Abstract
Coactivators are a diverse group of non-DNA binding proteins that induce structural changes in agonist-bound nuclear receptors (NRs) that are essential for NR-mediated transcriptional activation. Once bound, coactivators function to bridge enhancer binding proteins to the general transcription machinery, as well as to recruit secondary coactivators that modify promoter and enhancer chromatin in a manner permissive for transcriptional activation. In the following review article, we focus on one of the most in-depth studied families of coactivators, the steroid receptor coactivators (SRC) 1, 2, and 3. SRCs are widely implicated in NR-mediated diseases, especially in cancers, with the majority of studies focused on their roles in breast cancer. We highlight the relevant literature supporting the oncogenic activity of SRCs and their future as diagnostic and prognostic indicators. With much interest in the development of selective receptor modulators (SRMs), we focus on how these coactivators regulate the interactions between SRMs and their respective NRs; and, importantly, the influence that coactivators have on the functional output of SRMs. Furthermore, we speculate that coactivator-specific inhibitors could provide powerful, all-encompassing treatments that target multiple modes of oncogenic regulation in cancers resistant to typical anti-endocrine treatments.
Keywords: coactivators, nuclear receptors, cancer, steroid receptor modulators
1. Introduction to NR coactivators
The existence of coactivators was first speculated from early experiments that revealed ligand-bound NRs, alone, may not be sufficient to communicate with the general transcription machinery and induce transcriptional activation (Kim, 2008; Klein-Hitpass et al., 1990; Meyer et al., 1989). NRs, in general, are a large superfamily of proteins that bind as homo- or heterodimers to specific DNA elements in order to elicit transcriptional activation of target genes. Steroid nuclear receptors, members of class I NRs, are specifically recruited to gene promoters upon the binding of a high-affinity ligand to the respective NR, which induces conformational changes in the NR essential for its activity. However, in vitro transcription experiments using only purified NRs and basal transcription factors could not induce transcriptional activation on their own (Kim, 2008; Klein-Hitpass et al., 1990). Additionally, the fact that overexpression of one NR could inhibit the transactivation function of another NR indicated that multiple NRs may compete for essential factors (Meyer et al., 1989), which are now termed coactivators. The first coactivator, steroid receptor coactivator 1 (SRC-1), was identified and cloned in our laboratory in 1995 (Onate et al., 1995). SRC-1 overexpression enhances ligand-induced transcriptional activation by progesterone receptor (PR), estrogen receptor α (ERα), glucocorticoid receptor (GR), thyroid receptor (TR), and retinoid X receptor (RXR). Importantly, overexpression of SRC-1 overcomes ERα-induced squelching of PR.
In addition to SRC-1, over 300 coactivators have now been identified and are implicated in a wide-range of human diseases (Lanz, 2008; Xu et al., 2009; Yan J., 2008). Coactivators are strictly defined by their lack of DNA binding, differentiating coactivators from classic transcription factors. Initially, coactivators were defined as molecules that simply bridge NRs to the general transcription machinery. While this is a fundamental role of coactivators, they also modify chromatin within promoter and enhancer regions or recruit secondary coactivators (co-coactivators) that modify the chromatin in a manner that supports binding of enhancer regulatory proteins and general transcription factors (Figure 1), such as through histone acetylation and specific sites of histone methylation. These modifications are well-known to be associated with active transcription (Johnson and Barton, 2007). Moreover, recruited co-coactivators mediate all substeps of transcription, including elongation, RNA splicing, and termination (Lonard and O’Malley B, 2007).
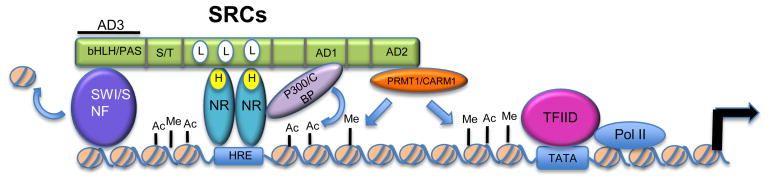
Figure 1. SRC-mediated coactivation of NRs.
SRC proteins are recruited to hormone bound NRs and bind through their LXXLL motifs, of which they have three. SRCs then recruit multiple secondary coactivator complexes that bind to their three activation domains (ADs). Three examples are shown: histone acetyltransferase, p300/CBP; histone methyltransferases, PRMT1 and CARM1; and chromatin remodeling complex, SWI/SNF. These secondary coactivators modify the chromatin and bridge the NR complex with the general transcription machinery to elicit transcriptional activation. SRCs (steroid receptor coactivators); bHLH/PAS (basic helix-loop-helix/Per-Arnt-Sim); S/T (serine/threonine –rich region); NR (nuclear receptor); Ac (acetylation); Me (methylation); HRE (hormone response element); L (LXXLL motifs).
True to the basis of Newton s 3rd law of motion, “for every action there is an equal and opposite reaction”, molecular counterparts to coactivators have been identified and coined corepressors. In contrast to coactivators, corepressors function by altering the chromatin structure of the promoter towards an inactive state. For example, corepressors SMRT (silencing mediator of retinoid and thyroid receptors) and NCOR (nuclear receptor corepressor) recruit and activate histone deacetylases, which orchestrate a transcriptionally repressive chromatin configuration [12, 13]. Corepressors were first discovered as regulators of class II NRs, such as thyroid hormone receptor (TR), peroxisome proliferator activated receptor (PPAR), and liver X receptor (LXR) (Baniahmad et al., 1995; Pace, 2008). These NRs constitutively bind DNA as a heterodimer with retinoid X receptor (RXR), and, in the absence of ligand, are bound by corepressors that actively inhibit transcription. The addition of ligand signals for a switch of corepressors for coactivators (Baniahmad et al., 1995; Glass and Rosenfeld, 2000). Thus, coactivators are essential for the transactivation function of NRs that are both recruited and constitutively bound to promoter and enhancer DNA.
The classification of coregulators into coactivators or corepressors is based on the general observations of their activity; however, it should be noted that in some instances coactivators can repress transcription and corepressors can activate transcription (Pace, 2008). For example, the coactivator SRC-2 was shown to function as a corepressor at the ERα-repressed TNFα promoter (Cvoro et al., 2006), and the corepressor SMRT coactivated TRα-driven transcription at a negative thyroid response element (Berghagen et al., 2002) and was found to be essential for full ERα transcriptional activity (Peterson et al., 2007). There are many plausible explanations for bimodal functions of coregulators, including the promoter context, which may alter the orientation of the NR-coregulator complex, influencing function. For example, SRC-2 activates GR-directed transcription when GR is bound directly to glucocorticoid response elements, but it represses GR tethered to DNA via activating protein 1 (AP-1) (Rogatsky et al., 2002).
2. Structural changes induced by coactivator-NR interactions
Unlike NRs, which have conserved structural domains, there are not universal structural motifs that define NR coactivators. NRs share structural homology in their central DNA binding domain and in their two transactivation domains (AF1 and AF2) (Nettles and Greene, 2005). AF1 is located within the N-terminal domain of NRs and activates transcription independent of ligand when the region is isolated, while AF2 is housed within the ligand-binding domain (LBD) of NRs that lies at the C-terminus. As its location indicates, AF2 activity is ligand-dependent. Crystal structure analyses of apo- and ligand-bound NRs have elucidated the conformational changes that occur upon ligand binding, which are essential for NR-mediated transcriptional ativation. The first crystal structure of an NR LBD was generated for apo-RXR and revealed 12 α-helices and a short β-turn (Bourguet et al., 2000). In the unliganded form, helices 10 and 11 are perpendicular to one another, jutting helix 11 inwards towards the ligand-binding pocket (LBP). Hydrophobic residues of helix 11 fill in the LBP and stabilize it. However, when ligand is present, helix 11 swings out and is in line with helix 10, opening up the hydrophobic LBP for the ligand to bind. Helix 12, which contains the AF2 domain, then folds inwards over the LBP (Bourguet et al., 2000; Wurtz et al., 1996). This conformational change is essential for ligand-induced transactivation function and coactivator recruitment.
While coactivators are a much more diverse group of proteins than NRs and lack such uniform structure, a motif within several coactivators, including the SRC family of coactivators, was identified by Heery et al. (Heery et al., 1997) and Torchia et al. (Torchia et al., 1997). This motif binds to the hydrophobic pocket created within helix 12 of the LBD upon ligand binding and is termed the LXXLL motif or NR box. Mutation of these residues inhibits the binding of SRC-1 to the ERα LBD in vitro and SRC-1-mediated activation of ERα in vivo (Heery et al., 1997). Several NRs have been cocrystalized with their cognate ligand and a short peptide comprising the LXXLL motif within the nuclear receptor interaction domain of coactivators, and they revealed that the peptide is stabilized by interactions between the leucine residues of the LXXLL motif and the hydrophobic groove of the LBP, as well as by hydrogen bonds between a lysine within helix 3 and a glutamate within the LXXLL motif located on helix 12 (Bourguet et al., 2000; Darimont et al., 1998; Gampe et al., 2000; Nolte et al., 1998; Shiau et al., 1998). A recently reported analysis of nuclear receptor coregulator motifs revealed that of 303 coregulators, 149 have at least one LXXLL motif; thus, while it is a common motif amongst coregulators, it is not universal (Lanz, 2008).
3. Steroid Receptor Coactivator (p160) family
3.1. Structural domains of SRC (p160) proteins
As mentioned previously, SRC-1 was the first identified NR coactivator and is the founding member of the SRC family. It was discovered in a yeast two-hybrid screen as a protein that interacts with the PR LBD (Onate et al., 1995). SRC-2, also known as GRIP1 (glucocorticoid receptor interacting protein 1) and TIF-2 (transcriptional intermediary factor-2), was found shortly afterwards as an interacting protein with various NRs (Hong et al., 1997; Hong et al., 1996; Voegel et al., 1996). SRC-3 was cloned by several different laboratories and, thus, has many other names, as well: p/CIP (p300/CBP cointegrator associated protein), RAC3 (RAR-associated coactivator 3), ACTR (activator of thyroid and retinoic acid receptor), AIB1 (amplified in breast cancer 1), and TRAM1 (thyroid receptor activator molecule 1) (Anzick et al., 1997; Chen et al., 1997; Li et al., 1997; Takeshita et al., 1997; Torchia et al., 1997). The SRC proteins are all approximately 160kDa in size and share 50–55% sequence similarity, with several structural domains conserved (Kim, 2008).
The N-terminus of SRCs contains a bHLH-PAS motif (basic helix loop helix- Per Arnt Sims) and is the most conserved domain within the family of proteins (75% similarity and 60% identity) (Kim, 2008). It is involved in several protein-protein interactions that recruit co-coactivators, such as CoCoA (coiled-coil coactivator), a protein that acts synergistically with CARM1 (coactivator-associated arginine methyltrasferase 1) and p300 to maximize transcriptional activation by NRs (Kim et al., 2003). The bHLH-PAS domain also contains a bipartite nuclear localization signal and is essential for proteasome-dependent turnover of the coactivators. Mutation of two key residues, K17 and K18, within this region prevented nuclear localization and showed decreased protein degradation in the presence of cyclohexamide (Li et al., 2007). The central region contains the nuclear receptor interaction domain (NRID). SRCs contain three α-helical LXXLL motifs essential for their interaction with NRs, and the sequences flanking these motifs are important for NR selectivity (Chang et al., 1999; Coulthard et al., 2003). The C-terminal half includes two activation domains: AD1 and AD2. Both of these ADs function by recruiting co-coactivators to promoter DNA that remodel and modify the chromatin in a manner permissive to active transcription (Kim, 2008; Xu et al., 2009). Figure 2 depicts some of the co-coactivators that interact with these regions; however, published emphasis has been granted to p300/CBP, which interacts with the AD1 domain, and CARM1 and PRMT1 (protein arginine N-methyltransferase 1), which interact with the AD2 domain. The C-terminal domain of SRC-1 and SRC-3 also contains weak HAT activity; however, the importance of this activity and its substrates are yet to be identified (Chen et al., 1997; Spencer et al., 1997).
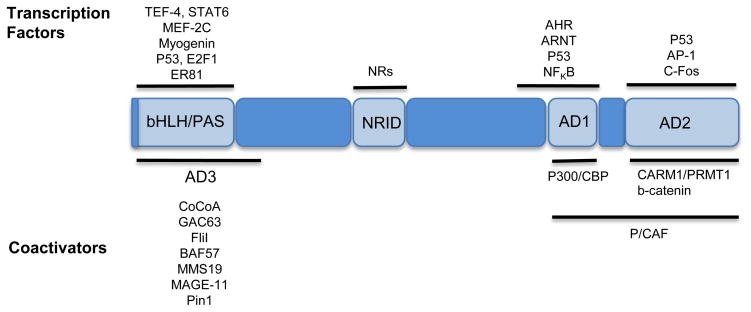
Figure 2. SRC-interacting proteins.
SRCs coactivate nuclear receptors (NRs), as well as numerous transcription factors. Once tethered to chromatin via these interactions, SRCs recruit a number of secondary coactivators that interact with its activation domains (ADs). This is a representative list of just some of SRC s interacting proteins, of which SRC-interacting domains have been mapped. Proteins are referenced and adapted from (Kim, 2008) (Yan J., 2008) (Xu et al., 2009) and as mentioned throughout the text.
3.2. Functions of SRCs
3.2.1. Recruitment of co-coactivators to the AD1 domain
NR-mediated gene transcription is an ordered, step-wise assembly of transcription factors and coactivators. Metivier et al. described the ordered recruitment of factors on the ERα gene target, pS2, in MCF7 cells, which occurs in a cyclical manner of gene transcription (Metivier et al., 2003). First, ERα binds in the presence of estrogens to estrogen response elements (EREs). Next, a host of coactivator protein complexes are recruited that function to modify and remodel chromatin and bridge the NR complex to the general transcription apparatus. SRCs are among the earliest proteins to be recruited once NRs are bound. SRCs then act as scaffolding proteins to recruit secondary or co-coactivators, such as p300, which binds to the AD1 region of SRCs (Dilworth and Chambon, 2001). In vitro transcription from chromatin-assembled templates revealed that the p300-interacting domain of SRC-1 is essential for SRC-1-mediated coactivation of PR. Additionally, the SRC-interacting domain of p300, but not its ERα-interacting region, is essential for p300-mediated coactivation of ERα (Kraus et al., 1999). These experiments, as well as structural and thermodynamic analysis of the interactions between SRC-3 and CBP/p300, support a step-wise assembly of factors where p160 coactivators are recruited first, followed by CBP/p300, which results in a high-affinity intermolecular complex (Demarest et al., 2002; Liu et al., 2001). CBP/p300 then, in turn, acetylates histones within the enhancer and promoter regions of the target gene to promote active transcription. Additionally, RNA helicase A (RHA) is recruited to the SRC-CBP/p300 complex and interacts with RNA polymerase II, bridging NRs with the general transcription machinery (Nakajima et al., 1997).
3.2.2. Recruitment of co-coactivators to the AD2 domain
The AD2 domain of SRCs is the site of recruitment for co-coactivators, such as CARM1 and PRMT1. CARM1 was first identified in a yeast two-hybrid screen using the AD2 region of SRC-2 as bait (Chen et al., 1999). This HMT (histone methyltransfease) specifically methylates histone H3 at arginines 2, 17, and 26. CBP-mediated acetylation of H3K18 promotes CARM1-mediated methylation of H3R17, suggesting an ordered recruitment of SRCs, CBP/p300, and then CARM1 (Daujat et al., 2002). CARM1, p300, and SRC-2 act synergistically together to enhance ERα-mediated gene transcription in transient transfection assays, and all three proteins are required for estradiol (E2)-induced transcription when low levels of ERα are present (Lee et al., 2002; Stallcup et al., 2003). PRMT1 also enhances NR-mediated transcription and functions synergistically with CARM1 (Koh et al., 2001; Stallcup et al., 2003); however, it has a different substrate than CARM1. PRMT1 methylates H4R3, and, interestingly, this event promotes p300/CBP-mediated acetylation on histone H4 (Lee et al., 2005; Wang et al., 2001). These data emphasize how the cross-talk of histone modifications is essential for optimal gene induction.
3.2.3. Recruitment of co-coactivators to the AD3 domain
The bHLH-PAS domain contains activation domain 3 (AD3). This region is a binding site for co-coactivators, such as CoCoA, GRIP1-associated coactivator 63 (GAC63), Flightless I (Fli-I), Brg1 and hBrm-associated factor 57 (BAF57), methyl methanesulfonate 19 (MMS19), and melanoma antigen gene protein-A11 (MAGE-11); all of which have been shown to interact with this region and enhance SRC-mediated activation of NR-directed transcription (Askew et al., 2009; Belandia et al., 2002; Chen et al., 2005; Kim et al., 2003; Kim et al., 2006; Lee et al., 2004; Wu et al., 2001). Many of these co-coactivators function synergistically with other co-coactivators that bind to the AD1 and AD2 domains of SRCs. For example, CoCoA acts synergistically with p300 and CARM1 in transient transfection assays (Kim et al., 2003). CoCoA may help stabilize the SRC-CBP/p300 complex, and it also associates with components of TFIID, bridging the SRC-CBP/p300 complex with the general transcription machinery (Kim, 2008). GAC63 functions synergistically with SRCs, CARM-1, and p300 (Chen et al., 2005), and Fli-I enhances SRC-2- and CARM-1-mediated coactivation (Lee et al., 2004). The coactivator, MAGE-11, is a novel AR (androgen receptor)- and SRC-interacting protein that increases the accessibility of SRCs to the AF2 domain of AR and is essential for SRC-mediated transcriptional activation of AR (Askew et al., 2009). Besides serving as histone modifying enzymes or protein bridges, coactivators promote chromatin remodeling. BAF57, a subunit of the ATP-dependent chromatin remodeling complex, SWI/SNF, binds to SRC-1 within its AD3 domain and potentiates SRC-mediated coactivation, presumably through chromatin remodeling (Belandia et al., 2002).
3.2.4. Coactivation of transcription factors
While the focus of this review is on the role of coactivators in NR-mediated transcription, we would be remiss not to mention that the SRC family of proteins also coactivate transcription factors. Transcription factors interacting with each of the three ADs of the SRCs have been identified, including myogenin to AD3, NF-KB to AD1, and AP-1 to AD2. SRCs coactivate these transcription factors in response to specific cellular signals, such as coactivation of NFKB in response to TNFα stimulation, (Wu et al., 2004) and coactivation of MEF2C and myogenin in response to skeletal muscle differentiation stimuli (Chen et al., 2000). Needless to say, SRCs play a role in a host of developmental and disease-related processes due to their diverse protein-protein interactions.
3.3. SRC-containing protein complexes
NRs function in large, multi-protein complexes whose components are directed and stabilized in the presence of ligand. Mass spectrometry analysis of proteins immunoprecipitated with an antibody targeting ERα in cells treated with vehicle or E2 revealed a relatively stable complex of 26 ERα-associated proteins (Lanz et al., 2010). Among these, E2-enrichment was greatest for SRC-3. SRCs, themselves, exist in dynamic complexes. An initial investigation into coactivator complexes reported that steady-state SRC complexes consist of six to ten stably associated protein and many more loosely-bound proteins (Jung et al., 2005; Lonard and O’Malley B, 2007). Like ERα, comparison of SRC-3 protein complexes in vehicle and E2-treated cells revealed different associating proteins (Lanz et al., 2010). In E2-treated cells, SRC-3 associates with a number of proteins involved in transcription and signal transduction. The association of these proteins likely varies dependent on the cellular context and function that the SRC protein is performing. Within these complexes, SRCs can be coded for specific functions through post-translational modifications (PTMs) induced by kinases, methyltransferases, and other protein-modifying enzymes associated with them (Feng et al., 2006; Wu et al., 2007; Wu et al., 2004), making SRCs extremely versatile.
3.4. SRCs in cancer
3.4.1. SRC-3
Coactivators, in general, are associated with a plethora of human diseases, including metabolic syndrome and a multitude of cancers (Lanz, 2008; Yan J., 2008; York and O’Malley, 2010). SRC-1 and SRC-3 are particularly overexpressed in breast cancer. NCOA3, the gene encoding SRC-3, is amplified in approximately 5–10% of breast cancers (Anzick et al., 1997; Bautista et al., 1998), while its mRNA is overexpressed in about 30–60% of cases (Anzick et al., 1997; Bouras et al., 2001; Zhao et al., 2003). Depending on the sampling population and its parameters, a variety of conclusions have been drawn as to SRC-3 s contribution to breast cancer and endocrine therapy treatment. In general, SRC-3 overexpression is associated with larger tumor size, higher tumor grade, and worse disease-free survival (Xu et al., 2009). Particularly in Her2/neu-positive tumors, SRC-3 overexpression is associated with tamoxifen resistance (Kirkegaard et al., 2007; Zhao et al., 2003). Thus, SRC-3 is a useful diagnostic and prognostic indicator for breast cancer.
In vivo mouse models have proven invaluable for assessing SRC-3 s role in breast cancer. A mouse mammary tumor virus (MMTV) LTR-driven SRC-3 transgenic mouse was created to specifically overexpress SRC-3 in the mammary tissue of mice (Torres-Arzayus et al., 2004). In this model, there was a significant increase in mammary tumors, as well as other tumors. Of the 145 tumors identified in the SRC-3 overexpressing mice, there were 48 mammary gland adenocarcinomas, 42 pituitary adenomas, 18 uterine leiomyosarcomas, 18 lung adenocarcinomas, and 6 or less of other types. These results are in contrast to the detection of only 3 lung and 2 pituitary gland tumors in the age-matched, wildtype mice. In contrast, loss of SRC-3 reduces tumor incidence induced in an MMTV-v-Ha-ras breast cancer mouse model (Kuang et al., 2004), an MMTV-PyMT (polyoma middle T oncoprotein) breast cancer mouse model (Qin et al., 2008), or induced by the chemical carcinogen DMBA (Kuang et al., 2005). Furthermore, the MMTV-PyMT SRC-3−/− mouse showed a decrease in breast tumor metastasis to the lung (Qin et al., 2008). SRC-3 promotes tumor metastasis by coactivating the Ets transcription factor, PEA3, which upregulates expression of the matrix metalloprotease 2 (MMP2) and MMP9 promoters. Another mouse model was made to look at the function of SRC-3 in Her2- overexpressing tumors. In line with findings that showed SRC-3 overexpresion correlates with Her2/neu levels, MMTV-ERBB2 mice heterozygous for SRC-3 showed delayed tumorigenesis compared with their SRC-3 wildtype counterparts.
SRC-3 overexpression correlates with the development and/or progression of several other cancers besides breast cancer, including lung cancer, prostate cancer, meningioma, colorectal cancer, and pancreatic cancer (Xu et al., 2009; Yan J., 2008). SRC-3 coactivates AR and is overexpressed in prostate cancer. Its expression correlates with worse tumor grade and worse disease-free survival (Gnanapragasam et al., 2001). It is also overexpressed, along with SRC-1 and SRC-2, in a majority of PR-positive meningiomas, 10–35% of colorectal cancers (Xie et al., 2005) and 6 of 9 tested pancreatic cancer cell lines (Ghadimi et al., 1999). Moreover, its expression increases throughout the progression of pancreatic cancer (Henke et al., 2004). Recently, there have been several publications highlighting the importance of SRC-3 in lung cancer. One study observed SRC-3 protein overexpression by immunohistochemistry (IHC) in 48.4% of non small cell lung sarcomas (NSCLS) and gene amplification by fluorescence in situ hybridization (FISH) in 8.2% of NSCLS (He et al., 2010). This same data set revealed a significant correlation between SRC-3 overexpression and ascending pathologic node stage, as well as shortened patient survival. Strikingly, these authors concluded that SRC-3 is the “most significant predictor for survival in multivariate analysis.” These findings are supported by a publication by Cai et al. that reported 27% of NSCLS tumors overexpress SRC-3 protein, and these patients have shorter overall and disease-free survival (Cai et al., 2010). Moreover, an analysis of NCOA3 copy number variation in lung and breast cancer cell lines revealed 25% of NSCLS, 35% of SCLC and 48% of breast cancer cell lines have gene amplification (Cai et al., 2010). Thus, SRC-3 is a bona-fide oncogene in multiple cancers; its influence likely expanding as more cancer types are explored.
3.4.2. SRC-1
Like SRC-3, SRC-1 expression positively correlates with breast tumorigenesis, with a reported increase in 19–29% of samples studied (Xu et al., 2009). Its expression, too, negatively correlates with disease-free survival, and positively with Her2 expression and tamoxifen resistance (Fleming et al., 2004a; Fleming et al., 2004b; Myers et al., 2004). An MMTV mouse model of breast cancer revealed that loss of SRC-1 does not affect tumor initiation or growth, but it significantly inhibits tumor metastasis to the lung (Wang et al., 2009). The mechanism of SRC-1 action, here, was shown to be due to loss of colony stimulating factor 1 (CSF-1), which recruits tumor-associated macrophages, and loss of PEA3-mediated TWIST expression, an epithelial-messenchymal transition (EMT)-promoting gene. In a study of 70 Her2-positive primary breast tumors, SRC-1 and the Ets transcription factor, PEA3, were significantly associated with tumor recurrence in a univariate analysis (Fleming et al., 2004b). In vitro experiments in breast cancer cell lines revealed that growth factors EGF and bFGF increase the association of Ets factors with their DNA binding elements and their association with both SRC-1 and SRC-3, leading to increased transcription (Myers et al., 2005) and yielding a potential pathway for SRC-1 and SRC-3-mediated gene activation independent of estrogen signaling. In addition to being amplified and/or overexpressed in breast cancers, high SRC-1 was reported in meningiomas and prostate cancer (Xu et al., 2009); therefore, assessing SRC-1 levels in patients with these cancers may be useful for prognostic purposes.
3.4.3. SRC-2
Unlike SRC-1 and SRC-3, there are few reports regarding a potential role for SRC-2 in oncogenesis. One study reported no significant change in SRC-2 protein levels between normal and malignant breast tissue (Hudelist et al., 2003); however another publication revealed a correlation between all three SRCs and cyclin D1 expression in ERα-positive breast tumors (Girault et al., 2003). Additionally, SRC-2 was reported to be high in 76% of meningiomas tested (Girault et al., 2003) and, along with SRC-1, in high-grade prostate tumors (Gregory et al., 2001). A fusion protein between SRC-2 and the transcription factor MOZ is frequently found in human acute myeloid leukemia (Troke et al., 2006). The AD1 domain of SRC-2, containing its interaction domain with CBP/p300, is fused with MOZ, and this fusion results in mislocalization of CBP from promyelocytic leukemia bodies. These initial studies give credence to the need for more investigation into potential roles for SRC-2 in oncogenesis.
4. Modulation of coactivator-NR interactions by SRMs
The identification of selective receptor modulators (SRMs) created much excitement for potential therapeutic drugs to treat cancers and other NR-mediated diseases. Indeed, the SRM, 4-hydroxytamoxifen (4HT), has had a great deal of success in the treatment of breast cancer (Jordan, 1992; Vogel et al., 2010). However, resistance to anti-endocrine therapies is an all too common problem. An initial clinical trial aimed at investigating the therapeutic effects of 4HT, the National Surgical Adjuvant Breast and Bowel Project (NSABP), found that women with breast cancer positive for ERα, but negative for axillary lymph nodes, benefited from 5 years of 4HT treatment. These women were then randomly selected to either receive placebo or continue 4HT treatment for 7 additional years. These results showed that women who took the placebo actually did slightly better than those receiving additional 4HT (Fisher et al., 2001). These data support the hypothesis that in some cellular contexts, anti-estrogens like 4HT can actually behave as ERα agonists when administered chronically. Agonistic effects of 4HT were previously implicated in studies analyzing the effects of 4HT treatment in the bone and uterus. Two years of 4HT treatment given to postmenopausal breast cancer patients had an estrogenic effect on the endometrium and the skeleton, increasing bone mass density (Jordan, 1992; Love et al., 1992; Wolf and Jordan, 1992). Raloxifene, another anti-estrogen drug, also displayed estrogenic effects in the bone but functioned as an anti-estrogen in the endometrium. These in vivo observations were supported by molecular work analyzing the activity of the transactivation domains of ERα on several promoters in the presence of estrogens or anti-estrogens, such as 4HT, and demonstrated 4HT-mediated, ERα-directed transcriptional activation of specific promoters (Tzukerman et al., 1994). The selective functional behavior of these drugs in different cellular contexts gave rise to the term selective estrogen receptor modulator (SERM).
4.1. Coactivator-NR interactions influenced by SERMs and SERDs
SERMs are classified as partial antagonists, as opposed to full antagonists such as ICI 182780, also known as fulvestrant. Both types of antagonists interfere with NR-coactivator interactions but by different mechanisms. Full or pure antagonists, such as ICI, create a disordered structure for helix 12 that cannot contact the LBD and form the coactivator docking site (Nettles and Greene, 2005; Pace, 2008). They also promote cytoplasmic localization of the receptor, immobilization within the nuclear matrix, and ubiquitin-mediated degradation of the NR via the proteasome system (Htun et al., 1999; Long and Nephew, 2006). These additional activities of full antagonists led to the term selective ER down-regulators (SERDs). Partial antagonists, or SERMs, such as raloxifene and 4HT, bind to the LBD and position helix 12 in such a manner that it occludes the coactivator binding site (Nettles and Greene, 2005). The structures of ERα-E2 and ERα-4HT revealed that they were both different from one another, as previously described and, importantly, from apo-receptor, indicating that neither 4HT nor raloxifene merely displaces estradiol from ERα and reverts the receptor back to its inactive state (Grasfeder, 2008; McDonnell et al., 1995). They create an entirely different structural conformation, which may be regulated in novel ways to promote agonist versus antagonist effects. Indeed, studies have shown an altered coactivator-NR structure in the presence of anti-estrogens, as opposed to estrogens. A peptide-based phage display screen probing changes in ERα conformation in the presence of buffer, E2, or 4HT revealed that different peptides are able to interact with ERα in the presence of different ligands, even when the ligands have the same agonistic or antagonistic effect (Chang et al., 1999; Norris et al., 1999; Paige et al., 1999; Wijayaratne et al., 1999). Thus, even if canonical coactivator sites are blocked with tamoxifen treatment, other sites may be available and provide a means for SERM-mediated transcriptional activation.
While these studies show that different ligands (E2 or 4HT) induce different structural conformations on ERα, it should be noted that not all ligands that promote divergent NR activity do so by creating alternate structural conformations. For example, the crystal structure of the PPARγ-RXRα complex bound to DNA, various ligands, and coactivator peptides was solved and revealed that rosiglitazone, a PPARγ agonist; GW9662, a suicide inhibitor of PPARγ; and BVT.13, a partial PPARγ agonist, all create similar PPARγ-RXRα structural conformations (Chandra et al., 2008). In this study, DNA was critical for the conformation of the PPARγ-RXRα structure, and the LBD influences DNA binding, and, thus, may influence the transcriptional activity of the NR by different ligands in this manner.
4.2. Coactivator levels and activity influence SRM agonist/antagonist activity
The pertinent question then became what factors are imposing the differential agonist and antagonist effects of SERM treatment. The O Malley laboratory provided some of the earliest evidence that the relative recruitment of coactivators or corepressors is instrumental towards the agonist/antagonist effects of SERMs (Smith et al., 1997). In HeLa cells, where 4HT is an agonist, overexpression of SRC-1 enhances 4HT s activity; whereas, overexpression of a corepressor such as SMRT, reduces 4HT-mediated stimulation (Smith et al., 1997). In MCF7 and T47D breast cancer cells, where 4HT acts as an ERα antagonist, corepressors were recruited to the repressed c-myc promoter, but coactivators were recruited to the same c-myc promoter in Ishikawa endometrial cells, where 4HT acts as an agonist (Shang and Brown, 2002). These data were recently corroborated by an analysis from Romano et al. that showed in all cellular contexts tested, when estradiol or 4HT was an activating stimulus, coactivators were recruited; while corepressors were recruited when 4HT acted as a repressing stimulus (Romano et al., 2010a). The question at hand now slightly shifts to what factors contribute towards the relative recruitment of coactivators versus corepressors. In the Brown lab study, it was found that SRC-1 is expressed at a much higher level in Ishikawa cells, where 4HT has agonist properties, than in MCF-7 cells, where it functions antagonistically. (Shang and Brown, 2002). Additionally, Romano et al. reported that overexpression of SRC-1 could convert tamoxifen from a transcriptionally repressive stimulus to an activating one, and overexpression of the corepressor NCoR could similarly reprogram tamoxifen-stimulated genes for transcriptional repression (Romano et al., 2010b). These functions of coactivators ring true for other NRs besides ERα. For example, overexpression of SRC-1 stimulates PR activity in HeLa cells treated with RU486- (a selective progesterone receptor modulator) (Liu et al., 2002). Likewise, low expression of the NR corepressor complex, NCoR/SMRT, is associated with better 4HT resistance (Lavinsky et al., 1998; Liu et al., 2002). Thus, the expression level of coactivators versus corepressors may determine whether a SERM has an agonist or antagonist effect.
Clinical data assessing coactivator levels in breast cancer samples from women treated with 4HT support this hypothesis. SRC-1 was scored as positive or negative by IHC in 52 human breast cancer tissue samples from patients treated for 5 years with 4HT (Fleming et al., 2004a). 92% of patients with disease recurrence were SRC-1-positive, compared with only 10% of the nonrecurrence patients being positive for SRC-1. Similar results were seen from an analysis of SRC-3 in 4HT-treated patients. Tumor specimens from 316 patients with axillary node-positive breast cancer treated with or without 4HT were scored for SRC-3 protein levels. This study found that high SRC-3 levels correlate with worse disease-free survival, indicative of 4HT resistance. The patients with the worst outcome were those with both SRC-3 and Her2 overexpression (Osborne et al., 2003). There are a few studies that did not show a statistical correlation between SRC-3 and overall disease recurrence; however, this is likely due to the cohort of samples collected and analyzed. In one such study, SRC-3, by itself, was not predictive of recurrence, but together with high Her2/3, it is predictive (Kirkegaard et al., 2007). Another data set by Dihge et al supports the hypothesis that SRC-3 may be most predictive when other factors are present to exacerbate a worse disease progression (Dihge et al., 2008). In this study of 297 breast cancer samples from patients treated with 4HT, SRC-3 expression was predictive for recurrence that occurs within the first 2 years but not long term at 5 years.
Tamoxifen treatment, itself, can raise coactivator levels, contributing towards resistance. The generation of SRC-1- and SRC-3- luciferase fusion proteins allowed for quantitative measurement of the relative changes in protein levels after treatment with E2, 4HT, or raloxifene and revealed a dramatic increase in protein expression with 4HT treatment (Lonard et al., 2004). Endogenous SRC-3 protein levels also increased in BT474 cells treated with 4HT (Su et al., 2008). Even with a low dose of tamoxifen, an increase in SRC-3 mRNA was seen (Haugan Moi et al., 2010). In fact, in this same study, an increase in all three SRCs was observed; however, the increase in SRC-3 was the most significant. Additionally, other signals may be induced in tamoxifen-resistant tumors, which either increase coactivator levels or their activity. In line with the correlation observed between SRC-3 and Her2 expression in breast cancers, Her2- overexpression stimulates phosphorylation of SRC-3 (Osborne et al., 2003). It is well established that phosphorylation of SRC-3 is essential for its function as an NR coactivator (Wu et al., 2002; Wu et al., 2004; Zheng et al., 2005). Moreover, knockdown of SRC-3 in Her2-amplified BT474 cells restores tamoxifen-induced inhibition of cell proliferation (Su et al., 2008).
Downstream of Her2 signaling lies the MAPK-activated Ets family of transcription factors. Ets factors are expressed in breast cancers and correlate with cancer progression and metastasis (Myers et al., 2005). Treatment of breast cancer cells with growth factors can increase the association between Ets factors and SRC-3 (Myers et al., 2005), as well as SRC-3 s association with ERα in 4HT-resistant MCF-7 cells (Zhao et al., 2009). In fact, knockdown of SRC-3 inhibited Her1 (EGFR)/Her2-induced cell growth and restored the sensitivity of 4HT-resistant cells to 4HT treatment. These studies and others suggest that breast cancers that are resistant to anti-endocrine therapies may have developed E2-independent mechanisms of cell growth. These mechanisms may include directing coactivators towards interaction with other factors besides NRs, such as Ets proteins (Myers et al., 2005) or E2F1 (Louie et al., 2004), allowing the cells to circumvent NRs altogether.
4.3. NR levels and post-translational modifications influence coregulator binding in the presence of SRMs
Signaling pathways that are upregulated in some cancers, such as the previously mentioned Her2 pathway, can modify coactivators or NRs to promote transcription in the presence of anti-estrogens. For example, activation of PKA (protein kinase A) promotes 4HT resistance through phosphorylation of ERα at S305. FRET analysis revealed that PKA-mediated ERα S305 phosphorylation alters the orientation between ERα and SRC-1 such that RNA polymerase II is recruited and transcription ensues (Zwart et al., 2007). PKA also induces phosphorylation of PR (Wagner et al., 1998) and AR, which blocks (Dotzlaw et al., 2002) corepressor recruitment and stimulates gene expression in the presence of their respective NR antagonists.
The expression level of NRs is another variable that can influence the agonist/antagonist activity of SRMs. A microarray-based profiling screen of prostate cancer xenograft models revealed a consistent increase in AR mRNA levels in models that had developed resistance to anti-androgen therapy (Chen et al., 2004). Overexpression of AR in LnCaP prostate cancer cells, mimicking the AR levels found in the anti-androgen resistant xenografts, converted the cells from SARM (selective androgen receptor modulator)- sensitive to resistant, as measured by their ability to grow in low androgen levels and in the presence of the anti-androgen, bicalutamide. In cells overexpressing AR, but not those lacking AR-overexpression, bicalutamide treatment resulted in the increased recruitment of SRC-1 and decreased recruitment of NCoR to the promoter of AR target genes, PSA and KLK-2 (Chen et al., 2004).
4.4. The promoter context may alter NR-coregulator interactions, influencing SRM effects
DNA, itself, has long been theorized to act as an allosteric regulator of transcription factors, such as NRs, (Lefstin and Yamamoto, 1998). DNA sequence influences NR binding, structural conformation, and the recruitment of coactivators and corepressors (Chandra et al., 2008; Heneghan et al., 2007; Lefstin and Yamamoto, 1998). Thus, the promoter context of NR-target genes can influence the effects of SRMs on NR activity. For example, 4HT and raloxifene treatments repress ERα bound to classical EREs in HeLa cells, but these ligands increase transcription when ERα is tethered to DNA through AP-1 (Webb et al., 1999). This pattern holds true in endometrial carcinoma cancer cells, as well. Genes, such as cathepsin D and EBAG9, whose promoters have classical EREs, are repressed by 4HT and raloxifene; however, 4HT activates transcription of c-myc and IGF-1 promoters that lack canonical EREs (Shang and Brown, 2002). As previously mentioned, SRC-2 activates GR-directed transcription when GR is bound directly to glucocorticoid response elements, but it represses GR tethered to DNA via activating protein 1 (AP-1) (Rogatsky et al., 2002). One potential explanation is that the promoter context regulates SRM s effects by altering the orientation of the NR in such a way that alters its interaction with coregulators or the activity of the coregulators. In summary, multiple variables, including coregulator and NR levels and activity, which can be influenced by various cancer-specific alterations in signaling pathways, as well as promoter-specific contexts may contribute to the differential agonist/antagonist effects of anti-endocrine therapies, such as tamoxifen (Figure 3).
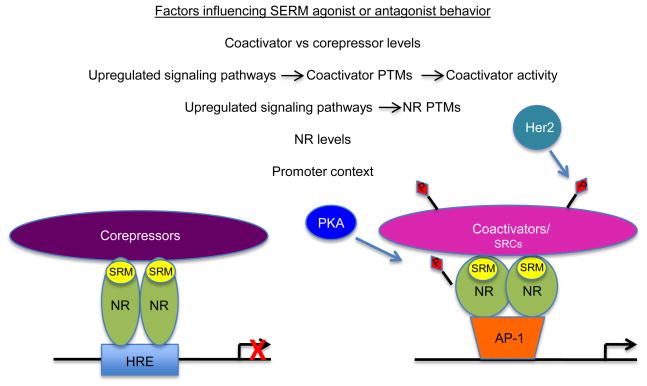
Figure 3. Multiple cellular factors influence coactivator-NR interactions in the presence of SRMs, affecting SRM functional effects on NR-mediated transcription.
Listed are several cellular variables, including NR and coactivator levels and activity, influenced by the differential upregulation of oncogenic signaling pathways, as well as gene-specific promoter contexts that determine whether a SRM will have agonist or antagonist effects. The cartoon demonstrates just some of these potential variables that can impact SRM antagonist/agonist activities and pathways that may be involved. Not all variable are likely to be in play at one time and are cancer, cell- and gene-dependent. SRM (selective receptor modulator); NR (nuclear receptor); HRE (hormone response element; AP-1 (activating protein 1); SRC (steroid receptor coactivator); PKA (protein kinase A); Her2 (human epidermal growth factor 2); P (phosphorylation).
5. Interrupting NR-coactivator interactions for therapeutic purposes
It is clear now that cancer cells can employ many mechanisms to circumvent SRMs that aim to silence NR activity. However, in each of these instances, once a cell becomes resistant to the anti-endocrine therapy, it still must be able to recruit coactivators in order to function. Thus, it stands to reason that interfering with NR-coactivator interactions should be very beneficial in treating NR-mediated diseases, such as cancer. Peptides identified in the previously mentioned phage-display screen can interfere with the agonistic activity of 4HT in resistant cells (Chang et al., 1999; Norris et al., 1999). These peptides, however, will require significant modifications to make them druggable, but some progress has been made in this area (Grasfeder, 2008). A peptide with a macrolactam ring was successful in preventing SRC-2 interaction with the TRβ/T3 complex (Geistlinger and Guy, 2001). Other peptide-derived antagonists can block the interaction of SRC-1 with ERα or ERβ, as well (Galande et al., 2004; Galande et al., 2005; Leduc et al., 2003). In addition to peptides, small molecule inhibitors (SMIs) have been investigated for potential therapeutic use. Rodriguez et al. demonstrated that a small molecule can target the coactivator binding pocket of ERα and displace a coactivator-derived peptide containing the LXXLL motif responsible for binding ERα (Rodriguez et al., 2004). Small molecules that inhibit coactivator binding to TRβ have also been discovered (Arnold et al., 2005). These SMIs were able to completely block TRβ-induced transcriptional activity. One potential problem with SMIs or peptide-based therapies that target NR-coactivator interactions is that the published literature has shown that overexpression of coactivators often circumvents the need for NRs to promote cell growth. As mentioned previously, SRC-3 interaction with other transcription factors, such as E2F1 and Ets factors, do not depend on an intact NR. Moreover, cells often upregulate other signaling pathways, such as EGFR and Her2, which can compensate for loss of an NR and promote coactivator function. In fact, in breast cancer ERα-negative cancers have a worse prognosis than ERα-positive cancers. Thus, SMIs that can directly bind to overexpressed coactivators and downregulate their activity or stability may be the preferred therapy of the future.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Arnold LA, Estebanez-Perpina E, Togashi M, Jouravel N, Shelat A, McReynolds AC, Mar E, Nguyen P, Baxter JD, Fletterick RJ, et al. Discovery of small molecule inhibitors of the interaction of the thyroid hormone receptor with transcriptional coregulators. J Biol Chem. 2005;280:43048–43055. doi: 10.1074/jbc.M506693200. [DOI] [PubMed] [Google Scholar]
- Askew EB, Bai S, Hnat AT, Minges JT, Wilson EM. Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators. J Biol Chem. 2009;284:34793–34808. doi: 10.1074/jbc.M109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad A, Leng X, Burris TP, Tsai SY, Tsai MJ, O’Malley BW. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista S, Valles H, Walker RL, Anzick S, Zeillinger R, Meltzer P, Theillet C. In breast cancer, amplification of the steroid receptor coactivator gene AIB1 is correlated with estrogen and progesterone receptor positivity. Clin Cancer Res. 1998;4:2925–2929. [PubMed] [Google Scholar]
- Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghagen H, Ragnhildstveit E, Krogsrud K, Thuestad G, Apriletti J, Saatcioglu F. Corepressor SMRT functions as a coactivator for thyroid hormone receptor T3Ralpha from a negative hormone response element. J Biol Chem. 2002;277:49517–49522. doi: 10.1074/jbc.M209546200. [DOI] [PubMed] [Google Scholar]
- Bouras T, Southey MC, Venter DJ. Overexpression of the steroid receptor coactivator AIB1 in breast cancer correlates with the absence of estrogen and progesterone receptors and positivity for p53 and HER2/neu. Cancer Res. 2001;61:903–907. [PubMed] [Google Scholar]
- Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- Cai D, Shames DS, Raso MG, Xie Y, Kim YH, Pollack JR, Girard L, Sullivan JP, Gao B, Peyton M, et al. Steroid receptor coactivator-3 expression in lung cancer and its role in the regulation of cancer cell survival and proliferation. Cancer Res. 2010;70:6477–6485. doi: 10.1158/0008-5472.CAN-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR-nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Norris JD, Gron H, Paige LA, Hamilton PT, Kenan DJ, Fowlkes D, McDonnell DP. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Molecular and cellular biology. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Kim JH, Stallcup MR. GAC63, a GRIP1-dependent nuclear receptor coactivator. Mol Cell Biol. 2005;25:5965–5972. doi: 10.1128/MCB.25.14.5965-5972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard VH, Matsuda S, Heery DM. An extended LXXLL motif sequence determines the nuclear receptor binding specificity of TRAP220. The Journal of biological chemistry. 2003;278:10942–10951. doi: 10.1074/jbc.M212950200. [DOI] [PubMed] [Google Scholar]
- Cvoro A, Tzagarakis-Foster C, Tatomer D, Paruthiyil S, Fox MS, Leitman DC. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol Cell. 2006;21:555–564. doi: 10.1016/j.molcel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- Dihge L, Bendahl PO, Grabau D, Isola J, Lovgren K, Ryden L, Ferno M. Epidermal growth factor receptor (EGFR) and the estrogen receptor modulator amplified in breast cancer (AIB1) for predicting clinical outcome after adjuvant tamoxifen in breast cancer. Breast Cancer Res Treat. 2008;109:255–262. doi: 10.1007/s10549-007-9645-1. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Dotzlaw H, Moehren U, Mink S, Cato AC, Iniguez Lluhi JA, Baniahmad A. The amino terminus of the human AR is target for corepressor action and antihormone agonism. Molecular endocrinology (Baltimore, Md. 2002;16:661–673. doi: 10.1210/mend.16.4.0798. [DOI] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Molecular and cellular biology. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B, Dignam J, Bryant J, Wolmark N. Five versus more than five years of tamoxifen for lymph node-negative breast cancer: updated findings from the National Surgical Adjuvant Breast and Bowel Project B-14 randomized trial. J Natl Cancer Inst. 2001;93:684–690. doi: 10.1093/jnci/93.9.684. [DOI] [PubMed] [Google Scholar]
- Fleming FJ, Hill AD, McDermott EW, O’Higgins NJ, Young LS. Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 2004a;89:375–383. doi: 10.1210/jc.2003-031048. [DOI] [PubMed] [Google Scholar]
- Fleming FJ, Myers E, Kelly G, Crotty TB, McDermott EW, O’Higgins NJ, Hill AD, Young LS. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J Clin Pathol. 2004b;57:1069–1074. doi: 10.1136/jcp.2004.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande AK, Bramlett KS, Burris TP, Wittliff JL, Spatola AF. Thioether side chain cyclization for helical peptide formation: inhibitors of estrogen receptor-coactivator interactions. J Pept Res. 2004;63:297–302. doi: 10.1111/j.1399-3011.2004.00152.x. [DOI] [PubMed] [Google Scholar]
- Galande AK, Bramlett KS, Trent JO, Burris TP, Wittliff JL, Spatola AF. Potent inhibitors of LXXLL-based protein-protein interactions. Chembiochem. 2005;6:1991–1998. doi: 10.1002/cbic.200500083. [DOI] [PubMed] [Google Scholar]
- Gampe RT, Jr, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- Geistlinger TR, Guy RK. An inhibitor of the interaction of thyroid hormone receptor beta and glucocorticoid interacting protein 1. J Am Chem Soc. 2001;123:1525–1526. doi: 10.1021/ja005549c. [DOI] [PubMed] [Google Scholar]
- Ghadimi BM, Schrock E, Walker RL, Wangsa D, Jauho A, Meltzer PS, Ried T. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am J Pathol. 1999;154:525–536. doi: 10.1016/S0002-9440(10)65298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault I, Lerebours F, Amarir S, Tozlu S, Tubiana-Hulin M, Lidereau R, Bieche I. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin Cancer Res. 2003;9:1259–1266. [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gnanapragasam VJ, Leung HY, Pulimood AS, Neal DE, Robson CN. Expression of RAC 3, a steroid hormone receptor co-activator in prostate cancer. Br J Cancer. 2001;85:1928–1936. doi: 10.1054/bjoc.2001.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasfeder LLaMDP. Nuclear Receptor Cofactor Interactions as Targets for New Drug Discovery. Singapore: World Scientific Publications; 2008. [Google Scholar]
- Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–4319. [PubMed] [Google Scholar]
- Haugan Moi LL, Hauglid Flageng M, Gandini S, Guerrieri-Gonzaga A, Bonanni B, Lazzeroni M, Gjerde J, Lien EA, DeCensi A, Mellgren G. Effect of low-dose tamoxifen on steroid receptor coactivator 3/amplified in breast cancer 1 in normal and malignant human breast tissue. Clin Cancer Res. 2010;16:2176–2186. doi: 10.1158/1078-0432.CCR-09-1859. [DOI] [PubMed] [Google Scholar]
- He LR, Zhao HY, Li BK, Zhang LJ, Liu MZ, Kung HF, Guan XY, Bian XW, Zeng YX, Xie D. Overexpression of AIB1 negatively affects survival of surgically resected non-small-cell lung cancer patients. Ann Oncol. 2010;21:1675–1681. doi: 10.1093/annonc/mdp592. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Coactivator assembly at the promoter: efficient recruitment of SRC2 is coupled to cooperative DNA binding by the progesterone receptor. Biochemistry. 2007;46:11023–11032. doi: 10.1021/bi700850v. [DOI] [PubMed] [Google Scholar]
- Henke RT, Haddad BR, Kim SE, Rone JD, Mani A, Jessup JM, Wellstein A, Maitra A, Riegel AT. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clin Cancer Res. 2004;10:6134–6142. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H, Holth LT, Walker D, Davie JR, Hager GL. Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell. 1999;10:471–486. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudelist G, Czerwenka K, Kubista E, Marton E, Pischinger K, Singer CF. Expression of sex steroid receptors and their co-factors in normal and malignant breast tissue: AIB1 is a carcinoma-specific co-activator. Breast Cancer Res Treat. 2003;78:193–204. doi: 10.1023/a:1022930710850. [DOI] [PubMed] [Google Scholar]
- Johnson AB, Barton MC. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat Res. 2007;618:149–162. doi: 10.1016/j.mrfmmm.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. The strategic use of antiestrogens to control the development and growth of breast cancer. Cancer. 1992;70:977–982. [PubMed] [Google Scholar]
- Jung SY, Malovannaya A, Wei J, O’Malley BW, Qin J. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol Endocrinol. 2005;19:2451–2465. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- Kim JH, Yang CK, Stallcup MR. Downstream signaling mechanism of the C-terminal activation domain of transcriptional coactivator CoCoA. Nucleic Acids Res. 2006;34:2736–2750. doi: 10.1093/nar/gkl361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JHaSMR. p160 Coactivators: Critical Mediators of Transcriptional Activation by Nuclear Receptors. Singaporte: World Scientific Publishing Co. Pte. Ltd; 2008. [Google Scholar]
- Kirkegaard T, McGlynn LM, Campbell FM, Muller S, Tovey SM, Dunne B, Nielsen KV, Cooke TG, Bartlett JM. Amplified in breast cancer 1 in human epidermal growth factor receptor - positive tumors of tamoxifen-treated breast cancer patients. Clin Cancer Res. 2007;13:1405–1411. doi: 10.1158/1078-0432.CCR-06-1933. [DOI] [PubMed] [Google Scholar]
- Klein-Hitpass L, Tsai SY, Weigel NL, Allan GF, Riley D, Rodriguez R, Schrader WT, Tsai MJ, O’Malley BW. The progesterone receptor stimulates cell-free transcription by enhancing the formation of a stable preinitiation complex. Cell. 1990;60:247–257. doi: 10.1016/0092-8674(90)90740-6. [DOI] [PubMed] [Google Scholar]
- Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Wang S, Medina D, O’Malley BW, Xu J. Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Zhang H, Lee AV, O’Malley BW, Xu J. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–1885. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, O’Malley BW. Global characterization of transcriptional impact of the SRC-3 coregulator. Mol Endocrinol. 2010;24:859–872. doi: 10.1210/me.2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, Lonard DM, O’Malley BW. Nuclear Receptor Coregulators in Human Diseases. Singapore: World Scientific Publishing Co; 2008. [Google Scholar]
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci U S A. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc AM, Trent JO, Wittliff JL, Bramlett KS, Briggs SL, Chirgadze NY, Wang Y, Burris TP, Spatola AF. Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions. Proc Natl Acad Sci U S A. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Li C, Wu RC, Amazit L, Tsai SY, Tsai MJ, O’Malley BW. Specific amino acid residues in the basic helix-loop-helix domain of SRC-3 are essential for its nuclear localization and proteasome-dependent turnover. Molecular and cellular biology. 2007;27:1296–1308. doi: 10.1128/MCB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Auboeuf D, Wong J, Chen JD, Tsai SY, Tsai MJ, O’Malley BW. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc Natl Acad Sci U S A. 2002;99:7940–7944. doi: 10.1073/pnas.122225699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc Natl Acad Sci U S A. 2001;98:12426–12431. doi: 10.1073/pnas.231474798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Molecular cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Tsai SY, O’Malley BW. Selective estrogen receptor modulators 4-hydroxytamoxifen and raloxifene impact the stability and function of SRC-1 and SRC-3 coactivator proteins. Molecular and cellular biology. 2004;24:14–24. doi: 10.1128/MCB.24.1.14-24.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Nephew KP. Fulvestrant (ICI 182,780)-dependent interacting proteins mediate immobilization and degradation of estrogen receptor-alpha. J Biol Chem. 2006;281:9607–9615. doi: 10.1074/jbc.M510809200. [DOI] [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Molecular and cellular biology. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love RR, Mazess RB, Barden HS, Epstein S, Newcomb PA, Jordan VC, Carbone PP, DeMets DL. Effects of tamoxifen on bone mineral density in postmenopausal women with breast cancer. N Engl J Med. 1992;326:852–856. doi: 10.1056/NEJM199203263261302. [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Clemm DL, Hermann T, Goldman ME, Pike JW. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Meyer ME, Gronemeyer H, Turcotte B, Bocquel MT, Tasset D, Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989;57:433–442. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- Myers E, Fleming FJ, Crotty TB, Kelly G, McDermott EW, O’Higgins NJ, Hill AD, Young LS. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br J Cancer. 2004;91:1687–1693. doi: 10.1038/sj.bjc.6602156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E, Hill AD, Kelly G, McDermott EW, O’Higgins NJ, Buggy Y, Young LS. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin Cancer Res. 2005;11:2111–2122. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annu Rev Physiol. 2005;67:309–333. doi: 10.1146/annurev.physiol.66.032802.154710. [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Norris JD, Paige LA, Christensen DJ, Chang CY, Huacani MR, Fan D, Hamilton PT, Fowlkes DM, McDonnell DP. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, Wong J, Allred DC, Clark GM, Schiff R. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- Pace MCaSCL. Coregulators as Determinants of Selective Receptor Modulator (SRM) Activity. Singapore: World Scientific Publishing Co., Pte, Ltd; 2008. [Google Scholar]
- Paige LA, Christensen DJ, Gron H, Norris JD, Gottlin EB, Padilla KM, Chang CY, Ballas LM, Hamilton PT, McDonnell DP, Fowlkes DM. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc Natl Acad Sci U S A. 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL. The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor alpha transcriptional activity. Mol Cell Biol. 2007;27:5933–5948. doi: 10.1128/MCB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O’Malley BW, Xu J. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Molecular and cellular biology. 2008;28:5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AL, Tamrazi A, Collins ML, Katzenellenbogen JA. Design, synthesis, and in vitro biological evaluation of small molecule inhibitors of estrogen receptor alpha coactivator binding. J Med Chem. 2004;47:600–611. doi: 10.1021/jm030404c. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci U S A. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A, Adriaens M, Kuenen S, Delvoux B, Dunselman G, Evelo C, Groothuis P. Identification of novel ER-alpha target genes in breast cancer cells: gene- and cell-selective co-regulator recruitment at target promoters determines the response to 17beta-estradiol and tamoxifen. Mol Cell Endocrinol. 2010a;314:90–100. doi: 10.1016/j.mce.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Romano A, Barca A, Kottra G, Daniel H, Storelli C, Verri T. Functional expression of SLC15 peptide transporters in rat thyroid follicular cells. Mol Cell Endocrinol. 2010b;315:174–181. doi: 10.1016/j.mce.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- Smith CL, Nawaz Z, O’Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85:139–145. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- Su Q, Hu S, Gao H, Ma R, Yang Q, Pan Z, Wang T, Li F. Role of AIB1 for tamoxifen resistance in estrogen receptor-positive breast cancer cells. Oncology. 2008;75:159–168. doi: 10.1159/000159267. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. The Journal of biological chemistry. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Troke PJ, Kindle KB, Collins HM, Heery DM. MOZ fusion proteins in acute myeloid leukaemia. Biochem Soc Symp. 2006:23–39. doi: 10.1042/bss0730023. [DOI] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, Bevers TB, Fehrenbacher L, Pajon ER, Wade JL, 3rd, et al. Update of the National Surgical Adjuvant Breast and Bowel Project Study of Tamoxifen and Raloxifene (STAR) P-2 Trial: Preventing breast cancer. Cancer Prev Res (Phila) 2010;3:696–706. doi: 10.1158/1940-6207.CAPR-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BL, Norris JD, Knotts TA, Weigel NL, McDonnell DP. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol Cell Biol. 1998;18:1369–1378. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wang S, Yuan Y, Liao L, Kuang SQ, Tien JC, O’Malley BW, Xu J. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci U S A. 2009;106:151–156. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA, Nilsson S, Kushner PJ. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13:1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- Wijayaratne AL, Nagel SC, Paige LA, Christensen DJ, Norris JD, Fowlkes DM, McDonnell DP. Comparative analyses of mechanistic differences among antiestrogens. Endocrinology. 1999;140:5828–5840. doi: 10.1210/endo.140.12.7164. [DOI] [PubMed] [Google Scholar]
- Wolf DM, Jordan VC. Gynecologic complications associated with long-term adjuvant tamoxifen therapy for breast cancer. Gynecol Oncol. 1992;45:118–128. doi: 10.1016/0090-8258(92)90273-l. [DOI] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O’Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O’Malley BW. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I kappa B kinase. Molecular and cellular biology. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Molecular cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Wu X, Li H, Chen JD. The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J Biol Chem. 2001;276:23962–23968. doi: 10.1074/jbc.M101041200. [DOI] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:206. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- Xie D, Sham JS, Zeng WF, Lin HL, Bi J, Che LH, Hu L, Zeng YX, Guan XY. Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol. 2005;36:777–783. doi: 10.1016/j.humpath.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. [Google Scholar]
- Yan J, TSY, Tsai M-J. A central Role of SRC-3/AIB1 in tumorigenesis. Singapore: World Scientific Publishing Co. Pte. Ltd; 2008. [Google Scholar]
- York B, O’Malley BW. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem. 2010;285:38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Yasui K, Lee CJ, Kurioka H, Hosokawa Y, Oka T, Inazawa J. Elevated expression levels of NCOA3, TOP1, and TFAP2C in breast tumors as predictors of poor prognosis. Cancer. 2003;98:18–23. doi: 10.1002/cncr.11482. [DOI] [PubMed] [Google Scholar]
- Zhao W, Zhang Q, Kang X, Jin S, Lou C. AIB1 is required for the acquisition of epithelial growth factor receptor-mediated tamoxifen resistance in breast cancer cells. Biochem Biophys Res Commun. 2009;380:699–704. doi: 10.1016/j.bbrc.2009.01.155. [DOI] [PubMed] [Google Scholar]
- Zheng FF, Wu RC, Smith CL, O’Malley BW. Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Molecular and cellular biology. 2005;25:8273–8284. doi: 10.1128/MCB.25.18.8273-8284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Griekspoor A, Berno V, Lakeman K, Jalink K, Mancini M, Neefjes J, Michalides R. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERalpha towards co-activator SRC-1. EMBO J. 2007;26:3534–3544. doi: 10.1038/sj.emboj.7601791. [DOI] [PMC free article] [PubMed] [Google Scholar]