplasmic contamination, because CamK is only evident in the whole tissue homogenate of SON. This supports the interpretation that the entire NK3 molecule translocates to the nucleus. The 711 AB shows enrichment of the 95kD band compared to blot in A. Pol II serves as loading control and demonstrates unintentional low loading of lane 2 as well as differential loading of lanes 3 and 4. Multiple samples of each brain region were prepared and all brain regions were analyzed on at least 2 blots.
Figure 7.
Comparison of whole tissue homogenates and nuclear fractions from tissue punches of substantia nigra (SN), lateral septum (LS), medial preoptic area (M), and ventral tegmental area (VT). As in Fig 6, tissue punches were pooled from 3–4 animals to obtain the nuclear fraction. Gels were run, blotted and probed as in Fig. 3 (e.g. sequential reaction with K2, 711, CamK, and Pol II ABs). A. Whole tissue homogenates probed with the N-terminal (K2) and 2nd extracellular loop (711) ABs. With the K2 AB the 3 bands seen in Figs 3 and 6 are evident in SN, LS, and VTA, but the 56–65kD band is low in MPOA compared to the fairly robust density of the 70kD and 95kd proteins. The 711 AB prominently labels the 70kD band in all of these brain regions, with differential expression reflecting differences in protein loading (see CamK blot). See Fig 8 for comparisons of relative abundance of NK3-ir bands across brain regions. B. Nuclear fractions from the same regions as in Part A. Whole tissue homogenate of SON is included for comparison (wS; lane 5). K2 AB: The 5665kD protein is prominent in all of these brain regions, and again, is not due to cytoplasmic contamination (see CamK picture). Again, this supports the interpretation that the entire NK3 molecule enters the nucleus in these brain regions. The 711 AB shows enrichment of the 95kD band. Pol II serves as loading control. Multiple samples of each brain region were prepared and all brain regions were analyzed on at least 2 blots.
In blots of whole tissue homogenates probed with the N-terminal AB (Figs 6A, 7A), all 3 of the NK3-R protein bands were more abundant in SON than in other brain regions (Fig 8A.). The glycosylated form of the receptor (e.g. the ~56–65kD protein band) was the most prominent form of NK3-R in most brain regions (e.g. SON, PVN, caudate, BNST, amygdala, and VTA (Fig 8B), but in lateral septum and MPOA, this form of NK3-R was low in spite of prominent ~70 and ~95kD protein bands (Fig 7A, Fig 8B). The ~70kD protein was evident in whole tissue preparations from all brain regions, and was detected by both the N-terminal and 2nd extracellular loop ABs. When corrected to CamK, it was most abundant in SON and VTA with lesser, but fairly uniform abundance in the other brain regions evaluated (Fig 8A). The ~95kD protein band was also distinctly labeled in whole tissue homogenates of all brain regions probed with both the N-terminal and the extracellular loop ABs (Figs 6A and 7A). These data demonstrate highest abundance of all 3 forms of the NK3-R in SON relative to the other brain areas examined, and also demonstrate that the predominant form of NK3-R differs across brain areas.
Figure 8.
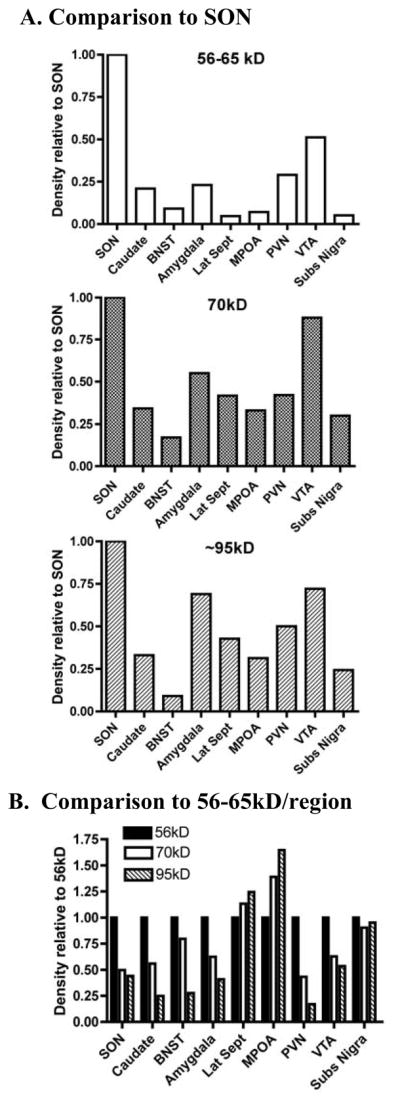
Densitometric analysis of blots shown in Figs 6A and 7A probed with the N-terminal (K2) AB. A. Comparison of the abundance of the 56–65kD, ~70kD, and ~95kD proteins in whole tissue homogenates from various brain regions to SON. Following normalization to CamK, each protein band was normalized to the density of that band in the SON lane on the same gel. All forms of NK3-R are more prominent in SON than any of the other brain regions analyzed. B. Comparison of the relative density of the ~70 and 95kD bands to the 56–65kD across brain regions. The 56–65kD form of NK3-R predominates in most brain regions, but in lateral septum and MPOA, the larger NK3-R forms predominate. The 3 forms of NK3-R are uniformly represented in substantia nigra.
In nuclear fractions from these same brain regions, the N-terminal AB labeled the 56–65kD protein in all brain regions analyzed (Figs 6B and 7B). This was in contrast to SON where the 56–65kD band was markedly less abundant in the nuclear fraction compared to the whole tissue homogenate (Fig 3A). As in SON, the 70kD band was not detected with either the N-terminal or the 2nd intracellular loop ABs in nuclear fractions from the other brain regions analyzed (Figs 6B and 7B). Both ABs recognized the 95kD band in the nuclear fraction, but the extracellular loop AB had much higher affinity for this form of NK3-R than the N-terminal AB. These observations demonstrate that the full length, glycosylated form of NK3-R can enter the nucleus, but the apparent 70kD NK3-R protein configuration does not enter the nucleus.
Following correction to the appropriate loading control, hydralazine did not consistently alter the abundance of any of the NK3 protein bands in brain regions other than SON.
Discussion
The current findings confirm that NK3-R translocates to the nucleus in SON and its translocation is activity induced. The results further demonstrate that: 1) the full length NK3-R molecule enters the nucleus; 2) NK3-R is glycosylated and also present in tight association with other cytosolic and nuclear proteins; 3) the predominant form of NK3-R differs between nuclear and non-nuclear compartments of the cell; 4) nuclear localization of NK3 occurs in multiple brain regions; and 5) the predominate form of NK3-R differs across brain regions.
Recognition of NK3-R-IR in the nucleoplasm of SON and PVN by ABs to the N-terminal region (K2), the 2nd extracellular loop (711), and the C-terminal tail [Novus Biologicals and K7 from J. Krause; (Howe et al., 2004; Haley and Flynn, 2006; Jensen et al., 2008)] demonstrates that the full-length NK3-R molecule enters the nucleus and provides strong evidence that the ABs are specific for NK3-R. The immunoblots presented here also demonstrate that both the N-terminal and 2nd extracellular loop ABs recognize NK3-R in multiple forms. The diffuse 56–65kD band represents NK3-R with variable N-linked glycosylation as evidenced by the band becoming less diffuse after incubation with PGNaseF to remove N-linked glycosylation moieties. The presence of NK3-R-IR associated with distinct protein bands larger than the predicted molecular weight for NK3-R (e.g. the 95kD bands) suggests that NK3-R remains tightly complexed with other proteins even under the denaturing conditions of SDS-PAGE. The presence of SDS-resistant protein complexes is consistent with the recognized role of chaperone proteins to mask hydrophobic regions of membrane proteins and thus maintain their solubility during trafficking between cellular compartments. Thus, although further work is required to identify the protein members of these NK3-R complexes, likely candidates include cytoplasmic heat shock proteins (Hsp) and nuclear histones. Members of the Hsp40 family have been identified as GPCR chaperone proteins involved in trafficking of GPCRs to the plasma membrane (Achour et al., 2008). The same or similar proteins may be involved in allowing NK3-R to exit the membrane and associate with importer proteins in order to enter the nucleus. In the nuclear compartment, NK3-R may be complexed with histones. In SON and PVN, immunoreactivity for NK3-R observed with either confocal or electron microscopy has a clustered appearance in the neuronal nuclei (Howe et al., 2004; Haley and Flynn, 2006; Jensen et al., 2008). The nuclear immunoreactivity for NK3-R reported in globus pallidus and VTA has a similar clustered appearance (Levesque et al., 2006; Lessard et al., 2009). This clustered appearance is consistent with localization of NK3-R to nuclear suborganelles some of which are rich in histones (Handwerger and Gall, 2006; Misteli, 2007). Histones form tetrameric complexes involved in DNA relaxation and transcription (Rouleau et al., 2004). Since the predicted size of a tetrameric histone/NK3-R complex is 96kD, enrichment of the ~95kD NK3-R protein complex in nuclear fractions is consistent with this being a histone/NK3-R complex. In contrast to enrichment of the ~95kD band in the nuclear fraction, the 70kD protein configuration is present in whole tissue homogenates, but not in the nucleus. The lower weight, glycosylated form of NK3-R is more prevalent in whole tissue homogenates than in nuclear fractions (e.g. Fig 3) and was the only form detected in membrane fractions (e.g. Fig 1).
Although the N-terminal and 2nd extracellular loop ABs both recognize the glycosylated and larger forms, they have differential affinity for these forms of the receptor. The N-terminal AB has greater affinity for the non-complexed, glycosylated form than the ~95kD complex while the 2nd-extracellular loop AB has greater affinity for the ~95kD nuclear complex than for the glycosylated form. This may reflect glycosylation- or protein complex-induced alterations in the configuration of the NK3-R molecule at the epitope region for each AB. The two ABs recognize the 70kD protein complex with similar affinity suggesting that the configuration and/or accessibility of both epitopes are maintained in this complex. The epitopes for the N-terminal and 2nd extracellular loop ABs are differentially obscured or recognizable in specific cell compartments: The N-terminal epitope is available in the 95kD complex in whole tissue homogenates of multiple brain regions, but in these same samples, the 2nd extracellular loop epitope is obscured (e.g. Figs 6A and 7A). In contrast, the 2nd extracellular loop epitope is strongly reactive in nuclear fractions, while the N-terminal epitope is only weakly available for AB binding (Figs 6B and 7B). These differences in receptor configuration may be required for or assist in directing the NK3-R functions specific to a given cellular compartment.
Confirmation that activation of NK3-R results in the appearance of NK3-R-IR in the nucleus in SON takes on even greater significance due to the wide distribution of NK3-R in the brain and the evidence that NK3-Rs are involved in a large number of important CNS functions. Specifically, NK3-R has been demonstrated in the retina, amygdala, ventral tegmental area (VTA), substantia nigra compacta (SNc), and nucleus tractus solitarius (NTS) as well as other regions (Stoessl and Hill, 1990; Stoessl, 1994; Ding et al., 1996; Shughrue et al., 1996; Yip and Chahl, 2000; Harrison et al., 2004). Thus, NK3-Rs are expressed in regions involved in mood disorders and motivated, compulsive, and addictive behaviors (amygdala and VTA). NK3-R agonists have been shown to suppress salt appetite and alcohol intake (Massi et al., 1990; Ciccocioppo et al., 1998; Flynn et al., 1999), and NK3-R antagonists produced anxiolytic and antidepressive reactions in gerbils (Salome et al., 2006). NK3-R antagonists blocked cocaine induced locomotor activity and anxiety behaviors in marmosets (De Souza Silva et al., 2006) while an NK3 agonist injected into SNc and VTA increased locomotor activity and rearing behavior in rats (Stoessl et al., 1991). Altered NK3-R expression occurs in a strain of hamsters with dystonia (Friedman et al., 2002), and a NK3-R antagonist reduces blood pressure in spontaneously hypertensive rats (Lessard et al., 2003). Nuclear NK3-R-IR has recently been shown ultrastructurally in globus pallidus and VTA neurons (Levesque et al., 2006; Lessard et al., 2009). In the current studies, NK3-R was detected in nuclear fractions from all brain areas examined. Since chronic alterations in responses characterize pathology of many of these CNS functions, altered gene expression is an attractive underlying mechanism, and therefore, the possibility that NK3-R activation directly regulates gene expression may be significant to understanding multiple brain functions and pathologies. Further work is required to determine if physiological perturbations alter nuclear NK3-R expression in other brain regions. Since the functions of these other brain areas are diverse, it is anticipated that the physiological signals initiating nuclear translocation in the various brain regions will be similarly diverse.
While a variety of signals may initiate the nuclear translocation of NK3-R, the final step is likely to involve docking with the importins (Jensen et al., 2010). This leaves the question of how the hydrophobic NK3-R protein is able to move to the cytoplasm to associate with importin. In some cases, such as with the epidermal growth factor receptor, ErbB-4, the protein is cleaved to generate a soluble, cytosolic fragment (Ni et al., 2001). This model would not appear to apply to NK3-R because the present data show that the full length NK3-R protein enters the nucleus. Rather, trafficking of the NK3-R may be similar to that for the fibroblast growth factor 1 receptor (FGFR1) and epidermal growth factor receptors: EGFR or ErbB-1, and ErbB-3. These receptors are translocated to the nucleus in their intact, full length form (Stachowiak et al., 1996; Lin et al., 2001; Offterdinger et al., 2002). In the case of EGFR it is internalized to the cytoplasm after ligand binding and trafficked to the endoplasmic reticulum (ER). Once in the ER the receptor is extracted from the lipid bilayer and moved to the cytosol by associating with the ERAD protein, Sec61 translocon (Liao and Carpenter, 2007). The model assumes that once retrotranslocated to the cytoplasm the receptor protein associates with chaperone proteins and/or importins that mask the hydrophobic region of the protein and enable transport into the nucleus (Reilly and Maher, 2001; Liao and Carpenter, 2007). Previous work shows that NK3-R associates with importin-B and this step is required for transport into the nucleus (Jensen et al., 2010). The full length NK3-R could be transported into the nucleus in a process like that described for EGFR.
The three forms of the NK3-R found in SON are present in the other brain regions examined. However, the relative expression of the three forms of NK3-R differs between regions. Specifically, in whole tissue homogenates, the glycosylated form is higher in SON than any other region, although it is also more prominent in amygdala and VTA than other areas and is the lowest form present in MPOA. The N-terminal AB recognizes the glycosylated form in nuclear fractions from several brain regions, but as in SON, the 70kD complex appears to be limited to non-nuclear compartments and although the 95kD complex recognized by the N-terminal AB is evident in several brain regions, the form recognized by the 2nd extracellular loop AB is markedly enriched in nuclear fractions from all areas. The differences in the predominant form of NK3-R detected in various brain regions suggests that NK3-R localization may be differentially regulated and therefore sub serve different primary functions depending on the region studied. Further studies are warranted to determine if the form of NK3-R in a particular region corresponds to differences in the functional roles of NK3-R.
Other membrane receptors including the GPCRs previously mentioned and several tyrosine kinase receptors (Kiefer et al., 1994; Stachowiak et al., 1996; Reilly and Maher, 2001) and cytokine receptors (Rycyzyn et al., 2000; Xiao et al., 2000; Zwaagstra et al., 2000; Jelaso and DeLong, 2005) have also been reported to localize to the nucleoplasm suggesting that information gained about nuclear localization and function of NK3-R may have broad physiological relevance. Nucleoplasm localization of the other GPCRs is cell type dependent, varies relative to its ligand-dependency, and in some cases has only been demonstrated in cultured cells or following transfection. To the best of our knowledge, nuclear localization of the NK3-R is the only example of ligand-initiated nuclear translocation in vivo.
Highlights.
NK3-R translocation to the nucleus is activity induced in SON
The full length NK3-R molecule enters the nucleus
NK3-R is glycosylated and tightly associated with cytosolic and nuclear proteins
Nuclear localization of NK3 occurs in multiple brain regions
The predominate form of NK3-R differs across brain regions.
Acknowledgments
Supported by NIH grants R21-NS059569 to CDS and RO1-NS57823 and P20-RR15640 to FWF.
Abbreviations
- AB
antibody
- BNST
bed nucleus of stria terminalis
- c
control
- C
caudate
- CamK
calcium calmodulin kinase
- h
hydralazine
- IR
immunoreactivity
- LS
lateral septum
- MPOA
medial preoptic area
- n
nuclear fraction
- NK3-R
neurokinin 3 receptor
- Pol II
RNA polymerase II
- PVN
hypothalamic paraventricular nucleus
- SN
substantia nigra
- SON
supraoptic nucleus
- VTA
ventral tegmental area
- w
whole tissue homogentate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achour L, Labbe-Jullie C, Scott MG, Marullo S. An escort for GPCRs: implications for regulation of receptor density at the cell surface. Trends Pharmacol Sci. 2008;29:528–535. doi: 10.1016/j.tips.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Buell G, Schulz MF, Arkinstall SJ, Mauary K, Missotten M, Adami N, Talabot F, Kawashima E. Molecular characterisation, expression and localisation of human neurokinin-3 receptor. FEBS Letters. 1992;299:90–95. doi: 10.1016/0014-5793(92)80107-r. [DOI] [PubMed] [Google Scholar]
- Ciccocioppo R, Panocka I, Polidori C, Froldi R, Angeletti S, Massi M. Mechanism of action for reduction of ethanol intake in rats by the tachykinin NK-3 receptor agonist aminosenktide. Pharmacol Biochem Behav. 1998;61:459–464. doi: 10.1016/s0091-3057(98)00090-2. [DOI] [PubMed] [Google Scholar]
- De Souza Silva MA, Mello EL, Jr, Muller CP, Jocham G, Maior RS, Huston JP, Tomaz C, Barros M. The tachykinin NK3 receptor antagonist SR142801 blocks the behavioral effects of cocaine in marmoset monkeys. Eur J Pharmacol. 2006;536:269–278. doi: 10.1016/j.ejphar.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Ding Y-Q, Shigemoto R, Takada M, Ohishi H, Nakanishi S, Mizuno N. Localization of the neuromedin K receptor (NK3) in the central nervous system of the rat. J Comp Neurol. 1996;364:290–310. doi: 10.1002/(SICI)1096-9861(19960108)364:2<290::AID-CNE8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Flynn FW, Smith ME, Beiber SL. Differential effects of intraventricular injections of Tachykinin NK1 and NK3 receptor agonists on normal and sham drinking of NaCl by sodium-deficient rats. BehavNeurosci. 1999;13:776–786. [PubMed] [Google Scholar]
- Friedman Y, Richter A, Raymond R, Loscher W, Nobrega JN. Regional decreases in NK-3, but not NK-1 tachykinin receptor binding in dystonic hamster (dt(sz)) brains. Neuroscience. 2002;112:639–645. doi: 10.1016/s0306-4522(02)00103-3. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved flourescence properties. JBiolChem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Haley GE, Flynn FW. Agonist and hypertonic saline-induced trafficking of the NK3-receptors on vasopressin neurons within the paraventricular nucleus of the hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1242–1250. doi: 10.1152/ajpregu.00773.2005. [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Harrison TA, Hoover DB, King MS. Distinct regional distributions of NK1 and NK3 neurokinin receptor immunoreactivity in rat brainstem gustatory centers. Brain Res Bull. 2004;63:7–17. doi: 10.1016/j.brainresbull.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Hay DWP, Giardina GAM, Griswold DE, Underwood DC, Kotzer C, Bush B, Potts W, Sandhu P, Lundberg D, Foley JJ, Schmidt DB, Martin LD, Kilian D, Legos JJ, Barone FC, Luttmann MA, Grugni M, Raveglia LF, Sarau HM. Nonpeptide tachykinin receptor antagonists. III. SB 235375, a low central nervous system-penetrant, potent and selective neurokinin-3 receptor antagonist, inhibits citric acid-induced cough and airways hyper-reactivity in guinea pigs. JPharmacolExpTher. 2002;300:314–323. doi: 10.1124/jpet.300.1.314. [DOI] [PubMed] [Google Scholar]
- Howe HE, Somponpun SJ, Sladek CD. Role of neurokinin 3 receptors in supraoptic vasopressin and oxytocin neurons. J Neurosci. 2004;24:10103–10110. doi: 10.1523/JNEUROSCI.3164-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans DA, Hassan G. Nuclear targeting by growth factors, cytokines, and their receptors: a role in signaling? Bioessays. 1998;20:400–411. doi: 10.1002/(SICI)1521-1878(199805)20:5<400::AID-BIES7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Jans DA, Hubner S. Regulation of protein transport to the nucleus: Central role of phosphorylation. PhysiolRev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- Jelaso AM, DeLong C. NGF and IL-1beta are co-localized in the developing nervous system of the frog, Xenopus laevis. Int J Dev Neurosci. 2005;23:575–586. doi: 10.1016/j.ijdevneu.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Jensen D, Zhang Z, Flynn FW. Trafficking of tachykinin neurokinin 3 receptor to nuclei of neurons in the paraventricular nucleus of the hypothalamus following osmotic challenge. Neuroscience. 2008;155:308–316. doi: 10.1016/j.neuroscience.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DD, Sundstrom K, Flynn FW. Expression of the nuclear transport protein importin ss-1 and its association with the neurokinin 3 receptor in the rat hypothalamus following acute hyperosmotic challenge. Neuroscience. 2010;170:1020–1027. doi: 10.1016/j.neuroscience.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer P, Acland P, Pappin D, Peters G, Dickson C. Competition between nuclear localization and secretory signals determines the subcellular fate of a single CUG-initiated form of FGF3. EMBO J. 1994;13:4126–4136. doi: 10.1002/j.1460-2075.1994.tb06730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey CG, Bussolati G, Bosco M, Kimura T, Pizzorno MD, Chernin MI, Cassoni P, Novak JF. Constitutive and ligand-induced nuclear localization of oxytocin receptor. J Cell Mol Med. 2007;11:96–110. doi: 10.1111/j.1582-4934.2007.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Lanca J, Cheng R, Nguyen T, Ji XD, Gobeil F, Jr, Chemtob S, George SR, O’Dowd BF. Agonist-independent nuclear localization of the apelin, angiotensin AT1, and bradykinin B2 receptors. JBiolChem. 2004;279:7901–7908. doi: 10.1074/jbc.M306377200. [DOI] [PubMed] [Google Scholar]
- Lessard A, Campos MM, Neugebauer W, Couture R. Implication of nigral tachykinin NK3 receptors in the maintenance of hypertension in spontaneously hypertensive rats: a pharmacologic and autoradiographic study. Br J Pharmacol. 2003;138:554–563. doi: 10.1038/sj.bjp.0705042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard A, Savard M, Gobeil F, Jr, Pierce JP, Pickel VM. The neurokinin-3 (NK3) and the neurokinin-1 (NK1) receptors are differentially targeted to mesocortical and mesolimbic projection neurons and to neuronal nuclei in the rat ventral tegmental area. Synapse. 2009;63:484–501. doi: 10.1002/syn.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M, Parent R, Parent A. Cellular and subcellular localization of neurokinin-1 and neurokinin-3 receptors in primate globus pallidus. Eur J Neurosci. 2006;23:2760–2772. doi: 10.1111/j.1460-9568.2006.04800.x. [DOI] [PubMed] [Google Scholar]
- Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–1072. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Massi M, Gentili L, Perfumi M, de Caro G, Schulkin J. Inhibition of salt appetite in the rat following injection of tachykinins into the medial amygdala. Brain Res. 1990;513:1–7. doi: 10.1016/0006-8993(90)91082-r. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Offterdinger M, Schofer C, Weipoltshammer K, Grunt TW. c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Steriotaxic Coordinates. New York, N.Y: Academic Press, Inc; 1982. p. 10003. [Google Scholar]
- Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Aubin RA, Poirier GG. Poly(ADP-ribosyl)ated chromatin domains: access granted. J Cell Sci. 2004;117:815–825. doi: 10.1242/jcs.01080. [DOI] [PubMed] [Google Scholar]
- Rycyzyn MA, Reilly SC, O’Malley K, Clevenger CV. Role of cyclophilin B in prolactin signal transduction and nuclear retrotranslocation. Mol Endocrinol. 2000;14:1175–1186. doi: 10.1210/mend.14.8.0508. [DOI] [PubMed] [Google Scholar]
- Salome N, Stemmelin J, Cohen C, Griebel G. Selective blockade of NK2 or NK3 receptors produces anxiolytic- and antidepressant-like effects in gerbils. Pharmacol Biochem Behav. 2006;83:533–539. doi: 10.1016/j.pbb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Shigemoto R, Yokota Y, Tsuchida K, Nakanishi S. Cloning and expression of a rat neuromedin K receptor cDNAs. JBiolChem. 1990;265:623–628. [PubMed] [Google Scholar]
- Shughrue PJ, Lane MV, Merchenthaler I. In situ hybridization analysis of the distribution of neurokinin-3 mRNA in the rat central nervous system. J Comp Neurol. 1996;372:395–414. doi: 10.1002/(SICI)1096-9861(19960826)372:3<395::AID-CNE5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Smith ME, Flynn FW. Distribution of Fos-like immunoreactivity within the rat brain following intraventricular injection of the selective NK3 receptor agonist, senktide. JCompNeurol. 2000;426:413–428. doi: 10.1002/1096-9861(20001023)426:3<413::aid-cne6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Song Z, Vijayaraghavan S, Sladek CD. Simultaneous exposure to ATP and phenylephrine induces a sustained elevation in the intracellular calcium concentration in supraoptic neurons. Am J Physiol Regul Integr Comp Physiol. 2006;291:R37–R45. doi: 10.1152/ajpregu.00718.2005. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK. Nuclear localization of functional FGF receptor 1 in human astrocytes suggests a novel mechanism for growth factor action. Brain Res Mol Brain Res. 1996;38:161–165. doi: 10.1016/0169-328x(96)00010-1. [DOI] [PubMed] [Google Scholar]
- Stoessl AJ. Localization of striatal and nigral tachykinin receptors in the rat. Brain Res. 1994;646:13–18. doi: 10.1016/0006-8993(94)90052-3. [DOI] [PubMed] [Google Scholar]
- Stoessl AJ, Hill DR. Autoradiographic visualization of NK-3 tachykinin binding sites in the rat brain, utilizing [3H]senktide. Brain Res. 1990;534:1–7. doi: 10.1016/0006-8993(90)90105-k. [DOI] [PubMed] [Google Scholar]
- Stoessl AJ, Szczutkowski E, Glenn B, Watson I. Behavioural effects of selective tachykinin agonists in midbrain dopamine regions. Brain Res. 1991;565:254–262. doi: 10.1016/0006-8993(91)91657-m. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Takahashi S, Zilliacus J. Interaction of the parathyroid hormone receptor with the 14-3-3 protein. Biochim Biophys Acta. 2003;1620:32–38. doi: 10.1016/s0304-4165(02)00503-2. [DOI] [PubMed] [Google Scholar]
- Wells A, Marti U. Signalling shortcuts: cell-surface receptors in the nucleus. Nature RevMol Cell Biol. 2002;3:1–6. doi: 10.1038/nrm905. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of Smad 3. J Biol Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- Yip J, Chahl LA. Localization of tachykinin receptors and Fos-like immunoreactivity induced by substance P in guinea-pig brain. Clin Exp Pharmacol Physiol. 2000;27:943–946. doi: 10.1046/j.1440-1681.2000.03366.x. [DOI] [PubMed] [Google Scholar]
- Zwaagstra JC, Guimond A, O’Connor-McCourt MD. Predominant intracellular localization of the type I transforming growth factor-beta receptor and increased nuclear accumulation after growth arrest. Exp Cell Res. 2000;258:121–134. doi: 10.1006/excr.2000.4905. [DOI] [PubMed] [Google Scholar]