Abstract
Cardiac valves originate from endocardial cushions (EC) formed by endothelial-to-mesenchymal transformation (EMT) during embryogenesis. The zinc-finger transcription factor Snai1 has previously been reported to be important for EMT during organogenesis, yet its role in early valve development has not been directly examined. In this study we show that Snai1 is highly expressed in endothelial, and newly transformed mesenchyme cells during EC development. Mice with targeted snai1 knockdown display hypocellular ECs at E10.5 associated with decreased expression of mesenchyme cell markers and downregulation of the matrix metalloproteinase (mmp) family member, mmp15. Snai1 overexpression studies in atrioventricular canal collagen I gel explants indicate that Snai1 is sufficient to promote mmp15 expression, cell transformation, and mesenchymal cell migration and invasion. However, treatment with the catalytically active form of MMP15 promotes cell motility, and not transformation. Further, we show that Snai1-mediated cell migration requires MMP activity, and caMMP15 treatment rescues attenuated migration defects observed in murine ECs following snai1 knockdown. Together, findings from this study reveal previously unappreciated mechanisms of Snai1 for the direct regulation of MMPs during EC development.
Keywords: Snai1, MMP15, endocardial cushion, heart valve, endothelial-to-mesenchymal transformation
Introduction
Development of heart valve structures is a complex process involving multiple molecular pathways that modulate cardiac morphogenesis (Combs and Yutzey, 2009; Srivastava, 2000). Alterations in these genetic networks during embryogenesis frequently lead to structural defects, malfunction and congenital heart disease (CHD) in approximately 1% of all live births (Hoffman and Kaplan, 2002). These commonly include valvular and septal defects that often require multiple corrective surgical procedures, or result in death in utero, childhood, or infancy (Lloyd-Jones et al., 2010). Despite the clinical significance, the intersecting regulatory pathways required for normal heart valve formation, and their genetic contributions to CHD are not complete. Understanding the structure-function relationship of genes important during embryonic valvulogenesis will provide insights into genetic causes of CHD associated with structural valve defects and dysfunction.
Heart valve formation is initiated in the atrioventricular canal (AVC), outflow tract (OFT) regions around embryonic day (E) 9.5 in the mouse and E3.0 in the chicken, following expansion of existing cardiac jelly and formation of endocardial cushions (ECs) (Combs and Yutzey, 2009; Person et al., 2005). ECs are reservoirs for a heterogeneous population of precursor cells that later remodel and give rise to the mature, and highly diversified heart valve structures (Lincoln et al., 2004). The cellular and molecular processes required for EC formation are critical to normal valve development and require endothelial-to-mesenchymal transformation (EMT) (Person et al., 2005). The EMT program involves several phenotypic changes of endothelial cells and is not restricted to ECs, but critical for the development of many tissues during embryogenesis (Barrallo-Gimeno and Nieto, 2005). In developing ECs, EMT initiation is characterized by delamination of a subset of endothelial cells overlying the future valve site following down regulation of adhesion molecules and lost cell-cell contacts (Person et al., 2005; Thiery et al., 2009). Subsequently, endothelial cells break away from their neighbors, transform into mesenchymal cells and acquire motile properties, allowing invasion and migration through the matrix-rich cardiac jelly (Person et al., 2005). Once migrated, these highly proliferative mesenchymal cells populate the ECs and form elongated valve primordia that continue to grow and elongate into mature leaflets and chordae tendinae in the AV valves and cusps in the OFT and pulmonic regions (Hinton et al., 2006; Lincoln et al., 2004; Lincoln et al., 2006b). Numerous signaling pathways have been shown to regulate EC EMT (Armstrong and Bischoff, 2004; Combs and Yutzey, 2009; Person et al., 2005). These include growth factors emanating from the adjacent myocardium including bone morphogenic proteins (Bmp) and transforming growth factors (Tgfβ), as well as more localized signaling from Wnt- and Notch-related pathways (Armstrong and Bischoff, 2004; Combs and Yutzey, 2009; Person et al., 2005). Targeted mutations in receptors, ligands or downstream mediators of these pathways have previously been shown to have detrimental effects on EC development and embryo survivability (Combs and Yutzey, 2009; Person et al., 2005), highlighting the importance of the EMT process for normal heart valve formation and cardiac function.
Members of the Snail family of transcription factors including snai1 (snail), and snai2 (slug) have previously been shown to play major roles in EMT in many developmental systems during embryogenesis, although direct roles for snai1 in the heart have not been examined (Cano et al., 2000; Niessen et al., 2008). Snai1−/− mutant mice die early in gestation due to defective gastrulation and mesoderm formation as cells fail to migrate because they cannot undergo EMT (Carver et al., 2001). Parallel studies have revealed that snai1 positively regulates EMT by directly repressing cell adhesion genes including E-cadherin (Batlle et al., 2000; Cano et al., 2000), claudins and occludins (Ikenouchi et al., 2003; Martinez-Estrada et al., 2006) and therefore cell-cell contacts are adversely maintained in snai1 mutant mice. In the absence of snai1, compensation by the homologous family member snai2 is not sufficient to rescue the lethal phenotype (Carver et al., 2001). Although snai2−/− mice are viable as a result of snai1 compensation (Jiang et al., 1998; Murray et al., 2007) and reports describe impaired cell migration and transformation in ECs (Niessen et al., 2008; Romano and Runyan, 1999). However, as one might expect, compensatory increases in snai1 expression are sufficient to rescue the reduced EC cellularization observed in embryonic snai2−/− mice (Niessen et al., 2008). Together, these studies highlight the importance of Snail family members during EMT and embryonic development.
In this study we aim to determine the direct role of Snai1 during EC formation using a mouse model with conditional reduced snai1 function and established in vitro systems. We demonstrate that Snai1 is highly expressed in overlying endothelial cells and transformed mesenchymal cells within the developing ECs, although expression is significantly downregulated during later, post-EMT stages of valvulogenesis. Mice with conditional heterozygous loss of snai1 in endothelial-derived cells (Tie2cre;Snai1fl/+) display fewer cells within the cardiac jelly of developing ECs, associated with reduced expression of mesenchymal gene markers. In addition, AVC regions from Tie2cre;Snai1fl/+ mice at E10.5, express significantly lower levels of the matrix metalloproteinase (mmp), mmp15 (MT2-MMP). Using the established collagen I gel explant assays (Bernanke and Markwald, 1982; Lincoln et al., 2006a; Runyan and Markwald, 1983), we show that treatment of AVC explants with adenovirus-targeting Snai1 (AdV-Snai1) promotes mmp15 expression and several EMT processes including cell transformation, invasion and migration. In contrast, treatment with a catalytically active MMP15 protein (caMMP15) promotes only cell motility. Further, pharmacological inhibition of MMP activity prevents Snai1-mediated mesenchyme cell migration, while caMMP15 treatment is sufficient to rescue attenuated migration phenotypes observed in AVC explants with targeted snai1 knockdown. Additional co-immunoprecipitation and luciferase assays indicate that Snai1 binds, and transctivates E-box-rich regions within mmp15. However, in the presence of the E2A family member, E47, Snai1 and E47 repress mmp15. Together, these studies support a direct role for Snai1 in EC development, and identify previously unappreciated mechanisms of mmp15 activity, regulated by snai1, for cell motility during EC EMT.
Materials and methods
Histological Analysis
Whole mouse and chicken embryos staged at embryonic day (E) 10.5 (mouse) and Hamburger Hamilton stage (HH St.) 14 (E2.5-3.0) were collected in 1× Phosphate Buffered Saline (PBS) and either fixed in 4% paraformaldehyde (PFA) overnight at 4°C or left unfixed. Fixed tissues were subsequently processed for paraffin embedding and 6μm tissue sections were cut as previously described (Lincoln et al., 2006a). Alternatively, unfixed tissue was immediately processed for frozen block cryoembedding (Lincoln et al., 2006a) and 12μm tissue sections were cut. For colorimetric and fluorescent immunohistochemistry, fixed tissue sections were processed (Lincoln et al., 2007) and incubated overnight at 4°C with primary antibodies against Snai1 (Abcam, 1:500), Smooth Muscle α-Actin (αSMA) (Invitrogen, 1:500), and Phospho-histone H3 (Upstate, 1:200). In contrast, unfixed frozen sections were post fixed in ice-cold acetone at −20° for 15 minutes, blocked in 5% bovine serum albumin/1xPBS for 30 minutes and incubated with anti-matrix metalloproteinase 15 (Mmp15) (Abcam, 1:200) for 2 hours at room temperature. Antigen retrieval was used for the detection of Snai1, Mmp15 and Phospho-histone H3 by boiling tissue sections in unmasking solution (Vector Labs) for 10 minutes and allowing to cool to room temperature prior to blocking. Detection using diaminobenzidine (DAB) was performed according to the manufacturer’s instructions (ABC staining system, Santa Cruz Biotechnology) and visualized on an Olympus BX51 microscope. Immunofluorescent staining was performed using appropriate secondary antibodies (Alexa-Fluor) and captured using Olympus Fluoview F-1000 confocal microscope. Alcian blue staining was performed on paraffin tissue sections from E10.5 and E13.5 Tie2cre;Snai1fl/+ embryos and counter-stained with nuclear fast red solution as previously reported (Lincoln et al., 2007).
Generation of Adenoviruses
Full-length FLAG-tagged mouse snai1 cDNA was generated by PCR amplification from E14.5 mouse limb template and ligated into the pShuttle-IRES-hrGFP-1 vector (Stratagene). In parallel, a control adenoviral construct was generated with no cDNA insert (AdV-GFP). Adenovirus generation and tittering was performed according to the manufacturer’s instructions using the AdEasy XL and AdEasy Viral Titer kits respectively (Stratagene). For AdV-Snai1 and AdV-GFP, infection efficiency was determined by counting the number of GFP-positive cells over the number of DAPI-positive nuclei in at least 5 microscopic fields (n=4). The AdV-cre was generated as previously described (Peacock et al., 2010) and snai1 knockdown in infected AVC explants from Snai1fl/fl mice was determined by qPCR using Taqman primers against mouse Snai1 (Applied Biosystems).
Endocardial Cushion Cell Culture
2D-Monolayer cell culture
AVC ECs were dissected from E4.5 white leghorn chicken eggs (Charles River Laboratories) and E11.5 Snai1fl/fl mouse embryos, trypsinized and cultured for 48 hours as a monolayer prior to treatment as described (Lincoln et al., 2006a).
3D-collagen gel I explant culture
AVC explants were collected from HH St. 14, and E5.0 chicks, and E11.5 Snai1fl/fl mouse embryos and placed lumen-side down on 3D-collagen I gels for 2 hours prior to treatment as previously described (Inai et al., 2008) (n=5-8).
Following initial culturing, monolayer cells or explants were infected with 3×106 and 2×107 PFU adenovirus respectively, expressing Snai1 (AdV-Snai1), Cre (AdV-Cre) (Peacock et al., 2010) or GFP (AdV-GFP) diluted in serum-free culture media (Lincoln et al., 2006a) for 2 hours. Following infection, fresh serum-free media was added and cells/explants were cultured for a further 48 hours (2D and 3D cultures mouse culture systems) or 72 hours (2D avian culture system). Alternatively, explants were treated with recombinant protein of the catalytic domain of MMP15 (caMMP15) (Chemicon/Millipore) at a final concentration of 1μg/mL in serum-free media for 36 hours (AVC 3D culture of E5.0 chicken embryos) or 48 hours (AVC 3D culture of HH St. 14 chicken embryos). For controls, explants were treated with serum-free media alone. For MMP inhibition studies, explants were subject to 2μg/mL, 10μg/mL and 20μg/mL GM6001 (Millipore) or dimethyl sulfoxide (DMSO) in the presence of either 2×107 PFU AdV-GFP or AdV-Snai1 for 36 hours. Following treatment, cells/explants were fixed in 4% PFA and subject to IHC or collected for mRNA extraction isolation (dissociated cell culture). To determine the area covered by migrating cells, 2D images were taken with an Olympus SZX7 and the total pixel counts of each cultured AVC explant was measured using ImagePro Plus software (n=5-8). To quantify the number of invading cells, immunostained 3-D collagen I gels were captured using an Olympus Fluoview F-1000 confocal microscope and Z-stack images were generated from multiple focal planes sections captured at 5μm intervals down to the furthest distance of invasion using Image J software. The fold change in cell invasion and maximal invading depth was calculated using ImagePro Plus software by counting the total pixel number and distance in Z-stacked images. Statistical significance over respective controls was determined using Student’s t-test (p<0.05) (n=5).
mRNA isolation, cDNA generation and quantitative PCR
Total mRNA was isolated from cultured monolayer EC cells as well as from E10.5 AVC regions from Tie2cre;Snai1fl/+ and Snai1fl/+ mice using TRIzol (Lincoln et al., 2007). 200-400ng of mRNA was used to generate cDNA using the high-capacity cDNA kit (Applied Biosystems) (Peacock et al., 2008). cDNA was subject to quantitative PCR amplification (StepOne Plus, Applied Biosystems) using specific primers targeting chicken vimentin (Forward: CCGACAGGATGTTGACAATG; Reverse: GGATGTGCTGTTCCTGGAGT), αSMA (Forward: CACCCAACTCTGCTGACTGA; Reverse: ACACCATCCCCAGAGTCAAG), VE-cadherin (Forward: ATCTCAGACAACGGCAATCC; Reverse: GAAAATTGCCACCAGTGCTT), claudin1 (Forward: GGAGGATGACCAGGTGAAGA; Reverse: TCTGGTGTTAACGGGTGTGA), fibronectin1 (Forward: CGTTCGTCTCACTGGCTACA; Reverse: ATTAATCCCGACACGACAGC), mmp15 (Forward: TGTCGGGGAACAACCTCTTC; Reverse: TTCTCCGTGTCCATCCACTG) and GAPDH (Forward: GGGTCTTATGACCACTGTCC; Reverse: GTAAGCTTCCCATTCAGCTCAG), or mouse fibronectin1 (Forward: GGTTGCCTTGCACGATGATA; Reverse: TGAGCTGAACACTGGGTGCT), αSMA (Forward: CTGACAGAGGCACCACTGAA; Reverse: CATCTCCAGAGTCCAGCACA), VE-cadherin (Forward: ACCGAGAGAAACAGGCTGAA; Reverse: AGACGGGGAAGTTGTCATTG), E-cadherin (Forward: CAAGGACAGCCTTCTTTTCG; Reverse: TGGACTTCAGCGTCACTTTG), snai1 (Forward: TGAGAAGCCATTCTCCTGCT; Reverse: GCCAGACTCTTGGTGCTTGT), mmp15 (Forward: GACCAGTATGGCCCCAACAT; Reverse: CCAATTGGCATGGGGTAGTT) and L7 (Forward: GAAGCTCATCTATGAGAAGGC; Reverse: AAGACGAAGGAGCTGCAGAAC). Also, Taqman probes (Applied Biosystems) were used to targeting mouse snai1. Alternatively, E10.5 mouse AVC cDNA was subject to SuperArray PCR analysis (PAMM-013C-12, SABiosciences) according to the manufacturer’s instructions (n=5). Following PCR analyses, the cycle count (Ct) was normalized to at least one housekeeping gene (GAPDH chicken, L7 mouse, the mean Ct of GAPDH, Gusb, Hsp90ab1) and the ΔCt and fold changes in experimental samples over controls were determined as described (Peacock et al., 2008). In addition, the mesenchymal to endothelial gene expression ratio in E10.5 mouse AVC cDNA was determined by subtracting the average ΔCt of endothelial (VE-cadherin, E-cadherin) from mesenchymal gene expression (fibronectin1, αSMA) (n=4). The ratio was further calculated based upon Power(2,−ΔΔCt). For absolute transcript calculation, the formula POWER(2, (40−ΔCt+CtL7) was used, with CtL7 denoting the average cycle count of L7 expression levels. Statistically significant differences in transcript levels were determined using student’s t-test on at least 3 independent experiments with p<0.05 considered significant after Student t-test analysis.
Generation of mice
Snai1fl/fl female mice (Murray et al., 2006) were bred with Tie2cre males (Kisanuki et al., 2001) to generate heterozygous offspring (Tie2cre;Snai1fl/+) and Cre negative Snai1fl/+ littermate controls at expected Mendelian ratios. Genotyping was performed by PCR as previously described (Carver et al., 2001; Lincoln et al., 2007; Murray et al., 2006). Timed embryonic staged litters of mice were collected at E10.5 and E13.5 from Snai1fl/fl female mice, counting day E0.5 by evidence of a copulation plug. Embryos were either fixed or AVC tissue (E10.5) was collected for mRNA extraction. Quantitation of the number of cells within the ECs from E10.5 and E13.5 Snai1fl/+ and Tie2cre;Snai1fl/+ mice was determined by counting the number of DAPI-positive nuclei in at least 3 tissue sections collected from 3-4 mutant and control mice.
Chromatin immunoprecipitation (ChIP)
Six canonical E-box consensus sites were identified within promoter region of the murine mmp15 gene (NC_000074.5; Chromosome: 8; Location: 8 D1; 8 45.5cM) and conservation between mouse, rat and human was determined using the basic local alignment search tool (NCBI blast). Snai1 binding to mmp15 was evaluated in whole E11.5 mouse embryonic hearts (8-10 hearts per sample, n=3). Protein/DNA complexes were cross-linked for 10 minutes in formaldehyde (Sigma) at a final concentration of 0.5%. Fixed tissue was lysed and sonicated three times for 10 seconds at 1 minute intervals (Ultrasonic cell disruptor; Microson). For ChIP, cell lysates were incubated with an antibody against Snai1 (6μg; Abcam) and incubated overnight at 4°C with gentle rocking. Immunoprecipitation with normal rabbit IgG was used as a negative control. ChIPs were performed according to the manufacturer’s instructions (EZ ChIP, MilliPore). Immunoprecipited and input DNA were subjected to qPCR using the following primers to amplify three E-box-rich regions within mmp15: region B (Forward: GCCACCACACCTGAATCTACTG; Reverse: GTGACTTGGGAAGCTAGGGTTG), region C (Forward: GAAGCAGGTGGATCTCTGTGAA; Reverse: TTGCCGGTTGTTCTAGCTGTAG) and region D (Forward: CCCCAGAGATAGGAGCTAAGCA; Reverse: CCTAACCCGAGGCTTCTCAGTA) Primers for region A were also included as a negative control as canonical E-box sites were not identified within this region: Forward: CCAGGAGTCCTAATCCCACACT; Reverse: TGCCCTCTACTGGTGA TTTCTG). Three independent ChIPs were performed and significant enrichment of E-box regions using the Snai1 antibody over IgG control as determined by qPCR, was determined by student’s t-test (p<0.05).
Dual Luciferase Assays
Luciferase constructs were generated using PCR to amplify three regions (B, C, D) of the mmp15 enhancer region (NCBI accession number NC_000074) from mouse tail genomic DNA template, using the following primers: pGL3-mmp15B, forward TAGGTACCATCCCCCTACGCAAGAAAAT and reverse TGAGCTCCTCCACTCTGGAGGCACACT (970bp). pGL3-mmp15C, forward TAGGTACCTCTCCCATGACCCTTTCATC and reverse TGAGCTCCCACTGGTGGACCTCTCTGT (1006bp). pGL3-mmp15D forward TAGGTACCGCGTGGGTGTGTAGGGTCTA and reverse TGAGCTCCCCAAACCGGG TGAGAAG (973bp). Amplified regions were gel purified and ligated into pGL3-basic using introduced Sac1 and Kpn1 (promega) sites. pcDNA-E47 and pcDNA-E12 plasmids were kindly provided by Dr. Nakamura (Funato et al., 2001).
Luciferase assays were performed in COS7 cells (ATCC) plated at 2×105 per well of a 24-well plate 24 hours prior to transfection with Lipofectamine reagent (Invitrogen) according to manufacturer’s instructions. 200ng of pGL3-mmp15B, pGL3-mmp15C, pGL3-mmp15D or empty pGL3, and 200ng of pcDNA-E47, pcDNA-E12 or empty pcDNA, were co-transfected into each well, along with 20ng of pGL4 (Renilla luciferase, Promega). All transfection were performed in 400μl OptiMem for 6 hours before the infection of either 3×106 PFU AdV-Snai1 or AdV-GFP. Cell lysates were collected 48 hours following treatment according to the manufacturer’s instructions for dual luciferase assays (Promega). Data is represented as an average percent of luciferase activity in cells infected with AdV-GFP and transfected with respective plasmids compared to AdV-GFP controls. All experiments were normalized to Renilla activity and transfected with equal concentrations of plasmid DNA (n=3-4).
Results
Reduced snail1 function in vivo leads to hypocellular endocardial cushions at E10.5
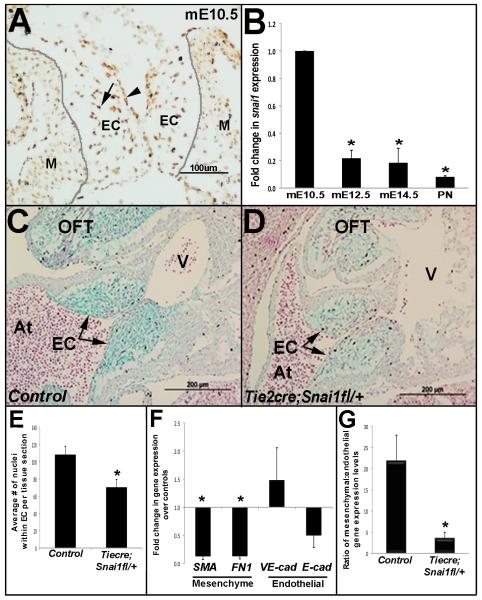
The expression pattern of Snai1 during endocardial cushion (EC) formation in the atrioventricular canal (AVC) was examined using immunohistochemistry (IHC) and quantitative PCR (qPCR). At E10.5, Snai1 expression is observed in endothelial cells overlaying the ECs (arrowhead, Fig. 1A), in addition to newly transformed mesenchyme cells within the cushions (arrow, Fig. 1A). Similar expression was observed in ECs within the outflow tract regions (data not shown). Nuclear expression is also detectable within the myocardium (M, Fig 1A). At the mRNA level, snai1 is most highly expressed in AVC regions at E10.5 when EMT is active, while expression is significantly reduced during post-EMT stages from E12.5 through post natal (Fig. 1B).
Figure 1. Tie2cre;Snai1fl/+ mice have reduced cell number in endocardial cushions at E10.5.
(A) Immunohistochemistry to show Snai1 localization in endothelial cells overlying the endocardial cushions (EC) (arrowhead) and transformed mesenchymal cells within the cushions (arrow). (B) Quantitative real-time PCR to show fold changes in snai1 expression in AVC regions isolated during differential stages of murine valve development. *p<0.05, statistical significance compared to E10.5, (n=4) (C, D) Alcian blue staining on tissue sections from E10.5 control snai1fl/+ (C) and Tie2cre;Snai1fl/+ (D) embryos to indicate atrioventricular canal and outflow tract (OFT) endocardial cushions (EC) (arrows, C-D), (n=3-4). (E) Quantitative analysis to show reduced number of nuclei within E10.5 AV ECs in Tie2cre;Snai1fl/+ mice compared to controls. (F) qPCR indicates significant decreases in expression of mesenchyme cell makers (SMA, fibronectin1) in Tie2cre;Snai1fl/+ embryos compared to the controls at E10.5. (G) The ratio of mesenchymal to endothelial gene expression is significantly increased in snai1 mutant mice, (n=4). *p<0.05, statistical significance compared to controls. At, atrium; V, ventricle; EC, endocardial cushion; AVC, atrioventricular canal; OFT, outflow tract; M, myocardium.
Although the role of Snai1 during EMT in many systems has been established, its role during EC formation has not been directly examined. To determine this, we used theTie2cre transgene to target loss of snai1 function in endothelial, and derived mesenchymal cells of the developing ECs in vivo (de Lange et al., 2004; Lincoln et al., 2004). Homozygous Tie2cre;Snai1fl/fl mice were never recovered after E9.5 and likely die due to vasculature defects as described following epiblast-specific snai1 deletion (Lomeli et al., 2009), However, heterozygous (Tie2cre;Snai1fl/+) mice are viable through adulthood and show a 50%±17% decrease in snai1 expression in AVC regions at E10.5. Upon histological examination, Tie2cre;Snai1fl/+ mice display significantly less cells within the AVC (Fig. 1C-E) and OFT (data not shown) ECs compared to littermate controls (Snai1fl/+) at E10.5. Insignificant differences in Phospho-histone H3 and cleaved caspase 3 expression suggest that hypocellular ECs observed in Tie2cre;Snai1fl/+ mice at E10.5 was not the result of changes in cell proliferation or apoptosis, respectively (data not shown, Fig. 2F). To determine changes in endothelial cell transformation in E10.5 Tie2cre;Snai1fl/+ mice, qPCR was used to quantitatively examine expression levels of mesenchymal (αSMA, fibronectin1) and endothelial (VE-cadherin, E-cadherin) cell markers in AVC regions isolated from Tie2cre;Snai1fl/+ and control mice. Expression of mesenchymal cell markers are significantly reduced in Tie2cre;Snai1fl/+ mice (SMA, −7.6-fold, fibronectin1, −7.4-fold), although insignificant changes were observed in endothelial markers (Fig. 1F). A consistent decrease in the ratio of mesenchymal to endothelial gene transcript levels confirms that observed decreases in mesenchymal cell marker expression was not a result of decreased cell number within ECs of Tie2cre;Snai1fl/+ mice (Fig. 1G).
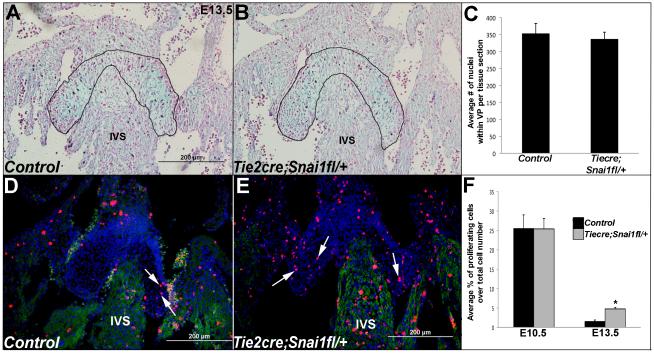
Figure 2. Proliferation is increased in endocardial cushions from Tie2cre;Snai1fl/+ mice at E13.5 and cell number is comparable with controls.
(A, B) Alcian blue staining on tissue sections from E13.5 Snai1fl/+ (control) and Tie2cre;Snai1fl/+ embryos to indicate AVC valve primordia structures (outlined). (C) Quantitative analysis to show comparable nuclei number within valve primordia of Tie2cre;Snai1fl/+ and control mice at E13.5. (D, E) Phospho-Histone H3 immunoreactivity to detect proliferating cells within valve primordia of Snai1fl/+ control (D) and Tie2cre;Snai1fl/+ (E) embryos at E13.5. (F) Percentage of Phospho-histone H3 positive cells within valve primordia from Tie2cre;Snai1fl/+ mice compared to controls at E10.5 and E13.5. n=4, *p<0.05, statistical significance compared to controls. IVS, interventricular septum.
Despite a significant decrease in the number of mesenchyme cells within ECs from Tie2cre;Snai1fl/+ mice at E10.5, by E13.5, cell number is comparable to controls (Fig. 2A-C). This is associated with increased Phospho-histone H3 immunoreactivity, indicative of cell proliferation in valve primordia from E13.5 Tie2cre;Snai1fl/+ mice compared to controls (Fig. 2D-F). Collectively, these findings demonstrate that reduced snai1 function leads to compromised EMT during early stages of EC development, however in association with increased proliferation, this phenotype recovers by later stages.
Mmp15 expression is reduced in atrioventricular canal regions from E10.5 Tie2cre;Snai1fl/+ mice
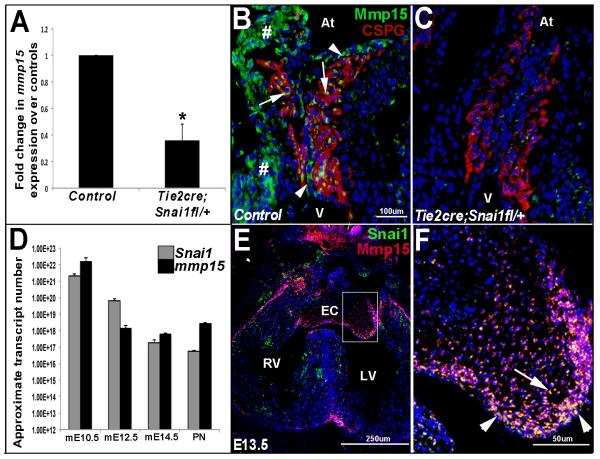
To identify downstream genes that may be affected by reduced snai1 function in developing ECs at E10.5, a gene expression screening approach was used. This high-throughout assay (Superarray) detects quantitative changes in expression of over 84 genes associated with the extracellular matrix (ECM) in AVC regions from E10.5 Tie2cre;Snai1fl/+ compared to Snai1fl/+ embryos. Table 1 indicates all the genes that were differentially expressed (p<0.05) in the Superarray analysis. The most significant difference was observed with mmp15 (MT2-MMP), a membrane bound matrix metalloproteinase family member decreased 5.3-fold in Tie2cre;Snai1fl/+ embryos (Fig. 3A). Interestingly, expression of other family members including mmps1, 2, 3, 7-14 were not affected (data not shown). Further IHC analysis shows reduced mmp15 protein expression in both endothelial (arrowheads, Fig. 3B, C) and mesenchymal cells (arrows, Fig. 3B,C) within ECs, highlighted by chondroitin sulfate proteoglycan immunoreactivity (red, Fig. 3B, C). To examine snai1 expression relative to mmp15 during valve development, qPCR was performed using cDNA from AVC regions at E10.5, E12.5, E14.5 and post natal stages (Fig. 3D). Approximate transcript levels of snai1 and mmp15 showed similar temporal expression patterns throughout valve development with transcript levels being highest at E10.5 and gradually declining to post natal stages. Consistently, co-localization studies show that snai1 and mmp15 are both expressed in endothelial (arrowhead, Figure 3F) and mesenchymal (arrow, Figure 3F) cells of the valves at E13.5 (Figs. 3E, F). Although due to the expected membrane localization of mmp15 and largely nuclear distribution of snai1, complete overlap is not always observed. Together, these studies suggest that snai1 and mmp15 are similarly expressed in developing valve structures and Snai1 function is required for mmp15 expression during EMT stages.
Table 1. Superarray analysis to show differential gene expression in E10.5 atrioventricular canal regions from Tie2cre;Snai1fl/+ mice.
Following analysis, gene expression was normalized to the mean cycle count of GAPDH, Gusb, Hsp90ab1. Fold changes are shown compared to snai1fl/+.
| GENE | Fold change | p value |
|---|---|---|
|
Emilin 1
(elastin microfibril interfacer 1) |
0.16±0.05 | 0.002 |
|
ltβ33
(Integrin- βeta 3) |
0.37±0.1 | 0.039 |
|
Lamc1
(laminin-gamma 1) |
0.39±0.09 | 0.030 |
| Mmp15 | 0.27±0.12 | 0.028 |
|
Sparc
(secreted protein, acidic, cysteine-rich) |
0.11 ±0.1 | 0.006 |
|
Spp1
(secreted phosphoprotein 1) |
0.14±0.13 | 0.023 |
|
Thbs1
(thrombospondin 1) |
0.08±0.1 | 0.022 |
|
Timp2
(tissue inhibitor of metalloproteinase 2) |
0.18±0.08 | 0.016 |
Figure 3. Mmp15 expression is reduced in endocardial cushions from Tie2cre;Snai1fl/+ mice at E10.5.
(A) Superarray qPCR analysis to show decreased mmp15 expression in AVC regions from E10.5 Tie2cre;Snai1fl/+ mice at E10.5 compared to snai1fl/+ controls. p<0.05, statistical significance compared to controls, (n=4). (B, C) Immunohistochemistry to show decreased mmp15 expression in endothelial (arrowheads) and mesenchymal (arrows) cells of the ECs in Tie2cre;snai1fl/+ mice (C) compared to controls (B). Red staining indicates chondroitin sulfate proteoglycan (CSPG) immunoreactivity to highlight the ECs (B, C), # indicates non-specific immunoreactivity in myocardium. (D) qPCR analysis to show approximate absolute transcript numbers of snai1 and mmp15 at mouse (m) embryonic stages (E) 10.5, 12.5 14.5 and post natal (PN) (n=4). (E, F) Co-localization studies reveal similar expression patterns between Snai1 (green) and Mmp15 (red) in endothelial (arrowheads) and mesenchyme (arrow) cells within the endocardial cushion at E13.5. EC, endocardial cushion; At, atria; V, ventricle; RV, right ventricle; LV, left ventricle.
Snai1, but not MMP15 is sufficient to initiate EMT and promote cell transformation in avian atrioventricular canal explants
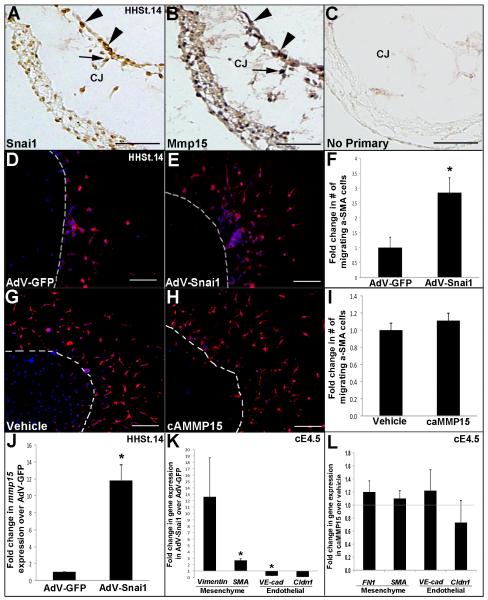
Understanding the mechanisms of EC EMT have been greatly enhanced by studies in the chick using collagen I gel explant systems (Bernanke and Markwald, 1982; Person et al., 2005). In the avian model, EC formation is initiated in the looped heart at HH St. 14, and by HH St.18, EMT is active in all cushion sets (Person et al., 2005). Worthy of mention, the EMT inductive signals in the chick appear conserved with the mouse system and include Bmp and Tgfβ signaling (Armstrong and Bischoff, 2004; Combs and Yutzey, 2009; Person et al., 2005). To investigate if Snai1 and MMP15 are sufficient to promote EMT in AVC endothelial cells, we employed a published in vitro collagen I gel explant assay (Bernanke and Markwald, 1982; Mjaatvedt et al., 1987; Runyan and Markwald, 1983). In this assay, HH St.14 AV canals are placed on a 3-dimensional (3D) rat-tail collagen I gel with the endothelial cell layer facing down, allowing cells to migrate onto the surface of the gel (Runyan and Markwald, 1983). With the myocardium intact, subsets of endothelial cells then invade the underlying gel as newly transformed mesenchymal cells (Inai et al., 2008; Mjaatvedt et al., 1987; Runyan and Markwald, 1983). To validate the use of the avian system in our study, IHC was performed to determine localization of Snai1 and Mmp15 in avian ECs at HHSt.14. As shown in Figures 4A and B, Snai1 and Mmp15 are similarly expressed in endothelial (Fig. 4A, B arrowheads) and mesenchyme (arrows) within the developing cushion. To determine if Snai1 or MMP15 promote cell transformation, HH St.14 AVC explants were subjected to Snai1 gain of function by infecting with a GFP-tagged adenovirus containing full-length FLAG-labeled mouse Snai1 (AdV-Snai1) (Fig. 4E), or a control virus with no insert (AdV-GFP) (Fig. 4D). Exogenous levels of murine Snai1 following adenoviral infection were confirmed by western blot to detect the FLAG epitope (data not shown). For overexpression experiments, the infection efficiency of the AdV-GFP and AdV-Snai1 adenoviruses was comparable with an average of 55.8%±2.29% cells being infected using AdV-GFP and 53.0%±6.45% with AdV-Snai1. To investigate the function of MMP15 during EC EMT, explants were alternatively treated with an exogenous human recombinant MMP15 catalytic domain that functions as a constitutively active form of MMP15 (caMMP15) (Rebustini et al., 2009). Following 48-hour treatment, explants were subjected to IHC to detect transformed αSMA-positive mesenchyme cells that migrate over the surface of the collagen gel away from the original explant site as (indicated by dotted white line) (Fig. 4D, E, G, H). Compared to AdV-GFP controls (Fig. 4D), AdV-Snai1 treatment (Fig. 4E) significantly increases the number of αSMA positive cells by over 2-fold (Fig. 4F), whereas compared to respective controls, caMMP15 does not promote EMT in HH St.14 AV canal explants (Fig. 4G-H). In addition, increased cell migration in response to AdV-Snai1 treatment was associated with increased mmp15 expression (11.83±1.82-fold), as determined by qPCR (Fig. 4J).
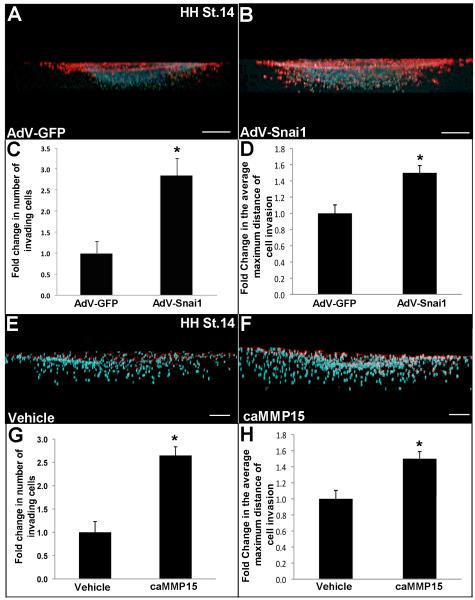
Figure 4. Snai1, but not MMP15 promotes initiation of endocardial cushion EMT and cell transformation.
(A, B) Immunohistochemistry to show Snai1 (A) and Mmp15 (B) expression in endothelial (arrowheads) and mesenchyme (arrows) cells of the developing EC in the chick at HH St.14. (C) No primary antibody control. Scale bar=50μm. (D-I) HH St.14 AVC explants on 3D collagen I gels were treated with AdV-GFP (D), AdV-Snai1 (E), vehicle (water) (G) or caMMP15 (H). Scale bar=200μm. (F, I) Graph to show fold change in the number of α-SMA positive cells (red) that have migrated away from the original explant site (dotted line) following treatments. Note increased α-SMA positive cells with AdV-Snai1 treatment. (J) qPCR to show increased mmp15 expression in AdV-Snai1 treated HH. St. 14 AVC explants compared to AdV-GFP controls. (K, L) qPCR to indicate changes in gene expression of mesenchymal (vimentin, SMA, fibronectin1 (FN1)) and endothelial (VE-cadherin (VE-cad) and Claudin1 (Cldn-1)) markers in cE4.5 dissociated endocardial cushion cells treated with AdV-Snai1 compared to AdV-GFP controls (K) or caMMP15 compared to vehicle (water) controls (L). Note the increase in mesenchymal gene expression and decrease in endothelial genes with AdV-Snai1, but not caMMP15 treatment. n=5-8, *p<0.05, compared to respective controls.
To support these studies, and determine if snai1 or MMP15 can promote cell transformation at later stages of EC development, ECs from HH St. 25-26 (cE4.5) embryos were removed from the adjacent myocardium, trypsinized and cultured as a 2-dimensional (2D) monolayer. This system allows for minimal cell-cell contact and therefore changes in transformation can be examined in a neutral environment (Lincoln et al., 2006a). Dissociated cells were similarly treated with AdV-Snail, AdV-GFP, caMMP15 or vehicle and following 72 hours, changes in expression of mesenchymal (vimentin, fibronectin, SMA) and endothelial (VE-cadherin, claudin1) cell markers were examined by qPCR (Fig. 4J, K). AdV-Snai1 treatment leads to increased expression of mesenchymal cell markers vimentin (+12.6-fold) and αSMA (+2.67-fold), and associated decreased expression of endothelial cell markers VE-cadherin (−4.0-fold) and claudin1 (−15.68-fold), compared to AdV-GFP controls (Fig. 4J). However, gene expression indicative of cell transformation was not affected with caMMP15 treatment compared to respective vehicle (water) controls (Fig. 4K). These data support a role of Snai1, but not MMP15, in promoting cell transformation during EC development.
Snai1 and MMP15 are sufficient to promote cell invasion and migration in developing endocardial cushions
For EC formation to occur, transformed cells from the overlying endothelial cell layer must invade the cardiac jelly and migrate to their destination within the expanding cushion (Person et al., 2005). To mimic EC cell invasion in vitro and examine if Snai1 and MMP15 play a role in this process, HH St. 14 AVC explants with minimal intact myocardium were placed on 3D collagen I gels and subjected to AdV-Snai1, AdV-GFP, caMMP15 or vehicle treatment. By reconstructing 2D stacks of Z series captured by confocal imaging, we observe that both AdV-Snai1 and caMMP15 treatments increase cell invasion (Fig. 5A-C, E-G), and the distance travelled by invading cells compared to respective controls (Fig. 5A, B, D, E, F, H). In addition, and consistent with observations in Figures 4J and K, the number of αSMA positive cells appears higher with AdV-Snai1, but not caMMP15 treatment. These findings suggest that in ECs, Snai1 and MMP15 play roles in promoting cell invasion during stages of EMT initiation.
Figure 5. AdV-Snai1 and caMMP15 treatments promote cell migration in HH St. 14 AVC explants.
(A, B, E, F) HH St.14 AVC explants were placed on 3D collagen I gels and treated with AdV-GFP (A), AdV-Snai1 (B), vehicle (water) (E) or caMMP15 (F). Scale bars in A, B=200μm, E, F=100μm. Confocal microscopy and Z-stack reconstruction (A, B, E, F) was used to determine the fold change in the number of cells invading the 3D collagen I gel (C, G), and the average maximum distance of invasion (D, H) in AdV-Snai1 (B) and caMMP15 (F) treated cultures compared to respective controls (A, E). Immunohistochemistry indicates α-SMA positive cells (red) and DAPI indicates nuclei (blue). Note increased invasion with AdV-Snai1 and caMMP15 treatments. n=5, *p<0.05, statistical significance compared to respective controls.
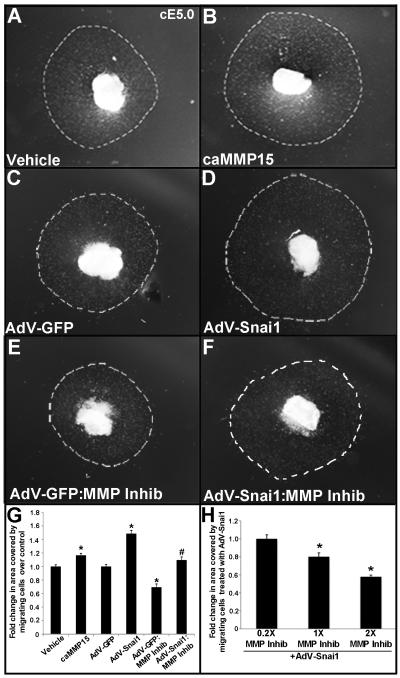
The sufficiency of Snai1 and MMP15 to promote migration of transformed mesenchyme cells within the cardiac jelly of developing ECs was examined using cE5.0 AVC explants on 3D collagen I gels. Similar to cell invasion assays, both caMMP15 and AdV-Snai1 treatments increase the area covered by migrating cells on the surface of the collage I gel, over respective controls (Figs. 6A-D, G). To further investigate the relationship between Snai1 and Mmps during cell migration, explants were treated with the pan-MMP inhibitor GM6001 (1x) (Fig. 6E-F) in the presence of AdV-GFP (Fig. 6E) or AdV-Snai1 (Fig. 6F). Compared to AdV-GFP controls (Fig. 6C), inhibition of MMP activity (AdV-GFP:MMP Inhib, Fig. 6E) significantly reduces the area covered by migrating cells on the gel surface. Further, MMP inhibition in the presence of AdV-Snai1 (Fig. 6F) attenuates the ability of AdV-Snai1 (Fig. 6D) to promote cell migration in a dose dependent manner (0.2x, 1x, 2x) (Fig. 6D compared to 6F, 6H). These observations suggest that Snai1-mediated cell migration in maturing ECs requires MMP activity.
Figure 6. Loss of MMP activity attenuates snai1-mediated mesenchyme cell migration in developing endocardial cushions in vitro.
(A-D) cE5.0 AVC collagen I explants treated with vehicle (water) (A), caMMP15 (B), AdV-GFP (C) AdV-Snai1 (D), or AdV-GFP (E) or AdV-Snai1 (F) in the presence of the MMP inhibitor (MMP Inhib) GM600. (G) Quantitation to show fold change in cell migration area on the surface of the collagen I gel (dotted line) following each treatment *p<0.05, statistical significnace compared to respective controls, #p<0.05, significance compared to AdV-Snai1. (H) Dose-response effects of GM6001 on cell migration in the presence of AdV-Snai1. n=5-8, *p<0.05 compared to 0.2x treatment.
caMMP15 treatment rescues attenuated mesenchyme cell migration in murine endocardial cushions with reduced snai1 function
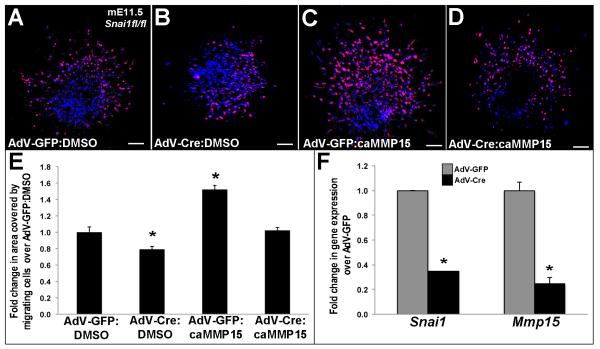
Tie2cre;Snai1fl/+ mice display defects in cell transformation (Fig. 1F). To determine if reduced snai1 function also affects cell migration, AVC regions isolated from E11.5 Snai1fl/fl mice were cultured on 3D collagen I gels and infected with AdV-Cre resulting in a 2.88-fold decrease in snai1 expression (Figure 7F). Compared to AdV-GFP controls (Fig. 7A), AdV-Cre treatment significantly reduces cell migration (Fig. 7A, B, E), associated with a 4-fold decrease in mmp15 (Figure 7F). However, cell migration defects are no longer observed when explants were co-treated with caMMP15 (Fig. 7B compared to 7D). Therefore suggesting that caMMP15 treatment is sufficient to rescue attenuated cell migration defects observed in ECs following reduced snai1 function.
Figure 7. caMMP15 treatment rescues attenuated cell migration defects observed in AVC explants with Snai1 knockdown.
(A-F) E11.5 AVC collagen I explants from snai1fl/fl mice treated with AdV-GFP:DMSO (A), AdV-Cre:DMSO (B), AdV-GFP:caMMP15 (C) or AdV-Cre:caMMP15 (D). (E) Quantification to show fold change in the area covered by migrating cells on the surface of the collagen I gel compared to AdV-GFP:DMSO treatment (*p<0.05). Note decreased migration with AdV-Cre:DMSO treatment compared to AdV-GFP:DMSO, and rescue with the addition of caMMP15 (AdV-Cre:caMMP15) to levels comparable with AdV-GFP:DMSO. (E) qPCR to show decreased Snai1 expressed with AdV-Cre infection in explants from snai1fl/fl mice, in addition to downregulation of mmp15. n=4-6, *p<0.05 compared to AdV-GFP).
Snai1 binds and regulates E-box-rich sequences within the promoter region of mmp15
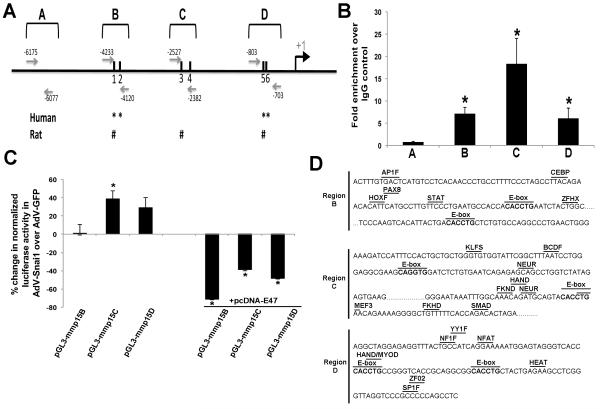
There are increasing data to suggest that Snai1 may regulate MMP function in several cell types to promote motility (Joseph et al., 2009; Ota et al., 2009; Rowe et al., 2009), however molecular interactions have not been reported. Snail family members regulate transcription via interactions with E-box binding sites (CANNTG) on target gene DNA. Within 6000bp upstream of the ATG transcription start site of the murine mmp15 gene, a total of six canonical E-box sites have been identified denoted within three regions (B-D), each containing two E-boxes (Fig. 8A). Of note, three of the six E-box sites are conserved with rat and four with human mmp15 promoter regions (indicated in Fig. 8A). Snai1 binding to regions B, C and/or D (Fig. 8A) in mmp15 was assessed using chromatin immunoprecipitation (ChIP) in whole E11.5 mouse hearts. Region A was included as a negative control as it does not contain canonical E-box sites. Cross-linked DNA/protein complexes immunoprecipitated with Snai1 antisera demonstrate significant enrichment of binding within B, C and D regions of mmp15 compared to IgG controls. In contrast, enrichment was not observed in region A (Fig. 8B). To further assess the functionality of E-boxes within regions B, C and D, luciferase assays were performed in COS-7 cells using pGL3 constructs containing ~1kb amplified sequences targeting each region (pGL3-mmp15B (970bp), pGL3-mmp15C (1006bp), pGL3-mmp15D (973bp)). 6 hours after transfection with respective pGL3-mmp15 constructs, cells were further infected with AdV-GFP or AdV-Snai1 (as used in Figures 4-6). Despite, enrichment observed by ChIP, AdV-Snai1 infection of cells transfected with pGL3-mmp15B had no significant effect on luciferase activity of Region B, compared to AdV-GFP:pGL3mmp15B controls (Figure 3C). Worthy of mention, significant luciferase activity was also not observed in AdV-infected cells co-transfected with empty vectors (data not shown). However, consistent with ChIP findings, AdV-Snai1 infection significantly increased transactivation of mmp15 region C (pGL3-mmp15C) (38.49%±8.73%) over AdV-GFP, with trends of activation also observed with Region D (pGL3-mmp15D)(29.0%±10.79%, p=0.3).
Figure 8. Snai1 binds and regulates E-box rich regions within the promoter region of mmp15 in whole mouse hearts.
(A) Schematic diagram to illustrate the location of six E-box sites (black solid bars) within three regions (B, C, D) of mmp15 at indicated base pair positions upstream of the ATG start site (+1). Region A is included as a negative control and does not contain canonical E-box sites. Grey arrows indicate the location of PCR primers used to determine enrichment in DNA/protein complexes following Snai1 pull down. *, # indicate conservation of E-box sites with human (*) and rat (#) sequences, n=3-4 (B) qPCR to show significant enrichment of Snai1 binding to regions B, C and D within the mmp15 promoter following co-immunprecipitation using whole E11.5 mouse hearts, over IgG controls. Note insignificant enrichment in region A that does not contain E-box sites. *p<0.05 compared to IgG controls (n=3). (C) Luciferase assays to show significant increased luciferase activity of pGL3-mmp15C in the presence of AdV-Snai1, compared to respective AdV-GFP controls. Co-transfection with pcDNA-E47, represses luciferase activity of pGL3-mmp15B, pGL3-mmp15C and pGL3-mmp15D in the presence of AdV-Snai1, but not AdV-GFP. Data is represented as a fold change in luciferase activity normalized to Renilla in AdV-Snai1 infected cells co-transfected with indicated plasmids compared to parallel AdV-GFP infected controls. n=3-4, *p<0.05. (D) Mmp15 sequence of Regions B, C and D to show predicted transcription factor binding sites, based on Genomatix analysis with predicted ‘matrix similarity’ >0.90. The core binding consensus sequences are indicated: AP1F, Activating Protein 1; CEBP, CCAAT/Enhancer Binding Protein; HOXF, Hox Family; PAX8, Pax 2/5/8 binding site; STAT, Signal transducer and activator of transcription 1; ZFHX, Two-handed zinc finger homeodomain transcription factors; KLFS, Krueppel like transcription factors; BCDF, Bicoid-like homeodomain transcription factors; NEUR, NeuroD, beta2, HLH domain; FKND, Fork head domain factor; HAND, Twist subfamily of bHLH factors; MEF3, myocyte-specific enhancing factor 3; SMAD, SMAD family; NF1F, nuclear factor 1; NFAT, Nuclear Factor of activated T-cells; YY1F, Activator/repressor binding to transcriptional initiation site; MyoD, myoblast determining factors; HEAT, Heat shock factors; SP1F, GC-Box factors SP1/GC; ZF02, C2H2 Zinc finger transcription factors 2.
Previous studies have shown that Snail and E2A (E47/E12) family members regulate activity of common target genes through interaction with specific E-box sites within the proximal promoter (Bolos et al., 2003; Cano et al., 2000; Peinado et al., 2004). To determine if E47 or E12 function effects Snai1-mediated transactivation of mmp15, luciferase assays were repeated in the presence of pcDNA-E47 or pcDNA-E12. Although E2A family members alone have been shown to act as transcriptional activators (Takahashi et al., 2004) and repressors (Batlle et al., 2000; Cano et al., 2000; Perez-Moreno et al., 2001), co-transfection of pGL3-mmp15B, pGL3-mmp15C or pGL3-mmp15D with pcDNA-E47 or pcDNA-E12 in the presence of AdV-GFP, had no effect on mmp15 activity (data not shown). However, co-transfection of pcDNA-E47 in the presence of AdV-Snai1 significantly repressed pGL3-mmp15B (−70.82%±-19.77%), pGL3-mmp15C (−39.09%±0.46%) and pGL3-mmp15D (−48.40%±-17.07%). Co-transfection with pcDNA-E12 had no significant effect on AdV-Snai1-mediated mmp15 activity. Collectively these studies reveal that Snai1 physically interacts and activates specific E-box-rich promoter regions within mmp15. Further, we provide evidence to suggest that E47 function plays an important role in Snai1-mediated regulation of mmp15.
Discussion
In developing ECs, the EMT program converts endothelial cells into migratory mesenchymal cells through sequential steps that require many phenotypic changes (Barrallo-Gimeno and Nieto, 2005; Thiery et al., 2009). The transcription factor Snai1 has previously been shown to be indispensible for EMT in many tissues during embryonic development and pathological processes (Nieto, 2009; Thiery et al., 2009). Despite snai1 expression correlating with EMT activity in developing valve structures (Liebner et al., 2004; Luna-Zurita et al.; Meadows et al., 2009; Nath et al., 2008; Wang et al., 2005), a direct role in EC formation has not previously been reported. This is likely due to the premature lethality of Snai1−/− embryos prior to cardiogenesis (Carver et al., 2001). To address this limitation, we performed in vivo studies using viable Tie2cre;Snai1fl/+ mice, in parallel with gain and loss of Snai1 function in AVC explants in vitro. The goal of this study was to use these previously established systems to reveal new insights into the role of Snai1 for cell transformation, invasion and migration in developing ECs in the embryonic heart.
Prior to EMT, Snai1 is highly expressed in endothelial cells of the AV canal and expression is maintained in this cell layer throughout EC formation and valvulogenesis (Fig. 1A, B, 4A) (G. Tao, Unpublished). During EMT, Snai1 expression is also observed in transformed mesenchyme cells, however expression is downregulated as these cells differentiate during remodeling stages (Fig. 1A, B, 4A) (Lincoln et al., 2006b). In this study we show that Snai1 is important in many sequential steps during EMT beginning with the early transformation of a subset of cardiac endothelial cells into mesenchyme cells, and proceeding mesenchyme cell invasion and migration. Further, we show that MMP activity is required for Snai1-mediated mesenchyme cell migration and more specifically, MMP15 activity is sufficient to rescue migratory phenotypes observed in ECs with targeted snai1 knockdown. Together, findings from this study suggest that Snai1 is an important regulator of EC formation and directly binds and regulates mmp15 to promote cell motility during this process.
Snai1-mediated signaling pathways in EMT have been extensively studied and therefore parallel mechanisms in the developing heart are considered. Several known upstream regulators of Snai1 are expressed during cardiogenesis (Luna-Zurita et al., 2010; Molin et al., 2003; Niessen et al., 2008; Thiery et al., 2009) and manipulation studies of Notch1, Tgfβs and Bmps, their receptors or intermediate signaling kinases affect snai1 expression in developing cushions (Luna-Zurita et al., 2010; Meadows et al., 2009; Wang et al., 2005). Therefore Snai1-induced EC EMT is likely initiated by these established signaling pathways in vivo. Downstream, we can speculate that Snai1 promotes EMT in developing cushions through mechanisms previously described in other non-cardiac tissues including repression of cell adhesion genes (Cano et al., 2000; Liebner et al., 2004; Meadows et al., 2009; Nath et al., 2008) and activation of mesenchymal markers (Batlle et al., 2000; Cano et al., 2000; Guaita et al., 2002; Ikenouchi et al., 2003; Person et al., 2005). In Tie2cre;Snai1fl/+ mice, it is likely that these signaling pathways are abrogated as a result of reduced snai1 function, leading to hypocellular ECs. In addition to attenuated transformation, it is plausible that deficient mesenchyme cell motility and/or imbalanced cell survival also contribute to this phenotype. Indeed, our data supports a positive role for Snai1 in cell invasion and migration in developing ECs, however changes in cell proliferation or apoptosis were not observed in Tie2cre;Snai1fl/+ mice at E10.5. In contrast, cell proliferation was enhanced at E13.5 and levels were sufficient to increase mesenchyme cell number to levels comparable with controls. Compensation of EC cellularity was similarly reported in Snai2−/− mice that exhibit hypocellular cushions at E9.5 but not E10.5, and attributed to increased snai1 expression (Niessen et al., 2008). However, increased expression of Snail1 family members were not detected in Tie2cre;Snai1fl/+ mice (data not shown), consistent with observations reported in snai1−/− mice (Carver et al., 2001). Therefore it is likely that alternative mechanisms promote the compensatory proliferative response in ECs from mice with reduced snai1 function.
In addition to developmental processes, Snai1-mediated EMT is active in many cancer cell lines and high expression levels correlative with increased cell migration and metastasis (Thiery et al., 2009; Wu and Zhou). A recent study suggested that this pathologic function of snai1 is mediated through induction of MT-MMPs, that when activated, recapitulate snai1’s ability to degrade basement membranes and promote cancer cell migration (Ota et al., 2009). In this study, we present several lines of evidence to suggest that in addition to disease, activated MT-MMPs also promote cell motility, but not transformation, in the embryonic heart during EC development. Further, we identify mmp15 (or MT2-MMP) as a direct target gene of Snai1, and a positive regulator of mesenchyme cell invasion and migration. This finding extends the current repertoire of known target genes regulated by Snai1 during EMT beyond cell transformation processes (Batlle et al., 2000; Cano et al., 2000; Guaita et al., 2002; Ikenouchi et al., 2003; Person et al., 2005).
In ECs from Tie2cre;Snai1fl/+ mice, mmp15 was the only MMP family member whose expression was affected by reduced snai1 function, although decreased trends of the MMP15 target gene mmp9 were also observed (data not shown). The widespread protein localization of mmp15 throughout the ECs suggests that this mmp family member may target ECM substrates in both endothelial and mesenchymal cells. Type IV collagen is a known substrate of MMP15 in basement membranes (Hotary et al., 2000; Rebustini et al., 2009) and its degradation in endothelial cells lining the AV canal and OFT regions is important for EMT and EC formation (Song et al., 2000). Therefore leading to the conclusion that mmp15 may facilitate endothelial cell delamination, independent of transformation in developing ECs. Mmp15’s function in mesenchymal cells is less speculative and its activity correlates with increased mesenchyme cell invasion and migration in AV canal explants. Like mmp15, other family members including mmp2 and 9 are also highly expressed in mesenchyme cells during EC EMT, where they facilitate ECM degradation to promote mesenchymal cell motility through the hyaluronan-rich cardiac jelly (Rupp et al., 2008; Shelton and Yutzey, 2007). The observation that Snai1 directly binds and activates mmp15 is consistent with a recent study showing that Snai2 (slug) binds and regulates activity of another family member, membrane-type 4 MMP (Huang et al., 2009). Findings from this current study have revealed a new role for snai1 as a transcriptional activator of mmp15 to promote cell migration during EC EMT.
The observation that Snai1 acts as a transcriptional activator of mmp15 is somewhat surprising as despite emerging evidence to support this function (Batlle et al., 2000; Cano et al., 2000; Guaita et al., 2002; Hwang et al., 2011; Ikenouchi et al., 2003; Person et al., 2005), Snai1 is more commonly known as a repressor (Nieto, 2009). In our system, Snai1 increases mmp15 transactivity by 38%, which could be considered a subtle, albeit significant, difference. As this study is one of the first to show direct transactivation of target genes by Snai1, it is not clear if the addition of co-activators (or removal of co-repressors) could increase mmp15 activity above this level. In contrast to AdV-Snai1 treatment, E47 alone has no effect on mmp15, yet E47 in the presence of AdV-Snai1, leads to repression. As previous studies have shown that Snai1 and E47 regulate common target genes through interaction with specific E-boxes (Bolos et al., 2003) (Peinado et al., 2004; Perez-Moreno et al., 2001), we speculate that overexpressed E47 binds with no functional effect, to prevent Snai1 interaction and transactivation of mmp15. This mechanism may also in part explain the subtle increase in mmp15 activity with AdV-Snai1 treatment: as endogenous E47 competes with Snai1 for E-box binding. This competition may also be dependent on differential binding affinities of Snail1 and E47 on the same target genes (Bolos et al., 2003). Although this might explain the lost activation of mmp15 by Snai1, the mechanism in which Snai1 and E47 cooperate to repress mmp15 is not clear, but likely requires recruitment of co-repressor machinery at the putative E-box sites. Our studies using the in vivo mouse and in vitro chick models, strongly suggest that Snai1 positively regulates mmp15 in these systems, therefore suggesting that endogenous E2A expression levels are low at this time. However, it remains unknown how Snai1-E2A work together to regulate mmp15 during post-EMT stages when cell motility is less active.
There is increasing evidence that adult heart valve disease has origins in embryonic development (Combs and Yutzey, 2009). Tie2cre;Snai1fl/+ mice display EC defects by E10.5, although heterozygous embryos recover by E13.5 and adult valve morphology is grossly indistinct from control animals. Therefore alternative approaches are needed to thoroughly examine the requirement of Snai1 during EC development, for adult cardiac function. Nonetheless this study provides new insights into the regulatory functions of Snai1 through mmp15, to form the pool of precursor cells within the ECs that later give rise to the diversified mature valve structures (Lincoln et al., 2004). Worthy of mention, snai1 expression is detectable at post-EMT stages in endothelial cells that overlay the maturing valve leaflets (G. Tao, Unpublished). It remains unknown why valve endothelial cells at these later stages do not undergo EMT in vivo, even in the presence of Snai1. As discussed above in the context of mmp15, this is likely dependent upon the maturation of VECs during valvulogenesis and therefore related to the absence (or presence) of some Snai1 targets or co-factors (Barrallo-Gimeno and Nieto, 2005). However, as treatment strategies for degenerative heart valve disease are limited, it would be beneficial to determine if snai1 can provide a genetic ‘cue’ to reactivate developmental EMT programs in post-natal valves as a means to maintain or replace the interstitial valve cell population during life. During later stages of valvulogenesis Snai1 may have additional functions, and based upon its repressive function on ECM genes (Seki et al., 2003) and positive regulation of mmp activity (this study), it is plausible that Snai1 is involved in valve remodeling (Lincoln et al., 2006b). Further, alterations in the regulation of Snai1 during this process may have implications in degenerative valve disease characterized by pathological remodeling and associated with increased MMP-2, -9 and -13 expression (Kaden et al., 2004; Rabkin et al., 2001; Soini et al., 2001). In conclusion, this current study has revealed new information into the role of snai1 during early heart valve formation that may provide insights into mechanisms of congenital heart valve disease that result from defects in EC formation.
*Highlights.
We examine the roles of Snai1 and MMP15 in EMT during endocardial cushion development
Snail is required for endothelial cell transformation, and mesenchyme cell invasion and migration during endocardial cushion EMT
MMP15 is sufficient to promote cell motility, but not transformation during endocardial cushion EMT
MMP activity is required for Snai1-mediated cell migration during endocardial cushion EMT
Increased MMP15 activity rescues cell migration defects in endocardial cushions following snai1 knockdown
Snai1 binds and regulates E-box rich regions within the mmp15 gene
Acknowledgements
We thank Harriet Hammond for editorial assistance, Dr. Nakamura for kindly providing DNA constructs, and Drs. Jacqueline Peacock, George McNamara and Yuhui Wen for scientific and technical support. This work was supported by NIH/NHLBI (1R01HL091878-JL) and AHA Predoctoral Fellowship (10PRE4360052-GT).
Grant Information: NIH/NHLBI R01HL91878 (JL)
AHA Predoctoral Fellowship 10PRE4360052 (GT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95:459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bernanke DH, Markwald RR. Migratory behavior of cardiac cushion tissue cells in a collagen-lattice culture system. Dev Biol. 1982;91:235–245. doi: 10.1016/0012-1606(82)90030-6. [DOI] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. Journal of cell science. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange FJ, Moorman AF, Anderson RH, Manner J, Soufan AT, de Gier-de Vries C, Schneider MD, Webb S, van den Hoff MJ, Christoffels VM. Lineage and morphogenetic analysis of the cardiac valves. Circ Res. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- Funato N, Ohtani K, Ohyama K, Kuroda T, Nakamura M. Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Molecular and cellular biology. 2001;21:7416–7428. doi: 10.1128/MCB.21.21.7416-7428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- Hinton RBJ, Lincoln J, Deutsch GH, Osinaka H, Manning PB, Benson DW, Yutzey KE. In preparation. 2006. Extracellular matrix remodeling and organization in developing and diseased aortic valves. [DOI] [PubMed] [Google Scholar]
- Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, Wu KJ, Yang MH. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 2009;11:1371–1382. doi: 10.1593/neo.91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Yang MH, Tsai ML, Lan HY, Su SH, Chang SC, Teng HW, Yang SH, Lan YT, Chiou SH, Wang HW. SNAIL Regulates Interleukin-8 Expression, Stem Cell-Like Activity, and Tumorigenicity of Human Colorectal Carcinoma Cells. Gastroenterology. 2011;141:279–291. e275. doi: 10.1053/j.gastro.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- Inai K, Norris RA, Hoffman S, Markwald RR, Sugi Y. BMP-2 induces cell migration and periostin expression during atrioventricular valvulogenesis. Dev Biol. 2008;315:383–396. doi: 10.1016/j.ydbio.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Joseph MJ, Dangi-Garimella S, Shields MA, Diamond ME, Sun L, Koblinski JE, Munshi HG. Slug is a downstream mediator of transforming growth factor-beta1-induced matrix metalloproteinase-9 expression and invasion of oral cancer cells. J Cell Biochem. 2009;108:726–736. doi: 10.1002/jcb.22309. [DOI] [PubMed] [Google Scholar]
- Kaden JJ, Vocke DC, Fischer CS, Grobholz R, Brueckmann M, Vahl CF, Hagl S, Haase KK, Dempfle CE, Borggrefe M. Expression and activity of matrix metalloproteinase-2 in calcific aortic stenosis. Z Kardiol. 2004;93:124–130. doi: 10.1007/s00392-004-1021-0. [DOI] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E. Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J Cell Biol. 2004;166:359–367. doi: 10.1083/jcb.200403050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev Dyn. 2004;230:239–250. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. BMP and FGF regulatory pathways control cell lineage diversification of heart valve precursor cells. Dev Biol. 2006a;292:292–302. doi: 10.1016/j.ydbio.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Kist R, Scherer G, Yutzey KE. Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol. 2007;305:120–132. doi: 10.1016/j.ydbio.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J, Lange AW, Yutzey KE. Hearts and bones: shared regulatory mechanisms in heart valve, cartilage, tendon, and bone development. Dev Biol. 2006b;294:292–302. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB, Ford E, Furie K, Gillespie C, Go A, Greenlund K, Haase N, Hailpern S, Ho PM, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott MM, Meigs J, Mozaffarian D, Mussolino M, Nichol G, Roger VL, Rosamond W, Sacco R, Sorlie P, Thom T, Wasserthiel-Smoller S, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Starling C, Gridley T. Epiblast-specific Snai1 deletion results in embryonic lethality due to multiple vascular defects. BMC Res Notes. 2009;2:22. doi: 10.1186/1756-0500-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest. 2010;120:3493–3507. doi: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Estrada OM, Culleres A, Soriano FX, Peinado H, Bolos V, Martinez FO, Reina M, Cano A, Fabre M, Vilaro S. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J. 2006;394:449–457. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows KN, Iyer S, Stevens MV, Wang D, Shechter S, Perruzzi C, Camenisch TD, Benjamin LE. Akt promotes Endocardial-Mesenchyme Transition. J Angiogenes Res. 2009;1:2. doi: 10.1186/2040-2384-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt CH, Lepera RC, Markwald RR. Myocardial specificity for initiating endothelial-mesenchymal cell transition in embryonic chick heart correlates with a particulate distribution of fibronectin. Dev Biol. 1987;119:59–67. doi: 10.1016/0012-1606(87)90206-5. [DOI] [PubMed] [Google Scholar]
- Molin DG, Bartram U, Van der Heiden K, Van Iperen L, Speer CP, Hierck BP, Poelmann RE, Gittenberger-de-Groot AC. Expression patterns of Tgfbeta1-3 associate with myocardialisation of the outflow tract and the development of the epicardium and the fibrous heart skeleton. Dev Dyn. 2003;227:431–444. doi: 10.1002/dvdy.10314. [DOI] [PubMed] [Google Scholar]
- Murray SA, Carver EA, Gridley T. Generation of a Snail1 (Snai1) conditional null allele. Genesis. 2006;44:7–11. doi: 10.1002/gene.20178. [DOI] [PubMed] [Google Scholar]
- Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- Nath AK, Brown RM, Michaud M, Sierra-Honigmann MR, Snyder M, Madri JA. Leptin affects endocardial cushion formation by modulating EMT and migration via Akt signaling cascades. J Cell Biol. 2008;181:367–380. doi: 10.1083/jcb.200708197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J Cell Biol. 2008;182:315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. Epithelial-Mesenchymal Transitions in development and disease: old views and new perspectives. Int J Dev Biol. 2009;53:1541–1547. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- Ota I, Li XY, Hu Y, Weiss SJ. Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc Natl Acad Sci U S A. 2009;106:20318–20323. doi: 10.1073/pnas.0910962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock JD, Levay AK, Gillaspie DB, Tao G, Lincoln J. Reduced sox9 function promotes heart valve calcification phenotypes in vivo. Circ Res. 2010;106:712–719. doi: 10.1161/CIRCRESAHA.109.213702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock JD, Lu Y, Koch M, Kadler KE, Lincoln J. Temporal and spatial expression of collagens during murine atrioventricular heart valve development and maintenance. Dev Dyn. 2008;237:3051–3058. doi: 10.1002/dvdy.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Marin F, Cubillo E, Stark HJ, Fusenig N, Nieto MA, Cano A. Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. Journal of cell science. 2004;117:2827–2839. doi: 10.1242/jcs.01145. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. The Journal of biological chemistry. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104:2525–2532. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, Pedchenko V, Hudson BG, Hoffman MP. MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell. 2009;17:482–493. doi: 10.1016/j.devcel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano LA, Runyan RB. Slug is a mediator of epithelial-mesenchymal cell transformation in the developing chicken heart. Dev Biol. 1999;212:243–254. doi: 10.1006/dbio.1999.9339. [DOI] [PubMed] [Google Scholar]
- Rowe RG, Li XY, Hu Y, Saunders TL, Virtanen I, Garcia de Herreros A, Becker KF, Ingvarsen S, Engelholm LH, Bommer GT, Fearon ER, Weiss SJ. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J Cell Biol. 2009;184:399–408. doi: 10.1083/jcb.200810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyan RB, Markwald RR. Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Dev Biol. 1983;95:108–114. doi: 10.1016/0012-1606(83)90010-6. [DOI] [PubMed] [Google Scholar]
- Rupp PA, Visconti RP, Czirok A, Cheresh DA, Little CD. Matrix metalloproteinase 2-integrin alpha(v)beta3 binding is required for mesenchymal cell invasive activity but not epithelial locomotion: a computational time-lapse study. Mol Biol Cell. 2008;19:5529–5540. doi: 10.1091/mbc.E07-05-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki K, Fujimori T, Savagner P, Hata A, Aikawa T, Ogata N, Nabeshima Y, Kaechoong L. Mouse Snail family transcription repressors regulate chondrocyte, extracellular matrix, type II collagen, and aggrecan. J Biol Chem. 2003;278:41862–41870. doi: 10.1074/jbc.M308336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev Biol. 2007;302:376–388. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soini Y, Satta J, Maatta M, Autio-Harmainen H. Expression of MMP2, MMP9, MT1-MMP, TIMP1, and TIMP2 mRNA in valvular lesions of the heart. J Pathol. 2001;194:225–231. doi: 10.1002/path.850. [DOI] [PubMed] [Google Scholar]
- Song W, Jackson K, McGuire PG. Degradation of type IV collagen by matrix metalloproteinases is an important step in the epithelial-mesenchymal transformation of the endocardial cushions. Dev Biol. 2000;227:606–617. doi: 10.1006/dbio.2000.9919. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Congenital heart defects: trapping the genetic culprits. Circ Res. 2000;86:917–918. doi: 10.1161/01.res.86.9.917. [DOI] [PubMed] [Google Scholar]
- Takahashi E, Funato N, Higashihori N, Hata Y, Gridley T, Nakamura M. Snail regulates p21(WAF/CIP1) expression in cooperation with E2A and Twist. Biochemical and biophysical research communications. 2004;325:1136–1144. doi: 10.1016/j.bbrc.2004.10.148. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Wang J, Sridurongrit S, Dudas M, Thomas P, Nagy A, Schneider MD, Epstein JA, Kaartinen V. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev Biol. 2005;286:299–310. doi: 10.1016/j.ydbio.2005.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. Snail: More than EMT. Cell Adh Migr. :4. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]