This study identifies CFL1 as a negative regulator of cuticle development in rice and Arabidopsis. CFL1 interacts with a class IV homeodomain-leucine zipper transcription factor to modulate the expression of two genes known to be involved in cuticle development.
Abstract
Plants have a chemically heterogeneous lipophilic layer, the cuticle, which protects them from biotic and abiotic stresses. The mechanisms that regulate cuticle development are poorly understood. We identified a rice (Oryza sativa) dominant curly leaf mutant, curly flag leaf1 (cfl1), and cloned CFL1, which encodes a WW domain protein. We overexpressed both rice and Arabidopsis CFL1 in Arabidopsis thaliana; these transgenic plants showed severely impaired cuticle development, similar to that in cfl1 rice. Reduced expression of At CFL1 resulted in reinforcement of cuticle structure. At CFL1 was predominantly expressed in specialized epidermal cells and in regions where dehiscence and abscission occur. Biochemical evidence showed that At CFL1 interacts with HDG1, a class IV homeodomain-leucine zipper transcription factor. Suppression of HDG1 function resulted in similar defective cuticle phenotypes in wild-type Arabidopsis but much alleviated phenotypes in At cfl1-1 mutants. The expression of two cuticle development-associated genes, BDG and FDH, was downregulated in At CFL1 overexpressor and HDG1 suppression plants. HDG1 binds to the cis-element L1 box, which exists in the regulatory regions of BDG and FDH. Our results suggest that rice and Arabidopsis CFL1 negatively regulate cuticle development by affecting the function of HDG1, which regulates the downstream genes BDG and FDH.
INTRODUCTION
Terrestrial plants are believed to have evolved from aquatic ancestors and have evolved a heterogeneous lipophilic covering layer, the cuticle, which prevents excessive water loss from plants and protects them against various biotic and abiotic stresses. The polyester layer covering the aerial parts of plants is mainly composed of cutin, with waxes embedded in the polymer matrix and deposited on its surface. Cutin is mainly composed of oxygenated C16 and C18 fatty acids cross-linked by ester bonds, which link the carboxyl group of one fatty acid to a primary or secondary hydroxyl group of another (Nawrath, 2002). Thus, the interesterified acyl chains form a polymeric network with a strongly hydrophobic character. Cuticular waxes are a complex mixture of very-long-chain fatty acids (VLCFAs) and their derivatives, such as very-long-chain alkanes, fatty alcohols, aldehydes, and ketones (Jenks et al., 2002). Together, cutin and waxes form the cuticle, which plays an important role in the control of nonstomatal water loss. The cuticle has also evolved to reflect or dissipate detrimental radiation and to resist invasion by insects and pathogens. Furthermore, the cuticle plays roles in plant development. For example, a properly synthesized cuticle is required for pollen–pistil interactions, and leaves with defectively developed cuticles show postgenital organ fusion (Pruitt et al., 2000; Sieber et al., 2000; Nawrath, 2006).
The cuticle changes continuously during plant development to maintain its balance between rigidity and flexibility (Suh et al., 2005). By using genetic, biochemical, and genomic approaches, many mutants with disruptions in genes involved in this process have been identified, and numerous genes have been cloned and characterized (Pollard et al., 2008; Samuels et al., 2008; Wallis and Browse, 2010). Cuticular wax is mainly composed of VLCFAs and their derivatives. These are produced via the actions of 3-ketoacyl-CoA synthases (KCSs), which sequentially add one two-carbon unit at a time to the C16 or C18 fatty acids that are produced in plastids via the de novo fatty acid synthesis pathway. To date, 21 members of the KCS family have been found in the Arabidopsis thaliana genome (Blacklock and Jaworski, 2006; Joubès et al., 2008). The functions of KCS1, KCS2, KCS6, KCS10, and KCS20 have been characterized in detail (Lolle et al., 1997; Millar et al., 1999; Todd et al., 1999; Yephremov et al., 1999; Fiebig et al., 2000; Pruitt et al., 2000). The 3-ketoacyl CoA reductase gene At KCR1 and enoyl reductase gene CER10, both of which encode enzymes that catalyze downstream reactions, have also been cloned (Zheng et al., 2005; Bach et al., 2008; Beaudoin et al., 2009). Through decarboxylation or acyl-reduction pathways, these VLCFA molecules are transformed into aldehydes, primary alcohols, alkanes, secondary alcohols, and ketones and then into wax esters (Jenks et al., 2002). Recent studies showed that cytochrome P450-dependent proteins are important for the production of cutin monomers. LACERATA (LCR) belongs to the CYP86A subfamily and is responsible for the synthesis of the hydroxy fatty acid components of cutin (Wellesen et al., 2001). CYP86A2, which encodes a monooxygenase, is also critical for cutin biosynthesis (Xiao et al., 2004). Transportation of hydrophobic molecules is also thought to be an important factor for cuticle formation. It was speculated that these components are transported by ATP binding cassette transporters to the plant surface through the plasma membrane and the cell wall (Kuromori et al., 2011; McFarlane et al., 2010; Bessire et al., 2011; Panikashvili et al., 2011).
Most Arabidopsis mutants with defective cuticles show decreased accumulation of one or more constituents of the cuticle. However, there are some defective-cuticle mutants or transgenic lines, for example, bdg, lcr, fdh, and 35S:WIN1, which show increased accumulation of cuticle constituents. BODYGUARD (BDG) encodes a protein in the α/β-hydrolase fold protein superfamily (Kurdyukov et al., 2006b), LCR encodes a cytochrome P450-dependent protein, and FIDDLEHEAD (FDH), also known as KCS10, encodes a KCS (Lolle et al., 1997; Yephremov et al., 1999; Wellesen et al., 2001). In both the fdh and lcr mutants, the amount of leaf residual-bound lipids and waxes increased (Voisin et al., 2009). WAX INDUCER1 (WIN1) is an ethylene response transcription factor and is reported to regulate wax and cutin biosynthesis (Broun et al., 2004; Kannangara et al., 2007). In addition to cuticular defects, these mutants also showed some other phenotypes (e.g., disturbed cell differentiation, altered organ morphology, and altered resistance to fungal attack) (Bessire et al., 2007; Tang et al., 2007). Recent studies in Arabidopsis showed that epidermal cells and the covering cuticle rely on each other for proper development. For example, epidermal cells produce molecules to construct the cuticle, whereas mutants with damaged epidermises, such as ale1, ale2, and acr4, usually have defective cuticles (Tanaka et al., 2001; Gifford et al., 2003; Watanabe et al., 2004; Tanaka et al., 2007). However, the development of epidermal cells is often perturbed in mutants that are defective in VLCFA biosynthesis, and it has been hypothesized that signals from the cuticle affect epidermal cell differentiation (Yephremov et al., 1999; Bird and Gray, 2003).
Although more and more genes with roles in cuticle biosynthesis have been described, few genes that regulate cuticle development have been reported, and the regulatory mechanisms remain unclear. To date, only a few genes, such as WIN1 and its two homologs, have been reported to regulate wax and cutin biosynthesis in Arabidopsis (Aharoni et al., 2004). In plants that overexpress WIN1, the expression of several genes involved in cutin and wax production were strongly affected. Long-chain acyl-CoA synthetase2 was identified as one of the possible direct targets of WIN1 (Kannangara et al., 2007). Homeodomain-leucine zipper (HD-ZIP) transcription factors are involved in regulation of cuticle development in maize (Zea mays) and tomato (Solanum lycopersicum) (Isaacson et al., 2009; Javelle et al., 2010). Since it is widely accepted that the cuticle structure is dynamic, there must be more regulatory genes involved in this process that need to be identified (Kannangara et al., 2007).
In this study, we identified CFL1 as one such regulatory gene by analyzing a dominant rice (Oryza sativa) mutant with a curly flag leaf phenotype. CFL1 encodes a WW domain protein, and there are two homologs of rice CFL1 in Arabidopsis: At CFL1 and At CFL2. Arabidopsis plants overexpressing At CFL1 displayed organ fusion phenotypes with decreased levels of epicuticular wax and defective cuticles. Loss-of-CFL1-function resulted in an enhancement of cuticle properties in Arabidopsis. Using the yeast two-hybrid system, we identified a protein that interacts with At CFL1, HDG1, a member of the class IV HD-ZIP transcription factor family, some members of which were previously shown to play an essential role in the development of the epidermis and its accessories (Nakamura et al., 2006). Suppression of HDG1 in wild-type plants by chimeric repression technology resulted in organ fusion and toluidine blue (TB) staining phenotypes similar to those of cauliflower mosaic virus (CaMV) 35S:At CFL1 plants, but the phenotypes were dramatically alleviated in the At cfl1-1 background. We found that HDG1 bound to the cis-element L1 box in the regulatory regions of two cuticle development-associated genes BDG and FDH. These data suggest that At CFL1 is possibly involved in cuticle development by affecting the function of HDG1, thus affecting the transcription of BDG and FDH. Our results suggest that this regulation is conserved in Arabidopsis and rice.
RESULTS
Curly Flag Leaf Mutant cfl1 Is a Dominant Mutant in Rice
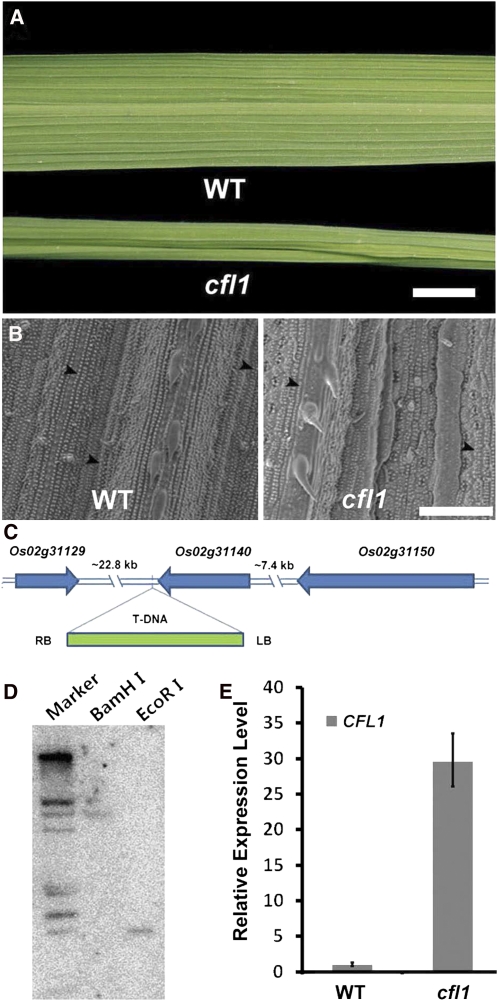
In rice, the flag leaf is essential for producing nutrients stored in grains (Yoshida, 1981), and the morphology and status of the flag leaf is critical to this process (Murchie et al., 1999). By screening a rice T-DNA insertion mutant collection, we identified a rice mutant with a curly flag leaf, which was therefore designated as curly flag leaf1 (cfl1), although later we found that all of its leaves were affected. Leaves of cfl1 mutants were almost indistinguishable from those of the wild type during vegetative growth. After tillering, cfl1 leaves became obviously curly, sometimes even cylindrical (Figure 1A). Scanning electron microscopy analysis showed that the surface of the epidermis of the cfl1 mutant was wrinkled and covered with less waxy papillae (WP) (Figure 1B, right panel), whereas wild-type leaves showed a smooth and well-organized epidermis with more WP (Figure 1B, left panel).
Figure 1.
Characterization of the Rice Mutant cfl1.
(A) Leaves of the wild type (WT) and homozygous cfl1 mutant at maximum tillering stage. Bar = 1 cm.
(B) Scanning electron micrograph of wild-type (left panel) and cfl1 (right panel) leaves at four-leaf stage. Arrowheads indicate WP on epidermis. Bar = 100 μm.
(C) Schematic of the genomic region flanking the T-DNA insertion site in cfl1. Arrows indicate the transcriptional orientation. LB, T-DNA left border; RB, T-DNA right border.
(D) DNA gel blot analysis of the cfl1 mutant using left border of T-DNA as probe. Rice genomic DNA was digested with BamHI (middle lane) or EcoRI (right lane). Note that there was only one band in each lane. Marker, λ DNA/EcoRI + HindIII digest.
(E) Relative expression level of rice CFL1 in the wild type and the cfl1 mutant. The expression level in the wild type is set to 1.0, and error bars represent the sd of three biological replicates.
Among the F2 offspring, the ratio of normal to curly leaved plants was ~1:3, suggesting that this mutation is dominant. Using thermal asymmetric interlaced (TAIL)-PCR, we identified a single T-DNA insertion in cfl1, which was located between Os02g31140 and Os02g31129, ~1.3 kb downstream of the Os02g31140 stop codon (Figure 1C). To clarify whether the T-DNA insertion cosegregates with the phenotype, we genotyped a T3 population of cfl1 mutants. Of 109 T3 plants, 27 lacked the T-DNA insertion, 26 were homozygous for the T-DNA insertion, and 56 were heterozygous for the T-DNA insertion (see Supplemental Figure 1 online). All plants with the T-DNA insertion showed curly leaf phenotypes, and the phenotype was more severe in the homozygous plants. All plants without the T-DNA insertion showed normal leaf phenotypes. These results suggest that cfl1 is a dominant mutant and indicate that the curly leaf phenotype is likely caused by this single T-DNA insertion. DNA gel blot analysis showed that there was indeed only a single T-DNA insertion in the cfl1 mutant, further supporting these conclusions (Figure 1D).
Next, we examined the expression levels of the four genes located within the region 25 kb upstream and downstream of the T-DNA insertion site. Quantitative real-time PCR analysis showed that, in 4-week-old rice leaves, the transcription level of Os02g31140 was increased, whereas expression of the other three neighboring genes (i.e., Os02g31129, Os02g31150, and Os02g31160) was not different from that in the wild type (Figure 1E). This result, together with the genetic data, suggests that the mutant phenotypes are caused by overexpression of Os02g31140, which was designated as CFL1.
Overexpression of CFL1 Is Responsible for the Curly Flag Leaf Phenotype
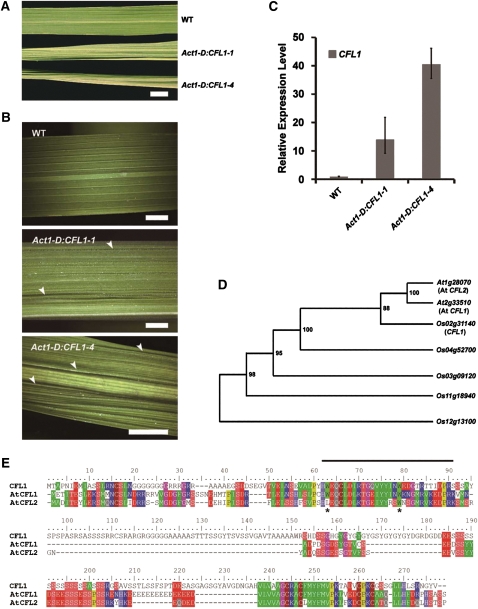
To clarify whether overexpression of CFL1 causes the curly flag leaf phenotype, we generated transgenic rice overexpressing CFL1 driven by an Actin 1 promoter and obtained more than 10 transgenic lines, most of which displayed the curly flag leaf phenotype. Two representative lines are shown in Figure 2A. The CFL1-overexpressing plants showed a crinkled epidermis, which differed from the smooth and well-organized epidermis of wild-type plants (Figure 2B). The severity of the crinkled epidermis phenotypes in 4-week-old rice leaves correlated with the expression levels of CFL1 demonstrated by quantitative real-time PCR analysis (Figure 2C). These results demonstrate that the curly flag leaf phenotype results from overexpression of CFL1.
Figure 2.
Characterization of CFL1 Structure and Function in Rice.
(A) Flag leaves of rice plants at maximum tillering stage, from top to bottom, the wild type (WT), Act1-D:CFL1-1, and Act1-D:CFL1-4. Act1-D represents the 5′ region fragment of rice actin 1 gene (Act1), which can drive constitutive expression of a gene in rice. Bar = 1 cm.
(B) Higher magnification of leaves presented in (B), from top to bottom, wild type, Act1-D:CFL1-1, and Act1-D:CFL1-4. Arrowheads indicate crinkles on epidermis. Bars = 5 mm.
(C) Relative expression level of CFL1 in wild-type, Act1-D:CFL1-1, and Act1-D:CFL1-4 plants by quantitative RT-PCR. The expression level in the wild type is set to 1.0, and error bars represent the sd of three biological replicates.
(D) Phylogenetic tree showing the predicted relationships between CFL1 homologs in Arabidopsis and rice. Numbers in the nodes indicate bootstrap values.
(E) Multiple sequence alignment of rice CFL1 and Arabidopsis CFL1 and CFL2. The WW domain is indicated with a line above the alignment. Two conserved Trp residues are indicated with stars.
CFL1, a single-copy gene in rice, encodes an unknown protein of 274 amino acid residues, with a WW domain located at its N terminus. There are two CFL1 orthologs in Arabidopsis, At2g33510 and At1g28070, designated as At CFL1 and At CFL2, respectively (Figure 2D; see Supplemental Data Set 1 online). Sequence alignment of the three proteins revealed another conserved motif at the C terminus, in addition to the WW domain (Figure 2E). Interestingly, At CFL2 appears to have lost the first W residue from the WW domain (Figure 2E).
Both At CFL1–Overexpressing Plants and Rice cfl1 Mutants Are Defective in Cuticle Development
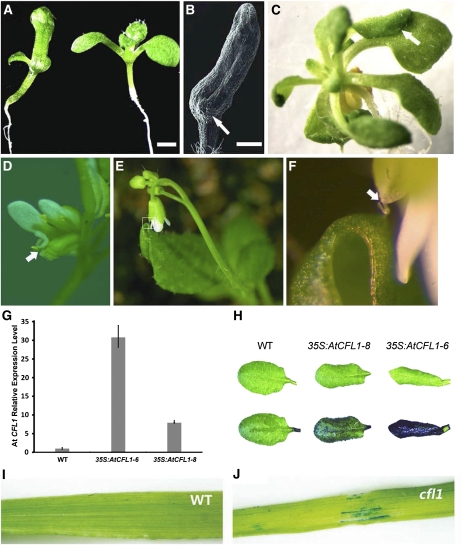
To investigate the function of CFL1, we first overexpressed rice CFL1 and Arabidopsis CFL1 and CFL2 in Arabidopsis. Plants overexpressing At CFL2 showed no obvious phenotypes compared with control plants; however, overexpression of either rice CFL1 or At CFL1 resulted in severe developmental defects. Here, we present only the At CFL1 overexpressor data. The most significant phenotype observed in the CaMV35S:AtCFL1 plants was organ fusion, including fused cotyledons, and fusions between leaves or sepals (Figures 3A to 3F). This organ fusion phenotype was observed in the T1 progeny of many At CFL1 overexpressor lines. This organ fusion did not occur between fully expanded leaves, not even when we clamped them together, suggesting that fusion probably occurs only in the primordia of young organs.
Figure 3.
Analysis of At CFL1 Overexpression Plants and rice cfl1 Plants.
(A) and (B) Cotyledon fusion in At CFL1 overexpression plants. Left, 10-d-old 35S:AtCFL1-6 seedling; right, wild-type seedling in (A). Scanning electron micrograph of fused cotyledon (B), arrow indicates emerging true leaf. Bars = 0.5 mm.
(C) to (F) Other organ fusions observed in At CFL1 overexpression plants, between rosette leaves (C), petal and carpel (D), and cauline leaf and sepal (E). (F) Higher magnification of boxed region in (E). Arrows in (C), (D), and (F) indicate organ fusion site.
(G) Relative expression levels of At CFL1 in overexpression and wild-type (WT) plants were measured with quantitative RT-PCR. The expression level in the wild type is set to 1.0, and error bars represent the sd of three biological replicates.
(H) Rosette leaves before (top) and after (bottom) staining with TB. From left to right, the wild type, 35S:AtCFL1-8, and 35S:AtCFL1-6, respectively.
(I) and (J) TB staining assay of rice cfl1. The wild-type leaf did not stain with TB (I), while patchy staining was observed in rice cfl1 leaves (J).
Because postgenital organ fusions are usually associated with cuticle defects, we used the TB assay (Tanaka et al., 2004) to examine whether there were any cuticular defects in the two overexpressor lines 35S:AtCFL1-6 and 35S:AtCFL1-8 (Figure 3G). Figure 3H shows that wild-type leaves with a normal cuticle repelled TB staining, whereas the leaves of 35S:AtCFL1-8 were partially stained by TB and those of 35S:AtCFL1-6 were completely stained by TB, suggesting that the cuticles in At CFL1 overexpressor plants were damaged. The intensity of the TB staining was correlated with the overexpression level of At CFL1 (Figure 3H). We also investigated whether the rice cfl1 mutant displayed defects in cuticle development by TB staining. As seen in At CFL1 overexpressing Arabidopsis plants, the leaves of rice cfl1 were also stained with TB, especially in areas where crinkles occurred (Figures 3I and 3J). This suggests that cuticle development is also affected in rice cfl1 mutants.
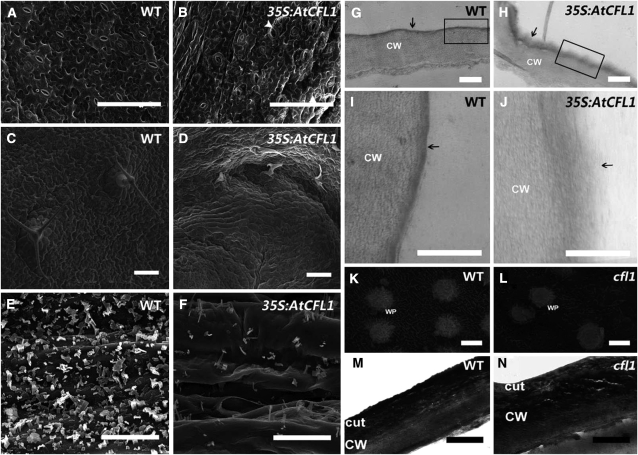
The Leaf Epidermal Surface Is Severely Altered in At CFL1 Overexpressing Plants and Rice cfl1 Mutants
To examine whether the leaf surface was altered in At CFL1–overexpressing plants and rice cfl1 mutants, we analyzed the first two true leaves using scanning electron microscopy. The abaxial surface of wild-type Arabidopsis leaves was smooth, reflecting the organized development of flattened layers of epidermal cells (Figure 4A). However, the epidermal cells of 35S:AtCFL1 plants were crinkled and deformed (Figure 4B). Paired stomata were observed on 35S:AtCFL1 plants (Figure 4B), resembling the phenotypes of the too many mouths and four lips mutants (Yang and Sack, 1995). The trichomes on 35S:AtCFL1 rosette leaves were misshapen and flattened, unlike those on wild-type rosette leaves, and were apparently arrested at earlier stages (Figures 4C and 4D). Next, we examined the epicuticular wax on inflorescence stems. The density of epicuticular wax crystals on the epidermal surface of 35S:AtCFL1-6 inflorescence stems was severely decreased (Figures 4E and 4F). The epicuticular wax on the wild-type inflorescence stem was mainly composed of columnar-shaped crystals, with a few structures of other shapes, including rods, vertical plates, and dendritic-like structures, whereas the epicuticular wax on the surface of the 35S:AtCFL1-6 inflorescence stem was mainly composed of rod- and tube-shaped structures (Figures 4E and 4F).
Figure 4.
Scanning Electron Microscopy and Transmission Electron Microscopy Analysis of At CFL1 Overexpression Plants and Rice cfl1 Mutants.
(A) and (B) Scanning electron micrograph of the abaxial surface of wild-type (A) and 35S:AtCFL1-6 (B) leaves. Arrowheads indicate paired stomata in (B).
(C) and (D) Scanning electron micrographs of the adaxial surface of wild-type (C) and 35S:AtCFL1-6 (D) leaves.
(E) and (F) Scanning electron micrographs of stems of the wild type (E) and 35S:AtCFL1-6 (F).
(G) to (J) Transmission electron micrographs of wild-type ([G] and [I]) and 35S:AtCFL1-6 ([H] and [J]) leaf epidermal cell sections. (I) and (J) are higher magnifications of boxed regions in (G) and (H), respectively; the cuticle is indicated with arrows.
(K) and (L) Scanning electron micrographs of leaves of wild-type (K) and cfl1 (L) plants at the four-leaf stage.
(M) and (N) Transmission electron micrographs of cuticle of the wild type (M) and cfl1 leaf (N).
Bars = 100 μm in (A) to (D), 10 μm in (E) and (F), 1 μm in (G) to (J), 2 μm in (K) and (L), and 1 μm in (M) and (N). CW, cell wall; cut, cuticle; WT, wild type.
To confirm that the cuticle was indeed affected, we examined the epidermis outer cell wall by transmission electron microscopy. Figures 4I and 4J show that the upper electron-dense membrane, representing the cuticle, covers a less dense layer that represents the cell wall. In the leaves of wild-type Arabidopsis plants, the cuticle is a continuous, condensed, and regular structure, the thin but electron-dense layer seen in Figures 4G and 4I. However, in 35S:AtCFL1-6 plants, the cuticle had a less osmiophilic membrane with reduced electron density (Figure 4H). The cuticle of 35S:AtCFL1-6 had a loose and fuzzy structure and was thicker than that of the wild type. The boundary between the cuticle and the cell wall was blurred in 35S:AtCFL1 plants, and the cuticle materials appeared to have invaded the cell wall (Figure 4J). Since the electron density signal is attributed to staining of lipid components by osmium tetroxide, the altered electron density and the deformed structure of the cuticle in 35S:AtCFL1-6 plants are probably a result of the change in the lipid components.
We then further examined the cuticle structure in rice cfl1 mutants. Unlike Arabidopsis, scanning electron microscopy analysis showed that the rice leaf epicuticular wax was mainly composed of vertical plate structures, with some WP scattered on the surface of the blade (Figure 4K). The wax on the leaf surface was markedly reduced in the rice cfl1 mutant (Figure 4L). In wild-type rice, the less dense layer representing the cell wall was covered by an electron-dense, continuous, uniform cuticle layer (Figure 4M). In cfl1 rice, this cuticle layer was less osmiophilic and more loose in appearance (Figure 4N), similar to the cuticle structure in 35S:AtCFL1 plants. Furthermore, a series of irregular layers was observed in the conjunction zone between the cuticle and cell wall, with some cavernous structures embedded in them (Figure 4N). These cavernous structures might represent a distortion of cell wall integrity due to inadequate protection by the defective cuticle in cfl1. These data suggest that both At CFL1–overexpressing plants and rice cfl1 mutants have impaired cuticle structure and composition and that CFL1 is possibly involved in cuticle development, the regulation of which is largely conserved in Arabidopsis and rice.
Cuticular Wax and Cutin Composition Are Severely Affected in 35S:AtCFL1 Plants
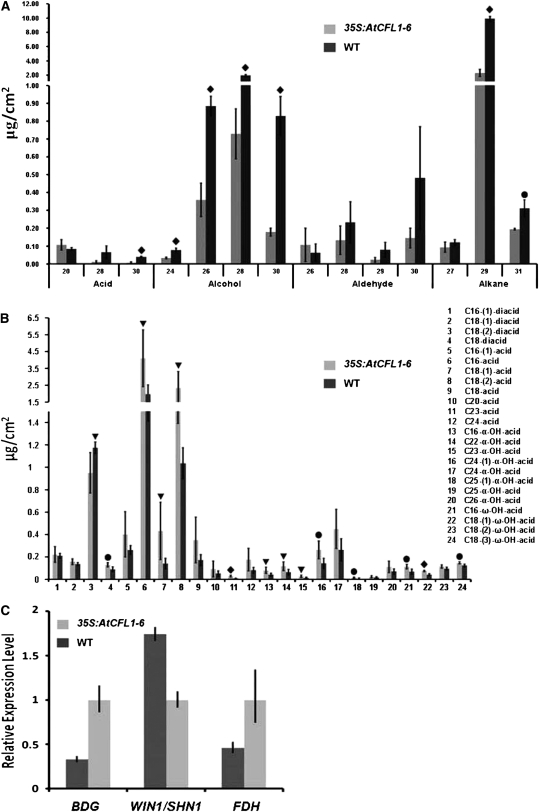
To further characterize the cuticle defects, we analyzed the cutin and wax composition of 35S:AtCFL1 and wild-type plants by gas chromatography–mass spectrometry (GC-MS) and gas chromatography–flame ionization detector (GC-FID). First, we compared the composition of wax between the stems of 35S:AtCFL1 and those of wild-type plants. In agreement with the observation that the amount of epicuticular wax crystals was reduced on the epidermis of the 35S:AtCFL1 inflorescence stem (Figures 4E and 4F), the GC-MS/GC-FID results showed that the total wax amount was significantly reduced to 7.762 ± 1.267 μg/cm2, less than one-third of the amount accumulated on the surface of wild-type stems (i.e., 25.776 ± 0.775 μg/cm2). Further analysis indicated that the amount of many components, such as C29 and C31 alkanes, C24, C26, C28, and C30 alcohols, and C30 fatty acid, was significantly reduced in the overexpression plants (Figure 5A).
Figure 5.
Cutin and Wax Composition and Gene Expression Analyses of Potential Target Genes.
(A) The cuticular wax composition of 35S:AtCFL1 and wild-type (WT) stems, in μg/cm2. Numbers indicate the chain length of substances arranged by substance classes. Each value is the mean ± sd of four replicates.
(B) Amounts of depolymerized cutin monomers in 35S:AtCFL1 and wild-type leaves, in μg/cm2. Each value is the mean ± sd of four replicates.
(C) Relative expression level of BDG, WIN1, and FDH in 35S:AtCFL1 and wild-type seedlings. The expression level of each gene in the wild type is set to 1.0, and error bars represent the sd of three biological replicates.
Analysis of the level of significance obtained with a Student's t test is marked by the following: triangle, P < 0.1; circle, P < 0.05; diamond, P < 0.01.
Then, we compared the composition of cutin monomers between the leaves of 35S:AtCFL1 and those of wild-type plants. We identified 24 compounds, 13 of which were significantly increased in 35S:AtCFL1. The amounts of C16, C18:1, C18:2, and C23 acids, which represent the major constituents of cutin fatty acid monomers in Arabidopsis, increased by 2.1-, 3.1-, 2.3-, and 3.8-fold, respectively, in 35S:AtCFL1. It is noteworthy that the amounts of most identified hydroxy acids, including five out of eight α-hydroxy acids and three out of four ω-hydroxy acids, were greater in 35S:AtCFL1 than in the wild type (Figure 5B).
Because loss-of-function mutants of either BDG, encoding an α/β-hydrolase fold protein, or FDH, encoding a KCS family protein, displayed similar phenotypes (i.e., organ fusion and altered cutin composition), we predicted that expression of BDG or FDH might be altered in At CFL1 overexpressor plants. Quantitative real-time PCR analysis showed that the expression level of both BDG and FDH was decreased in At CFL1 overexpressor plants. However, the expression of WIN1/SHN1, encoding an ethylene response transcription factor family member that causes a more permeable cuticle and increased amounts of major cutin monomers when overexpressed, was increased in 35S:AtCFL1 plants, consistent with the GC-MS results (Figure 5C). Our data suggest that the cuticular wax and cutin composition were severely affected by the overexpression of At CFL1, possibly through downregulation of BDG and FDH and/or upregulation of WIN1/SHN1.
At CFL1 Is Expressed in Cells Differentiated from Epidermal Cells
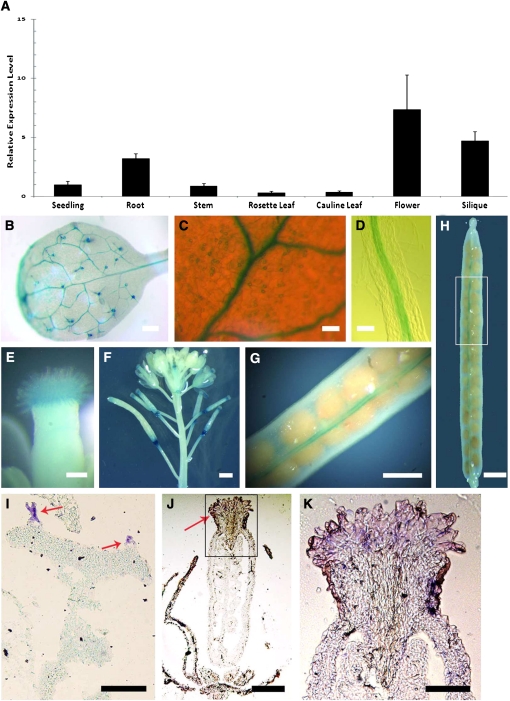
To investigate the relationship between At CFL1 and cuticular wax/cutin composition, we analyzed the expression pattern of At CFL1 in wild-type plants using quantitative RT-PCR. The expression of At CFL1 was detected in 10-d-old seedlings and in all organs of 6-week-old plants; the expression levels were relatively higher in roots, flowers, and siliques (Figure 6A). To further clarify the detailed expression pattern of At CFL1, we fused an ~1.6-kb fragment of the At CFL1 promoter to the Escherichia coli β-glucuronidase (GUS) reporter gene and transformed it into Arabidopsis. GUS activity was observed in trichomes (Figures 6B), guard cells (Figure 6C), root endodermis and central cylinder (Figure 6D), and stigmatic papillar cells (Figure 6E). When petals and sepals withered, we found strong GUS expression in the abscission zone at the bottom of the silique (Figure 6F). After that stage, At CFL1 expression became more prominent along the valve margin-replum boundary, where dehiscence and pod shatter occur (Figures 6G and 6H). These data suggest that the expression of At CFL1 is with abscission and dehiscence. RNA in situ hybridization analysis (Figures 6I to 6K) further confirmed the expression of At CFL1 in trichomes and stigmatic papillar cells. Interestingly, trichomes, guard cells, and stigmatic papillar cells are all differentiated from epidermal cells for particular functions (Lolle and Pruitt, 1999).
Figure 6.
Spatial Expression Pattern of At CFL1.
(A) Analysis of At CFL1 expression level in different organs by quantitative RT-PCR. The expression level in the seedling is set to 1.0, and error bars represent sd of three biological replicates.
(B) GUS-stained 5th true leaf of Arabidopsis.
(C) Magnification of a GUS-stained 5th true leaf of Arabidopsis.
(D) Longitudinal view of GUS-stained root from 10-d-old seedling.
(E) GUS-stained stigma.
(F) GUS-stained inflorescence stem with buds, flowers, and siliques.
(G) and (H) GUS-stained silique. (G) is a higher magnification of the boxed region in (H).
(I) to (K) Accumulation of At CFL1 transcripts, as visualized by in situ hybridization of the leaf tissues (I) and inflorescence ([J] and [K]). (K) is the higher magnification of the boxed region in (J). Arrows indicate trichomes in (I) and stigma in (J).
Bars = 1 mm in (B), (F), (H), and (I), 50 μm in (C), (D), and (K), 0.1 mm in (E), 20 μm in (K), and 0.5 mm in (G).
Loss of At CFL1 Function Results in Cuticle Biosynthesis in Trichomes in Arabidopsis
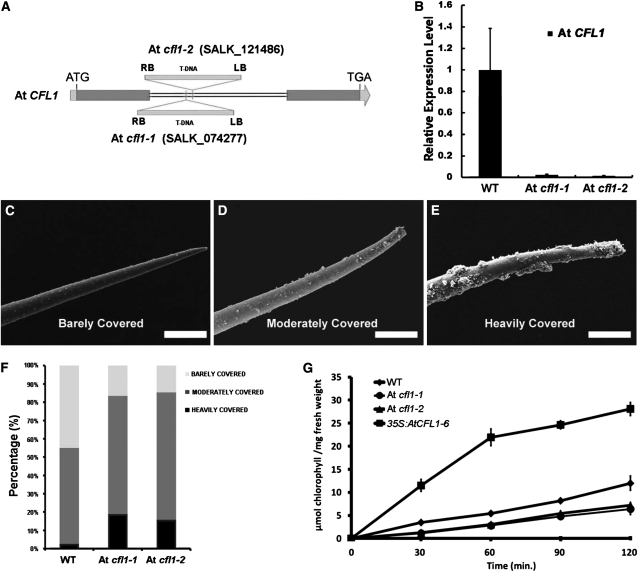
To define the physiological roles of At CFL1, we identified two SALK mutants (SALK_074277 and SALK_121486) from ABRC, later designated as At cfl1-1 and At cfl1-2, respectively. A single T-DNA is inserted in the only intron of At CFL1 in both At cfl1-1 and At cfl1-2 (Figure 7A). Quantitative RT-PCR analysis showed that the transcription of At CFL1 was decreased in At cfl1-1 and At cfl1-2 (Figure 7B). Although the morphological appearance of At cfl1 seedlings was similar to that of the wild type, scanning electron microscopy analysis showed that there was significantly more epicuticular wax on the surface of trichomes on the inflorescence stem of At cfl1-1 and At cfl1-2 (Figures 7C to 7F). However, the crystal density of the epicuticular wax on the epidermal surface was the same in wild-type and mutant plants (see Supplemental Figure 2 online). We grouped Arabidopsis trichomes into three types in terms of their epicuticular wax covering; barely, moderately, and heavily covered (Figures 7C to 7E). We analyzed 240 wild-type and 540 trichomes from the two mutants by scanning electron microscopy. The results showed that 64.7% of the trichomes were moderately wax covered and 16.5% were heavily covered in At cfl1-1, ~69.4% of the trichomes were moderately wax-covered and 15.8% were heavily covered in Arabidopsis At cfl1-2, whereas in the wild type, these percentages were 52.5 and 4.5%, respectively (Figure 7F). Because At CFL1 is specifically expressed in trichomes, it is reasonable to speculate that At CFL1 participates in cuticle development in trichomes.
Figure 7.
SALK T-DNA Insertion Mutant of At CFL1.
(A) T-DNA insertion sites in At CFL1. The arrow represents the trancriptional orientation of the gene. LB, T-DNA left border; RB, T-DNA right border. Blue boxes, exons; gray boxes, untranslated regions; double lines, introns.
(B) Relative expression level of At CFL1 in wild-type (WT), At cfl1-1 (SALK_074277), and At cfl1-2 (SALK_121486) plants. The expression level in the wild type is set to 1.0, and error bars represent the sd of three biological replicates.
(C) to (E) Scanning electron microscopy of Arabidopsis stem trichome barely covered (C), moderately covered (D), and heavily covered (E) with wax. Bars = 25 μm.
(F) Relative percentage of each kind of trichome in the wild type and SALK mutants.
(G) Chlorophyll leaching assays with mature rosette leaves of the wild type, SALK mutants, and 35S:AtCFL1-6 submerged in 80% ethanol for different time intervals. The results are from three independent assays.
To confirm these findings, we conducted a chlorophyll leaching assay to test whether the cuticular membrane properties were altered in At cfl1 mutants. The aerial parts of wild-type, At cfl1-1, and At cfl1-2 plants were submerged in 80% ethanol for different periods of time, and the chlorophyll concentration was then determined. The results showed that chlorophyll was extracted significantly more slowly from the At cfl1 mutants than from wild-type plants (Figure 7G), indicating that the cuticle of At cfl1 mutants is more resistant to ethanol. These data suggest that the cuticular membrane properties were enhanced in At cfl1-1 and At cfl1-2.
At CFL1 May Interact with HDG1
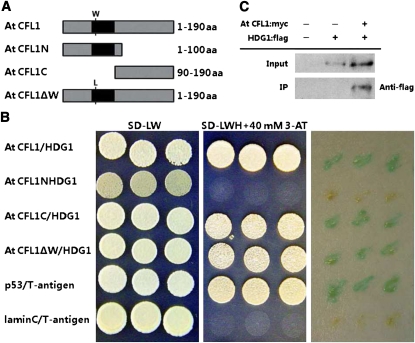
To investigate how At CFL1 is associated with cuticle biosynthesis in Arabidopsis, we performed a yeast two-hybrid system assay using full-length At CFL1 as bait. We screened 3.4 million colonies from a normalized cDNA library constructed from 14-d-old Arabidopsis seedlings and identified seven clones that were positive for HIS3 and LacZ expression, three of which were found to carry HDG1 (see Supplemental Table 2 online). HDG1 is a member of the class IV HD-ZIP gene family, which has 16 members in Arabidopsis (Nakamura et al., 2006). Some gene members of this family, such as GLABRA2 (GL2), ANTHOCYANINLESS2 (ANL2), PROTODERMAL FACTOR2 (PDF2), and ARABIDOPSIS THALIANA MERISTEM LAYER1, are involved in the development of the epidermis or epidermis-associated structures, such as trichomes and root hairs (Rerie et al., 1994; Kubo et al., 1999; Abe et al., 2003; Ohashi et al., 2003). To determine which domain of At CFL1 is responsible for its interaction with HDG1, we generated two deletions, the N-terminal 100 amino acid residues (At CFL1N) and the C-terminal 90–amino acid residues (At CFL1C), and a point mutation that substituted the first conserved Trp residue in the WW domain with a Leu residue (At CFL1ΔW), and then tested their abilities to interact with HDG1 (Figure 8A). The results showed that At CFL1C and At CFL1ΔW could interact with HDG1, whereas At CFL1N could not (Figure 8B), suggesting that the C terminus is responsible for the At CFL1-HDG1 interaction and that the first conserved Trp residue in the WW domain is not important for the interaction. In the C terminus, we found an unknown motif that is conserved between At CFL1 and its homolog At CFL2, but its exact function needs to be further investigated.
Figure 8.
Interaction between At CFL1 and HDG1.
(A) Schematic representation of the peptides used for yeast two-hybrid interaction assay. From top to bottom, the diagram represents the full sequence of At CFL1, N-terminal peptide of At CFL1, C-terminal peptide of At CFL1, and full sequence of At CFL1 with one amino acid (aa) mutated from Trp (W) to Leu (L).
(B) Yeast two-hybrid interaction analysis of At CFL1, At CFL1N, At CFL1C, and At CFL1ΔW with HDG1. Transformed yeast strains were plated on SD-LW and SD-LWH medium and colony-lift filter assays performed with X-gal, from left to right, respectively. Interaction between p53 and T-antigen was used as a positive control and interaction between lamin C and T-antigen as a negative control.
(C) Immunoprecipitation assay between Arabidopsis HDG1 and At CFL1 in vivo. Lane 1, no input; lane 2, HDG1:flag; lane 3, At CFL1:myc and HDG1:flag.
To further confirm the At CFL1-HDG1 interaction, we performed a coimmunoprecipitation (Co-IP) experiment. Myc-tagged At CFL1 and FLAG-tagged HDG1 were coexpressed in leaves of Nicotiana benthamiana, and then total proteins were extracted for Co-IP and immunoblot analyses. The results showed that HDG1-FLAG coimmunoprecipitated with myc-At CFL1 (Figure 8C), suggesting that HDG1 interacts with At CFL1 in vivo.
HDG1 Chimeric Repressor Plants Resemble the TB Staining and Organ Fusion Phenotypes of 35S:AtCFL1 Plants
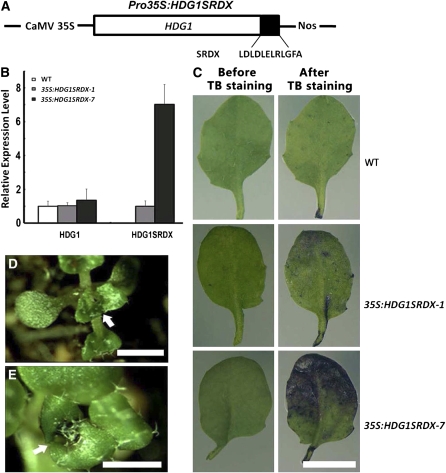
To further confirm this interaction genetically, we first examined the phenotypes of the hdg1 mutants from SALK (SALK_062171 and SALK_147739) but found no abnormal phenotypes, possibly due to functional redundancy within the HD-ZIP IV gene family. This is consistent with the report that the double mutant of HDG1 and its closest homolog ANL2 exhibited no additional abnormal phenotypes to those reported for ANL2 (Nakamura et al., 2006). Therefore, we used chimeric repressor silencing technology to knock down expression of HDG1 by fusing it with the EAR motif repression domain driven by the CaMV 35S promoter (Figure 9A). The chimeric construct was designated HDG1SRDX and transformed into wild-type Arabidopsis. We chose two transgenic lines, 35S:HDG1SRDX-1 and 35S:HDG1SRDX-7, for further analyses. Quantitative RT-PCR analysis showed that, in 2-week-old seedlings, endogenous HDG1 was expressed in the two transgenic lines at the same level as in the wild type and that HDG1SRDX transcripts were more abundant in 35S:HDG1SRDX-7 than in 35S:HDG1SRDX-1 (Figure 9B). When leaves of the two transgenic lines were stained using the TB staining assay, those of 35S:HDG1SRDX-1 were partially stained by TB, while those of 35S:HDG1SRDX-7 were strongly stained (Figure 9C). The finding that the intensity of TB staining was positively correlated with HDG1SRDX expression level suggests that the phenotypes of the 35S:HDG1SRDX plants are not likely due to cosuppression effects on HDG1. Interestingly, we also observed organ fusion between leaves of these transgenic plants, resembling that seen with 35S:AtCFL1 plants (Figures 9D and 9E). These data suggest that the HDG1 chimeric repressor plants are also defective in cuticle development.
Figure 9.
Overexpression of the Chimeric HDG1 Repressors Result in Cuticle Defects.
(A) Schematic representation of the construct used for expression of the chimeric HDG1 repressors. CaMV 35S, SRDX, and NOS represent the CaMV 35S promoter, the repression domain of 12 amino acids, and the NOS terminator sequence, respectively.
(B) The relative expression level of HDG1SRDX transgene and endogenous HDG1 in 35S:HDG1SRDX plants. The expression levels of HDG1 in the wild type (WT) and HDG1SRDX in 35S:HDG1SRDX-1 are set to 1.0, and error bars represent sd of three biological replicates.
(C) TB staining assay of rosette leaves. From top to bottom, the wild type, 35S:HDG1SRDX-1, and 35S:HDG1SRDX-7. Bars = 1 cm.
(D) and (E) Fusion of leaves observed in 35S:HDG1SRDX plants. Arrows indicate sites of organ fusion. Bars = 1 cm.
At CFL1 Is Epistatic to HDG1
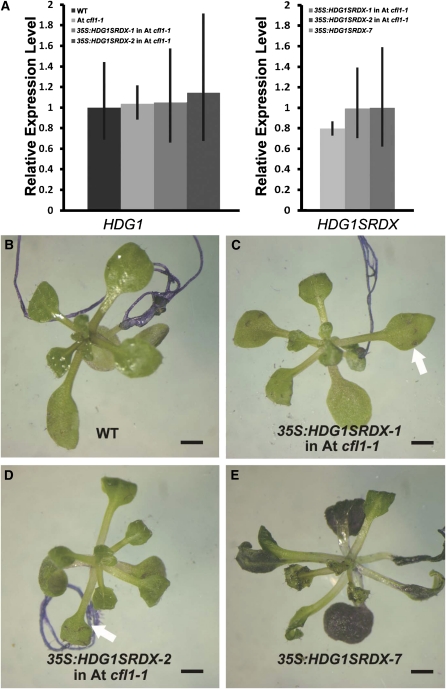
To clarify the genetic relationship between HDG1 and At CFL1, we transformed the HDG1SRDX construct into the At cfl1-1 mutant. We chose two transgenic lines that exhibited a similar level of HDG1SRDX transcript as those in the 35S:HDG1SRDX-7 plants (Figure 10A) and applied a TB staining assay. We found that TB staining intensity was dramatically alleviated in the 35S:HDG1SRDX plants with At cfl1-1 background compared with that in the 35S:HDG1SRDX-7 plants (Figures 10B to 10E), suggesting that the cuticle development defect in the wild-type plants caused by the HDG1 chimeric repressor was largely rescued in the At cfl1-1 mutant. These data suggest that At CFL1 and HDG1 are involved in the same genetic pathway to regulate cuticle development and that the function of HDG1 is possibly dependent on the presence of At CFL1.
Figure 10.
Overexpression of the Chimeric HDG1 Repressor in At cfl1-1 Rescued the Cuticle Defects Observed in 35S:HDG1SRDX-7 Plants.
(A) The relative expression level of HDG1SRDX transgene (right) and endogenous HDG1 (left) in 35S:HDG1SRDX plants in a wild-type (WT) and At cfl1-1 mutant background. The expression levels of HDG1 in the wild type and HDG1SRDX in 35S:HDG1SRDX-7 are set to 1.0, and error bars represent sd of three biological replicates.
(B) to (E) TB staining assay of seedlings. Wild-type seedlings did not stain with TB (B) and 35S:HDG1SRDX-7 plants did stain (E), while the staining was much alleviated in the At cfl1-1 mutant ([C] and [D]). Bar = 0.5 cm.
HDG1 Binds to the L1 Box and BDG and FDH May Be Target Genes of HDG1
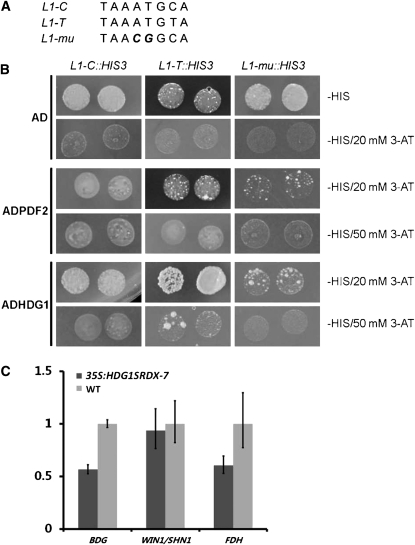
At ML1 and PDF2 bind to the L1 box 5′-TAAATGYA-3′ (Abe et al., 2001, 2003). Since HDG1 belongs to the same subgroup in the HD-ZIP family, we performed a yeast one-hybrid assay to test whether HDG1 could bind to the L1 box. We synthesized three DNA fragments for the assay: two were identical to the consensus sequence, 5′-TAAATGCA-3′ (as L1-C) and 5′-TAAATGTA-3′ (as L1-T), while the other contained two point mutations, 5′-TAACGGCA-3′ (as L1-mu) (Figure 11A). The results showed that the positive control PDF2 bound to both L1-C and L1-T sequences when selected with either 20 or 50 mM 3-aminotriazol (3-AT) but did not bind to the Li-mu sequence (Figure 11B). HDG1 bound to both L1-C and L1-T sequences when selected with 20 mM 3-AT, but it did not bind to the L1-T sequence when selected with 50 mM 3-AT (Figure 11B). This slight difference in binding specificity is consistent with the idea that the function of HDG1 has diversified from those of other family members, such as PDF2.
Figure 11.
HDG1 Binds to the L1-Box.
(A) Sequences of target DNA. Italic letters in third line indicate substitutions.
(B) Interaction of HDG1 with the sequences. AD with no fusion protein was used as negative control and ADPDF2 as positive control.
(C) Relative expression level of BDG, WIN1, and FDH in 35S:HDG1SRDX-7 and wild-type seedlings. The expression level of each gene in the wild type (WT) is set to 1.0, and error bars represent sd of three biological replicates.
Since, as mentioned above, genes such as BDG, FDH, and WIN1/SHN1 were reported to play important roles in cuticle development, we scanned the promoter sequence of those genes with PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/) and identified two L1-boxes, located at ~700 and 270 bp upstream of the start codon of BDG and FDH, respectively, but did not detect an L1-box in the promoter region of WIN1/SHN1 (Higo et al., 1999). Quantitative real-time PCR analysis showed that BDG and FDH were downregulated in HDG1 chimeric repressor plants as they were in At CFL1 overexpression plants, but WIN1/SHN1 was not changed significantly (Figure 11C). These data support the idea that BDG and/or FDH are direct targets of HDG1.
In conclusion, our data suggest that At CFL1 negatively regulates cuticle development in both rice and Arabidopsis by regulating the activity of HDG1, which regulates the downstream genes BDG and FDH.
DISCUSSION
In this work, we present evidence that rice and Arabidopsis CFL1 are involved in the regulation of cuticle development. We show that At CFL1 negatively regulates cuticle development by directly interacting with and modulating the activity of HDG1. CFL1 belongs to a novel class of plant-specific genes that encode WW domain–containing proteins and is conserved in many plant species, including sorghum (Sorghum bicolor), maize, poplar (Populus tremula), grape (Vitis vinifera), and soybean (Glycine max). No CFL1 homologs have yet been reported in moss or algae; both moss and algae are poikilohydric organisms where a water-impermeable cuticle is not needed or is even nonexistent. Whether lack of the CFL1-mediated regulatory pathway is associated with the lack of a water-impermeable cuticle in moss and algae needs further investigation.
CFL1 is a WW domain protein. In yeast and vertebrates, the WW domain binds proteins containing particular Pro motifs. For example, the WW domain of Yes-associated protein binds to the SH3 domain of the Yes oncoprotein via a Pro-rich region (Chen and Sudol, 1995; Sudol et al., 1995). The WW domain is also frequently associated with proteins involved in signal transduction; for example, in the rat transcription factor activator FE65, the WW domain coincides with its activator region (Ermekova et al., 1997; Meiyappan et al., 2007). In Arabidopsis, there are 13 genes that encode proteins containing a WW domain, most of which are functionally unidentified. FLOWERING LOCUS C (FCA) and SUPPRESSOR OF ACTIN9 (SAC9) are the few exceptions. FCA is involved in the promotion of the transition of the vegetative meristem to reproductive development, whereas SAC9, encoding a phosphoinositide phosphatase, has a role in stress responses (Macknight et al., 1997; Williams et al., 2005). The WW domains of CFL1 differ from typical WW domains, in that the spacing between the three conserved residues, Trp, Trp, and Pro, is shorter (Sudol et al., 1995). Because deletion of the WW domain or mutation of the conserved Trp residue did not affect the interaction between At CFL1 and HDG1, it is possible that At CFL1 may interact with other proteins to implement its function through this atypical WW domain.
Similar to some cer mutants and most previously isolated cuticular defective mutants, 35S:AtCFL1 plants displayed enhanced TB staining, increased chlorophyll leaching, and decreased amounts of epicuticular wax crystals in stems. However, more cutin accumulated in the epidermis of the leaves. This is consistent with other reports: for example, Arabidopsis plants overexpressing WIN1/SHN1 and its homolog exhibited phenotypes such as glossy leaves, increased drought tolerance, and increased cuticle permeability to ethanol, but the amounts of some major constituents of cutin were substantially increased (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). Loss of function of either FDH or LCR, two genes involved in cutin and wax biosynthesis, resulted in plants with defective cuticles, but the amounts of some major cutin components were increased in the mutants (Voisin et al., 2009). Another example is BDG; loss of BDG function resulted in a series of morphological and cuticular defects, but the amount of residual bound lipids and wax accumulation were increased in leaves of bdg. That 35S:AtCFL1 plants showed increases in the amounts of 13 of the 24 compounds found in cutin suggests that cuticle-defective phenotypes may not be caused by the decreased amount of cutin composition. The altered structure and changed composition of the cuticle and the status of epidermal cells may also contribute to the cuticle-defective phenotypes.
HDG1 belongs to the class IV HD-ZIP family (also known as the HD-GL2 family after the first identified member, GL2) (Rerie et al., 1994). GL2 is required for the specification of root hairless cells and for trichome outgrowth from the shoot epidermis (Ohashi et al., 2003). The functions of other genes in this class are also closely associated with the epidermis. For example, ANL2 regulates the accumulation of anthocyanin in subepidermal and epidermal cells in the shoot, and extra cells were produced between the epidermal and cortical layer in the roots of anl2 plants (Kubo et al., 1999). The surface of the pdf2 atml1 double mutant appeared to lack an epidermis (Abe et al., 2003). There is increasing evidence that HD-ZIP IV transcription factors have roles in plant cuticle development. Recently, a gene annotated as a homolog of ANL2 was identified in tomato, and mutation of this gene altered the cutin content (Isaacson et al., 2009). Moreover, it was reported that overexpression of the HD-ZIP IV transcription factor Outer Cell Layer1 in maize affected the expression level of several genes involved in lipid metabolism and cuticle development, in some cases directly (Javelle et al., 2010). Our data suggest that the function of HDG1 is also associated with epidermis development and cuticle development. Neither overexpression nor mutation of HDG1 resulted in developmental defects, suggesting that functional redundancy exists among the members of this class. Consistent with this idea, the double mutant of HDG1 and ANL2, the closest homolog of HDG1, produced no observable morphological changes (Nakamura et al., 2006). The relationships among members of the class IV HD-ZIP gene family are intriguing, since there are 16 members in this class, but most members have overlapping but distinct roles in epidermis development in Arabidopsis, similar to class III members of this gene family (Prigge et al., 2005).
The relationship between At CFL1 and HDG1 is very interesting. Our data showed that At CFL1 was able to interact with HDG1 and that HDG1 chimeric repressor plants resemble 35S:AtCFL1 plants in terms of the TB staining and organ fusion phenotypes. The transcription level of HDG1 was not changed in At cfl1-1 mutants nor in At CFL1 overexpression plants. HDG1 activity does not appear to feed back on its own expression because we found that HDG1 expression level was not significantly changed in HDG1 chimeric repressor plants and that no L1-box was found in the promoter of HDG1. Our genetic data also showed that the cuticle development defects resulting from the HDG1 chimeric repressor were more severe in wild-type plants than in the At cfl1-1 mutant, suggesting that the inhibitory function of HDG1 was At CFL1 dependent. All these biochemical and genetic data indicate that At CFL1 probably serves as an enhancer of HDG1 activity inhibition and that this regulation happens through protein–protein interaction rather than through changes in gene transcription.
Recently, HDG1 was reported to possess a possible EAR motif in its N-terminal region, with the help of a combination of novel algorithms (Kagale et al., 2010). However, it seems that this EAR motif does not inhibit the activity of HDG1 because overexpression of HDG1 in wild-type Arabidopsis plants did not result in similar phenotypes as 35S:AtCFL1 or 35S:HDG1SRDX plants. One possible explanation is that the EAR motif itself is too short (i.e., five to six amino acid residues) to implement function and that the proper functioning of the EAR motif requires the flanking sequences and/or tertiary structure or assistance by some unknown cofactors. Because the EAR motif is in the middle of HDG1 rather than at the C terminus (Kagale et al., 2010), it is also possible that this EAR motif is not well situated to suppress HDG1 activity efficiently or that the suppression requires the presence of At CFL1.
In the expression pattern analysis, we found strong GUS expression driven by the At CFL1 promoter in places where abscission and dehiscence occurred. This expression pattern resembles those of the SHINE transcription factor genes (Broun et al., 2004; Kannangara et al., 2007). Abscission and dehiscence are two physiological processes that involve cell separation in which specific layers are preprogrammed to differentiate and distinguish from neighboring cells. In these processes, formation of an anatomically different protective layer is required, often with deposition of heterogeneous lipophilic materials (e.g., suberin). The expression pattern coincidence of At CFL1 and the SHINE clade transcription factor genes suggests that cuticle modification plays an important role in abscission and dehiscence. However, it is interesting to note that the expression level of WIN1/SHN1 was not changed in the HDG1 overexpressor plants or in 35S:HDG1SRDX plants, suggesting that WIN1/SHN1 is probably not regulated by HDG1. Taking into consideration that the expression level of WIN1/SHN1 changed significantly in 35S:AtCFL1 plants and that no L1-box was found in the promoter, introns, and 3′-untranslated region of WIN1/SHN1, it is possible that At CFL1 regulates the expression of WIN1 through a route other than HDG1. It is therefore reasonable to conclude that At CFL1 plays a central role in regulating cuticle development in the abscission and dehiscence regions by directly interacting with and regulating HDG1 and/or by regulating WIN1/SHN1. It seems that the regulation of cuticle development is much more complicated than expected, and it would be interesting to investigate the specific factors involved in the At CFL1–mediated regulation of WIN1/SHN1.
The epidermis is the most important interface between plants and the environment. Many mutants with cuticle defects also show a defective epidermis and vice versa. We showed that At CFL1 not only had a role in the correct formation of the cuticle but also played a role in epidermis development. Although regulation of the downstream cutin biosynthesis genes by At CFL1 and HDG1 is reasonably acceptable, an alternative explanation does exist and that is that the status of epidermal cells could change, resulting in changes in the composition and structure of the cuticle. In addition, it was proposed that small molecular derivatives from the cuticle could affect epidermal cell differentiation. Hence, At CFL1 is also possibly involved in regulating the status of epidermal cells. The composition and structure of the cuticle that covers the epidermis varies temporally and spatially and should be compatible with environmental factors and developmental stages. Therefore, the regulation of cuticle development and epidermis development is extremely complicated. Nevertheless, the CFL1-mediated regulation pathway is pivotal to both cuticle and epidermis development, and additional environmental or developmental cues probably determine the final direction of the pathway. The phenotypic similarity between rice cfl1 mutants and 35S:AtCFL1 overexpressor plants suggests that the regulatory pathway for cuticle development likely evolved before the divergence of monocots and dicots and remains highly conserved.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia-0) and rice var Zhonghua 11 (Oryza sativa ssp japonica) were used as the wild type. The rice mutant cfl1 was obtained from the Shanghai T-DNA Insertion Population generated by Hong-Wei Xue (Fu et al., 2009). Rice plants were grown in the field or in the greenhouse at 30°C/25°C day/night cycles. The Arabidopsis T-DNA insertion mutant SALK_074277 was obtained from the ABRC, and the left border of the T-DNA was confirmed by sequencing. Arabidopsis plants were grown on half-strength Murashige and Skoog medium, with pH adjusted to 5.7 with 1 mol/L KOH, 0.6% (w/v) phytoagar containing 1% Suc, or on soil in the greenhouse under long-day conditions (16-h-light/8-h-dark cycle) at 22 to 23°C.
TAIL-PCR and DNA Gel Blot Analyses
The flanking sequence of the T-DNA insertion in cfl1 was determined by TAIL-PCR using the specific and arbitrary degenerate primers as described (Qin et al., 2003). Genomic DNA was extracted from wild-type and cfl1 plants, and 15 μg of each sample was used for DNA gel blots as described (Wu et al., 2003).
Vector Construction and Transformation
The open reading frames of CFL1 were amplified from rice cDNA with primers CFL1-F and CFL1-R and from Arabidopsis with primers AtCFL1-F and AtCFL1-R, respectively, by RT-PCR. The Arabidopsis CFL1 promoter was amplified from genomic DNA with primers Pro-AtCFL1-F and Pro-AtCFL1-R. These products were cloned into the EcoRV site of pBluescript SK+ (pBS) (designated pBS-CFL1, pBS-AtCFL1, and pBS-ProAtCFL1, respectively) and sequenced.
The CFL1 overexpression construct was generated by ligation of the XhoI-SpeI fragment from pBS-CFL1 and the SalI-SpeI fragment from the pAP1380 plasmid. The pAP1380 construct was generated by cloning the EcoRV-SmaI fragment from pActin1-D into the SmaI site of pCAMBIA 1380 (McElroy et al., 1990; McElroy et al., 1991). 35S:AtCFL1 was constructed by ligation of the XhoI-SpeI fragment from pBS-AtCFL1 and the XhoI-SpeI fragment from pJim19. The SalI-BglII fragment of pBS-ProAtCFL1 was cloned into pCAMBIA 1381Xa (Cambia) that had been digested with SalI-BglII to fuse it with the GUS reporter gene.
For the chimeric repressor construct, the HDG1 coding sequence was amplified from Arabidopsis cDNA by RT-PCR with primers HDG1SRDX-F and HDG1SRDX-R and cloned into the EcoRV site of pBS (designated pBS-HDG1SRDX). The 35S:HDG1SRDX was generated by ligation of the following DNA fragments: the BglII-SpeI fragment from pBS-HDG1SRDX, the PstI-BglII 35S promoter fragment from pCAMBIA1302, and the PstI-BglII fragment from pCSRDX (Guo et al., 2009).
Constructs were transformed into Agrobacterium tumefaciens GV3101/pMP90 using the freeze-thaw procedure and plant transformation was conducted as previously reported (Qin et al., 2005).
Microscopy
The fourth leaf of rice and the first pair of true leaves of Arabidopsis were excised for imaging the epidermal surface by scanning electron microscopy. Samples were fixed and dehydrated as described (Guo et al., 2009). The samples were CO2 critical point dried for 3 h (Hitachi critical point dryer, HCP-2; Hitachi Koki) and coated with gold powder, then examined under a scanning electron microscope (Hitachi S3000N). To see the epicuticular wax particles on the surface, Arabidopsis inflorescence stems and rice leaves were mounted directly and observed.
For transmission electron microscopy, rice and Arabidopsis leaves were collected and fixed in 2% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 6.8, for 3 h at room temperature and then for 24 h at 4°C. Samples were washed three times with 0.1 M sodium phosphate buffer and refixed in 1% OSO4 for 1 h and then dehydrated by a series of graded acetone. Samples were then embedded in a complete resin mixture (Spi-chem Spurr) and incubated at 70°C for 9 h before they were sectioned using a Leica EM UC6 ultramicrotome and stained with uranyl acetate and lead stain solutions. Sections were observed with a JEM-100CX-II (JEOL).
Total RNA Isolation and Quantitative RT-PCR Analysis
Total RNA was extracted from frozen material using TRIzol reagent and then treated with RNase-free DNase (TaKaRa) to eliminate genomic DNA. Total RNA was reverse transcribed using the Superscript II RT kit (Invitrogen), and the cDNA was used as the template for RT-PCR or quantitative RT-PCR as described (Liu et al., 2008).
Real-time PCR was performed as described (Xing et al., 2007) on an MJ Research thermocycler using SYBR Green Realtime PCR Master Mix (TOYOBO). Primer sequences are listed in Supplemental Table 1 online. Gene expression values were standardized to the expression level of TUB2. For each sample, at least three replications were performed in one experiment. The relative expression level of each gene was calculated using the cycle threshold (CT) 2−ΔΔCT method (Livak and Schmittgen, 2001).
In Situ Hybridization
In situ hybridization experiments were performed as previously described with some modifications (Liu et al., 2008). Arabidopsis inflorescence tissues were collected from 40-d-old wild-type plants, whereas leaves and roots were collected from 14-d-old seedlings. A 534-bp fragment that included the second exon and 3′-untranslated region of At CFL1, which was amplified with the primer CFL1-IS-F and CFL1-IS-R, was used as the probe.
Staining with TB and Chlorophyll Leaching
Arabidopsis plants were grown on culture medium for 3 to 4 weeks before being stained as described (Tanaka et al., 2004). Rosette leaves from wild-type and mutant plants were submerged in 0.05% solution of TB at room temperature for 2 min for Arabidopsis or 10 min for rice. Leaves were rinsed with tap water before being photographed.
Rosette leaves of 4-week-old Arabidopsis plants were used for the chlorophyll-leaching assay. Rosette leaves were weighed and submerged in 30 mL of 80% ethanol at room temperature in the dark with gentle shaking. Four-hundred microliters of the extract solution was removed from each sample every 30 min over a 2-h period. Samples were measured at 664 and 647 nm, and calculations were performed as described (Lolle et al., 1997).
Analysis of Residual Bound Lipids and Wax
Composition analyses of cuticular lipid polyesters and wax were performed as described (Kurdyukov et al., 2006a, 2006b).
Yeast Two-Hybrid Screening and Co-IP Assay
The full-length cDNA of At CFL1 was cloned into the pGBKT7 vector (Clontech) and transformed into yeast strain AH109. The resulting yeast cells were transformed with plasmids from a cDNA library constructed from 14-d-old Arabidopsis seedlings. Transformants (~3.4 × 106) were screened on medium that lacked His but contained 10 mM of 3-AT. Surviving clones were assayed for LacZ activity. The prey plasmids were isolated from these HIS3+ LacZ+ yeast clones and transformed into Escherichia coli and then sequenced for confirmation.
Tobacco (Nicotiana benthamiana) leaves were used for the Co-IP assay. Individual Agrobacterium colonies transformed with the appropriate constructs were grown at 30°C for 20 h in 50 mL medium with 20 μM acetosyringone. The bacteria were pelleted by centrifugation, resuspended in infiltration medium (10 mM MgCl2/10 mM MES, pH 5.7/150 μM acetosyringone) to OD600 = 0.5 to 1.0, and incubated at room temperature for ~3 h. The solution was then injected into tobacco leaves by a syringe through an incision. Tobacco leaves expressing At CFL1:myc and HDG1:flag were ground in IP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM PMSF, and 0.1% Nonidet P-40) and centrifuged at 1200 rpm for 10 min. Supernatant (600 μL) was incubated with anti-myc antibody coupled to Protein A sepharose beads (Amersham) for 30 min, and the beads were washed three times with IP buffer. Then bound proteins were eluted with 2× SDS buffer. All procedures were performed on ice.
Yeast One-Hybrid Assays
For the yeast one-hybrid assay, a yeast reporter strain was prepared. Oligonucleotide primer pairs (Eco-L1C-Xba and C-Eco-L1C-Xba; Eco-L1T-Xba and C-Eco-L1T-Xba) containing three tandem repeated L1 box sequences were annealed and inserted into the EcoRI and XbaI sites upstream of the HIS3 minimal promoter in pHISi-1 (Clontech) to form pHISi-1-L1CG and pHISi-1-L1TA, respectively. Mutated oligonucleotide pairs Eco-L1m-Xba and C-Eco-L1m-Xba were also used to form pHISi-1L1m. After linearization at the XhoI site, each of the plasmids was integrated in the his3 locus of the YM4271 host strain to generate new reporter strains. The full-length cDNA of HDG1 and PDF2 was cloned into the pDEST22 vector (Invitrogen) and transformed into the reporter strains. The transformed yeast cells were grown on medium that lacked His but contained 0, 20, or 50 mM of 3-AT. Growth status of the yeast was observed to determine the binding activity.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, Rice Genome Annotation Project, or GenBank/EMBL databases under the following accession numbers: CFL1 (Os02g31140), At CFL1 (At2g33510), At CFL2 (At1g28070), HDG1 (At3g61150), PDF2 (At4g04890), WIN1/SHN1(At1g15360), BDG (At1g64670), and FDH (At2g26250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Linkage Analysis of the T-DNA Insertion and Curly Leaf Phenotype.
Supplemental Figure 2. Scanning Electron Micrograph of the Stems of the Wild Type and At cfl1-1.
Supplemental Table 1. Primer Information for Genes Used in This Study.
Supplemental Table 2. Seven Positive Clones Identified from Yeast Two-Hybrid Analysis.
Supplemental Data Set 1. The Amino Acid Sequence Alignment Used to Generate the Phylogeny of CFL and Its Homolog Proteins.
Acknowledgments
We thank Hongwei Xue (Institute of Plant Physiology and Ecology, CAS) for providing the rice cfl1 mutant and Sheila McCormick (University of California, Berkeley, CA), Matthew Terry (University of Southampton), and Xing-Wang Deng (Yale University) for helpful suggestions and valuable discussions. The work was supported by the National Hi-Tech R&D Program of China (Grant 2006AA10A101) and the National Natural Science Foundation of China (Grant 30625002 to L.-J.Q.) and partially supported by the 111 Project. Financial support by the Deutsche Forschungsgemeinshaft to L.S. is also acknowledged.
AUTHOR CONTRIBUTIONS
R.W. and L.-J.Q. designed research. F.W. and L.S. performed research on cutin and wax composition analysis. R.W., S.L., S.H., C.Y., and G.Q. performed the other parts of the research. R.W., L.-J.Q., and H.G. analyzed data and wrote the article.
References
- Abe M., Katsumata H., Komeda Y., Takahashi T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 [DOI] [PubMed] [Google Scholar]
- Abe M., Takahashi T., Komeda Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26: 487–494 [DOI] [PubMed] [Google Scholar]
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach L., et al. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 105: 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F., Wu X., Li F., Haslam R.P., Markham J.E., Zheng H., Napier J.A., Kunst L. (2009). Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire M., Borel S., Fabre G., Carraça L., Efremova N., Yephremov A., Cao Y., Jetter R., Jacquat A.-C., Métraux J.-P., Nawrath C. (2011). A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire M., Chassot C., Jacquat A.C., Humphry M., Borel S., Petétot J.M.C., Métraux J.P., Nawrath C. (2007). A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird S.M., Gray J.E. (2003). Signals from the cuticle affect epidermal cell differentiation. New Phytol. 157: 9–23 [DOI] [PubMed] [Google Scholar]
- Blacklock B.J., Jaworski J.G. (2006). Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem. Biophys. Res. Commun. 346: 583–590 [DOI] [PubMed] [Google Scholar]
- Broun P., Poindexter P., Osborne E., Jiang C.-Z., Riechmann J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.I., Sudol M. (1995). The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 92: 7819–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermekova K.S., Zambrano N., Linn H., Minopoli G., Gertler F., Russo T., Sudol M. (1997). The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J. Biol. Chem. 272: 32869–32877 [DOI] [PubMed] [Google Scholar]
- Fiebig A., Mayfield J.A., Miley N.L., Chau S., Fischer R.L., Preuss D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F.-F., Ye R., Xu S.-P., Xue H.-W. (2009). Studies on rice seed quality through analysis of a large-scale T-DNA insertion population. Cell Res. 19: 380–391 [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean S., Ingram G.C. (2003). The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130: 4249–4258 [DOI] [PubMed] [Google Scholar]
- Guo Y., Qin G., Gu H., Qu L.-J. (2009). Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21: 3518–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T., Kosma D.K., Matas A.J., Buda G.J., He Y., Yu B., Pravitasari A., Batteas J.D., Stark R.E., Jenks M.A., Rose J.K.C. (2009). Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J. 60: 363–377 [DOI] [PubMed] [Google Scholar]
- Javelle M., Vernoud V., Depege-Fargeix N., Arnould C., Oursel D., Domergue F., Sarda X., Rogowsky P.M. (2010). Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 154: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M.A., Eigenbrode S.D., Lemieux B. (2002). Cuticular waxes of Arabidopsis. In The Arabidopsis Book 1: e0016, doi/10.1199/tab.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J., Raffaele S., Bourdenx B., Garcia C., Laroche-Traineau J., Moreau P., Domergue F., Lessire R. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara R., Branigan C., Liu Y., Penfield T., Rao V., Mouille G., Höfte H., Pauly M., Riechmann J.L., Broun P. (2007). The transcription factor WIN1/SHN1 regulates Cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Peeters A.J.M., Aarts M.G.M., Pereira A., Koornneef M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov S., Faust A., Nawrath C., Bär S., Voisin D., Efremova N., Franke R., Schreiber L., Saedler H., Métraux J.-P., Yephremov A. (2006b). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov S., Faust A., Trenkamp S., Bär S., Franke R., Efremova N., Tietjen K., Schreiber L., Saedler H., Yephremov A. (2006a). Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224: 315–329 [DOI] [PubMed] [Google Scholar]
- Kuromori T., Ito T., Sugimoto E., Shinozaki K. (June 20, 2011). Arabidopsis mutant of AtABCG26, an ABC transporter gene, is defective in pollen maturation. J. Plant Physiol. http://dx.doi.org/10.1016/j.jplph.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Liu J., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lolle S.J., Berlyn G.P., Engstrom E.M., Krolikowski K.A., Reiter W.-D., Pruitt R.E. (1997). Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev. Biol. 189: 311–321 [DOI] [PubMed] [Google Scholar]
- Lolle S.J., Pruitt R.E. (1999). Epidermal cell interactions: A case for local talk. Trends Plant Sci. 4: 14–20 [DOI] [PubMed] [Google Scholar]
- Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., Dean C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89: 737–745 [DOI] [PubMed] [Google Scholar]
- McElroy D., Blowers A.D., Jenes B., Wu R. (1991). Construction of expression vectors based on the rice actin 1 (Act1) 5′ region for use in monocot transformation. Mol. Gen. Genet. 231: 150–160 [DOI] [PubMed] [Google Scholar]
- McElroy D., Zhang W., Cao J., Wu R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H.E., Shin J.J.H., Bird D.A., Samuels A.L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiyappan M., Birrane G., Ladias J.A.A. (2007). Structural basis for polyproline recognition by the FE65 WW domain. J. Mol. Biol. 372: 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Clemens S., Zachgo S., Giblin E.M., Taylor D.C., Kunst L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie E.H., Chen Yz., Hubbart S., Peng S., Horton P. (1999). Interactions between senescence and leaf orientation determine in situ patterns of photosynthesis and photoinhibition in field-grown rice. Plant Physiol. 119: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K.T., Takahashi T. (2006). Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 141: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. (2002). The biopolymers cutin and suberin. In The Arabidopsis Book 1: e0021, doi/10.1199/tab.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. (2006). Unraveling the complex network of cuticular structure and function. Curr. Opin. Plant Biol. 9: 281–287 [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Panikashvili D., Shi J.X., Schreiber L., Aharoni A. (2011). The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol. 190: 113–124 [DOI] [PubMed] [Google Scholar]
- Pollard M., Beisson F., Li Y., Ohlrogge J.B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R.E., Vielle-Calzada J.-P., Ploense S.E., Grossniklaus U., Lolle S.J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.-J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., et al. (2003). Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Sci. 165: 941–949 [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Sieber P., Schorderet M., Ryser U., Buchala A., Kolattukudy P., Métraux J.-P., Nawrath C. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12: 721–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M., Bork P., Einbond A., Kastury K., Druck T., Negrini M., Huebner K., Lehman D. (1995). Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 270: 14733–14741 [DOI] [PubMed] [Google Scholar]
- Suh M.C., Samuels A.L., Jetter R., Kunst L., Pollard M., Ohlrogge J., Beisson F. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Onouchi H., Kondo M., Hara-Nishimura I., Nishimura M., Machida C., Machida Y. (2001). A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128: 4681–4689 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Tanaka H., Machida C., Watanabe M., Machida Y. (2004). A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Watanabe M., Sasabe M., Hiroe T., Tanaka T., Tsukaya H., Ikezaki M., Machida C., Machida Y. (2007). Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 134: 1643–1652 [DOI] [PubMed] [Google Scholar]
- Tang D., Simonich M.T., Innes R.W. (2007). Mutations in LACS2, a long-chain acyl-coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol. 144: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J., Post-Beittenmiller D., Jaworski J.G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17: 119–130 [DOI] [PubMed] [Google Scholar]
- Voisin D., Nawrath C., Kurdyukov S., Franke R.B., Reina-Pinto J.J., Efremova N., Will I., Schreiber L., Yephremov A. (2009). Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet. 5: e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J.G., Browse J. (2010). Lipid biochemists salute the genome. Plant J. 61: 1092–1106 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Tanaka H., Watanabe D., Machida C., Machida Y. (2004). The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 39: 298–308 [DOI] [PubMed] [Google Scholar]
- Wellesen K., Durst F., Pinot F., Benveniste I., Nettesheim K., Wisman E., Steiner-Lange S., Saedler H., Yephremov A. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω -hydroxylation in development. Proc. Natl. Acad. Sci. USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.E., Torabinejad J., Cohick E., Parker K., Drake E.J., Thompson J.E., Hortter M., Dewald D.B. (2005). Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol. 138: 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fan Z., Guo L., Li Y., Zhang W., Qu L.-J., Chen Z. (2003). Over-expression of an Arabidopsis δ-OAT gene enhances salt and drought tolerance in transgenic rice. Chin. Sci. Bull. 48: 2594–2600 [Google Scholar]
- Xiao F.M., Goodwin S.M., Xiao Y.M., Sun Z.Y., Baker D., Tang X.Y., Jenks M.A., Zhou J.M. (2004). Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Qin G., Shi Y., Ma Z., Chen Z., Gu H., Qu L.J. (2007). GAMT2 encodes a methyltransferase of gibberellic acid that is involved in seed maturation and germination in Arabidopsis. J. Integr. Plant Biol. 49: 368–381 [Google Scholar]
- Yang M., Sack F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A., Wisman E., Huijser P., Huijser C., Wellesen K., Saedler H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. (1981). Physiological analysis of rice yield. In Fundamentals of Rice Crop Science, Sheehy J.E., Mitchell P.L., Hardy B., (Makita City, Philippines: International Rice Research Institute; ), pp. 231–251 [Google Scholar]
- Zheng H., Rowland O., Kunst L. (2005). Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]