Abstract
Gene conversion, a form of concerted evolution, bears enormous potential to shape the trajectory of sequence and functional divergence of gene paralogs subsequent to duplication events. fog-2, a sex-determination gene unique to Caenorhabditis elegans and implicated in the origin of hermaphroditism in this species, resulted from the duplication of ftr-1, an upstream gene of unknown function. Synonymous sequence divergence in regions of fog-2 and ftr-1 (excluding recent gene conversion tracts) suggests that the duplication occurred 46 million generations ago. Gene conversion between fog-2 and ftr-1 was previously discovered in experimental fog-2 knockout lines of C. elegans, whereby hermaphroditism was restored in mutant obligately outcrossing male–female populations. We analyzed DNA-sequence variation in fog-2 and ftr-1 within 40 isolates of C. elegans from diverse geographic locations in order to evaluate the contribution of gene conversion to genetic variation in the two gene paralogs. The analysis shows that gene conversion contributes significantly to DNA-sequence diversity in fog-2 and ftr-1 (22% and 34%, respectively) and may have the potential to alter sexual phenotypes in natural populations. A radical amino acid change in a conserved region of the F-box domain of fog-2 was found in natural isolates of C. elegans with significantly lower fecundity. We hypothesize that the lowered fecundity is due to reduced masculinization and less sperm production and that amino acid replacement substitutions and gene conversion in fog-2 may contribute significantly to variation in the degree of inbreeding and outcrossing in natural populations.
Keywords: gene conversion, gene duplication, intraspecific variation, sex determination, Caenorhabditis elegans
Introduction
Gene conversion, the nonreciprocal exchange of genetic material between genes, is facilitated by high levels of sequence identity between DNA sequences and has the dual effect of homogenizing intergenic sequences while increasing intragenic variation (King 1998; Nielsen et al. 2003). This may be especially important in the evolution of new genes as exceptionally high identity between newly duplicated genes may cause them to be particularly susceptible to gene conversion, leading to the maintenance of low sequence divergence between duplicate pairs or concerted evolution (Ohta 1983; Teshima and Innan 2004). Yet, gene conversion can also increase variation within genes by generating different alleles in natural populations (Innan 2003a). Here, we analyzed DNA-sequence variation in natural populations of Caenorhabditis elegans in two paralogous genes, fog-2 and ftr-1, for which gene conversion had previously been detected in laboratory populations (Katju et al. 2008).
Caenorhabditis elegans predominantly reproduces by hermaphroditic self-fertilization, though rare outcrossing with males does occur. Hermaphroditism has evolved convergently in two Caenorhabditis, C. elegans and C. briggsae (Cho et al. 2004; Kiontke et al. 2004; Hill et al. 2006). The gene fog-2 unique to C. elegans encodes a protein involved in the hermaphroditic spermatogenesis pathway and may have played a major role in C. elegans sex evolution (Schedl and Kimble 1988; Nayak et al. 2005). It is thought to have originated from a postspeciation partial duplication of ftr-1, a gene of unknown function located on chromosome V, approximately 800 bp upstream of fog-2 (Nayak et al. 2005; Katju et al. 2008). The FOG-2 and FTR-1 proteins encoded by the paralogs are structurally similar in that both are members of the FTR (fog 2–related) gene family, a family of proteins containing an N-terminal F-box domain and a C-terminal FOG-2 Homology (FTH) domain, also called Domain of Unknown Function 38 (DUF38) (Kipreos and Pagano 2000). In FOG-2, the DUF38/FTH domain of the protein forms a complex with GLD-1, a translational repressor that binds to the mRNA transcript of the feminizing gene tra-2, repressing feminization and promoting hermaphrodite spermatogenesis (Schedl and Kimble 1988; Clifford et al. 2000). The C-terminus of the FOG-2 protein, where GLD-1 interaction occurs, shows much lower sequence identity to FTR-1 than other regions. Notably, FOG-2 binding to GLD-1 is eliminated with the deletion of the last 64 amino acids of the protein (Nayak et al. 2005). fog-2 appears to have arisen from a partial duplication of ftr-1 followed by recruitment of surrounding genomic sequence in the downstream region to create an uninterrupted reading frame, as the 3′ end of the gene contains a 134-bp sequence completely unique to fog-2 (Katju and Lynch 2006; Katju et al. 2008). The recruitment of this unique sequence in the 3′ end of fog-2 may have facilitated neofunctionalization after duplication. This idea is bolstered by the observation that the unique region of fog-2 includes the last 16 amino acids of the DUF38/FTH domain, where GLD-1 binding occurs (Nayak et al. 2005).
The fog-2 and ftr-1 paralogs provide a useful model for studying the effects of gene conversion on genetic variation for several reasons. Firstly, fog-2 and ftr-1 exhibit qualities of genes that are more likely to undergo conversion in the C. elegans genome. In C. elegans, gene conversion occurs most often in gene duplicates that are members of multigene families, physically close, oriented in the same direction, and highly similar in sequence (Semple and Wolfe 1999). fog-2 and ftr-1 are members of the FTR gene family, are located less than 1 kb apart on chromosome V, and are oriented in the same direction. In addition, there exists a high level of sequence conservation (84% similarity) between fog-2 and ftr-1, with several segments averaging approximately 50 bp in length that have retained complete identity. Secondly, the incidence of gene conversion between fog-2 and ftr-1 has been established. In a recent study that used strains with a fog-2 nonsense mutation to create male–female lines, spontaneous reversions to hermaphroditism were the result of gene conversion by ftr-1 restoring functionality of fog-2 (Katju et al. 2008). Furthermore, the presence of regions of complete sequence identity between the genes suggests fixation of past gene-conversion events (Katju et al. 2008). Finally, the evolution of the fog-2 gene is also of interest because of its role in sex determination and the possibility that sequence variation in fog-2 may play an important role in the degree of self-fertilization and outcrossing in the wild.
Materials and Methods
Preparation of Genomic DNA
Forty isolate populations of C. elegans from widespread geographic locales were obtained from the Caenorhabditis Genetics Center (table 1). The majority of these strains are natural isolates, with some laboratory strains. Each isolate was grown on agarose with Escherichia coli HB101. Worms were collected using a 4-ml wash of M9 salts (Wood 1988), then centrifuged for 30 s at 3,000 rpm. The supernatant was extracted, and the worms were resuspended in 1-ml M9. After 30 s, worms were centrifuged for 30 s at 3,000 rpm and resuspended in 1-ml Tris-EDTA-NaCl (TEN) solution. The worms were stored for extraction in 50-μl TEN at −20 °C. To extract genomic DNA, standard methods for proteinase K and phenol–chloroform extraction were used. Purified DNA was stored in 50 μl molecular grade water at −20 °C.
Table 1.
List of 40 Isolates of Caenorhabditis elegans and Their Geographic Origins Comprising This Population-Genetic Study.
| Isolate | Origin | fog-2 Allelea | ftr-1 Alleleb | Haplotypec |
| AB1 | Adelaide, Australia | 2 | 2 | b |
| AB2 | Adelaide, Australia | 3 | 4 | d |
| AB3 | Adelaide, Australia | 4 | 4 | e |
| AB4 | Adelaide, Australia | 4 | 4 | e |
| CB3192 | Altadena, CA | 5 | 5 | f |
| CB3196 | Altadena, CA | 3 | 4 | d |
| CB3197 | Altadena, CA | 4 | 4 | e |
| CB3198 | Pasadena, CA | 4 | 6 | g |
| CB3199 | Pasadena, CA | 4 | 4 | e |
| CB4851 | Bergerac, France | 5 | 1 | h |
| CB4852 | Bristol, United Kingdom | 4 | 4 | e |
| CB4853 | Altadena, CA | 4 | 4 | e |
| CB4854 | Altadena, CA | 6 | 4 | i |
| CB4855 | Palo Alto, CA | 4 | 4 | e |
| CB4856 | Hawaii, United States | 2 | 3 | c |
| CB4857 | Claremont, CA | 4 | 4 | e |
| CB4858 | Pasadena, CA | 3 | 4 | d |
| CB4932 | Taunton, United Kingdom | 2 | 2 | b |
| DH424 | El Prieto Canyon, CA | 5 | 7 | j |
| DR1344 | Subclone of Bergerac BO | 5 | 1 | h |
| DR1345 | Claremont, CA | 4 | 4 | e |
| DR1346 | Claremont, CA | 5 | 1 | h |
| DR1347 | Claremont, CA | 5 | 1 | h |
| DR1348 | Claremont, CA | 5 | 1 | h |
| DR1349 | Pasadena, CA | 5 | 4 | k |
| DR1350 | Pasadena, CA | 4 | 4 | e |
| JU258 | Madeira, Portugal | 4 | 4 | e |
| JU262 | Le Blanc, France | 4 | 4 | e |
| JU263 | Le Blanc, France | 4 | 4 | e |
| KR314 | Vancouver, Canada | 4 | 4 | e |
| LSJ1 | Berkeley, CA | 5 | 1 | h |
| N2 | Ancestral, Bristol, United Kingdom | 1 | 1 | a |
| PB303 | Isopod association | 4 | 8 | l |
| PB306 | Isopod association | 4 | 4 | e |
| PS2025 | Altadena, CA | 7 | 9 | m |
| RC301 | Freiburg, Germany | 8 | 8 | n |
| RW6999 | Subclone of Bergerac | 5 | 1 | h |
| RW7000 | Bergerac, France | 5 | 1 | h |
| TR388 | Madison, WI | 5 | 10 | o |
| TR389 | Madison, WI | 5 | 11 | p |
fog-2 allele identification number found in each isolate. N2 is assigned as 1 and AB1, CB4856, and CB4932 as 2, with all other numbers assigned alphabetically.
ftr-1 allele identification number found in each isolate. N2 is assigned as 1, AB1 and CB4932 as 2, and CB4856 as 3, with all other numbers assigned alphabetically.
Haplotype identification letter found in each isolate. N2 is assigned as a, AB1 and CB4932 as b, and CB4856 as c, with all other numbers assigned alphabetically.
Polymerase chain reaction (PCR) and Sequencing
DNA was diluted to 20 ng/μl in preparation for PCR amplification. The sequences of fog-2 and ftr-1 for the N2 laboratory strain were retrieved from the C. elegans genome deposited in WormBase (http://www.wormbase.org, release WS150, November 30, 2005, gene ids 00001482 and 00013756, respectively). Unique primers were designed to anneal to the 5′ and 3′ regions of each gene (table 2). These primer sets were used in PCR amplification and returned DNA fragments encompassing the entire coding region of fog-2 and ftr-1, corresponding to ∼1.4 and 1.6 kb in length, respectively. For PCR amplification of both genes, a touchdown thermocycling protocol was used. This consisted of denaturation at 94 °C for 2 min, followed by 9 cycles of 94 °C for 45 s, 68 °C (−1 °C) for 45 s, and 72 °C for 15 s, followed by 32 cycles of 94 °C for 45 s, 58 °C for 45 s, and 72 °C for 15 s. The length of the DNA fragments was checked using gel electrophoresis. DNA was extracted from the gel using the illustra GFX PCR DNA and Gel Band Purification Kit (manufacturer's protocol, GE Healthcare).
Table 2.
Primer Sequences for PCR Amplification and Sequencing of fog-2 and ftr-1.
| Primer | Sequence (5′ to 3′) |
| fog-2 F2 | TCGTCTCTTCACATGAAGCTTC |
| fog-2 F4 | TGAGGGACGTGTATGCGAAGA |
| fog-2 R1 | AGACGACCTGGCCCTTTTGT |
| fog-2 R3 | CTGGATTTCTTGAAGACATCGAA |
| fog-2 UF1 | GGTACTTGCTCGAAAAATGCGT |
| ftr-1 DR1 | GGTATTGAGAACAAAATCGAGCAG |
| ftr-1 F2 | CACATGGTCTTTTATGATGCTCCC |
| ftr-1 F3 | GTAGATGCGATGATATTCCC |
| ftr-1 F4 | CCATCGCTGTTTGAAATGC |
| ftr-1 R2 | GTCATTGATTGCCCTGGAGATC |
| ftr-1 R3 | GTAGATGCGATGATATTCCC |
| ftr-1 R4 | GTGATGTACCTTGATTGTATTGC |
| ftr-1 UF1 | CCTAGGACTACTGTAGAAGGTACGC |
Sequencing and Alignment
DNA was sequenced using the Big Dye Terminator v3.1 Cycle Sequencing Kit and protocol (Applied Biosystems, Foster City, CA). To obtain sequence data for the full coding region of each gene, primers were designed to anneal to various regions of the gene (table 2). Resulting sequences were assembled and proofed using Sequencher 4.8 software (Gene Codes, Ann Arbor, MI) and manually aligned against the N2 sequences for both fog-2 and ftr-1 using the Se-Al 2.0a11 program (Andrew Rambaut, University of Edinburgh). The sequences have been deposited in Genbank under the accession numbers GU736201-GU736292.
Data Analysis
Sequence variation was analyzed using MEGA (Kumar et al. 2008), DnaSP (Librado and Rozas 2009), and TASSEL software (Bradbury et al. 2007). Gene-conversion tracts were identified using GENECONV software that employs a statistical test to detect gene conversion (Sawyer 1999). To examine the relative contribution of gene conversion to the parameters measured, regions of suspected gene conversion were removed from the data set and the calculations repeated. Nucleotide diversity, Tajima's D (Tajima 1989), nonsynonymous to synonymous ratios, and linkage disequilibrium were analyzed for both genes with and without gene conversion. For all sliding window analyses, both a 50-nt window slid in 10-nt increments and a 100-nt window slid in 25-nt increments were used.
Association between fog-2 Allele and Brood Size
Given the direct role of fog-2 in spermatogenesis in C. elegans hermaphrodites, it is reasonable to expect a contribution of fog-2 genotype to fecundity, and hence fitness, within this species. We used published data on isolate-specific brood size from Hodgkin and Doniach (1997) and tested for an association, between fecundity and the particular genotype at fog-2 using a Wilcoxon two-sample test.
Results
Sequence Divergence between fog-2 and ftr-1
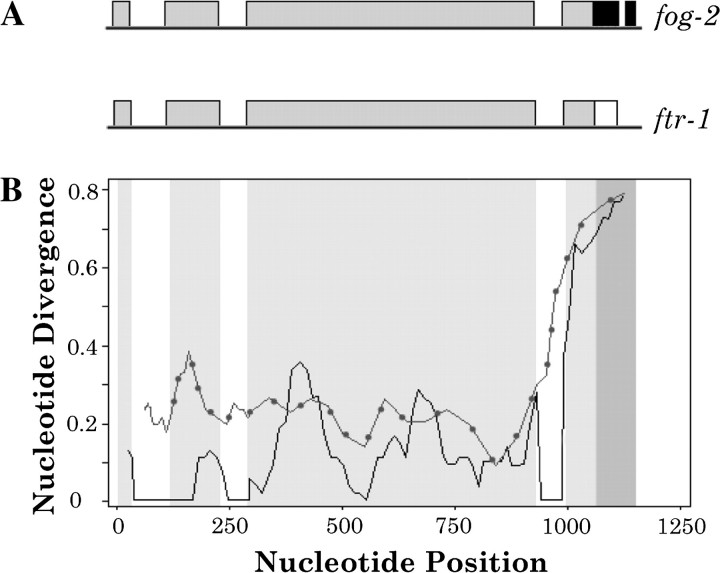
The total sequence divergence between the N2 laboratory strain sequences of fog-2 and ftr-1 was calculated to be 0.16 (including introns and excluding the unique regions at the 3′ end in both genes). The results from a sliding window analysis of divergence between the two paralogs are shown in figure 1. Nucleotide diversity at both terminal ends in each intron takes a noticeable dip, suggesting purifying selection at intron splice junctions. Synonymous site divergence was calculated between fog-2 and ftr-1 after removing the nonhomologous region at the 3′ end as well as a section where indels had altered the reading frame between the two paralogs. Synonymous and nonsynonymous site divergence between fog-2 and ftr-1 is 0.17 and 0.13, respectively. The KA/KS ratio is 0.81. We were interested in using the synonymous divergence to estimate an approximate age of the fog-2/ftr-1 duplication. However, fog-2 and ftr-1 include regions of complete sequence identity that are strongly suggestive of a history of gene conversion between the two genes, which would tend to make the duplication erroneously appear evolutionarily more recent than its true age (Katju et al. 2008). We therefore calculated the synonymous site divergence after removing three regions that were statistically significant in a previous analysis of gene conversion between the genes. The KS value for fog-2/ftr-1 excluding the converted regions is 0.25. The base substitution rate in C. elegans has been estimated to be 2.7 × 10−9 per site per generation (Denver et al. 2009). The sequence divergence of neutral sites between two DNA sequences is 2 μt, where μ is the mutation rate and t is time. This suggests that the duplication occurred 46 million generations ago. In a previous analysis of the age of hermaphroditism in C. elegans, the authors assumed that on average, wild populations of C. elegans had six generations per year (Cutter et al. 2008). Using these assumptions for the sake of comparison to Cutter et al.’s (2008) analysis, the fog-2/ftr-1 duplication occurred 7.2 Ma. However, Cutter et al. (2008) used base substitution rates that are 3-fold higher than the most recent estimates from high-throughput sequencing (Denver et al. 2009). If we employ the previous estimate of the base substitution rate of 9 × 10−9 per site per generation (Denver et al. 2004), the time since the fog-2/ftr-1 duplication is 2.3 Ma, compared with Cutter et al.’s (2008) estimate of 4 Ma as an upper boundary for the origin of hermaphroditism in C. elegans.
FIG. 1.
(A) Gene structure of fog-2 and ftr-1. Shaded rectangles represent exons and horizontal lines represent introns and flanking regions. Regions of homology between the genes are indicated by similar shading. The figure is drawn to scale. (B) Sliding window of intergenic nucleotide divergence between fog-2 and ftr-1 of the N2 strain. Nucleotide position in the alignment (x axis) is plotted against KA and KS, sequence divergence (y axis). KA is shown by a solid black line, and KS is shown by a dotted gray line. A 50-nt window was slid in 10-nt increments. The highly divergent region towards the 3’ end corresponds to the nonhomologous recruited sequence in fog-2. Two regions of complete identity exist (∼300 and 600 bp).
Sequence Variation in fog-2 and ftr-1
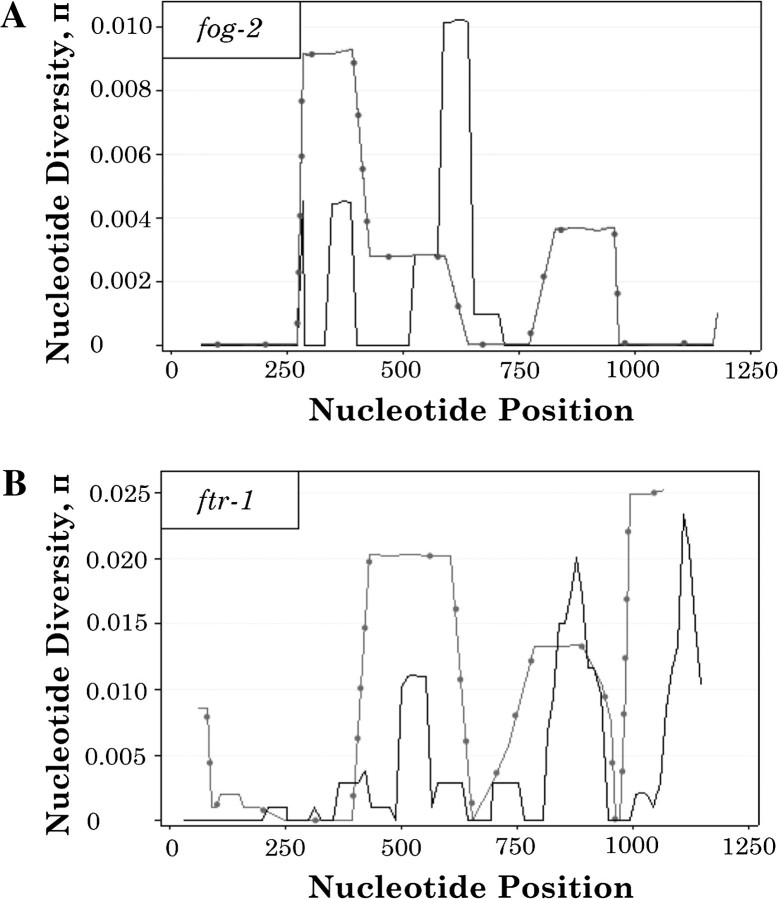
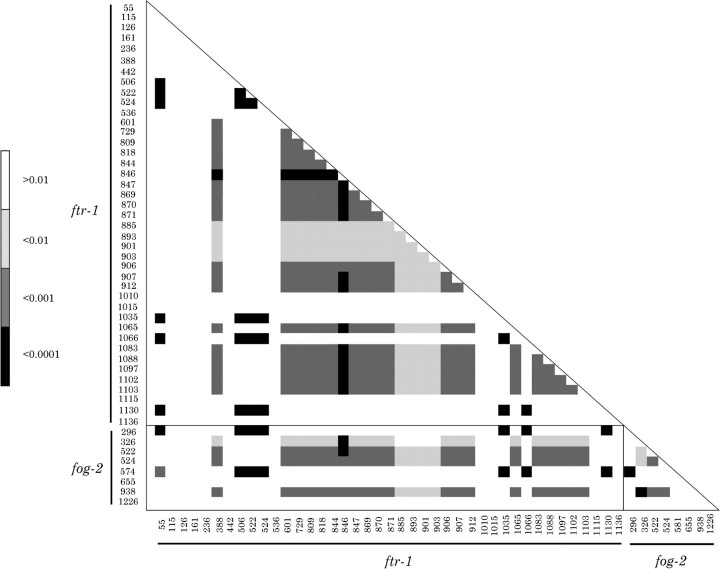
The number of distinct alleles in fog-2 and ftr-1 in the sample is 8 and 11, respectively, and the total number of haplotypes is 16 (table 1). The number of segregating sites in fog-2 and ftr-1 is 8 and 41, respectively. Nucleotide diversity as the average number of nucleotide differences per site, π, was also calculated for both fog-2 and ftr-1 (table 3), and sliding windows of intragenic π were computed separately for each paralog (fig. 2). The measure of π is higher for ftr-1 than fog-2 in all cases. Nucleotide diversity in introns is greater than that in exons for fog-2, but the opposite pattern is observed in ftr-1. Nucleotide diversity was further classified with respect to synonymous and nonsynonymous sites (table 3). In fog-2, the nucleotide diversity is similar at synonymous and nonsynonymous sites (0.0015 and 0.0013, respectively). Nucleotide diversity is 3-fold higher at synonymous sites than at nonsynonymous sites in ftr-1 (0.014 and 0.005, respectively), which would suggest purifying selection. For both genes, nucleotide diversity at synonymous sites, or πS, are within the range found previously for C. elegans genes (Cutter 2006), with πS at the low end of the range in fog-2 and the high end of the range in ftr-1. Tajima's D was calculated for both genes under the same conditions described above (table 3). None of the results reported here are statistically significant. The McDonald–Kreitman test of adaptive protein evolution (McDonald and Kreitman 1991) between the two paralogs found no significant deviation from neutral evolution. Analysis of polymorphic sites in fog-2 and ftr-1 revealed extensive linkage disequilibrium between and within genes (fig. 3). In fact, of 946 pairwise linkage disequilibrium measures, D′ was less than 1 in only 12 instances.
Table 3.
Measures of Nucleotide Diversity and Selection for fog-2 and ftr-1 in Isolate Populations of Caenorhabditis elegans.
| Gene | Sequence Data | πa | πAb | πSc | πA/πSd | θA/θSe | Df | DSg |
| fog-2 | Introns and exons | 0.0015 | −0.171 | −0.575 | ||||
| Introns only | 0.0017 | 1.406 | ||||||
| Exons only | 0.0014 | 0.0013 | 0.0015 | 0.83 | 0.59 | −0.579 | −0.246 | |
| Without gene conversion | 0.0011 | 0.0011 | 0.0009 | 1.24 | 0.88 | −0.344 | ||
| ftr-1 | Introns and exons | 0.0061 | −0.898 | 0.394 | ||||
| Introns only | 0.0025 | −0.913 | ||||||
| Exons only | 0.0070 | 0.0050 | 0.0136 | 0.37 | 0.71 | −0.843 | −0.022 | |
| Without gene conversionh | 0.0046 | 0.0035 | 0.0087 | 0.41 | 0.76 | −1.313 | ||
| Without gene conversioni | 0.0040 | 0.0023 | 0.0100 | 0.23 | 0.70 | −0.163 |
Nucleotide diversity.
Nucleotide diversity at nonsynonymous sites.
Nucleotide diversity at synonymous sites.
Ratio of diversity at nonsynonymous sites to diversity at synonymous sites.
Ratio of the value of Watterson's θ at nonsynonymous and synonymous sites.
Tajima's D, a measure of selection in which deviation from zero indicates nonneutral evolution (Tajima 1989). Negative values for D are indicative of purifying selection or recent population expansion.
Tajima's D at sites unlikely to be under selection (introns and silent sites in exons).
Refers to exclusion of the gene-conversion tract shown in figure 4B.
Refers to exclusion of the gene-conversion tracts shown in figure 4B and C.
FIG. 2.
Sliding windows of intragenic nucleotide diversity for fog-2 and ftr-1 in natural isolate populations. Nucleotide position in the alignment (x axis) is plotted against π, intragenic nucleotide diversity (y axis). A 50-nt window was slid in 10-nt increments. Note that the scales for π are different on both graphs. fog-2 shows much lower π values overall. (A) Nucleotide diversity between natural isolates in fog-2. The peak at 500 bp in fog-2 corresponds to the gene conversion event identified by GENECONV between 507 and 599 bp and disappears when the gene-conversion event is excluded from the analysis. (B) Nucleotide diversity between natural isolates in ftr-1. The peak at 500 bp in ftr-1 corresponds to the gene-conversion event identified by GENECONV between 498 and 599 bp and also disappears when the gene-conversion event is excluded from the analysis. The peak from 800 to 950 bp corresponds to the putative recombinant allele identified in ftr-1. The removal of this allele from the analysis greatly reduces π in this region. Additionally, the peak spanning 1,000–1,150 bp has features indicative of gene conversion involving a more divergent locus.
FIG. 3.
Linkage disequilibrium in fog-2 and ftr-1 within C. elegans populations. The paralogs are differentiated by black lines. Numbers indicate nucleotide position in the alignment of each paralog, with indels introduced in the fog-2 alignment in order to show positional homology with ftr-1. Shading indicates significance by Fischer's exact test, with light shading corresponding to P < 0.01, medium shading to P < 0.001 and dark shading to P < 0.0001. The gene conversion event detected in ftr-1 consists of the three polymorphisms at 506, 522, and 524 bp in the ftr-1 alignment, whereas the gene conversion event in fog-2 consists of the points at 522 and 524 bp in the fog-2 alignment. The ftr-1 recombinant allele consists of all polymorphisms from 601 to 912 bp in the ftr-1 alignment.
In order to address the possibility that strains isolated from the same geographic site may be clonal replicates of one another, we identified 11 strains that were indistinguishable at the fog-2 and ftr-1 loci from others isolated from the same location. We conducted the analyses with 1) inclusion and 2) exclusion of these strains with identical haplotypes. Table 3 represents the results when these 11 strains were included. Estimates of overall nucleotide diversity were marginally altered, with the exclusion of these strains (π changed by −0.02% to 0.13%; πA changed by 0.01–0.13%; and πs changed by 0.001–0.17%).
Contribution of Gene Conversion to Sequence Diversity
Evidence of gene conversion was found in both fog-2 and ftr-1, with each gene containing one allele matching the expected pattern of gene conversion (fig. 4). A statistical test for gene conversion implemented in GENECONV detected gene conversion in the third exon of fog-2 (range from 507 to 599 bp in the alignment, P = 0.001). This gene-conversion tract consists of 75 bp that are identical between fog-2 and ftr-1 as well as a shared polymorphism between the two genes immediately upstream of the identical region. This particular fog-2 allele with N2 ftr-1 rather than fog-2 nucleotides is present in 3 of 40 isolates (7.5%): AB1, CB4856, and CB4932 (fog-2 allele 2 in table 1 and fig. 4A). The minimum and maximum lengths of the gene conversion tract are 3 and 93 bp, respectively. The same region in ftr-1 (range from 498 to 599 bp in the alignment; P = 0.002) contains 3 bp matching N2 fog-2 rather than ftr-1 in 20 of 40 isolates (50%): AB2, AB3, AB4, CB3196, CB3197, CB3199, CB4852, CB4853, CB4854, CB4855, CB4857, CB4858, DR1345, DR1349, DR1350, JU258, JU262, JU263, KR314, and PB306 (ftr-1 allele 4 in table 1). The minimum and maximum lengths of the gene-conversion tract are 19 and 102 bp, respectively. Two of the 3 bp matching fog-2 in the converted ftr-1 allele aligned with the converted nucleotides matching ftr-1 in the converted fog-2 allele (from 522 to 524 bp in the alignment; see fig. 4A and B). These two sites can be described as shared polymorphic sites or sites where both of the corresponding sites in the genes are polymorphic (Innan 2003a). These conversion events are not shared between isolates, that is, isolates with the N2 ftr-1 nucleotides in fog-2 do not contain N2 fog-2 sequences in ftr-1 and vice versa.
FIG. 4.
Nucleotide sequence alignments showing possible gene-conversion and recombination events in fog-2 and ftr-1. Numbers at the top represent base position in the alignment. The uppermost sequence is the N2 laboratory-strain sequence of the gene in question and is depicted in white. Dots represent nucleotides identical to the N2 sequence. The middle sequence represents the converted or recombinant allele. The bottom sequence indicates the N2 laboratory-strain sequence of the paralogous gene and is shaded in light gray. (A) Gene conversion in fog-2. The region of gene conversion in the fog-2 allele (middle sequence) is shown in gray to indicate sequence similarity with the N2 ftr-1 gene sequence. The minimum gene-conversion tract length of 3 bp is shown by the boxed region. The region surrounding the converted nucleotides is identical between fog-2 and ftr-1 over a 93-bp stretch (the maximum length of the gene-conversion tract). (B) Gene conversion in ftr-1. The region of gene conversion in the ftr-1 allele (middle sequence) is shown in light gray to indicate sequence similarity with the N2 fog-2 gene sequence. The minimum gene-conversion tract length of 19 bp is shown by the boxed region. The region surrounding the converted nucleotides is identical between fog-2 and ftr-1 over a 102-bp stretch (the maximum length of the gene conversion tract). The two converted nucleotides at 522 and 524 bp are the same 2 nt that have undergone conversion in fog-2. (C) The middle sequence represents an allele possibly generated by recombination with fog-2 or other FTR family sequence. Only part of the allele, which covers a 294-bp range of the gene, is shown. Dark gray shading is used to represent this allele, as it does not fully match either fog-2 or ftr-1. Note that the detected allele matches fog-2 rather than ftr-1 for short stretches, as indicated by the boxed regions in C but contains a number of unique polymorphisms that do not match either gene. The boxed region from 892 to 906 bp was identified as gene conversion by GENECONV.
Another ftr-1 allele was found in three isolates (AB1, CB4856, and CB4932; ftr-1 alleles 2 and 3 in table 1 and fig. 4C). This allele was highly divergent over 103 bp of the gene (range from 809 to 912 bp in the alignment). It matched fog-2 in many regions but was diverged from both fog-2 and ftr-1 in others such that neither gene conversion nor point mutation alone seem likely mechanisms to account for its formation. A possible cause could be multiple recombination events with fog-2 and other paralogs in the FTR gene family. However, a search using the basic local alignment search tool (BLAST) against the C. elegans genome found no exact matches for those regions of the allele differing from both fog-2 and ftr-1. To further test the possibility that recombination with other paralogs in the FTR family yielded this chimeric allele, we identified three paralogs closely related to fog-2 and ftr-1 that also lie in genomic proximity (fbxa-113, fbxa-114, and fbxa-115). We then identified all known single nucleotide polymorphisms (SNPs) for these paralogs in the Hawaiian strain, CB4856, which is one of the strains that harbors the divergent ftr-1 allele and is the only strain for which genomewide SNP data are currently available. However, none of the SNP haplotypes found in CB4856 matched the divergent regions of the chimeric allele.
The GENECONV test detected gene conversion within a 14-bp region of this allele in AB1 and CB4932 (range from 892 to 906 bp in the alignment, P = 0.010). This tract contained 4 bp matching fog-2 rather than ftr-1 and was flanked on both sides by polymorphisms unique to AB1 and CB4932. The sequence of CB4856 was intermediate in this area in that it contained only 2 of the 4 bp matching fog-2. Interestingly, this unusual allele and the fog-2 gene conversion tract were shared between the same three isolates, AB1, CB4856, and CB4932. These strains are geographically distinct: AB1 was isolated from Adelaide, Australia; CB4856 from Hawaii, United States; and CB4932 from Taunton, United Kingdom. In addition, they are phylogenetically distinct (Denver et al. 2003; Cutter 2006).
To look at the relative contribution of the two detected gene conversion events as well as the suspected recombinant allele in ftr-1 to population diversity between isolates, all analyses were repeated with the detected gene-conversion events removed from the data set (table 3). An appreciable proportion of π in both genes can be attributed to the observed gene conversion and recombination events. The exclusion of gene-conversion tracts in fog-2 and ftr-1 removes 22% and 34% of the nucleotide diversity from the genes, respectively.
Contribution of fog-2 Genotype to Fecundity
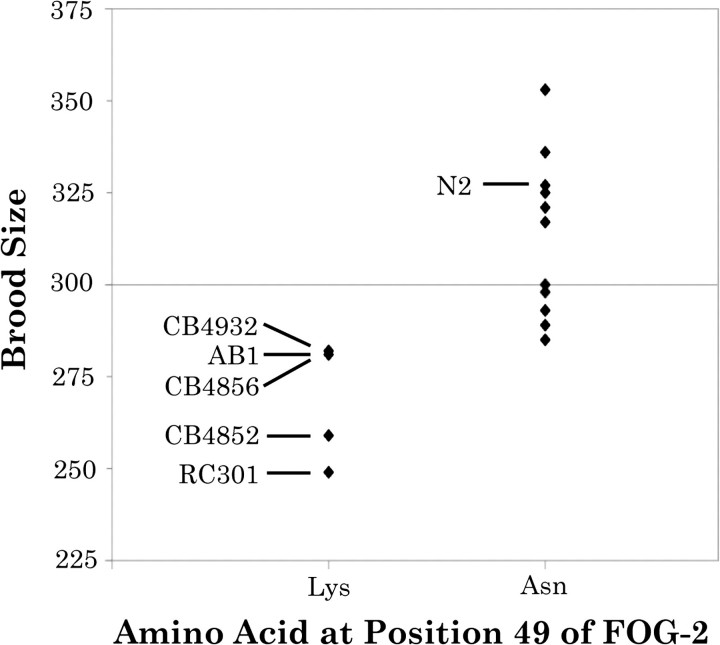
fog-2 contains four nonsynonymous polymorphisms. To explore the possibility that these nonsynonymous mutations may have phenotypic effects, we utilized data from a previous study of fecundity in natural isolates. Hodgkin and Doniach (1997) measured the number of viable progeny (brood size) in 19 of the 40 strains comprising this study. In their analyses, hermaphrodites of most strains produced approximately 300 progeny via self-fertilization. Three strains with active Tc1 elements had particularly small brood sizes that the authors ascribed to the detrimental effects of a large population of selfish elements, given that these strains produced a larger number of dead eggs relative to other strains with silenced Tc1 elements. To avoid confounding the deleterious effects of transposon activity with fog-2 genotype, we excluded these three strains (CB4851, DH424, and RW7000) from our analysis. The average brood size of the remaining 16 strains is 300. The five strains with the smallest brood sizes (AB1, CB4852, CB4856, CB4932, and RC301) share a radical amino acid replacement between lysine and asparagine at position 49, located at the center of a well-conserved region in the F-box domain. Three of these five strains (AB1, CB4856, and CB4932) also possess the ftr-1 converted fog-2 allele, including a second nonsynonymous substitution, shown in figure 4A. Strains with an asparagine at position 49 have, on average, a 14% smaller brood size than strains with lysine at the same position (fig. 5). The difference between the two genotypes (asparagine vs. lysine at position 49) is significant (Wilcoxon two-sample test, z = −3.0, P = 0.003).
FIG. 5.
Average brood size as determined by Hodgkin and Doniach (1997) for a subset of 15 of the 40 strains used in this study. To the left and right are brood sizes for strains with asparagine and lysine at position 49 of FOG-2, respectively.
Discussion
The two genes in this study, fog-2 and ftr-1, appear to have arisen from tandem duplication after the divergence of the C. elegans lineage from its most closely related congenerics (Nayak et al. 2005; Katju et al. 2008). FOG-2 is required for spermatogenesis in C. elegans hermaphrodites and the evidence suggests that the C-terminal end of the protein is essential for its function (Nayak et al. 2005). The last 30 amino acids in the FOG-2 protein do not have sequence similarity to other ftr genes, nor do the 3′ untranslated region and flanking sequence show sequence similarity between fog-2 and ftr-1, suggesting partial tandem duplication with recruitment as the mode of duplication (Katju and Lynch 2006; Katju et al. 2008).
A study employing relaxation of selection on optimal codon usage to estimate the time since C. elegans evolved hermaphroditism concluded that it was unlikely to have evolved more than 4 Ma (Cutter et al. 2008). Using the same assumptions about mutation rates and generation time, synonymous sequence divergence between ftr-1 and fog-2 suggests that the duplication resulting in the origin of fog-2 occurred approximately 2.3 Ma. This is concordant with the estimate of 4 Ma as the upper boundary for the evolution of hermaphroditism (Cutter et al. 2008) and suggests that the duplication of fog-2 occurred around the same time as the evolution of hermaphroditism. Further, hermaphroditism in C. elegans vastly postdated its evolutionary divergence from its congeneric species, C. briggsae and C. remanei. This suggests that the lineage leading to C. elegans, after its split from C. briggsae and C. remanei, most likely existed as a gonochoristic obligate male–female outcrosser for the majority of its evolutionary past, with the origin of hermaphroditism being a relatively recent derived character state. However, using a more recent estimate of mutation rate from whole-genome sequencing of mutation accumulation lines, the origin of fog-2 dates to 7.2 Ma.
These estimates come with important caveats. Foremost, the abundance of information relating to the cellular, developmental and genetic mechanisms in C. elegans is in stark contrast to the scant data available regarding the ecology and natural history of the species. Hence, if the average number of generations in the wild is significantly different from the value assumed in our analyses, the date of origin for fog-2 and the evolution of hermaphroditism in C. elegans will be concomitantly altered. Another caveat is concerned with the initial period following the duplication event. Although we removed regions of the genes from the analysis that have undergone recent gene conversion, this does not correct for a period of unknown length during which sequence divergence between the copies may have been limited by frequent gene conversion.
Of the gene duplicates that were the focus of this study, only fog-2 has a known function, namely, the regulation of spermatogenesis in C. elegans hermaphrodites. Loss-of-function mutations in fog-2 disrupt spermatogenesis in hermaphrodites, effectively turning them into females. This phenotype has been used to generate experimental populations of obligate outcrossers to examine the role of sex in evolution, as well as in other experiments that are more appropriately conducted with outcrossing rather than largely self-fertilizing populations (Chasnov and Chow 2002; Stewart and Phillips 2002; Cutter 2005; Manoel et al. 2007; Katju et al. 2008; Morran et al. 2009). Hermaphrodites usually outcompete obligate outcrossers that contain mutations in fog-2 or other loci involved in C. elegans sex determination (Chasnov and Chow 2002; Stewart and Phillips 2002), even under experimental conditions imposing a high mutational load when outcrossing is expected to be more beneficial (Cutter 2005; Manoel et al. 2007). However, it has been recently shown that increased mutation rates, as well as adaptation to novel environments, favors outcrossing worms (Morran et al. 2009).
The levels of intraspecific variation in fog-2 and ftr-1 observed are typical to C. elegans genes (Cutter 2006; Jovelin et al. 2009). Variation is several times higher in ftr-1, as is the level of polymorphism potentially due to gene conversion. Our results show that 22% and 34% of the nucleotide diversity in fog-2 and ftr-1 is due to gene conversion. A short sequence containing shared polymorphism between fog-2 and ftr-1 is in the same area as the previously detected gene conversion events in laboratory populations (Katju et al. 2008). The area containing evidence of gene conversion in the wild, as well as in the experimental populations, is immediately upstream of a stretch of DNA that harbors the most extensive sequence identity between the two genes (100% sequence identity for 75 bp; Katju et al. 2008). It is likely that the reason why shared polymorphism and other signs of contributions from gene conversion to polymorphism in fog-2 and ftr-1 is relatively limited is that, for most of the length of the genes, DNA divergence is close to 20%. This level of divergence would hinder the formation of heterologous duplex intermediates in gene conversion.
Strains AB1, CB4856, and CB4932 share fog-2 gene conversion alleles and a highly divergent ftr-1 allele (fig. 4). This divergent ftr-1 allele follows the mosaic pattern generally attributed to gene conversion or other recombination events (Betrán et al. 1997, fig. 4). Clusters of nucleotides that are identical to fog-2 are flanked by other differences between the two sequences, and it is possible that recombination events responsible for this pattern involved FTR genes other than fog-2. However, this donor FTR locus is not in the sequenced N2 genome nor is it present in the most closely related F-box paralogs in the CB4856 strain. It is possible that the F-box gene family to which fog-2 and ftr-1 belong is in some flux and that the donor loci have been lost.
It has long been supposed that gene conversion could present a potential obstacle to the evolution of novel function in a duplicate gene, as frequent gene conversion with the ancestral gene could eliminate divergence as it arises (Walsh 1987; Teshima and Innan 2004). However, in the case of fog-2, the partial nature of the gene-duplication event may have considerably ameliorated this problem. If the nonhomologous recruited sequence at the 3′ end (fig. 1) is even partially responsible for the GLD–1 binding ability of FOG-2, gene conversion with ftr-1 would be less able to disrupt fog-2 neofunctionalization as the genes do not have high enough identity in this region to erroneously line up during DNA replication. In C. elegans, this type of structural heterogeneity immediately following duplication may not be an unusual means to circumvent the problem of gene conversion, as approximately half of the young gene duplicates in C. elegans originate from partial or chimeric duplications (Katju and Lynch 2003, 2006).
Gene conversion can contribute substantially to nucleotide diversity in duplicated genes (Innan 2003b). One study has found evidence that gene conversion accounted for 81 of 82 polymorphisms between paralogs with 96% identity in Plasmodium falciparum (Nielsen et al. 2003). In another study, a region of the human genome that experienced repeated conversion events was shown to have nucleotide diversity levels higher than most of the genome (Bosch et al. 2004). Gene conversion is also known to contribute directly to phenotypic diversity, such as gene conversion between the red and green human opsin genes, which is one of the causes of blue cone monochromacy, a form of color blindness (Reyners et al. 1995).
Caenorhabditis elegans hermaphrodites normally do not produce enough sperm to fertilize all their eggs, and variation in brood size is therefore more likely to be due to sperm limitation and efficiency of fertilization rather than the amount of eggs produced. We have found that experimental fog-2 strains sometimes revert to hermaphroditism via gene conversion with ftr-1, an upstream paralog of fog-2 (Katju et al. 2008). If gene conversion between ftr-1 and fog-2 is common in the wild, it has the potential to shape genetic variation at these loci in natural populations, thereby modifying the number of sperm produced by hermaphrodites. This could have important consequences for the degree of inbreeding versus outcrossing in natural populations of C. elegans (Katju et al. 2008). There is a statistical association between a radical amino acid replacement in a conserved area of the F-box domain and brood size as calculated by Hodgkin and Doniach (1997). The alleles sharing this mutation were from relatively unrelated strains isolated from diverse geographic locations (Denver et al. 2003; Cutter 2006). However, given pervasive linkage disequilibrium in natural populations of C. elegans, this association could be due to linked alleles and not the mutation in question and additional experiments will be needed to test this suggested relationship between fog-2 genotype and fecundity. Previous results have shown that deletion of the C-terminal domain has the largest consequences for the function of FOG-2 (Nayak et al. 2005). However, based on our results, we hypothesize that substitutions in other parts of the gene can modulate the degree of self-fertilization in the wild. In the light of new evidence that shows that outcrossing is favored during adaptation and high mutation rates (Morran et al. 2009), different fog-2 alleles could be selected for under different conditions. Thus, gene conversion could play an important role in both generating diversity in fog-2 as well as facilitating back mutations or reversals.
Acknowledgments
All nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This manuscript was improved by the comments of two anonymous reviewers. H.S.R. was supported by an NSF DEB-0731350 UnO (Undergraduate Opportunities) assistantship. This research was facilitated by start-up funds from the University of New Mexico to Vaishali Katju.
References
- Betrán E, Rozas J, Navarro A, Barbadilla A. The estimation of the number and length distribution of gene conversion tracts from population DNA sequence data. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Hurles ME, Navarro A, Jobling MA. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Chasnov JR, Chow KL. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics. 2002;160:983–994. doi: 10.1093/genetics/160.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Jin SW, Cohen A, Ellis RE. A phylogeny of the Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Mutation and the experimental evolution of outcrossing in Caenorhbditis elegans. J Evol Biol. 2005;18:27–34. doi: 10.1111/j.1420-9101.2004.00804.x. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–184. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Wasmuth JD, Washington NL. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics. 2008;178:2093–2104. doi: 10.1534/genetics.107.085787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, et al. (11 co-authors) A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci U S A. 2009;106:16310–16314. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Thomas WK. Phylogenetics in Caenorhabditis elegans: an analysis of divergence and outcrossing. Mol Biol Evol. 2003;20:393–400. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 2006;10:531–538. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H. The coalescent and infinite-site model of a small multigene family. Genetics. 2003a;163:803–810. doi: 10.1093/genetics/163.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H. A two-locus gene conversion model with selection and its application to the human RHCE and RHD genes. Proc Natl Acad Sci U S A. 2003b;100:8793–8798. doi: 10.1073/pnas.1031592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Dunham JP, Sung FS, Phillips PC. High nucleotide divergence in developmental regulatory genes contrasts with structural elements of olfactory pathways in Caenorhabditis. Genetics. 2009;181:1387–1397. doi: 10.1534/genetics.107.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, LaBeau EM, Lipinski KJ, Bergthorsson U. Sex change by gene conversion in a Caenorhabditis elegans fog-2 mutant. Genetics. 2008;180:669–672. doi: 10.1534/genetics.108.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Lynch M. On the formation of novel genes by duplication in the Caenorhabditis elegans genome. Mol Biol Evol. 2006;23:1056–1067. doi: 10.1093/molbev/msj114. [DOI] [PubMed] [Google Scholar]
- King LM. The role of gene conversion in determining sequence variation and divergence in the Est-5 family in Drosophila pseudoobscura. Genetics. 1998;148:305–315. doi: 10.1093/genetics/148.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DHA. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci U S A. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:3001–3007. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Manoel D, Carvalho S, Phillips PC, Teotónio H. Selection against males in Caenorhabditis elegans under two mutational treatments. Proc R Soc B. 2007;274:417–424. doi: 10.1098/rspb.2006.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favor outcrossing over self-fertilization. Nature. 2009;462:350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:57–71. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KM, Kasper J, Choi M, Befrod T, Kristiansen K, With DF, Volkman SK, Lozovsky ER, Hartl DL. Gene conversion as a source of nucleotide diversity in Plasmodium falciparum. Mol Biol Evol. 2003;20:726–734. doi: 10.1093/molbev/msg076. [DOI] [PubMed] [Google Scholar]
- Ohta T. On the evolution of multigene families. Theor Pop Biol. 1983;23:216–240. doi: 10.1016/0040-5809(83)90015-1. [DOI] [PubMed] [Google Scholar]
- Reyners E, Van Thienen M, Meire F, De Boulle K, Devries K, Kesteljn P, Willems PJ. Gene conversion between red and defective green opsin gene in blue cone monochromacy. Genomics. 1995;29:323–328. doi: 10.1006/geno.1995.9998. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. GENECONV: a computer package for the statistical detection of gene conversion. St Louis (MO): Washington University; 1999. http://www.math.wustl.edu/∼sawyer. [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;139:579–606. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple C, Wolfe KH. Gene duplication and gene conversion in the Caenorhabditis elegans genome. J Mol Evol. 1999;48:555–564. doi: 10.1007/pl00006498. [DOI] [PubMed] [Google Scholar]
- Stewart AD, Phillips PC. Selection and maintenance of androdioecy in Caenorhabditis elegans. Genetics. 2002;160:975–982. doi: 10.1093/genetics/160.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Innan H. The effect of gene conversion on the divergence between duplicated genes. Genetics. 2004;166:1553–1560. doi: 10.1534/genetics.166.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JB. Sequence-dependent gene conversion: can duplicated genes diverge fast enough to escape conversion? Genetics. 1987;117:543–557. doi: 10.1093/genetics/117.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. The nematode C. elegans. New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]