Abstract
Cells exposed to stimuli exhibit a wide range of responses ensuring phenotypic variability across the population. Such single cell behavior is often examined by flow cytometry; however, gating procedures typically employed to select a small subpopulation of cells with similar morphological characteristics make it difficult, even impossible, to quantitatively compare cells across a large variety of experimental conditions because these conditions can lead to profound morphological variations. To overcome these limitations, we developed a regression approach to correct for variability in fluorescence intensity due to differences in cell size and granularity without discarding any of the cells, which gating ipso facto does. This approach enables quantitative studies of cellular heterogeneity and transcriptional noise in high-throughput experiments involving thousands of samples. We used this approach to analyze a library of yeast knockout strains and reveal genes required for the population to establish a bimodal response to oleic acid induction. We identify a group of epigenetic regulators and nucleoporins that, by maintaining an ‘unresponsive population,’ may provide the population with the advantage of diversified bet hedging.
Keywords: flow cytometry, high-throughput experiments, statistical regression model, transcriptional noise
Introduction
Cells in a genetically identical population do not necessarily behave similarly when exposed to a particular condition. Phenotypic variation within populations is often observed and can be ascribed to stochastic variations in gene expression (Elowitz et al, 2002). Upstream signaling fluctuations or low concentrations of molecules governing gene expression lead to inherently stochastic expression patterns that can be augmented or mitigated by gene regulatory network structures (Ratushny et al, 2008; Cagatay et al, 2009), organization of chromatin (Raser and O’Shea, 2004) or epigenetic regulation (Raj and van Oudenaarden, 2008). Such population variation has been evolutionarily tuned for each gene and can confer an advantage to a population by enabling it to produce multiple phenotypes and to hedge its bets in case of a change of the environmental cues (Acar et al, 2008). This mechanism is particularly relevant for the cellular response to stress, which involves highly committed structural remodeling (Ozbudak et al, 2002; Maamar et al, 2007; Suel et al, 2007).
Flow cytometry is an excellent technique to measure such single cell behaviors within large populations of cells, and has been extensively used to analyze transcriptional noise (Newman et al, 2006). However, quantitative comparisons of such behaviors spanning large numbers of samples, involving large knockout libraries and time point measurements, have thus far not been carried out. The reason, ironically, has not been the inability to generate very large-scale data sets in a high-throughput manner, but rather the absence of appropriate analytical methods to perform such quantitative comparisons in the face of substantial variability in cellular physical characteristics that confound the quantification of fluorescence. The primary confounding characteristics are cell volume and cell granularity, which are correlated with forward scattered light (FSC) and side scattered light (SSC) measurements, respectively. For example, bigger cells may show an apparent increased fluorescence. Notwithstanding the fact that FSC and SSC measurements are useful for a variety of phenotyping purposes, including delineation of dead cells and determination of cell types, the cellular physical variability potentially masks the true heterogeneity of expression in a sample due to biological noise (Newman et al, 2006). Thus, minimizing the source of fluorescence variance in a population due to the physical characteristics of the cells is critical to performing quantitative fluorescence-based comparisons in flow cytometry experiments and revealing expression-based phenotypic variation. A common method to reduce this variability relies on the selection of a subgroup of cells with similar physical characteristics. This is accomplished by creating a ‘gate’ in the FSC/SSC two-dimensional space and discarding all the cells that fall outside of the gate. Assuming that the gate is restrictive enough, the encapsulated cells will be morphologically homogeneous and hence show a reduced variability in their fluorescence. Different, but often used applications of gating are to discriminate subpopulations within complex samples, to remove auto-fluorescence background or to separate dead cells. For these purposes, a less restrictive gate is commonly used.
To remove as much morphological variability as possible, it is often considered that the gate should be as small as possible. However, in practice, choosing a minimal gate dramatically reduces the sample size and essentially ignores the ungated cell population, masking potentially relevant biological events and biasing results. In addition, gating creates a dilemma when comparing multiple biological samples: one must choose a single gate for all the samples that is representative for each individual sample. This is a time-consuming and user-dependent (i.e., subjective) process that may become impossible in high-throughput screens, even with automated gating procedures. Indeed, the variety of experimental conditions in a typical high-throughput experiment, such as differences in environmental stimuli, genetic perturbations and time points, will inherently lead to a large diversity in cellular morphology across such conditions, and therefore, to no or only little overlap between samples in the FSC/SSC two-dimensional space. This, in turn, makes it impossible to choose a gate that contains sufficient data for each sample while being able to reduce the variability introduced by cell size and granularity.
To address this need, we have developed a regression model that uses all the cells in a biological sample to normalize cell size and granularity effects on fluorescence across multiple samples. This approach avoids the subjectivity associated with selecting a gate, dramatically increases the sample size available for analysis, and enables systematic quantitative comparisons of large-scale data sets. Using several experiments, we aim to establish these advantages of the regression model compared to gating for the purpose of removing the variability in fluorescence due to morphological characteristics. It is important to point out, however, that the regression model does not substitute gating in the cases where a specific population needs to be selected from a complex sample. Delineating subpopulations can be performed by manual gating using dedicated flow cytometry software or using automated procedures (Lo et al, 2008; Pyne et al, 2009) and most frequently not only based on FSC and SSC, but also using FL channels that measure cell-type-specific markers. In these cases, the regression model is a powerful complement that can be used either before gating to remove the effect of cell morphology leading to a better separation between subpopulations (see analysis of multiplex samples in the Results section) or afterwards to remove the morphology-associated variation in fluorescence.
We first demonstrate the effectiveness of the regression model in the analysis of cellular heterogeneity in the galactose response of yeast and its utility for studying population variability in the context of high-throughput screening of a yeast deletion strain library. Second, we highlight one specific application in the deconvolution of a mixed sample of fluorescently bar-coded mammalian cells, which enables multiplexing analysis, demonstrating the generality of the regression framework. Finally, we have applied the method to a large compendium of yeast flow cytometry data consisting of time series of Pot1p–GFP expression during a carbon source shift from glucose to oleate and back to glucose on a miniarray of 148 mutant strains carrying deletions of all non-essential chromatin regulators and nucleoporins in yeast. Cells undergoing this carbon shift change substantially in morphology. Traditional gating precludes proper analysis due to the lack of overlap in the FSC/SSC two-dimensional space between different mutants and time points. Our results unveil new modes of regulation at an epigenetic level of Pot1p expression and point to the genes implicated in this regulation. Thus, the regression-based model not only serves as a useful and practical alternative or complement to gating, but also enables heretofore impossible large-scale systems biology studies to be carried out with flow cytometric data.
Results
Compensating for the variability due to cell size and cell granularity using regression
The regression model takes as input the raw flow cytometry data of a set of biological samples; each sample is assumed to consist of the two scatter measurements (FSC and SSC) and one fluorescence measurement (FL) for a number of cells (or events). The model outputs the FL intensities compensated for FSC and SSC.
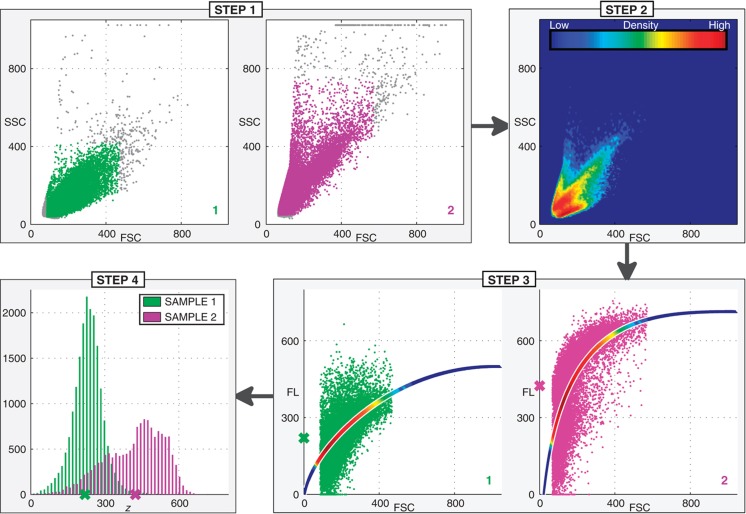
The procedure follows four steps, which are graphically depicted in Figure 1 and mathematically described in the Materials and methods section and in more detail in Supplementary Figures S1–S6. To explain the rationale behind the regression model, we describe the different steps in analogy to gating. In gating, one selects both the size (or shape) of the gate and the position of the gate in the two-dimensional FSC/SSC space. Choosing the size determines how many cells (and thus how much variability) is retained, whereas choosing the position of the gate determines the average cell size and granularity of the retained cells. When different biological samples are compared, the same gate (position and shape) is used to enable a quantitative comparison of the intensity and variation in fluorescence between the samples. Below, we outline the four steps of the regression procedure and explain how they relate to gating.
Figure 1.
Compensating for the effect of cell size and cell granularity using regression. In this example, the experiment consists of two biological samples (sample 1 and sample 2). During preprocessing in step 1, spurious events (depicted in gray) are discarded. In step 2, the FSC and SSC measurements are used to estimate the density of cells in the two-dimensional FSC/SSC space. The regression model of FL on FSC and SSC for each sample is indicated by the colored lines in step 3. (For visualization purposes, only the FSC is depicted as an independent variable. The SSC is also an independent variable and the actual regression model represents a surface, not a curve.) The average fluorescence intensity for each sample is computed by evaluating the regression model across the complete two-dimensional FSC/SSC space and weighting each location in this space by its corresponding density (estimated in step 2) before averaging. The colors within the regression lines indicate the weights and are directly related to the colors of the density estimate in step 2. The average fluorescence values are indicated by the green and purple cross on the y axis for samples 1 and 2, respectively. Step 4 depicts a histogram of the fluorescence intensities compensated for the effect of cell size and cell granularity (as measured by FSC and SSC, respectively). The values are obtained as the residuals (distances from the regression model) offset by the average fluorescence intensity.
Step 1 Preprocessing. This is a standard preprocessing step in the analysis of flow cytometry data to remove spurious events.
Step 2 The overall density of the cells across all biological samples in the two-dimensional FSC/SSC space is estimated. This density will be used in step 4 to compute the average fluorescence intensity for each sample. In analogy to gating, where the same gate is used for all samples, we will use this one density for all samples. In contrast to gating, where only a subset of cells with specific morphological properties is selected as determined by the position of the gate, we use a distribution across the entire two-dimensional FSC/SSC space, which represents the ‘average cell’ in terms of morphological features across all biological samples.
Step 3 A regression model of FL on FSC and SSC is applied to each sample independently. The distribution of the residuals (or regression errors) represents the variability in fluorescence that is not due to cell size and cell granularity. In gating, this is the variability in fluorescence of the cells that are left in the gate, given that the gate is small enough to assume that these cells are morphologically uniform.
Step 4 To compute the fluorescence intensities compensated for FSC and SSC, the residuals (which are centered around zero) are offset by the sample-specific average fluorescence intensity. This intensity is computed by evaluating the (sample-specific) regression model across the two-dimensional FSC/SSC space weighted by the (sample-unspecific) density estimated in step 2. In analogy to gating, the average fluorescence intensity is the average of the fluorescence values of the cells in the gate. Since the same gate is used for all samples, the fluorescence intensities can be compared between samples. Likewise in this procedure, we use the (same) sample-unspecific density to compute the average fluorescence intensity for each sample.
The use of the (sample-unspecific) density to compute the average fluorescence intensity can be seen as a normalization procedure that enables the direct comparison between the average fluorescence intensities of all biological samples in the experiment. It should be noted that when additional samples are added to the experiment, the density in step 2 will have to be recomputed, and the resulting average fluorescence intensities will change. Thus, the average fluorescence intensity of a biological sample is not an absolute measure of fluorescence, but should be interpreted relative to the average intensities of the other samples in the experiment. This is similar to gene expression levels, which are normalized across a set of microarray measurements.
Regions in the two-dimensional FSC/SSC space with many cells, that is, high-density areas, will have a larger influence on the average fluorescence intensity. Therefore, the regression model and standard gating will have very similar average fluorescence intensities, when the position of the gate is chosen in a high-density area. Obviously, this is often the case as one does not normally place a gate in regions with few cells.
The regression model is implemented in MATLAB and available as Supplementary Information and at http://code.google.com/p/flowregressionmodel/.
Cellular heterogeneity in the galactose response of yeast
In order to compare the regression approach with a traditional gating procedure, a previously published data set was reanalyzed (see Materials and methods, Dataset 1) (Ramsey et al, 2006). In this paper, the variability of the expression of GAL1 was quantified in a wild-type (WT) strain and in a mutant strain in which two feedback loops controlling GAL1 expression had been disabled.
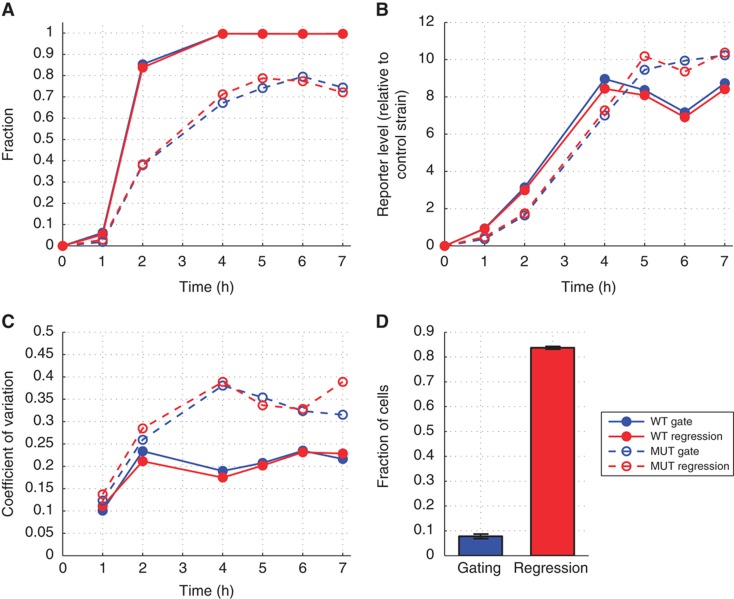
The data set was reanalyzed as described in the paper as well as using the regression model. The histograms of the fluorescence obtained by both methods showed a remarkable similarity (Supplementary Figures S7 and S8). Statistics calculated in the original paper were compared with those obtained by the regression model. The fraction of responding cells obtained with both methods was practically identical, indicating that the regression model accurately differentiated the two (i.e., responding and non-responding) populations of the biological sample (Figure 2A). The mean intensity for all cells was also consistent between the two methods (Figure 2B). Finally, the coefficients of variation (CV) remained very comparable among all conditions, showing that the regression model removed the variation due to FSC/SSC at least as well as the gating method (Figure 2C).
Figure 2.
Analysis of Gal1–GFP in response to galactose in WT and mutant strains. (A) Fraction of responding cells over time after a galactose induction at t=0. (B) Mean intensity of all cells. (C) CV of active cells. (D) Fraction of cells gated or used for the regression model. All results show the average of three replicates. Error bars have been omitted for clarity, but they are similar between the two methods. Panels (A, B) correspond to Figure 2 of Ramsey et al (2006).
The only significant difference between the two approaches was the number of cells used to calculate these statistics. Using a gating approach, only 7.8±0.9% of the original cells were taken into account (even 3.5% in one case). In contrast, the regression model used 84±0.6% (Figure 2D) (15±0.4% cells were removed due to preprocessing). The relevance of this difference in sample size is exemplified in one extreme case (time=1 h, replicate 1, mutant), where the CV was calculated from only 72 cells due to the restrictive gating, producing a much less reliable statistic compared with the regression model which used 1080 cells.
The regression model as a tool to study population variability
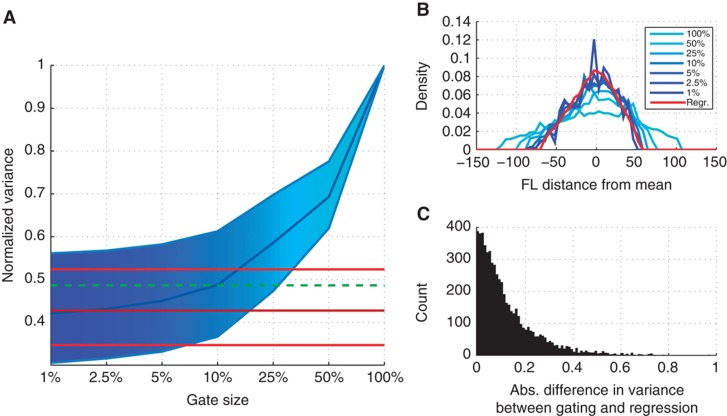
To evaluate the regression model in the task of removing the variability in fluorescence intensities due to cell size and cell granularity, the method was compared with standard gating when applied to a high-throughput screening of Pot1p–GFP expression, containing 5883 biological samples (see Materials and methods, Dataset 2). Gating was performed for each biological sample individually. First, the center of a circular gate was set as the location with the highest density in the two-dimensional FSC/SSC space for that individual sample. Then, the radius of the circular gate was chosen such that the gate contained 1, 2.5, 5, 10, 25, 50 or 100% of the cells in that sample. The variance of the FL for each of these seven different gates was normalized by dividing by the overall variance of that sample (100% of the cells). Next, the variance of the FL was analyzed in a similar way after applying the regression model to each of the 5883 biological samples.
The experiment showed that as the size of the gate was reduced, the variance tended to converge to a constant value (Figure 3A). This observation agrees with Newman et al (2006), who demonstrated a relationship between the size of the gate and the variance in the fluorescence, but only to a certain size of the gate. When the gate is small enough, the FSC/SSC-dependent variability is virtually removed, and when the gate is made even smaller, the variability remains constant, since it is no longer affected by cell size and granularity. The variance component of the regression model agrees well with the ‘converged’ gate variance, indicating that the algorithm successfully removes the effect of cell size and cell granularity (as does the smallest gate), but the regression model does so without significantly reducing the sample size. We compared our regression model with a much simpler version, including only linear FSC and SSC terms. In contrast to our regression model, the simple linear model is not a suitable alternative to gating as it fails to remove as much FL variance as the ‘converged’ gate. See Supplementary Figure S6 for a detailed comparison between different regression models.
Figure 3.
Comparing the variance components between gating and regression. (A) The blue polygon represents the median and the interquartile range of the variance of the fluorescence after gating with different sizes of the gate (x axis). The red lines indicate the median and the interquartile range of the variance of the fluorescence after regression. (The latter median and interquartile values are constant values that are not dependent on the x axis, since the regression model uses all data points; they are depicted as lines for representation only). The green dashed line indicates the median of the variance of the fluorescence after regression with a simple linear model, including only linear effects of FSC and SSC. (B) Distribution of the mean-subtracted fluorescence intensities based on gates with different sizes (shades of blue) and the regression model (red) for one biological sample (n=4469). The distribution is computed by normalized histogram binning using 50 bins and is represented by a continuous line that connects the centers of the histogram (C) Histogram of the difference in variance between the regression model and the ‘converged’ gate across all biological samples.
Figure 3B shows that the variance (width of the distribution) decreases with the gate size and is comparable between the smaller gates and the regression-based variance. However, for small gate sizes the distribution is very spiky (noisy), which is due to the small number of cells in the gate used to estimate the distribution. In a follow-up experiment, described in Supplementary Information, a random sampling strategy was used to reduce the number of cells in the biological samples of this data set. This experiment demonstrated that the regression model yielded much better estimates of the CV and the FL distribution (using the complete biological sample as a ‘ground truth’), and that at least 10 times fewer cells are needed to obtain the same accuracy of these statistics (Supplementary Figure S9; Supplementary Tables T1 and T2). It is important to note that the regression model produced reliable estimates in situations where the gating approach would be ineffective, thus demonstrating its considerable advantage in applications where only few cells can be analyzed.
Figure 3C shows a histogram representing the difference in variance between the regression model and the ‘converged’ gating for each individual sample. The variance of the ‘converged’ gate was computed by taking the median of the variance for gates with 1, 2.5 and 5% of the cells. For more than half of the biological samples, the difference in variance was <0.1 (10% of the total variance). To further investigate the difference in variance components between gating and regression, a more principled experimental approach based on comparing the expression of two reporters under the control of the same promoter using a two-color assay was carried out (Elowitz et al, 2002) (see Materials and methods, Dataset 3). This analysis, which is described in detail in Supplementary Information and corresponding Supplementary Figure S10, again demonstrated that the regression model provides an excellent estimate of transcriptional noise (extrinsic and intrinsic) without compromising statistical power.
Dissecting bar-coded flow cytometry data using the regression model
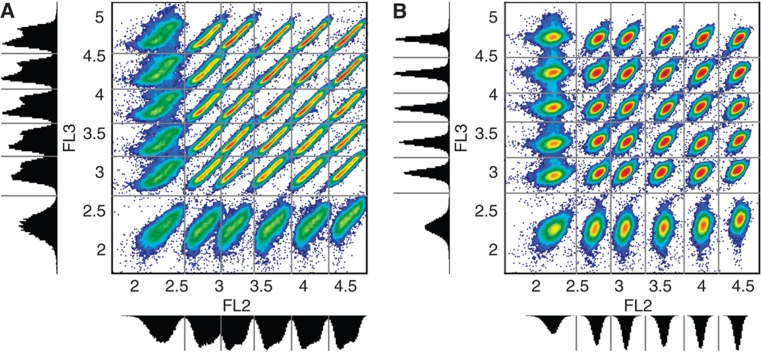
Another application of the regression model is found in fluorescent cell bar coding that (by multiplexing) can drastically reduce antibody consumption and acquisition time (Krutzik and Nolan, 2006). Each sample in such a data set is a mixture of different fluorescently bar-coded populations corresponding to different experimental conditions or time points. A flow cytometry data set of mammalian cells consisting of 36 (6 × 6) fluorescently bar-coded subpopulations was analyzed (see Materials and methods, Dataset 4). These subpopulations can be distinguished in the two-dimensional space defined by the FL2 and FL3 channels by dividing this space up into rectangular areas using a thresholding scheme. This is referred to as ‘forward deconvolution’ (Krutzik and Nolan, 2006) (Figure 4A). However, the smear in these populations due to the effect of cell morphology on fluorescence leads to overlapping subpopulations, making it difficult to clearly distinguish population boundaries. When the regression model is applied, this smear is substantially reduced, producing more coherent subpopulations that are easier to separate (Figure 4B). On average, the CV of FL2 and FL3 in these 36 rectangular areas is halved when the regression model is applied. The more coherent subpopulations enable automated algorithms that can separate the subpopulations in a reliable way. In Supplementary Information, a mixture modeling approach is outlined that quantifies (and clearly shows) the higher separability between bar-coded samples (Supplementary Figures S13–S15).
Figure 4.
Scatter plots of bar-coded samples. (A) Density plot of the FL2 and FL3 raw data with their corresponding histograms of a mixture of 6 × 6 populations stained with different concentrations of PACblu-NHS (in FL2) and Alexa 488-NHS (in FL3). The gray lines (minima in the histograms) separate the 36 populations. Colors represent density of cells with a gradation from red (more dense) to blue (less dense). This figure is similar to Figure 3A in Krutzik and Nolan (2006). (B) Identical to (A), except the FL2 and FL3 data are compensated for cell size and granularity using the regression model.
Comparing biological samples without overlap in cell size and granularity
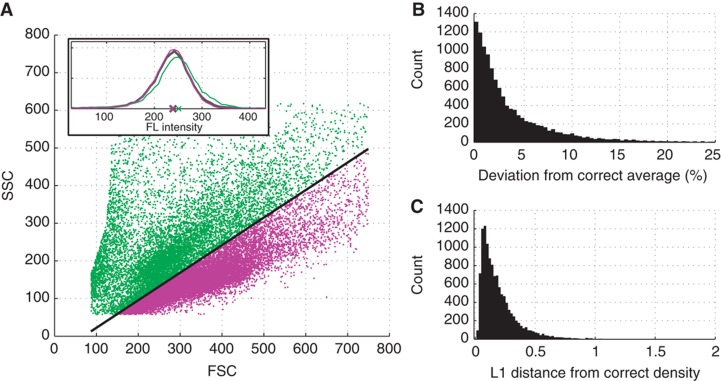
Most flow cytometry studies do not focus on a single biological sample, but compare a number of them. In such cases, a single gate must be selected in order to compare across all biological samples. Otherwise, it would not be clear to what extent the measured fluorescent intensity is affected by cell size and granularity rather than by the experimental condition of interest. However, it might be impossible to find a gate that is small enough to remove the effect of cell size and granularity and at the same time contains enough cells for each of the biological samples to accurately compute statistics. For example, in Dataset 2, the smallest square gate that would contain 750 (2.5–5%) of the cells in every biological sample is, in fact, so large that on an average the gate retains 46% of the cells in a biological sample. It was previously observed that the gate should retain only between 1–5% of the cells in order to successfully remove the effect of cell size and granularity. Even if only 100 (∼0.5%) cells per sample are required, the resulting gate still retains 15% of the cells on average. Thus, choosing a single gate for all the samples essentially precludes reliable analysis. The regression model is not hampered by the different physical characteristics of the cells across biological samples; indeed, even biological samples that do not exhibit any overlap are comparable. This is so because monotonicity constraints ensure that the regression surface, that is, the function of FSC and SSC to approximate FL, has a stable behavior in areas of the FSC/SSC space where there are no or only few data points (i.e. cells) (see Materials and methods and Supplementary Information).
To test this assumption, each of the 5883 biological samples was split into two samples using the first principal component in the two-dimensional FSC/SSC space (Figure 5A). Such a pair of samples was then treated as one experiment and analyzed using the regression model. The fluorescence intensities compensated for the SSC and FSC for these two samples were then compared with those of the original non-split sample using two metrics. First, the difference in average fluorescence intensity between the two samples and the ‘correct’ original sample was measured (Figure 5B). Second, the differences between the distributions of the fluorescence intensities of the split and non-split samples were measured (Figure 5C). This difference was defined as the L1 (i.e. absolute) distance between the probability density functions. The L1 distance is between 0 (when the distributions are identical) and 2 (when the distributions are completely different). Overall, for 90% of the biological samples, the average fluorescence intensities of the two halved counterparts differed <10% from the ‘correct’ intensity computed on the whole sample, with most samples (i.e., the median) differing <2.5%. Also, the distribution of the FL intensities of the halved parts agrees well with that of the complete sample. This analysis demonstrates the utility of the approach for analyzing multiple biological samples where gating would preclude comparative analyses.
Figure 5.
Comparing biological samples without overlap in the SSC/FSC space. (A) The cells of one biological sample in the FSC/SSC space are split up into two parts (green and magenta) using the first principal component axis (black line). Inlay: the distributions of the fluorescence densities obtained from the regression model. The green and magenta lines represent the two ‘halved’ samples, while the black line represents the distribution of the complete sample. The crosses represent the means (i.e. the average fluorescence intensities). In this case, the mean of the magenta and green distribution differ by 0.6 and 5.0% from the mean of the whole sample and 0.06 and 0.23 in terms of absolute difference in density, respectively. (B) Histogram of the deviation between the correct average (of the non-split sample) and the ‘halved’ samples across all biological samples. (C) Histogram of the L1 (absolute) difference in density between the ‘halved’ samples and the whole sample across all biological samples. These absolute differences range from 0 (identical densities) to 2 (completely different densities).
Analysis of the epigenetic regulation of POT1 expression under the carbon shift from glucose to oleate and back to glucose
POT1 encodes 3-ketoacyl-CoA thiolase required for β-oxidation metabolism. Under glucose conditions, it is repressed and its expression is highly induced when cells use fatty acids as a carbon source (Einerhand et al, 1991; Igual et al, 1992). Its transcriptional network has been well characterized (Smith et al, 2006; Ratushny et al, 2008), but its characteristic bimodal profile of expression during oleate induction indicates a more complex transcriptional behavior that has yet to be explained. Some recent studies point to epigenetic regulation mediated by the histone variant H2A.Z (Htz1) (Wan et al, 2009). In addition, the POT1 gene changes its localization relative to the nuclear periphery during oleate induction, a behavior previously shown for highly expressed genes (Casolari et al, 2004; Cabal et al, 2006; Capelson et al, 2010; Kalverda et al, 2010) and subtelomeric chromosomal loci (Galy et al, 2000), opening the possibility of transcriptional control associated with the nuclear periphery or nuclear pore complex (NPC) (see Materials and methods and Supplementary Figure S16). Thus, we aimed to study the effects of chromatin remodeling factors and nucleoporins on expression behavior at a population level.
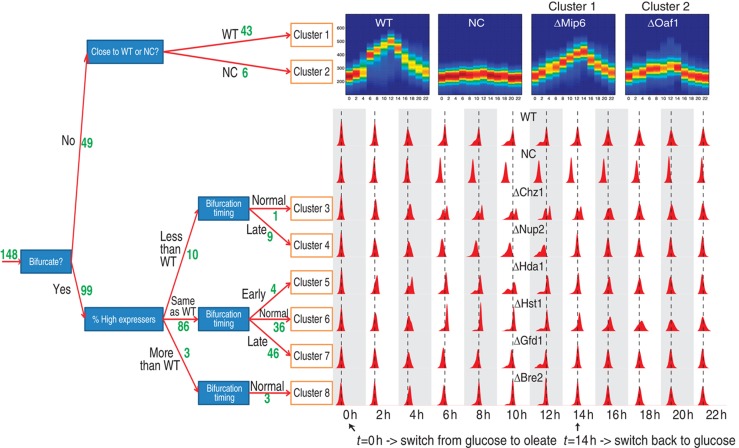
We monitored Pot1p–GFP expression by flow cytometry during a cycle of induction and repression on a miniarray of 148 strains carrying mutations for non-essential chromatin modifiers and nucleoporins, along with six identical WT strains and six negative controls (NCs) (see Materials and methods, Dataset 2). After the regression model was applied to all biological samples in this data set, replicates were combined and a Gaussian mixture model was fit to each time point for each strain separately using the EM approach described in Song et al (2010). The resulting model represents Pot1p–GFP expression as either one Gaussian distribution (unimodal population) or two Gaussian distributions (bimodal distribution). The WT strains consistently showed bimodality at time points 6, 8, 10, 12 h after the carbon shift from glucose to oleate, indicating a clear bifurcation event around 6 h after which two different subpopulations can be recognized, one with higher expression, the other with lower expression. At 14 h (i.e. 2 h after the shift back to glucose) the two subpopulations merged again into a unimodal population. We used a rule-based clustering procedure to group the 148 mutant strains according to their expression behavior over time relative to WT. This grouping was based on four properties: (1) whether the bifurcation event occurred, and if so, (2) the time points of bimodality and (3) the relative population sizes of the high and low expressers, and if no bifurcation occurred (4) the overall similarity of the expression to WT. See Materials and methods for more details on this procedure. This led to eight characteristic profiles and are summarized in Table I and Figure 6.
Table 1. Deletion strains clustered based on their Pot1p–GFP expression behavior during the carbon source shift.
| Cluster no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bifurcate? | No | No | Yes | Yes | Yes | Yes | Yes | Yes | ||||||
| Closer to WT or NC? | WT | NC | ||||||||||||
| % High expressers | Less than WT | Less than WT | Same as WT | Same as WT | Same as WT | More than WT | ||||||||
| Bifurcation onset time | Same as WT | Late | Early | Same as WT | Late | Same as WT | ||||||||
| Clusters are characterized by the four properties indicated beneath the cluster number. First, deletion strains are divided into a group that showed no bifurcation along the time series (49 strains) and a group that did show bifurcation at some time point(s) (99 strains). The 49 ‘non-bifurcated strains’ were further split up into a group that was close to WT (43 strains, cluster 1) or to negative control samples (NC) (6 strains, cluster 2) in terms of their expression profile. The 99 ‘bifurcated strains’ were split up into three groups according to the population sizes of high and low expressers with respect to WT: (1) strains for which the high expressing population was much smaller than the percentage of high expressers in WT, (2) strains with a larger population of high expressers than WT and (3) strains with the same percentage of high expressers as WT. A further dissection was made based on the onset time of bifurcation, because many deletion strains (59) showed an earlier (4) or later (55) onset of bifurcation. This resulted in clusters 3–8, with 1, 9, 4, 36, 46 and 3 strains, respectively. | ||||||||||||||
| Acs1 | Leo1 | Sir4 | Adr1 | Chz1 | Bre1 | Hda1 | Dot1 | Mak31 | Spt7 | Ahc1 | Lge1 | Sas5 | Bre2 | |
| Aim4 | Mdm20 | Snf12 | Htl1 | Hfi1 | Hos2 | Eaf1 | Nat1 | Vps72 | Asm4 | Mlp1 | Set2 | Gis1 | ||
| Arp5 | Mip6 | Snf5 | Oaf1 | Hos3 | Stb2 | Eaf6 | Nat3 | Yaf9 | Chd1 | Mlp2 | Set3 | Nat5 | ||
| Arp6 | Nto1 | Snf6 | Pip2 | Htz1 | Vps71 | Eaf7 | Nup42 | Ydl089w | Dyn2 | Nap1 | Sgf11 | |||
| Asf1 | Nup120 | Snt1 | Swc3 | Ies1 | Gtt3 | Pex3 | Gfd1 | Nat4 | Snf11 | |||||
| Cdc73 | Nup133 | Spt20 | Vps75 | Ngg1 | Hst1 | Pml39 | Hat1 | Nhp10 | Snf2 | |||||
| Eaf3 | Nup84 | Swc5 | Nup2 | Ioc2 | Rad54 | Hat2 | Nup100 | Snl1 | ||||||
| Eaf5 | Paf1 | Swi3 | Sas2 | Ioc3 | Rco1 | Hda3 | Nup170 | Spp1 | ||||||
| Esc1 | Rph1 | Swr1 | Sir1 | Ioc4 | Rtf1 | Hos1 | Nup188 | Sum1 | ||||||
| Fpr4 | Rsc2 | Taf14 | Isw1 | Rxt2 | Hpa2 | Nup60 | Swd1 | |||||||
| Gcn5 | Sdc1 | Yng1 | Isw2 | Rxt3 | Hpa3 | Oaf3 | Swd3 | |||||||
| Hda2 | Sds3 | Itc1 | Sap30 | Hst2 | Pho23 | Ubp8 | ||||||||
| Hsl7 | Sgf73 | Mad1 | Sas3 | Hst4 | Pom152 | Uip3 | ||||||||
| Ies3 | Sif2 | Mad2 | Shg1 | Jhd1 | Rad6 | Ypr174c | ||||||||
| Kap114 | Sin3 | Mak10 | Spt3 | Kap120 | Rpd3 | |||||||||
| Kap122 | Sir3 | Mak3 | Spt5 | Kap123 | Sas4 | |||||||||
Figure 6.
Decision tree divides the 148 deletion strains into 8 clusters. The green numbers near the red arrows indicate the number of deletion strains in the branch of the tree. For clusters 1 and 2, a heatmap representation of the expression over time is shown for one gene in each cluster. For clusters 3–8, a histogram representation is shown of one gene per cluster. The dashed lines are aligned to the mean of the WT distribution for unimodal time points. For the bimodal WT time points (i.e., 6, 8 10 and 12 h), the lines are aligned to the mean of the high expressing distribution.
Among the 148 mutants, 36 had a profile similar to WT (cluster 6) and 46 showed only a delay in the response, while keeping a similar intensity and distribution shape (cluster 7). Mutants in these two clusters represent 55% of the total and were considered as non-relevant for POT1 regulation. Two other clusters identified enhanced phenotypes: four mutants had an earlier response than WT (cluster 5) and three mutants had a larger subpopulation of high expressers than WT (cluster 8). We considered these mutants as attenuators of POT1 transcriptional regulation. Only one mutant (Δchz1) had a smaller subpopulation of high expressers and the same bifurcation onset time as WT (cluster 3), while nine other mutants with a small subpopulation of high expressers bifurcated later (cluster 4). These data are consistent with the previous results, wherein genome-wide microarray analysis showed that deletion of CHZ1 can dramatically affect the induction of oleate-responsive genes (Wan et al, 2009).
A total of 49 deletion strains did not exhibit any bimodality over the time course. Interestingly, 12 of the 148 deletion strains were reported to grow slower on oleate (Smith et al, 2006), and 8 of these are found in this set of 49. Within these 49 strains, one group showed a profile close to the untagged NC (cluster 2). This group included mutants of transcription factors implicated in POT1 expression (Oaf1, Pip2 and Adr1). This was an expected result and served as an additional control to validate the assay. The mutant strains Δswc1, Δhtl1 and Δvps75 were also in this cluster. These genes may also play a fundamental, but as of yet, unknown role in POT1 expression. More striking is the profile that showed a loss of the characteristic bimodal expression of POT1 while maintaining similar levels of expression as WT (cluster 1). There are 43 mutants in this group, representing 28% of the total. This phenotype is interesting, as it has been associated with adaptation to rapidly changing environments, where cells committed to a change might have an adaptation disadvantage if this change is only transient (Acar et al, 2008). Remarkably, genes in this cluster are significantly enriched for mutual synthetic lethal interactions and negative genetic effects, while the genes in the WT clusters 6 and 7 were underrepresented for these and other protein–protein interactions (Supplementary Table T3).
Between the bimodal WT response and the lack of bimodality for cluster 1, we can place the mutants of cluster 4 that maintained the bimodality, albeit after a delay and only with a small percentage of high expressers. Interestingly, this group included Htz1, which has previously been reported to be involved in the oleate response (Wan et al, 2009). One hint of the possible mechanism implicated in this behavior is the fact that many genes in cluster 1 are related to the histone variant Htz1: Kap114 imports Htz1 into the nucleus (Straube et al, 2010); Swc5, Swr1 and Arp6 are parts of the Swr1 complex, implicated in Htz1 chromatin binding (Wu et al, 2009); Cdc73 and Paf1, as parts of the Paf1 complex, have also been shown to have a genetic interaction with Htz1. Finally, Snf5, Snf6, Snf12, Swi3 and Taf14 are parts of the SWI/SNF complex and important for Htz1 binding to chromatin. Interestingly, the three main components of Nup84 complex (Nup84, Nup120 and Nup133) were also present in cluster 1. These proteins are implicated in gene recruitment to the nuclear periphery (a process termed reverse recruitment) (Menon et al, 2005). POT1 follows this mechanism of activation (Supplementary Figure S16), indicating a possible relationship between promoter nuclear localization and bimodal behavior of expression at the population level.
Discussion
We present a novel automated methodology that compensates for the effect of cell morphology on flow cytometry data, and thereby enables a quantitative analysis of high-throughput flow cytometry data. The algorithm normalizes the effect of the physical characteristics of cell size and cell granularity on the fluorescence intensity, thereby enabling the analysis of fluorescence intensities (protein abundance) in the presence of different morphological characteristics of cells in a population. In contrast to traditional gating, which discards the large majority of cells, the regression model retains all cells and thereby provides more accurate statistics, higher consistency across replicates and the ability to handle biological samples that contain far fewer cells (at least 10-fold), allowing for faster and cheaper data acquisition. This is relevant when one is looking for rare cells (e.g., stem cells), or when performing high-throughput screens where only a few hundred cells per experimental condition are being assayed.
The fact that the regression model uses a much larger fraction of cells in a biological sample points to an important feature of the method, namely that it provides fluorescence information across the complete population of cells in the biological sample. A traditional gating approach, on the other hand, reports the behavior of the cells with the specific physical properties (cell size and granularity) that were used to define the gate. In particular, when biological function is correlated with morphological characteristics, for example, cell-cycle-dependent genes (Supplementary Figures S11 and S12), the choice of the gate has a profound influence on the observed fluorescence, potentially (and inadvertently) leading to subjective and biased data analysis. However, if biological function is correlated with morphological characteristics, the regression model would remove this biological effect on fluorescence. Batenchuk et al (2011) present a methodology to reduce extrinsic transcriptional noise using a large gate followed by a cell morphology binning approach, which might be promising in such a scenario. A detailed discussion of this topic is found in Supplementary Information. Related to this point is the fact that flow cytometry experiments are in general difficult to reproduce, since there is no easy and formal way to supply a description of the gate, which is often manually drawn using a flow cytometry software package. The regression model, by avoiding the gate altogether, affords a much greater degree of reproducibility. However, it should again be pointed out that in many applications, the goal of gating is not only to remove morphology-associated variation in fluorescence (which the regression model accomplishes), but also to delineate or characterize subpopulations (e.g., removing dead cells), especially for complex mixtures containing different cell types. In such cases, an initial gating procedure of some sort is still necessary with the regression model becoming a powerful complement. Especially when subpopulations do not behave uniformly in terms of the relationship between fluorescence and morphological characteristics (as assumed by the regression model), initial delineation of subpopulations is essential.
Flow cytometry experiments are rapidly growing in size using high-throughput technologies. Researchers often desire to follow the protein expression behavior across different conditions and time points for large collections of cell types, strains or perturbed cells. These different experimental and genetic conditions can lead to samples with widely differing morphological characteristics, making it more difficult or even impossible to choose a proper gate. The regression model overcomes this by enabling the direct comparison of samples even if their cells do not share similar characteristics in terms of cell size and granularity. As we have shown, the regression model ensures that different biological samples are directly comparable to one another, even in the case where there is no overlap whatsoever between the cells in the FSC/SSC two-dimensional space in two or more biological samples. This is accomplished by extrapolation of the average fluorescence intensity across cell sizes and granularities that were not present in the actual sample. Monotonicity constraints were introduced into the regression model to guarantee stable behavior of these extrapolated values. Although in theory, this cannot guarantee the validity of the obtained extrapolated intensities, in practice, this approach worked exceptionally well in all experiments and (large) data sets examined. This makes the regression model a suitable systems biology tool to analyze large (high-throughput) flow cytometry data sets containing hundreds or thousands of biological samples in a highly automated manner.
The algorithm also proved valuable for dissecting bar-coded flow cytometry data from a human cell line, demonstrating its widespread utility. Indeed, the flexibility of the regression model, due to the inclusion of non-linear terms, is apparent from the fact that two different types of staining are successfully modeled: cytoplasmic staining (GFP), where the correlation between fluorescence and forward scatter depends on cell volume, and surface staining (bar coding), where this correlation depends on surface area. Further, the universality of the method was established by applying it to data sets from different organisms and laboratories.
Finally, we have used this methodology to analyze the effect of chromatin remodeling proteins on POT1 expression and variability during a carbon shift from glucose to oleate and back to glucose. Yeast cells change in size and morphology during this carbon shift, making the analysis impossible by traditional gating. Pot1p–GFP shows a clear bimodal pattern; that is, during the carbon shift there is a bifurcation event, after which two different subpopulations can be recognized; one with a higher expression, the other with low expression. This indicates that only a fraction of the cells in the population can achieve activation under oleate induction. This behavior has been previously ascribed to transcriptional network architecture (Ramsey et al, 2006), but the results presented here together with previous observations (Ratushny et al, 2008) indicate that in the case of POT1, the effect is also mediated by chromatin modifiers. In particular, Htz1 appears to play an important role in controlling this bimodal behavior. Specifically, deletions of HTZ1 and some of its major effectors (in either its nuclear transport or its chromatin-binding functions) showed either no bimodality or a delayed bimodality with only a low percentage of high expressers.
Several nucleoporins were included in the miniarray because an increasing amount of data suggest that gene activity is linked to physical position within the nucleus and the NPC may provide a means for genes to be recruited to the periphery and either promote gene activation or repression. The precise role(s) of the NPC in these dynamic, complex (and potentially physically distinct) activities operating at the nuclear periphery remain to be elucidated. POT1 is localized to the nuclear periphery coincident with activation (Supplementary Figure S16). This behavior is shared with other highly expressed genes and, in particular genes within subtelomeric regions (POT1 is located near a subtelomeric region (∼40 kb from left telomere of Chr IX)) (Casolari et al, 2004; Cabal et al, 2006; Brickner et al, 2007). This movement is important for robust transcriptional activation and has been termed reverse recruitment. This activity requires the seven-member Nup84 complex. Only three members of this complex were included in the miniarray, yet all three (Nup84, Nup120 and Nup133) were found in the non-bimodal cluster 1 leading to the hypothesis that subtle differences in POT1 promoter localization can also be the cause of the divergent fate of cells exposed to oleate. In addition, Nup2 has been associated with peripheral localization of genes during their activation (Brickner et al, 2007). It is also suggested that this recruitment promotes Htz1-mediated epigenetic memory. Independent studies have found that Nup2 and Htz1 are functionally linked in chromatin function through boundary activity (separating active from repressed chromatin) (Ishii et al, 2002; Dilworth et al, 2005). Remarkably, Nup2 was found in the same nine-member cluster (cluster 4) as Htz1. Taken together, these results suggest that the analysis performed here is sufficiently precise to reveal functional relationships among proteins of different families.
We hypothesize that the transitions between chromatin activity states mediated by Nup2, Htz1 and/or Nup84 complex may have been evolutionarily selected to create a bimodal profile to facilitate adaptation to non-predictable environments, where a commitment to oleate metabolism implies a high risk if the change in carbon source is transient. Biological advantages derived from this phenotype are related to the high investment that cells need to make in order to adapt to oleate as a carbon source. This not only requires expressing new genes, but also commits cells to major structural changes, such as creating new peroxisomes. Maintaining a heterogeneous population, especially in cases of highly committed responses, can be advantageous as it leaves a fraction of the population able to respond quickly upon a switch back to the original conditions (Acar et al, 2008). This study provides the first thorough analysis of this phenomenon in the oleate response in yeast leading to the identification of several chromatin modifiers, and NPC components required to maintain population variability during the transcriptional response.
Materials and methods
General description of flow cytometry data
Raw flow cytometry data consist of two scatter measurements (FSC and SSC) and one or more fluorescence channels (FL) for each measured cell (or event) in a biological sample. FSC and SSC correspond to cell size and cell granularity, respectively. Here, only one fluorescence channel is assumed. However, this method can be applied to each fluorescence channel independently if there is more than one. Further, the goal is to analyze multiple biological samples, say, N samples. For each sample n (with n=1,…, N) the FSC is denoted as column vector xnFSC, the SSC as column vector xnSSC and the FL as column vector xnFL. Within each biological sample, these three column vectors have the same length, which is the number of measured cells in that sample, denoted by sn.
Compensating for the variability due to cell size and cell granularity using regression
The approach follows four steps (graphically depicted in Figure 1). The MATLAB scripts for these steps are found in Supplementary information and MATLAB Code and at http://code.google.com/p/flowregressionmodel/. Specifically, step 1: FC_preprocess.m, step 2: selectdensescatterarea_ cont.m, step 3 and 4: doregress_constrained.m.
Step 1: Preprocessing
The raw data are processed to eliminate instrument errors and outliers as described in Newman et al (2006):
The first and last 0.2 s of data are removed to minimize errors due to uneven sample flow through the cytometer.
All FL, FSC and SSC data with minimal or maximal values are removed, as these values are saturated and thus undefined.
The bottom and top 5% of the FSC and SSC data are excluded to limit the influence of cellular debris and aggregated cells.
This step is performed for each biological sample independently and leads to the removal of 10–20% of the cells, resulting in somewhat lowered values of the sn's.
Step 2: Determining the density of cells in the two-dimensional FSC/SSC space
For each biological sample n, a two-dimensional density function fn(u) is estimated, where u is a two-dimensional variable that represents a location in FSC/SSC space, that is, u=[uFSC
uSSC]. fn(u) is estimated from xnFSC and xnSSC using a bivariate kernel density estimator based on the linear diffusion process described in Botev et al (2010) (MATLAB file exchange 17204, kde2d, Zdravko Botev, University of Queensland). fn(u) is computed for all grid points of an equally spaced grid of 256 by 256 that covers the complete FSC/SSC space. We denote the locations of the grid points of this grid by the set G. After 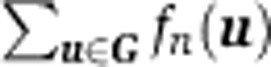 is normalized to one for each biological sample n, the N densities are averaged to construct the overall density function f(u),
is normalized to one for each biological sample n, the N densities are averaged to construct the overall density function f(u),
 |
The main reason for averaging the N densities instead of computing one density for all cells in all samples is that we want to give each sample the same weight, that is, samples with fewer cells should not be considered less important. Note that 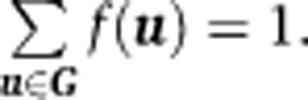 .
.
Step 3: Regressing the fluorescence values on the FSC and SSC measurements
For each biological sample n, the following linear least-squares regression problem is solved
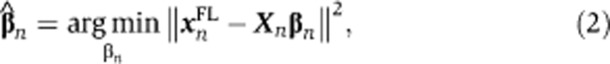 |
where 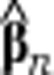 is a column vector of regression coefficients to be estimated and Xn is the design matrix:
is a column vector of regression coefficients to be estimated and Xn is the design matrix:
where 1 represents the intercept, that is, a column vector of sn ones. Thus, the model itself is linear, yet it includes non-linear terms and interaction effects in the design matrix. We consider this design matrix as a generic model containing all terms that are potentially necessary to model the often complex relationship between cell morphology and fluorescence. Experiments with various flow cytometry data sets demonstrated that higher-order polynomial (interaction) effects do not often form significant predictors. However, the MATLAB code (Supplementary Information) allows one to easily add these or any other terms if necessary. Since nine parameters (eight regression coefficients and the intercept) are estimated using (in most cases) thousands of data points, overfitting of the regression coefficients is not an issue.
Since two variables (i.e., the FSC in xnFSC and the SSC in xnSSC) are used to predict the FL (in xnFL), the resulting regression model can be represented by a regression surface, that is, a (non-linear) function of two variables. The regression problem is solved under the constraint that this regression surface is monotone across the two-dimensional FSC/SSC space. This constraint leads to stable behavior of the regression surface even in areas of the FSC/SSC space where there are no data points (i.e., cells) (Supplementary Figures S1–S5). See Supplementary Information for a complete description of the constrained regression model and visualizations of the regression surface. In Supplementary Information, we also describe a detailed comparison of different regression models demonstrating the benefit of the monotonicity constraints and complex design matrix (Supplementary Figure S6).
The regression step can also be performed on log-transformed FL data, which is more appropriate when the FL measurement data are recorded on a linear scale instead of the more common logarithmic scale.
Step 4: Computing the fluorescence intensities compensated for the effect of the SSC and FSC
Computation of the fluorescence intensities compensated SSC and FSC requires two ingredients: the sample-specific average fluorescence intensity and the sample-specific residuals (or regression errors).
For each biological sample n, the average fluorescence intensity an is computed as the average height of the regression surface across the two-dimensional FSC/SSC space weighted by the density f(u). The height of the regression surface at location u, hn(u), is given by
 |
where U is found by substituting location u into the design matrix:
To compute the average fluorescence intensity an, hn(u) is not simply the average across all grid points, but is weighted by f(u), that is,
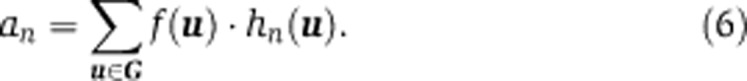 |
Note that the weights f(u) are not dependent on n, and that high-density areas have more weight in determining an, while areas without any cells in any biological sample do not contribute to the average fluorescence at all.
The variability in fluorescence that is not due to cell size and granularity is given by the residuals (or regression errors) rn,
 |
In order to obtain the fluorescence intensities compensated for the SSC and FSC, denoted by vector zn, the residuals rn, which form a distribution centered around zero, are offset by the average fluorescence intensity an:
In analogy to gating, an is the average fluorescence intensity of the cells in the gate and rn represents the cell-specific deviation from this average. However, in contrast to gating, zn has length sn, that is, the compensated fluorescence intensity is computed for all cells in the biological sample.
Compensating for the effect of cell size and cell granularity using gating
After preprocessing (step 1), an area is defined in the two-dimensional FSC/SSC space. This area is called the gate. The gate is the same for all N biological samples. All cells outside of the gate are discarded. For the cells inside the gate, the FL intensities are stored in column vector gn; that is, elements of gn form a subset of xnFL and comprise the cells inside the gate. Commonly, the shape of the gate is circular, ellipsoidal or (rotated) rectangular. In general, the length of gn (the number of cells in the gate) is much smaller than sn.
Dataset descriptions
Dataset 1: Gal data
A control data set was obtained from the paper ‘Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast’ (Ramsey et al, 2006). In particular, the time-course experiment described in Figures 2 and 3 of that paper corresponding to data acquired 1, 2, 4, 5, 6 and 7 h after galactose induction with the galactose response marker Gal1 tagged with GFP at its C-terminus was reanalyzed. Data were obtained from the WT and a strain in which the upstream regulators of GAL1, GAL3 and GAL80 were constitutively expressed under the CYC1 promoter. A non-tagged control strain was used to set the background and to normalize fluorescence intensities.
The data were analyzed in two steps as described in the paper: (1) cells were gated in the FSC channel with a gate centered at the mean FSC of the NC strain with a width of 0.1 standard deviations or alternatively analyzed with the regression model and (2) only cells with a fluorescence intensity three times higher than the mean of the non-tagged control strain were considered induced and used to calculate the CV.
Dataset 2: Pot1p–GFP time series
In all, 155 unique deletion strains, expressing the Pot1p–GFP chimera in a BY4741 strain background, from Invitrogen (Carlsbad, CA), were retrieved from a library previously constructed in our laboratory (Saleem et al, 2008). Cells were cultured in 96-well plates at 30°C for 12 h in YPBD (0.3% yeast extract, 0.5% peptone, 0.5% potassium phosphate buffer, pH 6.0, 2% glucose) to mid-logarithmic phase. Samples were then pelleted, washed and induced for 12 h in YPBO (0.5% Tween 40 and 0.2% oleic acid). Induction data were acquired at 0 (before YPBO induction), 2, 4, 6, 8, 10 and 12 h after induction or upon repression. In the latter case, cells were pelleted after 12 h in YPBO, washed and incubated in YPBD for 12 h more and data were collected at 2, 4, 6, 8, 10 and 12 h time points. The experiment was repeated for three biological replicates.
To proceed to flow cytometry analysis, 50 μl of each sample were removed, washed, fixed in 3.7% formaldehyde for 5 min and re-suspended in PBS. Samples were immediately analyzed with a FACSCalibur (BD Biosciences) using the following parameters: FSC: E0, 1.87 Amp linear scale; SSC: 660 V, 1.6 Amp linear scale; FL1: 650 V logarithmic scale. Cells were loaded onto the FACSCalibur using the high-throughput sampler (BD Biosciences). The high-throughput sampler was run in standard mode using a 96-well flat-bottomed plate and was set to sample 20 000 events or 50 μl at a rate of 2 μl/s. Only files containing >1000 cells were accepted. The final number of samples recorded was 5883 discarding 6% of the samples.
Samples were then preprocessed as described in step 1 of the regression model. Auto-fluorescence was not normalized as NCs were used as an indicator of non-responsive mutants in further analysis.
See MATLAB script example_standard.m in Supplementary information and MATLAB Code for an example of applying the regression model to these data. The complete Pot1 dataset can be found at http://code.google.com/p/flowregressionmodel/.
Dataset 3: Two-color assay
To construct a strain expressing the Pot1-mCherry chimera, the POT1 open reading frame was tagged in a BY4741 strain background, from Invitrogen (Carlsbad, CA) at its 3′ end through homologous recombination with a PCR-based strategy in frame with the sequence encoding Discosoma sp red fluorescent protein (Shaner et al, 2004). Proper genomic integration was confirmed by PCR. Two-color diploid strains were obtained by mating the resulting strain with a BY4742 (Invitrogen) derivative strain carrying Pot1–GFP (previously generated in the laboratory (Saleem et al, 2008)) obtaining a diploid strain carrying both chimeras under the same promoter and chromatin environment. Single clones were selected and cultured at 30°C for 12 h in YPD (1% yeast extract, 2% peptone, 2% glucose) and then induced for 12 h with YPBO. Samples were immediately analyzed with FACSAria (BD Biosciences). Flow was set at 2 μl/s. FSC and SSC were measured linearly and set at 77V and 280 V, respectively. Pot1-mCherry was measured at Texas Red Channel (630/22 nm, 566 V, linear), Pot1–GFP was measured at FITC channel (530/30 nm, 358 V, linear). In all, 50 000 events were monitored. The mean expression at the Texas Red channel was normalized to be equal to the mean in the FITC channel.
The regression model was applied independently for each fluorescent channel. Gates were circular, centered at the value with highest density in the two-dimensional FSC/SSC space and with a radius such that gates included 1, 2.5, 5, 10, 25, 50 or 100% of the cells.
Dataset 4: Bar-coded mammalian cells
A 6 × 6 fluorescently bar-coded sample published in Krutzik and Nolan (2006) was used. This sample, as described in Figure 3 and Materials and methods section of that paper, contains 36 samples of U937 cells (a well-established cell line originated from human histiocytic lymphoma) labeled with six concentrations of Pacific Blue-NHS (0, 0.15, 0.6, 2.5, 10 and 40 μg/ml and/or Alexa 488-NHS (0, 0.07, 0.3, 1.3, 5 or 20 μg/ml). The data were obtained via the Cytobank portal (https://www.cytobank.org). Following the same methodology as in the original paper, the sample was analyzed with the regression model after gating to remove spurious events.
See MATLAB script example_barcoded.m in Supplementary information and MATLAB Code for an example of applying the regression model to these barcoded data.
Data storage and handling
Data obtained from FACSCalibur were stored in FSC 2.0 format. Each parameter was described by 1024 channels uniformly distributed along the range of acquisition. FSC and SSC were recorded on a linear scale. FL1 was recorded and stored on a logarithmic scale. Data obtained with FACSAria were stored in FSC 3.0 format. Each parameter was described by 262 143 channels uniformly distributed along the range of acquisition. FSC and SSC were recorded in linear scale. FL1 and FL2 were recorded on a logarithmic scale but stored on a linear scale. FSC files where imported into MATLAB using fca_readfcs (file exchange 9608, Laszlo Balkay, University of Debrecen, PET Center).
Rule-based clustering of deletion strains
Of the 5883 biological samples of Dataset 2, 3663 were used in clustering: Although triplicates were measured, the third replicate seemed unreliable and was discarded. For 7 of the 155 deletion strains, <1000 cells were detected for more than half of the time points. These strains were discarded, leaving 148 deletion strains used in clustering. Additionally, there were six identical WT strains and six NCs. The Pot1p–GFP expression values were obtained as the fluorescent intensity values compensated for the effect of the SSC and FSC using the regression model. A Gaussian mixture model was fit to each time point for each strain separately using the EM approach described in Song et al (2010). This approach takes expression densities as input. The density of Pot1p–GFP expression for each biological sample was estimated using the one-dimensional version of the kernel density estimator described above. Then, for each strain and each time point, the duplicates were combined by averaging the two densities. The EM fitting procedure of Song et al uses cross-validation to determine the number of Gaussian components (1 or 2). If the mean log-likelihood (over the cross-validation folds) of the two-component model was 4 standard deviations greater than the mean log-likelihood of the one-component model, the two-component model was selected. Otherwise, the fluorescent intensity values were modeled using one Gaussian distribution. Thus, after applying the EM procedure, each time point of each strain is represented by either one or two Gaussian distributions.
The WT strains consistently showed bimodality at time points 6, 8, 10 and 12 h. First, we divided the deletion strains into two groups; one group that showed no bifurcation along the time series (49 strains) and the other group that did show bifurcation at some time point(s) (99 strains). The 49 ‘non-bifurcated strains’ were further split up into a group that was close to WT (43 strains) or to NC (6 strains) in terms of their expression profile (by using a simple least-squares distance criterion applied to the means of the Gaussian distributions). For the 99 ‘bifurcated strains,’ we analyzed the population sizes of high and low expressers. For WT, the high expressing subpopulation comprised 75±6% of the total population. The bifurcated strains were divided into three groups: (1) strains for which the high expressing population comprised <63% (two standard deviations lower than WT) of the total (10 strains), (2) strains for which the high expressing population comprised >88% (two standard deviations higher than WT) of the total (3 strains) and (3) strains with higher expressers between 63 and 88% (86 strains). Of the 99 bifurcated strains, only four (Δahc1, Δhda3, Δloc2 and Δshg1) showed bimodality as late as 14 h. However, many deletion strains (59) showed an earlier (4) or later (55) onset of bifurcation. This provided a further dissection of the bifurcated strains. The clustering is found in Table I and graphically depicted in Figure 6.
Nuclear localization of POT1
Yeast cells expressing a LacI–GFP and Nup49p–GFP were tagged with a 256 lacop array upstream from the POT1 genomic loci (Supplementary Figure S16). POT1 promoter nuclear position was scored within three zones, with zone 1 being directly on the nuclear periphery, zone 2 to being in proximity to but seperated from the nuclear periphery and zone 3 being in the center of the nucleus. Cells were grown to ∼1 × 107 cells/ml cultures in YPD and then washed and transferred to oleate-containing media (SCIM) for 6 h at 30°C. The average percentage of GFP-dots in each zone was tallied at 0 h (YPD), 1, 4 and 6 h into oleate induction and averaged over two biological replications where n⩾100.
Supplementary Material
Supplementary figures S1–16, Supplementary tables S1–3
Acknowledgments
We thank Peter Krutzik for providing the bar coded flow cytometry data and for help with analysis of this data set. We thank Hector Rovira and Jake Lin for setting up the Google Code site. This research was supported by National Institutes of Health grants R01 GM075152, R01 GM072855 and GM076547. Support to YW from The National Natural Science Foundation of China (31071146), and Excellent Young Teachers Program of Southeast University (3231001201) is also acknowledged. We thank the anonymous reviewers for their detailed and helpful comments.
Author contributions: TAK and OR devised the method and wrote the manuscript. TAK implemented the method. OR and YW performed the experiments. GPN wrote the manuscript and provided expertise in working with the bar-coded data. JDA and IS supervised the project and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acar M, Mettetal JT, van Oudenaarden A (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40: 471–475 [DOI] [PubMed] [Google Scholar]
- Batenchuk C, St-Pierre S, Tepliakova L, Adiga S, Szuto A, Kabbani N, Bell JC, Baetz K, Kærn M (2011) Chromosomal position effects are linked to sir2-mediated variation in transcriptional burst size. Biophys J 100: L56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botev Z, Grotowski J, Kroese D (2010) Kernel density estimation via diffusion. Ann Stat 38: 2916–2957 [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH (2007) H2A. Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U (2006) SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Cagatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Suel GM (2009) Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell 139: 512–522 [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW (2010) Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD (2005) The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol 171: 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einerhand AW, Voorn-Brouwer TM, Erdmann R, Kunau WH, Tabak HF (1991) Regulation of transcription of the gene coding for peroxisomal 3-oxoacyl-CoA thiolase of Saccharomyces cerevisiae. Eur J Biochem 200: 113–122 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403: 108–112 [DOI] [PubMed] [Google Scholar]
- Igual JC, Gonzalez-Bosch C, Franco L, Perez-Ortin JE (1992) The POT1 gene for yeast peroxisomal thiolase is subject to three different mechanisms of regulation. Mol Microbiol 6: 1867–1875 [DOI] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK (2002) Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562 [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M (2010) Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371 [DOI] [PubMed] [Google Scholar]
- Krutzik P, Nolan G (2006) Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods 3: 361. [DOI] [PubMed] [Google Scholar]
- Lo K, Brinkman RR, Gottardo R (2008) Automated gating of flow cytometry data via robust model-based clustering. Cytometry A 73: 321–332 [DOI] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D (2007) Noise in gene expression determines cell fate in Bacillus subtilis. Science 317: 526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM (2005) Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Pyne S, Hu X, Wang K, Rossin E, Lin TI, Maier LM, Baecher-Allan C, McLachlan GJ, Tamayo P, Hafler DA, De Jager PL, Mesirov JP (2009) Automated high-dimensional flow cytometric data analysis. Proc Natl Acad Sci USA 106: 8519–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Smith JJ, Orrell D, Marelli M, Petersen TW, de Atauri P, Bolouri H, Aitchison JD (2006) Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat Genet 38: 1082–1087 [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny AV, Ramsey SA, Roda O, Wan Y, Smith JJ, Aitchison JD (2008) Control of transcriptional variability by overlapping feed-forward regulatory motifs. Biophys J 95: 3715–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem RA, Knoblach B, Mast FD, Smith JJ, Boyle J, Dobson CM, Long-O’Donnell R, Rachubinski RA, Aitchison JD (2008) Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. J Cell Biol 181: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Smith JJ, Sydorskyy Y, Marelli M, Hwang D, Bolouri H, Rachubinski RA, Aitchison JD (2006) Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol Syst Biol 2: 2006.0009; DOI: 10.1038/msb4100051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Phenix H, Abedi V, Scott M, Ingalls BP, Kaern M, Perkins TJ (2010) Estimating the stochastic bifurcation structure of cellular networks. PLoS Comput Biol 6: e1000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube K, Blackwell J Jr, Pemberton L (2010) Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic 11: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB (2007) Tunability and noise dependence in differentiation dynamics. Science 315: 1716–1719 [DOI] [PubMed] [Google Scholar]
- Wan Y, Saleem R, Ratushny A, Roda O, Smith J, Lin CH, Chiang JH, Aitchison J (2009) The role of the histone variant H2A.Z/Htz1p on TBP recruitment, chromatin dynamics and regulated expression at oleate-responsive genes. Mol Cell Biol 29: 2346–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Wu CH, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, Wu C (2009) N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem 284: 6200–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1–16, Supplementary tables S1–3