Abstract
Self/non-self discrimination is a fundamental requirement of life. Endogenous peptides presented by major histocompatibility complex class I (MHC I) molecules represent the essence of self for CD8 T lymphocytes. These MHC I peptides (MIPs) are collectively referred to as the immunopeptidome. From a systems-level perspective, very little is known about the origin, composition and plasticity of the immunopeptidome. Here, we show that the immunopeptidome, and therefore the nature of the immune self, is plastic and moulded by cellular metabolic activity. By using a quantitative high-throughput mass spectrometry-based approach, we found that altering cellular metabolism via the inhibition of the mammalian target of rapamycin results in dynamic changes in the cell surface MIPs landscape. Moreover, we provide systems-level evidence that the immunopeptidome projects at the cell surface a representation of biochemical networks and metabolic events regulated at multiple levels inside the cell. Our findings open up new perspectives in systems immunology and predictive biology. Indeed, predicting variations in the immunopeptidome in response to cell-intrinsic and -extrinsic factors could be relevant to the rational design of immunotherapeutic interventions.
Keywords: biochemical network, major histocompatibility complex, mTOR, plasticity, transcriptome
Introduction
While unicellular eukaryotes primarily employ self/non-self discrimination to avoid self-mating, multicellular organisms use self/non-self discrimination primarily in immune defense (Boehm, 2006). Failure to respond to non-self can lead to death from infection whereas untoward response to self paves the way to autoimmunity. Peptides presented by major histocompatibility complex class I (MHC I) molecules represent the essence of self for CD8 T lymphocytes (Rammensee et al, 1993; Klein et al, 2009). These MHC I-associated peptides (MIPs) regulate all key events that occur during the lifetime of CD8 T cells in the thymus and the periphery (Goldrath and Bevan, 1999; Klein et al, 2009). MIPs are presented at the surface of most nucleated cells in jawed vertebrates and are collectively referred to as the MHC I immunopeptidome (Istrail et al, 2004).
Classic reductionist approaches have established the broad outlines of the MIP processing system and shown that MIPs derive from degradation of endogenous proteins by the proteasome and other peptidases (Yewdell et al, 2003; Hammer et al, 2007). They have also highlighted the complexity of the MIP repertoire. Thus, MHC I molecules present peptides encoded not only in the primary open reading frames but also those encoded in alternate reading frames (Starck and Shastri, 2011). Moreover, progresses in mass spectrometry (MS) have allowed increasingly sophisticated and comprehensive large-scale analyses of MIPs (Mester et al, 2011). Large-scale analyses have yielded unprecedented insights into the peptide specificities and motifs of MHC molecules and the diversity of the MIP repertoire (Hunt et al, 1992; Engelhard et al, 1993; Bonner et al, 2002; Weinzierl et al, 2008). They have also demonstrated that MIPs derive from all cell compartments and that the MIP repertoire can be modified by neoplastic transformation (Schirle et al, 2000; Hickman et al, 2004; Weinzierl et al, 2007; Fortier et al, 2008). Nevertheless, we still know very little about the genesis and molecular composition of the immunopeptidome: why do proteins such as STT3B yield abundant MIPs while others do not (Perreault, 2010)? MS studies have also revealed that the immunopeptidome is not a random sample of the proteome: many abundant proteins do not generate MIPs, while some low-abundance proteins generate large amounts of MIPs (Milner et al, 2006). Furthermore, large-scale analyses have yielded conflicting results on the relation between the transcriptome and the MIP repertoire (Weinzierl et al, 2007; Fortier et al, 2008; Mester et al, 2011). Therefore, further systematic studies based on high-throughput technologies and integrative approaches are needed in order to elucidate the mechanisms that mould the immunopeptidome. In-depth mechanistic understanding of the immunopeptidome biogenesis would allow prediction of its molecular composition and would therefore be highly relevant to the development of immunotherapies (Zarling et al, 2006; Sette and Rappuoli, 2010).
A key unresolved question is whether the immunopeptidome of a particular cell is plastic and affected by its metabolic state. This issue is of fundamental importance because cells targeted by CD8 T lymphocytes (i.e. infected and neoplastic cells) are metabolically perturbed by intracellular parasite/viruses or various transforming events. We addressed this question by studying the immunopeptidome of a mouse lymphoma cell line (EL4) treated with the classic mammalian target of rapamycin (mTOR) inhibitor, rapamycin. We selected this model because rapamycin is an exquisitely specific inhibitor of the serine/threonine kinase mTOR, and because mTOR is a central regulator of cellular homeostasis through its involvement in promoting anabolic and inhibiting catabolic processes (Sengupta et al, 2010). Furthermore, mTOR signaling is regulated in numerous physiological and pathological conditions in all cell types, including neoplastic and immune cells (Araki et al, 2009; Caron et al, 2010; Sengupta et al, 2010).
Results
The effect of rapamycin on mTOR signaling in EL4 cells
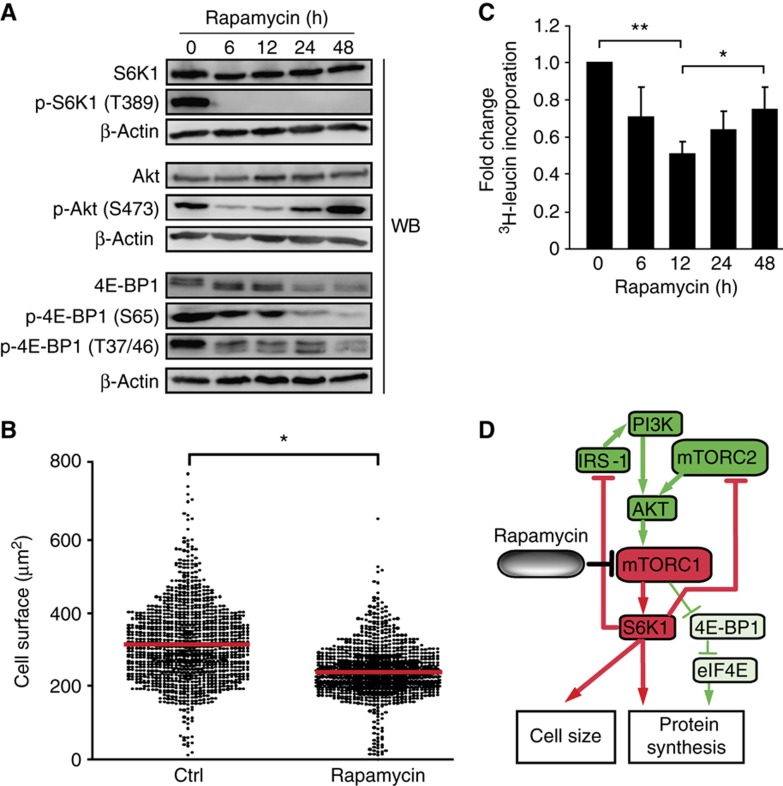
As a prelude to our peptidomic studies, we first assessed the effects of rapamycin on EL4 cells. The first mTOR complex, mTORC1, phosphorylates both S6K1 and 4E-BP1 (Dowling et al, 2010). Recent studies have shown that rapamycin, previously thought to completely inhibit mTORC1 activity, differentially affects 4E-BP1 and S6K1 (Choo et al, 2008; Choo and Blenis, 2009; Thoreen et al, 2009). While rapamycin treatment completely and sustainably inhibits S6K1 activation, it partly and variably inhibits 4E-BP1 phosphorylation (Choo et al, 2008). To evaluate how rapamycin affected mTORC1 signaling in EL4 cells, we treated EL4 cells with rapamycin for different time durations up to 48 h. As expected, Thr-389 on S6K1 was fully dephosphorylated upon rapamycin treatment, correlating with a decrease in cell size (Figure 1A and B). In contrast, Ser-65 and Thr-37/46 on 4E-BP1 were partially dephosphorylated by rapamycin (Figure 1A). As previously reported, we also noted that abundance of 4E-BP1 was decreased after 24 h of rapamycin treatment (Figure 1A) (Dilling et al, 2002). Because 4E-BP1 is critically involved in cap-dependent translation via regulation of eIF4E, these results suggested that translation was maintained in EL4 cells in the presence of rapamycin. Accordingly, we observed that protein synthesis decreased during the first 12 h of rapamycin treatment, but progressively recovered thereafter (Figure 1C). Prolonged rapamycin treatment for up to 24 h has also been shown to inhibit the assembly of mTORC2, the second mTOR complex (Sarbassov et al, 2006). Here, we observed that mTORC2-mediated phosphorylation of AKT at Ser-473 was transiently decreased but recovered and was increased after treatment for 48 h. Thus, rapamycin treatment for 48 h inhibited mTORC1, but activated mTORC2 in EL4 cells. Collectively, these results showed that rapamycin-mediated mTORC1 inhibition perturbed protein synthesis and cell size by differentially inhibiting S6K1 and 4E-BP1 in EL4 cells (Figure 1D). Of direct relevance to our analyses of the MIP landscape, rapamycin treatment did not affect expression levels of key proteins involved in the MHC I antigen processing and presentation pathway (Supplementary Figure S1).
Figure 1.
Rapamycin differentially inhibits S6K1 versus 4E-BP1 in EL4 cells. Cells were treated with 20 ng/ml of rapamycin for the indicated time periods. (A) Levels of the indicated proteins were determined by western blotting. β-Actin served as a loading control. Data are representative of three independent experiments. (B) EL4 cells were treated or not (ctrl) with rapamycin for 48 h. One thousand cells were counted for each condition. Cell size was measured by light microscopy. The red line corresponds to the average cell size. Data are representative of three independent experiments. *P<0.0005 (Student's t-test). (C) Relative protein synthesis was measured by [3H]-leucine incorporation. Data (mean±s.d.) are representative of three independent experiments. *P<0.01, **P<0.005 (Student's t-test). (D) Model for effects of rapamycin-mediated mTORC1 inhibition in EL4 cells. Proteins and reactions were curated from the literature. Color code is based on the results in Figure 1 after 48 h of rapamycin treatment. Activation and inhibition of components/reactions are depicted in green and red, respectively. Partial inactivation is represented by the thinner green lines.
High-throughput MS-based studies unveil the plasticity of the immunopeptidome
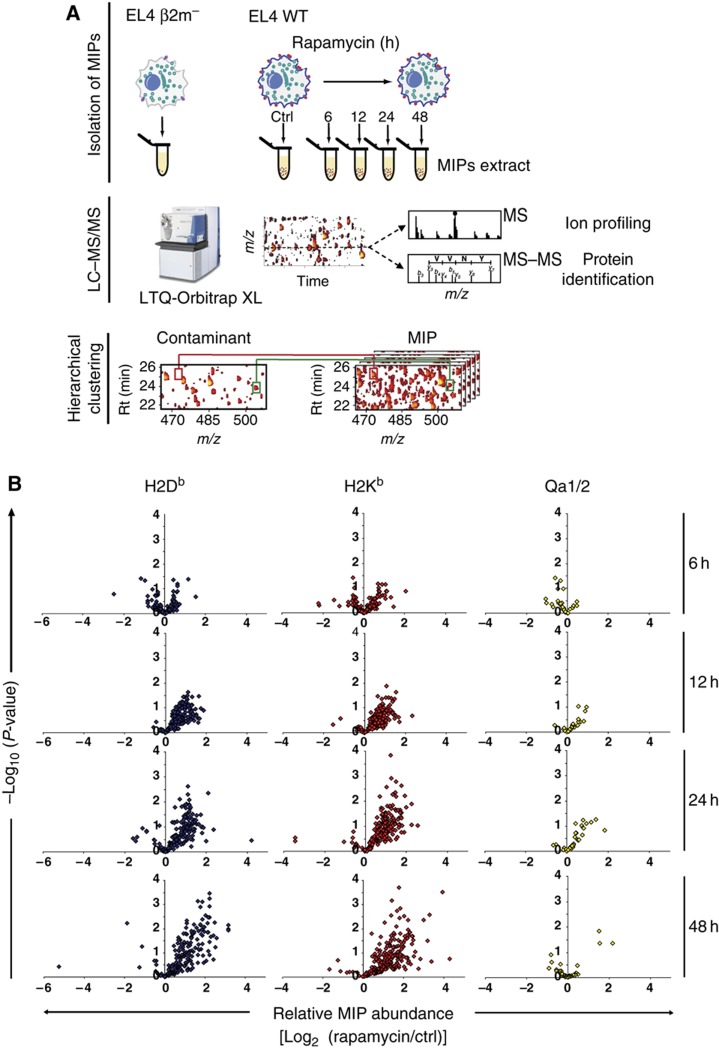
The plasticity of the immunopeptidome can only be estimated by systems-level analyses (Benoist et al, 2006; Germain et al, 2011), and MS with high accuracy analyzers is the most comprehensive and versatile tool in large-scale proteomics (Yates et al, 2009). We therefore used a high-throughput MS-based approach (Fortier et al, 2008; de Verteuil et al, 2010) to profile in a time-sequential manner the abundance of MIPs presented by EL4 cells exposed to rapamycin for up to 48 h (Figure 2A; Supplementary Information). Subtraction of ‘contaminant peptides’ eluted from β2m mutant EL4 cells allowed specific identification of genuine MIPs (Supplementary Table S1) (Fortier et al, 2008; de Verteuil et al, 2010). This resulted in identification and quantification of the relative abundance of 422 unique MIPs associated to different MHC I allelic products (Figure 2B; Supplementary Table S2). The salient finding was that the abundance of most peptides presented by classical MHC Ia molecules (H2Db and H2Kb) increased over time after 12–48 h of rapamycin treatment (Figure 2B). In contrast, the abundance of most peptides associated to non-classical MHC Ib molecules (Qa1 and Qa2) was not augmented after treatment with rapamycin for 48 h. Accordingly, exposure to rapamycin led to an increase in cell surface levels of H2Db and H2Kb but not Qa1 and Qa2 (Supplementary Figure S2B). Clustering analysis of the time-series data sets revealed that the abundance of numerous MIPs increased progressively in response to rapamycin (Supplementary Figure S2A). In general, the dynamic range of this response was modest with a median fold change in MIP abundance of 2.1 relative to untreated cells (calculated from Supplementary Table S2). Nonetheless, a subset of 222 MIPs showed fold changes between 2 and 15, including 6 MIPs detected exclusively on rapamycin-treated cells (Supplementary Table S2). Perturbation of cellular metabolic activity with rapamycin therefore unveiled the plasticity of the immunopeptidome, by increasing both the abundance and the diversity of MIPs at the cell surface in a time-dependent manner.
Figure 2.
Rapamycin increases the abundance of MIPs presented by MHC Ia molecules. (A) Experimental design for identification and relative quantification of MIPs. EL4 cells were incubated with 20 ng/ml of rapamycin for 0 h (ctrl), 6, 12, 24 and 48 h. Cells from all experimental conditions were harvested simultaneously. MIPs from rapamycin-treated and ctrl EL4 cells were isolated by mild acid elution and analyzed by nanoLC–MS–MS/MS. Heat maps displaying m/z, retention time and abundance were generated. A logarithmic intensity scale distinguishes between low (dark red) and high (bright yellow) abundance species. Analysis of β2m- mutant EL4 cells allows discrimination of genuine MIPs from contaminants (Fortier et al, 2008; de Verteuil et al, 2010). Examples of peptides that were differentially expressed (red line) or not (green line) between WT and β2m- mutant EL4 cells are highlighted in the boxes. (B) Volcano plots show the relative abundance of 416 H2b-associated peptides identified in three biological replicates at each time point. Six MIPs were detected uniquely after 48 h of rapamycin treatment.
mTOR inhibition induces functionally coherent changes in the transcriptome and the immunopeptidome
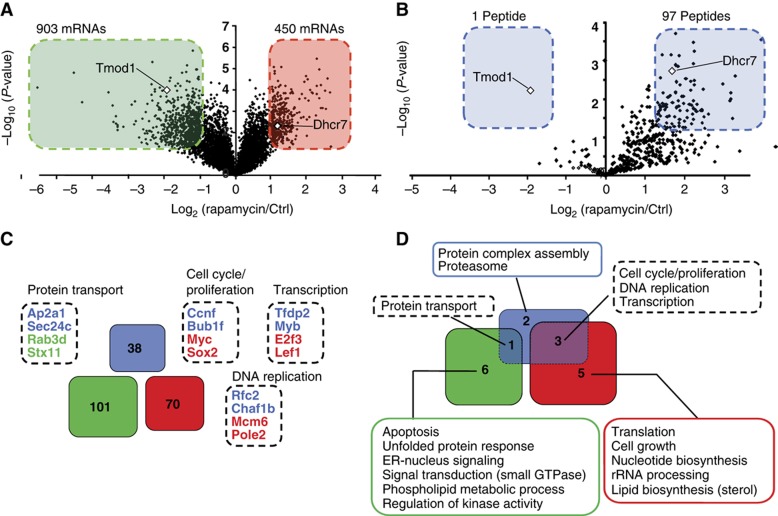
Whether the immunopeptidome accurately projects intracellular metabolic changes to the cell surface is a fundamental question that has not been addressed yet. More precisely, we asked herein whether variations in the immunopeptidome reflected perturbations of rapamycin-sensitive metabolic processes regulated at the transcript level. To address this question, we first defined the impact of rapamycin on the transcriptome of EL4 cells. Using NimbleGen MM8 385K microarrays, we compared the transcriptome of untreated EL4 cells to that of EL4 cells treated with rapamycin for 48 h. A total of 1353 differentially expressed transcripts were identified (fold change >2.0; P<0.05), which represents about 3% of transcripts in EL4 cells (Figure 3A). Among these, 903 and 450 mRNAs were under- and overexpressed, respectively. These genes are hereafter referred to as differentially expressed genes (DEGs). We next compared the 1353 DEGs with 98 unique source genes coding for the most differentially expressed MIPs (DEMs) (fold change >2.5; P<0.05: as described in Fortier et al, 2008) (Figure 3B; Supplementary Table S2). Based on microarray gene expression data, only two DEM source genes (Tmod1 and Dhcr7) were found to be differentially expressed at the transcript level (Figure 3A and B). Interestingly, the only DEM that was less abundant was also found to be downregulated at the transcript level. Thus, these results indicate that only a minority of DEMs originate from DEGs.
Figure 3.
Rapamycin-mediated mTOR inhibition induces functionally coherent changes in the transcriptome and the immunopeptidome. (A) Volcano plot representation of the relative abundance of 42 586 transcripts after 48 h of rapamycin treatment. Transcripts were considered to be differentially expressed when the fold difference in abundance was >2.0 (P<0.05). Transcripts over- and underexpressed in rapamycin-treated cells relative to ctrl cells are depicted in the red and green boxes, respectively. (B) Volcano plot representation of the relative abundance of 422 MIPs after 48 h of rapamycin treatment. MIPs that were the most differentially expressed (fold difference relative to untreated cells >2.5; P<0.05; fold difference and P-value based on Fortier et al, 2008) are highlighted in the blue boxes. (C, D) GO enrichment analyses were performed for DEM source genes and DEGs. In all, 38 DEM source genes (blue) were associated to 6 significantly enriched cellular processes. In all, 101 (green) and 70 (red) genes coding for under- and overexpressed mRNAs were associated to 7 and 8 significantly enriched cellular processes, respectively. (C) Venn diagram showing no overlap between DEM source genes and DEGs. Dashed boxes show representative genes that contributed to enrichment of four cellular processes in both DEM source genes and DEGs. (D) Venn diagram showing functional overlap between cellular processes overrepresented in DEM source genes and DEGs. The four cellular processes overrepresented in both DEM source genes and DEGs are listed in the dashed boxes.
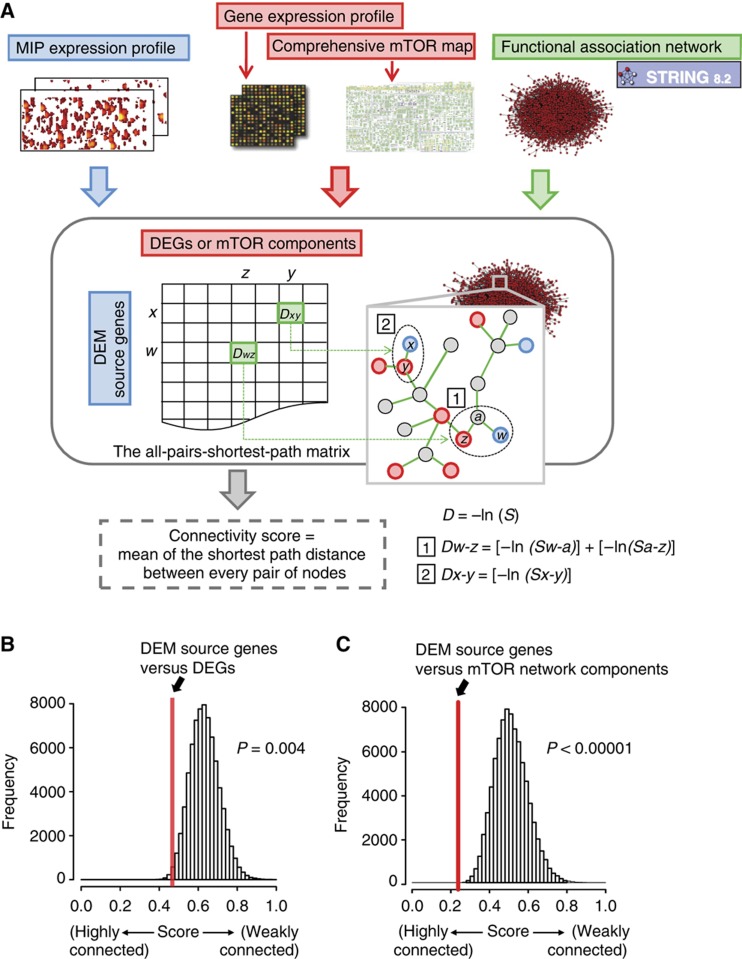
Analysis of Gene Ontology (GO) annotations revealed that 101 and 70 genes (Figure 3C; Supplementary Figure S3B and C) coding for under- and overexpressed mRNAs were associated to 7 and 8 (Figure 3D) significantly enriched cellular processes, respectively (P<0.05) (Supplementary Table S3). GO term analysis on the 98 DEM source genes also revealed a significant (P<0.05) enrichment for 6 cellular processes (Figure 3D) implicating 38 DEM source genes (Figure 3C; Supplementary Figure S3A; Supplementary Table S3). Analysis of DEGs and DEM source genes involved in enriched cellular processes yielded several findings. First, there was no overlap between the 171 DEGs and the 38 DEM source genes (Figure 3C). Second, cross-comparison of the overrepresented functional groups identified four distinct cellular processes (protein transport, cell cycle/proliferation, DNA replication and transcription) that were enriched in both DEGs and DEM source genes (Figure 3D). In order to determine whether this result implies a significant functional relationship between transcriptome and immunopeptidome variations, we developed an all-pairs-shortest-path matrix, which scores the functional connectivity between two lists of genes (Figure 4A). Using this matrix, we first calculated a connectivity score between the 171 DEGs and the 38 DEM source genes identified above. Then, bootstrapping was used as a statistical sampling method to calculate control connectivity scores from 105 sets of 38 randomly selected MIP source genes from a database of 891 unique source genes encoding H2b-associated peptides (Supplementary Table S4). These analyses revealed that the 38 DEM source genes were tightly interconnected to the 171 DEGs (bootstrapping; P=0.004) (Figure 4B). Hence, while most DEMs did not originate directly from DEGs, they do originate from genes that are tightly functionally connected to DEGs. In other words, this systems-level analysis demonstrates that rapamycin-mediated mTOR inhibition induces functionally coherent changes in the transcriptome and the immunopeptidome. Given the tremendous complexity of the transcriptome, this finding suggests that the immunopeptidome of the cell is more complex than anticipated and that its plasticity might be extensive.
Figure 4.
DEM source genes are tightly connected to transcriptomic changes and the mTOR network. (A) An all-pairs-shortest-path matrix was developed by using computed scores (S) in the STRING database (http://string-db.org/). The all-pairs-shortest-path matrix was used to calculate functional associations between (1) DEM source genes and DEGs and (2) DEM source genes and mTOR network components. Each functional association in the all-pairs-shortest-path matrix was transformed into a distance (D). The matrix shows DEM source genes (rows), DEGs or mTOR network components (columns), and the shortest path distance between every pair of nodes (genes/proteins) in the association network (e.g. Dw-z and Dx-y). A connectivity score corresponds to the mean of the shortest path distance between every pair of nodes in a given matrix. (B, C) The all-pairs-shortest-path matrix was used to calculate functional connectivity scores. The red lines represent the connectivity score between DEM source genes and DEGs (B), and between DEM source genes and mTOR network components (C). A bootstrap procedure was used to calculate control connectivity scores represented by the Gaussian distributions.
DEMs arise from biochemical networks connected to mTOR
The above results suggested that DEMs originated from genes connected to the mTOR network. However, considering that mTOR has a pervasive role in protein synthesis and degradation (Caron et al, 2010), we could not discard the possibility that DEMs originated from some non-specific generic effect of rapamycin on protein metabolism. In the latter case, DEM-coding genes would not be tightly connected to the mTOR network. Hence, in order to further evaluate the relationship between DEMs and mTOR, we first conducted an analysis on the 98 DEM source genes (Figure 3B; Supplementary Table S2) using the interaction network database STITCH (Kuhn et al, 2010). This analysis uncovered a network containing 30 DEM source genes that were interconnected and organized within discrete functional modules (Supplementary Figure S4). Strikingly, the network included the chemicals rapamycin and everolimus (rapamycin analog) in addition to components (e.g. Rictor, Sgk1) and modules (e.g. mTOR signaling, translation, lipid biosynthesis) known to be directly regulated by mTOR (Caron et al, 2010). Therefore, we reasoned that rapamycin-mediated changes in the immunopeptidome might originate from genes that are very closely connected to components of the mTOR signaling network. To systematically evaluate this assumption, we measured the connectivity score between the 30 DEM source genes and components extracted from a comprehensive map of the mTOR interactome and signaling network (Supplementary Table S5 based on Caron et al, 2010). By using the all-pair-shortest-path matrix described above, we calculated that the 30 DEM source genes were strongly interconnected to the mTOR network components relative to random assignments (bootstrapping; P<10−5) (Figure 4C). This systems-level analysis demonstrates that a substantial fraction of rapamycin-induced variations in the immunopeptidome arise from biochemical networks whose components are highly connected to the target of rapamycin (i.e. mTOR). Thus, our data reinforce the notion that the immunopeptidome projects a functional representation of intracellular metabolic changes to the cell surface. Further immunopeptidomic studies are needed to evaluate whether the mTOR network regulates the repertoire of MIPs in other contexts.
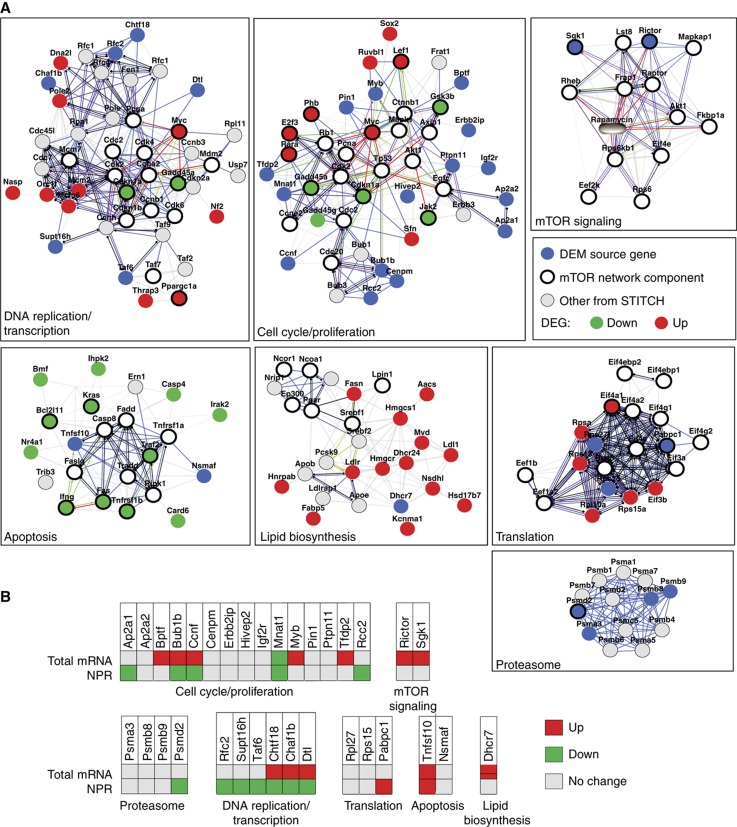
To visualize the organization of relationships between DEM source genes, the transcriptome and the mTOR network, we integrated in a global network the mTOR interactome and signaling network (Supplementary Table S5 based on Caron et al, 2010), the 98 DEM source genes (Figure 3B; Supplementary Table S2) and the 171 DEGs (Figure 3C; Supplementary Figure S3B and C). We found that 33 DEM source genes were constituents of 7 discrete functional subnetworks (Figure 5A). All modules included components of the mTOR network. Integration of microarray data revealed that DEGs were found in 5 out of the 7 modules containing DEM source genes. Notably, two modules contained DEM source genes but no DEGs: the proteasome and the core mTOR signaling modules. Overall, 82% of the DEM source genes (27/33) were localized within subnetworks transcriptionally regulated by DEGs. Collectively, these data indicate that rapamycin-induced variations in the immunopeptidome arise mostly from transcriptionally perturbed subnetworks that are connected to components of the mTOR network.
Figure 5.
DEM source genes are regulated at multiple layers within specific mTOR subnetworks. (A) The STITCH database (http://stitch.embl.de/) was used to generate a network from protein–protein interactions and functional associations involving identified DEM source genes, DEGs and mTOR network components. From the total network, subnetworks of DNA replication/transcription, cell cycle/proliferation, apoptosis, lipid biosynthesis, translation, proteasome and mTOR signaling were extracted. Legend for functional associations (edges) is depicted in Supplementary Figure S4. (B) EL4 cells were treated or not with rapamycin for 48 h. Total mRNA and NPR levels of the DEM source genes depicted in (A) were assessed by quantitative real-time PCR (see also Supplementary Table S6).
The immunopeptidome integrates events occurring at the transcriptional and co- or post-translational level
Generation of MIPs exploits generic cellular processes including transcription, translation and protein degradation (Yewdell et al, 2003; Fortier et al, 2008; de Verteuil et al, 2010). However, the relative contribution of these different regulatory processes in moulding the immunopeptidome remains elusive. Here, we first sought to determine whether the 33 DEM source genes depicted in Figure 5A were translationally and/or transcriptionally regulated in response to rapamycin. After 48 h of rapamycin treatment, polysome-associated RNA as well as total RNA from both rapamycin-treated and -untreated cells were analyzed using quantitative real-time PCR. Translation of a large number of genes is reduced following rapamycin treatment (Grolleau et al, 2002). Accordingly, analysis of normalized polysomal mRNAs (NPR) (see Materials and methods) showed that, in rapamycin-treated cells, ribosomal loading efficiency was decreased for 36% (12/33), and increased for 6% (2/33) of the DEM source mRNAs (Figure 5B). In contrast, we observed that 36% (12/33) of the source genes were upregulated at the transcript level. Two points can be made from these data. First, while transcriptional activation could explain the increased abundance of about 36% of MIPs in rapamycin-treated cells, the role of translational activation appears negligible. As a corollary, increased levels of 20/33 MIPs can be ascribed to neither enhanced transcription nor translation of their source genes. From this, we infer that increased MIP levels on rapamycin-treated cells are caused mainly by co- and/or post-translational mechanisms.
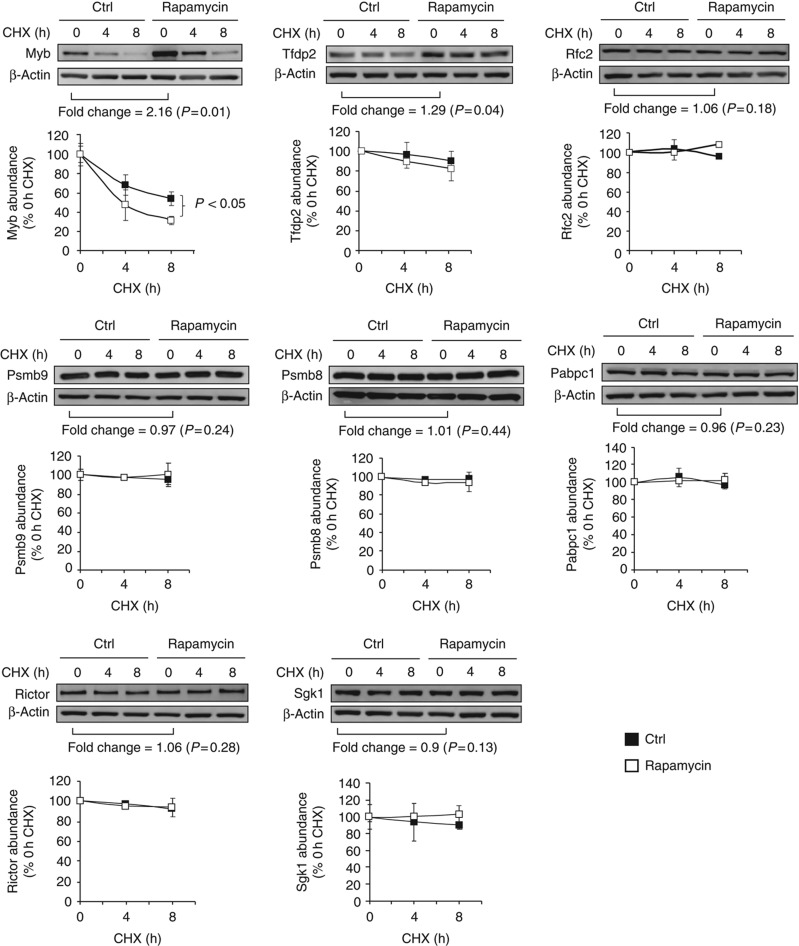
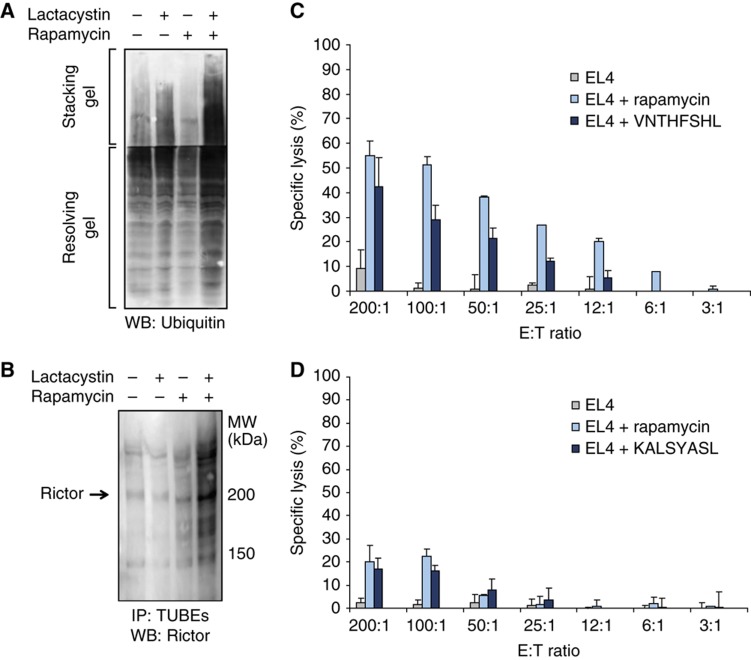
In order to probe the role of co- and/or post-translational mechanisms, we first assessed the abundance and stability of eight DEM source proteins by western blotting on total lysates from EL4 cells treated or not with rapamycin and cycloheximide (Figure 6; Supplementary Table S6). The abundance of two proteins was increased (Myb and Tfdp2) while that of the other six proteins was unchanged (Rfc2, Psmb9, Psmb8, Pabpc1, Rictor and Sgk1). Notably, the eight DEM source proteins were relatively stable with a half-life ⩾8 h in the presence or absence of rapamycin. Myb was the only DEM source protein degraded more rapidly (two-fold; half-life from 8 to 4 h) in the presence of rapamycin. These data strongly suggest that MIP levels do not correlate with the abundance or stability of the native form of DEM source proteins. However, up to 30% of proteins are so rapidly degraded (with a mean t1/2 of ∼10 min) by the ubiquitin-proteasome that they escape detection in standard cycloheximide chase experiments (Qian et al, 2006). Most of these rapidly degraded proteins are defective ribosomal products (DRiPs) resulting from imperfections of protein synthesis or assembly (Yewdell and Nicchitta, 2006). Accordingly, most protein species have two half-lives: a longer one for the well-conformed native protein and a short one for DRiPs. Importantly, MIPs are generated at higher efficiency from rapidly degraded proteins than from old proteins (Yewdell et al, 2003; Yewdell and Nicchitta, 2006). Western blotting experiments on whole EL4 cell extracts revealed that rapamycin increased the amount of proteasomal substrates, that is, ubiquitinated proteins that accumulate in lactacystin-treated cells (Figure 7A). We focused on Rictor, a DEM source protein that is a component of the mTOR complex 2, to evaluate more specifically whether rapamycin enhances proteasomal substrates generation from DEM source proteins. Rictor is coding for the KALSYASL peptide, which was exclusively detected after 48 h of rapamycin treatment (Supplementary Table S2). EL4 cells were treated or not with rapamycin for 48 h, with or without lactacystin for the last 8 h. In the presence of lactacystin, we observed a substantial accumulation of polyubiquitinated Rictor proteins in rapamycin-treated cells relative to untreated cells (Figure 7B). This result indicates that more polyubiquitinated Rictor proteins are subjected to proteasome-mediated degradation upon rapamycin treatment. Because steady-state levels of the native Rictor protein were not affected by rapamycin (Figure 6), we infer that the overabundant Rictor MIP derives from a rapidly degraded pool of Rictor and not from the long-lived native form of Rictor. Collectively, our data indicate that changes in the immunopeptidome integrate events occurring at the transcriptional and co- or post-translational level. Furthermore, consistent with the fact that mTOR links protein quality and quantity control (Qian et al, 2010), our work suggests that enhanced generation of rapidly degraded proteasomal substrates is instrumental to rapamycin-induced changes in the immunopeptidome. Altogether, our results show that the immunopeptidome projects at the cell surface a representation of biochemical networks and metabolic events regulated at multiple levels inside the cell.
Figure 6.
Relative abundance and stability of eight DEM source proteins in the presence or absence of rapamycin. EL4 cells were treated or not (ctrl) with 20 ng/ml of rapamycin for 48 h. Levels of DEM source proteins were determined by western blotting and quantified by densitometry. Fold change indicates the rapamycin/ctrl protein abundance ratio. To estimate the half-life of DEM source proteins, cells were treated with cycloheximide (CHX) for 0, 4 and 8 h in the presence and absence of rapamycin. Results in the graphs are expressed as a percentage of the remaining protein abundance in CHX-treated cells relative to CHX-untreated cells. β-Actin served as a loading control. One representative western blot out of three is shown for individual proteins (P-value: Student's t-test). Source data is available for this figure in the Supplementry Information.
Figure 7.
Rapamycin-treated cells contain increased levels of proteasomal substrates and express antigenic MIPs. (A, B) EL4 cells were treated or not with 20 ng/ml rapamycin for 48 h, combined or not with 10 μM of lactacystin for the last 8 h. (A) Accumulation of ubiquitinated proteins was measured on total cell lysates by western blotting. (B) Polyubiquitinated proteins from total cell lysates were isolated with TUBEs and levels of polyubiquitinated Rictor were then determined by western blotting. One representative western blot out of three is shown. (C, D) Mice were immunized with DCs coated with VNTHFSHL (C) or KALSYASL (D) peptide. Splenocytes from primed mice were tested for cytotoxic activity against CFSE-labeled target EL4 cells at different E/T ratios. EL4 cells coated with VNTHFSHL or KALSYASL were used as positive control. Data represent the mean±s.d. for three mice per group. Source data is available for this figure in the Supplementry Information.
MIPs appearing de novo on rapamycin-treated cells elicit cytotoxic T-cell responses
Finally, we wished to determine whether MIPs that were exclusively detected on rapamycin-treated cells could be antigenic. To test this, we selected the KALSYASL and the VNTHFSHL peptide, which are encoded by Rictor and Ttc21b, respectively (Supplementary Table S2). We immunized C57BL/6 mice with dendritic cells (DCs) coated with KALSYASL or VNTHFSHL synthetic peptide. Splenocytes from immunized mice did not kill untreated EL4 cells, but they showed specific cytotoxicity for EL4 cells treated with rapamycin or EL4 cells coated with the peptide used for T-cell priming (Figure 7C and D; Supplementary Figure S5). Further studies are needed to evaluate (1) the proportion of novel or overabundant MIPs that are antigenic and (2) whether particular pathways and cellular processes generate MIPs that are more likely to be antigenic. Nevertheless, the antigenicity of KALSYASL and VNTHFSHL is consistent with the notion that the immune system is tolerant to MIPs expressed at physiological levels but can mount biologically relevant immune responses toward self MIPs present in excessive amounts (Schild et al, 1990).
Discussion
Early proteomic studies were conducted with analyzers whose sensitivity (dynamic range) and accuracy were orders of magnitude inferior to that of MS analyzers that are now available (Depontieu et al, 2009; Yates et al, 2009; Nilsson et al, 2010). As a result, early studies on the immunopeptidome identified only the more abundant MIPs. Those MIPs were found to derive from highly abundant housekeeping proteins that are common to many cell types (Marrack et al, 1993; Hughes and Hughes, 1995; Barnea et al, 2002; Engelhard et al, 2002). More recently, high-throughput MS-based analyses have shown that the immunopeptidome conceals a cell-type-specific signature. Thus, though the immunopeptidome of DCs and thymocytes partially overlap, no less than 40% of their MIPs are cell-type specific (Fortier et al, 2008; de Verteuil et al, 2010). The present study demonstrates that perturbation of a single signaling pathway can lead to significant changes in the composition of the immunopeptidome. More specifically, our work shows that dynamic changes of mTOR signals are reflected in MHC I presentation of numerous peptides associated with the mTOR interactome and its signaling network. An important question is whether changes in the MIP repertoire induced by rapamycin would be found in response to any type of cell stress. Two elements strongly support our contention that rapamycin-induced changes in the immunopeptidome are connected to the mTOR network. First, we found that the connectivity between DEM source genes and mTOR network components was amazingly strong (P<10−5). A bootstrap procedure (500 000 iterations) failed to reveal a single set of peptide source genes that were so tightly connected to component of the mTOR network (Figure 4C). Second, we have previously evaluated the impact of other types of cell stress (tunicamycin, palmitate or glucose deprivation) on expression of MHC–peptide complexes at the surface EL4 cells (the cell line used the present study) (Granados et al, 2009). We found that cell stress induced by tunicamycin, palmitate and glucose deprivation led to a decrease in the generation of MIPs and in the expression of MHC I molecule. These effects are the exact opposite of what we found upon treatment of EL4 cells with rapamycin: overexpression of MHC I molecules (Supplementary Figure S2B) and increased abundance of cell surface MIPs (Figure 2B). In our opinion, it is therefore sound to conclude that rapamycin-induced variations in the immunopeptidome (1) arise from biochemical networks whose components are highly connected to the target of rapamycin (i.e. mTOR) and (2) are not unspecific changes found with any type of cell stress. However, mTOR senses and integrates multiple environmental cues and mTOR network components interact with many signaling pathways, such as the Wnt pathway (Inoki et al, 2006; Caron et al, 2010). Therefore, further studies will be needed to determine whether perturbation of mTOR by other agents or in other contexts would bring the same changes in the MIP repertoire as those induced by rapamycin.
An important implication for the immune system is that, at the peptide level, the immune self is plastic and its molecular composition is influenced by the cell's metabolic activity. We found that variations in the immunopeptidome functionally reflected perturbations of rapamycin-sensitive metabolic processes regulated in the transcriptome. More precisely, we observed that changes in the MIP repertoire originate from genes that are very closely functionally connected to DEGs. We observed that only 2% of DEMs did originate directly from DEGs as measured by DNA microarray experiments. In contrast, quantitative PCR revealed that 36% of the DEM source genes were slightly upregulated at the transcript level. In fact, eight DEM source genes (Bptf, Bub1b, Ccnf, Chaf1b, Dtl, Mnat1, Myb and Tfdp2) in Figure 3 come from DEGs that were identified by quantitative PCR (fold change between 1.2 and 3.1; Supplementary Table S6) but missed by microarrays. Differences in the sensitivity of both techniques are likely to explain this discrepancy. Indeed, whereas microarrays permit genome-wide profiling of mRNA expression levels, traditional quantitative PCR is by far more sensitive (Germain et al, 2011). Therefore, genome-wide quantitative PCR would have revealed a more significant overlap between DEM source genes and DEGs. Although the overlap between DEM source genes and DEGs was underestimated from microarray data, altogether our data indicate that changes in MIP abundance originate mostly from post-transcriptional mechanisms. Furthermore, data integration analysis revealed that 82% of DEMs arised from subnetworks that were transcriptionally reorganized in response to rapamycin. Thus, modulation in the composition of the immunopeptidome might originate predominantly from gene-regulatory subnetworks that are transcriptionally perturbed in response to multiple cellular stresses (i.e. pathogen infection (Kumar et al, 2010)). In other words, genes involved in transcriptionally perturbed subnetworks, although not necessarily differentially expressed at the transcript level, might participate in modulating the composition of the immunopeptidome. Consequently, identification of components within transcriptionally perturbed subnetworks might enable prediction of peptide source genes modulating the immunopeptidome's composition.
mTOR integrates environmental cues in terms of nutrients and growth factors. Therefore, it is reasonable to infer that cell-extrinsic factors modulate the repertoire of self peptides presented by MHC I. In addition, our results suggest that the peptide MHC I repertoire is a functional, if not a direct, representation of the transcriptional state of a cell. Together, that means the nature of the immune self is much more complex than anticipated. Moreover, the notion that MIPs presented specifically on metabolically stressed cells can be antigenic (e.g. KALSYASL and VNTHFSHL) could have several implications that need to be explored: immune responses against metabolic stress-associated MIPs could contribute to elimination of infected or transformed cells (Gleimer and Parham, 2003), but might also elicit autoimmunity (Todd et al, 2008). In keeping with this hypothesis, some self MIPs were found to be upregulated on infected cells, and some of them were immunogenic (Herberts et al, 2003; Hickman et al, 2003; Ovsyannikova et al, 2005).
From a more general perspective, our data provide a proof of concept that acquisition and integration of large-scale, quantitative biological data from multiple regulatory layers (i.e. -omics) may enable prediction of variations in the composition of the immunopeptidome. The notion that the immunopeptidome is plastic and conveys at the cell surface an integrative view of cellular protein metabolism opens up new perspectives in systems immunology and predictive biology (Benoist et al, 2006; Germain et al, 2011). The immune system has co-evolved with pathogens and positive selection imposed by pathogens has hastened the evolution of immune genes and thereby increased the complexity of the immune system (Waterston et al, 2002; Hedrick, 2004). Given the complexity of the immune system and the quintessential importance of self/non-self discrimination, it is imperative to further develop and exploit systems-level quantitative methods that will enable modeling of the immunopeptidome's plasticity. Such systems-oriented approaches will help to simulate and predict how cell-autonomous and -extrinsic elements modulate the composition of the immunopeptidome in health and disease.
Materials and methods
Cell culture, flow cytometry and determination of cell size
EL4 cell lines were cultured as previously reported (Fortier et al, 2008). Rapamycin, lactacystin and cycloheximide were purchased from Sigma-Aldrich. For MHC I labeling, cells were stained with anti-H2Db (B22–249.R1; Cedarlane), anti-H2Kb (Y3; ATCC), anti-Qa2 (clone 1-1-2; BD Pharmingen), anti-Qa1b (6A8.6F10.1A6) antibodies and analyzed on a BD LSR II flow cytometer using FACSDiva software (BD Biosciences) (Meunier et al, 2005). Cell number and cell size were measured by light microscopy using an Axio Imager microscope (Zeiss) and the Northern Eclipse software (Empix Imaging Inc.).
Western blotting and isolation of polyubiquitinated proteins
EL4 cells were harvested and lysed in RIPA buffer, samples were resolved by SDS–PAGE and immunoblotted with the following antibodies: ERAAP (gift from N Shastri, University of California, Berkeley, CA); Tapasin (gift from TH Hansen, Washington University School of Medicine, St Louis, MO); Lmp7, Lmp2, β1, β5, ERp57, Sgk1, β2 m, PDI (Abcam); Calnexin, Calreticulin, β-actin (Sigma); PA28, S6K1, p-S6K1 (Thr-389), 4E-BP1, p-4E-BP1 (Ser-65), p-4E-BP1 (Thr-37/46), α-tubulin, Rictor, Akt, p-Akt (S473), Ubiquitin (Cell Signaling); Myb (Upstate, Millipore); Mecl1, β2 (Biomol International); Rfc2 (Proteintech Group, PTG); Tfdp2 (Proteintech Group, PTG); Pabpc1 (Cell Signaling); H2Kb/H2Db (2G5; Thermo Scientific); Tap1, Tap2 (Santa Cruz Biotechnology). Isolation of polyubiquitinated proteins was achieved by using Tandem Ubiquitin Binding Entities (TUBEs) (LifeSensors, Inc.) according to the manufacturer's instruction.
Metabolic radiolabeling
To measure protein synthesis, EL4 cells were cultured in the absence or presence of rapamycin (20 ng/ml) for 0 h (ctrl), 6, 12, 24 and 48 h. During the last hour, cells were transferred to 96-well plates (0.5 × 106 cells/well, 12 replicates per conditions) and [3H]-leucine (10 μCi/ml) (Perkin-Elmer) was added to each well. At the end of the culture period, cells were harvested with a Cell Harvester (Perkin-Elmer) on a fiber glass filter (Packard) for washing. Filters were dried for 24 h and 20 μl of scintillation liquid per well (MicroScint-O, Perkin-Elmer) was added. Radioactivity incorporation was determined with the TopCount-NXT microplate scintillation and luminescence counter (Packard).
Peptide extraction and MS analyses
EL4 cells were treated with 20 ng/ml of rapamycin for 0, 6, 12, 24 and 48 h. MIPs were then isolated from EL4 cells as described previously using mild acid elution (Fortier et al, 2008; de Verteuil et al, 2010). Three biological replicates (5 × 108 EL4 cells per replicate) were prepared. MIPs obtained after acid elution were separated using an off-line 1100 series binary LC system (Agilent Technologies) to remove contaminating species. Peptides were loaded on a homemade strong cation exchange (SCX) column (0.3 mm internal diameter × 50 mm length) packed with SCX bulk material (Polysulfoethyl ATM, PolyLC). Peptides were fractionated with a gradient of 0–25% B after 25 min, 25–60% B after 35 min (solvent A=5 mmol/l ammonium formate, 15% acetonitrile, pH 3.0; solvent B=2 mol/l ammonium formate, 15% acetonitrile, pH 3.0). MIPs were collected in five consecutive fractions and brought to dryness using a speedvac. MIP fractions were resuspended in 2% aqueous acetonitrile (0.2% formic acid) and analyzed by nanoLC–MS/MS on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) (Fortier et al, 2008). Full mass spectra were acquired with the Orbitrap analyzer operated at a resolving power of 60 000 (at m/z 400) and collision-activated dissociation tandem mass spectra were acquired in data-dependent mode with the quadrupole linear ion trap analyzer. Mass calibration used an internal lock mass (protonated (Si(CH3)2O))6; m/z 445.12057) and typically provided mass accuracy within 5 p.p.m. for all nanoLC–MS/MS experiments.
MS/MS sequencing, hierarchical clustering and identification of MIP source proteins
Peptidomic data were analyzed using Xcalibur software and peak lists were generated using Mascot distiller (version 2.1.1, http://www.matrixscience.com). Database searches were performed against the International Protein Index mouse database (version 3.23 containing 51 536 sequences and 24 497 860 residues) using Mascot (version 2.2, http://www.matrixscience.com) with a mass precursor tolerance of ±0.05 Da and a fragment tolerance of ±0.5 Da. Searches were performed without enzyme specificity and a variable modification of oxidized Met. All search results were filtered using an MHC motif filter based on the predicted mouse MHC I allele motifs (Supplementary Information). Raw data files were converted to peptide maps comprising m/z values, charge state, retention time and intensity for all detected ions above a threshold of 10 000 counts using in-house software (Mass Sense) (Fortier et al, 2008; de Verteuil et al, 2010). Peptide maps were aligned and clustered together to profile the abundance of Mascot identified peptides using hierarchical clustering with criteria based on m/z and time tolerance (±0.02 m/z and ±1 min). This resulted in a list of non-redundant peptide clusters for all replicates of all samples to be compared. Peptides found in eluates from β2m mutant EL4 cells were considered ‘contaminant peptides’. Subtraction of contaminant peptides eluted from untreated or rapamycin treated (for 48 h) β2m EL4 cells allowed specific identification of genuine MIPs (Fortier et al, 2008; de Verteuil et al, 2010). MIPs were further inspected for mass accuracy and MS/MS spectra were validated manually. The Sidekick resource (http://www.bioinfo.iric.ca/sidekick/Main) was used to identify MIP source proteins. The list of MIPs reported in the present work has been provided to The Immune Epitope Database and Analysis Resource (http://beta.immuneepitope.org/) (Vita et al, 2010). MS/MS of all peptide identifications are available at http://www.thibault.iric.ca/proteoconnections (Courcelles et al, 2011) under the project name: MHC I immunopeptidome, and were submitted to the PRIDE database (Vizcaíno et al, 2010) under the accession numbers: 18855, 18856.
MIP classification and abundance profiling
Each MIP listed in Supplementary Tables S1 and S2 was classified according to restriction size and binding motif (Fortier et al, 2008; de Verteuil et al, 2010). The raw ratios for the time profiles of detected MIPs were then clustered using the TIGR MultiExperiment Viewer (TMEV) software. Hierarchical clustering with Euclidean distances and average linkage was used.
Microarray analysis
EL4 cells were treated or not with 20 ng/ml of rapamycin for 48 h. Three biological replicates were prepared. Total RNA was extracted from EL4 cells with TRIzol RNA reagent (Invitrogen) as instructed by the manufacturer. Samples were purified using DNase (Qiagen) and the RNeasy Mini kit (Qiagen), and the overall quality was analyzed with the 2100 Bioanalyzer (Agilent Technologies). Purified RNA (10 μg/sample) was hybridized on MM8 385K NimbleGen chips according to the manufacturer's instruction. Arrays were scanned using a GenePix4000B scanner (Axon Instruments) at 5 μm resolution. Data were extracted and normalized using the NimbleScan 2.4 extraction software (NimbleGen Systems). Further microarray analyses were performed using GeneSpring GX 7.3.1. The complete microarray data sets have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-3007.
Isolation of total and polysomal RNA for quantitative real-time PCR
EL4 cells were treated or not with 20 ng/ml of rapamycin for 48 h. Three biological replicates were prepared. Total RNA was extracted as described above. Polysomal RNA was extracted from 2 × 108 EL4 cells as described previously (Rajasekhar et al, 2003). Gene expression level was determined using primer and probe sets from Universal ProbeLibrary (https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). Primer sequences are given in Supplementary Table S7. PCR reactions for 384-well plate format were performed as described in Baron et al (2007). All reactions were run in triplicate, and the mean values were used for quantification. The mouse Hprt1 and Tbp genes were used as endogenous controls for total RNA and polysomal RNA, respectively. In order to obtain an unbiased profile of the normalized ribosomal loading efficiencies for cells, we calculated the net polysomal mRNA to total mRNA ratios. Such data are presented in the manuscript as NPR (Rajasekhar et al, 2003).
GO enrichment and connectivity score
The Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource (Peters and Sette, 2007) was used to identify significantly enriched GO terms associated to DEM source genes and DEGs from untreated and rapamycin-treated EL4 cells. To calculate connectivity scores between two lists of nodes (genes/proteins), computed scores from STRING (http://string-db.org/) (Szklarczyk et al, 2011) were used. By using computed scores in STRING, functional associations between two lists of nodes were integrated into an all-pairs-shortest-path distance matrix. Each functional association in the all-pairs-shortest-path matrix was transformed into a ‘distance’ (D), defined as –ln(S), where S is the computed STRING score. The all-pairs-shortest-path matrix contains the length of the shortest path (distance) between every pair of nodes in the network. A connectivity score was then obtained by calculating the mean of the shortest path distance between every pair of nodes in a given matrix.
STITCH network
The 98 DEM source genes highlighted in Figure 3B were used to build the DEM network (Supplementary Figure S4). The 98 source genes encoding DEMs, the 171 DEGs classified within the enriched GO terms (P<0.05) in Supplementary Figure S3B and C and the mTOR network components in Supplementary Table S5 were used to build the global rapamycin-regulatory network. Subnetworks (Figure 5A) were manually extracted based on the modular organization of the total network (Hartwell et al, 1999; Rives and Galitski, 2003). Each of these molecules was individually searched in the STITCH (http://stitch.embl.de/) (Kuhn et al, 2010) database for its functional associations. The parameters used to elucidate the network were as follows: 40 additional nodes, a network depth=1, and interactions with a minimum STITCH combined score of 0.400. Functional associations identified for all of the molecules were then imported into Adobe Illustrator as a Scalable Vector Graphics image (action view configuration).
In vitro cytotoxicity assay
Bone marrow-derived DCs were generated as previously described (Fortier et al, 2008; de Verteuil et al, 2010). On day 9 of culture, peptides were added at a concentration of 2 × 10−6 M and incubated with DCs for 3 h at 37 °C. For mouse immunization, 106 peptide-pulsed DCs were injected i.v. in C57BL/6 females at day 0 and day 7. On day 14, splenocytes were harvested from the spleens of immunized mice and depleted of red blood cells using 0.83% NH4Cl. Cells were plated at 5 × 106 cells/well in 24-well plates and restimulated with 2 × 10−6 M peptide at 37 °C. After 6 days, cytotoxicity was evaluated by a 4-h CFSE-based assay. The percentage of specific lysis was calculated as follows: (number of remaining CFSE+ cells after incubation of target cells alone – number of remaining CFSE+ cells after incubation with effector cells)/number of CFSE+ cells after incubation of target cells alone × 100.
Supplementary Material
Acknowledgments
We thank Raphaëlle Lambert and Mathieu Courcelles (IRIC, Montreal, Canada) for technical support. We also thank Stephen W Michnick (Université de Montréal, Montreal, Canada) and Valeria de Azcoitia for comments on the manuscript. This work was supported by grants from the Canadian Institutes for Health Research (MOP 42384) and the Canadian Cancer Society (019475). EC and KV are supported by training grants from the Cole Foundation. M-HF was supported by a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ). CP, PT and PPR are supported by the Canada Research Chairs Program. IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and the FRSQ.
Author contributions: EC designed the study, carried out the experiments, analyzed the data, prepared the figures and wrote the first draft of the manuscript. KV and M-PH carried out the experiments and prepared the figures. M-HF carried out the experiments and the MS analyses. J-PL, AB and GV carried out the bioinformatics analysis. CP and PT designed the study, analyzed the data, discussed the results and wrote the manuscript. SL and PPR contributed to the study design, discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R (2009) mTOR regulates memory CD8 T-cell differentiation. Nature 460: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea E, Beer I, Patoka R, Ziv T, Kessler O, Tzehoval E, Eisenbach L, Zavazava N, Admon A (2002) Analysis of endogenous peptides bound by soluble MHC class I molecules: a novel approach for identifying tumor-specific antigens. Eur J Immunol 32: 213–222 [DOI] [PubMed] [Google Scholar]
- Baron C, Somogyi R, Greller LD, Rineau V, Wilkinson P, Cho CR, Cameron MJ, Kelvin DJ, Chagnon P, Roy DC, Busque L, Sékaly RP, Perreault C (2007) Prediction of graft-versus-host disease in humans by donor gene expression profiling. PLoS Med 4: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C, Germain RN, Mathis D (2006) A plaidoyer for ‘systems immunology’. Immunol Rev 210: 229–234 [DOI] [PubMed] [Google Scholar]
- Boehm T (2006) Quality control in self/nonself discrimination. Cell 125: 845–858 [DOI] [PubMed] [Google Scholar]
- Bonner PL, Lill JR, Hill S, Creaser CS, Rees RC (2002) Electrospray mass spectrometry for the identification of MHC class I-associated peptides expressed on cancer cells. J Immunol Methods 262: 5–19 [DOI] [PubMed] [Google Scholar]
- Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H (2010) A comprehensive map of the mTOR signaling pathway. Mol Syst Biol 6: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Blenis J (2009) Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle 8: 567–572 [DOI] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J (2008) Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA 105: 17414–17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelles M, Lemieux S, Voisin L, Meloche S, Thibault P (2011) ProteoConnections: a bioinformatics platform to facilitate proteome and phosphoproteome analyses. Proteomics 11: 2654–2671 [DOI] [PubMed] [Google Scholar]
- de Verteuil D, Muratore-Schroeder TL, Granados DP, Fortier MH, Hardy MP, Bramoullé A, Caron E, Vincent K, Mader S, Lemieux S, Thibault P, Perreault C (2010) Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Mol Cell Proteomics 9: 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depontieu FR, Qian J, Zarling AL, McMiller TL, Salay TM, Norris A, English AM, Shabanowitz J, Engelhard VH, Hunt DF, Topalian SL (2009) Identification of tumor-associated, MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc Natl Acad Sci USA 106: 12073–12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilling MB, Germain GS, Dudkin L, Jayaraman AL, Zhang X, Harwood FC, Houghton PJ (2002) 4E-binding proteins, the suppressors of eukaryotic initiation factor 4E, are down-regulated in cells with acquired or intrinsic resistance to rapamycin. J Biol Chem 277: 13907–13917 [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhard V, Brickner A, Zarling A (2002) Insights into antigen processing gained by direct analysis of the naturally processed class I MHC associated peptide repertoire. Mol Immunol 39: 127. [DOI] [PubMed] [Google Scholar]
- Engelhard VH, Appella E, Benjamin DC, Bodnar WM, Cox AL, Chen Y, Henderson RA, Huczko EL, Michel H, Sakaguichi K (1993) Mass spectrometric analysis of peptides associated with the human class I MHC molecules HLA-A2.1 and HLA-B7 and identification of structural features that determine binding. Chem Immunol 57: 39–62 [PubMed] [Google Scholar]
- Fortier MH, Caron E, Hardy MP, Voisin G, Lemieux S, Perreault C, Thibault P (2008) The MHC class I peptide repertoire is molded by the transcriptome. J Exp Med 205: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, Meier-Schellersheim M, Nita-Lazar A, Fraser ID (2011) Systems biology in immunology: a computational modeling perspective. Annu Rev Immunol 29: 527–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleimer M, Parham P (2003) Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity 19: 469–477 [DOI] [PubMed] [Google Scholar]
- Goldrath AW, Bevan MJ (1999) Selecting and maintaining a diverse T-cell repertoire. Nature 402: 255–262 [DOI] [PubMed] [Google Scholar]
- Granados DP, Tanguay PL, Hardy MP, Caron E, De Verteuil D, Meloche S, Perreault C (2009) ER stress affects processing of MHC class I-associated peptides. BMC Immunol 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolleau A, Bowman J, Pradet-Balade B, Puravs E, Hanash S, Garcia-Sanz JA, Beretta L (2002) Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J Biol Chem 277: 22175–22184 [DOI] [PubMed] [Google Scholar]
- Hammer GE, Kanaseki T, Shastri N (2007) The final touches make perfect the peptide-MHC class I repertoire. Immunity 26: 397–406 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW (1999) From molecular to modular cell biology. Nature 402: C47–C52 [DOI] [PubMed] [Google Scholar]
- Hedrick SM (2004) The acquired immune system; a vantage from beneath. Immunity 21: 607–615 [DOI] [PubMed] [Google Scholar]
- Herberts CA, van Gaans-van den Brink J, van der Heeft E, van Wijk M, Hoekman J, Jaye A, Poelen MC, Boog CJ, Roholl PJ, Whittle H, de Jong AP, van Els CA (2003) Autoreactivity against induced or upregulated abundant self-peptides in HLA-A*0201 following measles virus infection. Hum Immunol 64: 44–55 [DOI] [PubMed] [Google Scholar]
- Hickman HD, Luis AD, Bardet W, Buchli R, Battson CL, Shearer MH, Jackson KW, Kennedy RC, Hildebrand WH (2003) Cutting Edge: Class I presentation of host peptides following HIV infection. J Immunol 171: 22–26 [DOI] [PubMed] [Google Scholar]
- Hickman HD, Luis AD, Buchli R, Few SR, Sathiamurthy M, VanGundy RS, Giberson CF, Hildebrand WH (2004) Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol 172: 2944–2952 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MK (1995) Self peptides bound by HLA class I molecules are derived from highly conserved regions of a set of evolutionarily conserved proteins. Immunogenetics 41: 257–262 [DOI] [PubMed] [Google Scholar]
- Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH (1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255: 1261–1263 [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, Macdougald OA, You M, Williams BO, Guan KL (2006) TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126: 955–968 [DOI] [PubMed] [Google Scholar]
- Istrail S, Florea L, Halldorsson BV, Kohlbacher O, Schwartz RS, Yap VB, Yewdell JW, Hoffman SL (2004) Comparative immunopeptidomics of humans and their pathogens. Proc Natl Acad Sci USA 101: 13268–13272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L, Hinterberger M, Wirnsberger G, Kyewski B (2009) Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol 9: 833–844 [DOI] [PubMed] [Google Scholar]
- Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von MC, Jensen LJ, Beyer A, Bork P (2010) STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res 38: D552–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV (2010) Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140: 731–743 [DOI] [PubMed] [Google Scholar]
- Marrack P, Ignatowicz L, Kappler JW, Boymel J, Freed JH (1993) Comparison of peptides bound to spleen and thymus class II. J Exp Med 178: 2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester G, Hoffmann V, Stevanovic S (2011) Insights into MHC class I antigen processing gained from large-scale analysis of class I ligands. Cell Mol Life Sci 68: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier MC, Delisle JS, Bergeron J, Rineau V, Baron C, Perreault C (2005) T cells targeted against a single minor histocompatibility antigen can cure solid tumors. Nat Med 11: 1222–1229 [DOI] [PubMed] [Google Scholar]
- Milner E, Barnea E, Beer I, Admon A (2006) The turnover kinetics of MHC peptides of human cancer cells. Mol Cell Proteomics 5: 357–365 [DOI] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR III, Bairoch A, Bergeron JJ (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods 7: 681–685 [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Johnson KL, Naylor S, Poland GA (2005) Identification of HLA-DRB1-bound self-peptides following measles virus infection. J Immunol Methods 297: 153–167 [DOI] [PubMed] [Google Scholar]
- Perreault C (2010) The origin and role of major histocompatibility complex class I-associated self peptides. Prog Mol Biol Transl Sci 92: 41–60 [DOI] [PubMed] [Google Scholar]
- Peters B, Sette A (2007) Integrating epitope data into the emerging web of biomedical knowledge resources. Nat Rev Immunol 7: 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian SB, Princiotta MF, Bennink JR, Yewdell JW (2006) Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem 281: 392–400 [DOI] [PubMed] [Google Scholar]
- Qian SB, Zhang X, Sun J, Bennink JR, Yewdell JW, Patterson C (2010) mTORC1 links protein quality and quantity control by sensing chaperone availability. J Biol Chem 285: 27385–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC (2003) Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell 12: 889–901 [DOI] [PubMed] [Google Scholar]
- Rammensee HG, Falk K, Rotzschke O (1993) Peptides naturally presented by MHC class I molecules. Annu Rev Immunol 11: 213–244 [DOI] [PubMed] [Google Scholar]
- Rives AW, Galitski T (2003) Modular organization of cellular networks. Proc Natl Acad Sci USA 100: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Schild H, Rotzschke O, Kalbacher H, Rammensee HG (1990) Limit of T cell tolerance to self proteins by peptide presentation. Science 247: 1587–1589 [DOI] [PubMed] [Google Scholar]
- Schirle M, Keilholz W, Weber B, Gouttefangeas C, Dumrese T, Becker HD, Stevanovic S, Rammensee HG (2000) Identification of tumor-associated MHC class I ligands by a novel T cell-independent approach. Eur J Immunol 30: 2216–2225 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Rappuoli R (2010) Reverse vaccinology: developing vaccines in the era of genomics. Immunity 33: 530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Shastri N (2011) Non-conventional sources of peptides presented by MHC class I. Cell Mol Life Sci 68: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von MC (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH (2008) The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol 8: 663–674 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B (2010) The immune epitope database 2.0. Nucleic Acids Res 38: D854–D862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Weinzierl AO, Lemmel C, Schoor O, Muller M, Kruger T, Wernet D, Hennenlotter J, Stenzl A, Klingel K, Rammensee HG, Stevanovic S (2007) Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol Cell Proteomics 6: 102–113 [DOI] [PubMed] [Google Scholar]
- Weinzierl AO, Maurer D, Altenberend F, Schneiderhan-Marra N, Klingel K, Schoor O, Wernet D, Joos T, Rammensee HG, Stevanovic S (2008) A cryptic vascular endothelial growth factor T-cell epitope: identification and characterization by mass spectrometry and T-cell assays. Cancer Res 68: 2447–2454 [DOI] [PubMed] [Google Scholar]
- Yates JR, Ruse CI, Nakorchevsky A (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng 11: 49–79 [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Nicchitta CV (2006) The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol 27: 368–373 [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Reits E, Neefjes J (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nature Rev Immunol 3: 952–961 [DOI] [PubMed] [Google Scholar]
- Zarling AL, Polefrone JM, Evans AM, Mikesh LM, Shabanowitz J, Lewis ST, Engelhard VH, Hunt DF (2006) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci USA 103: 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.