This work characterizes a hypomorphic allele of ELF3, delineating a functional domain needed to inhibit phytochrome action at ELF3. These findings are consistent with ELF3 being a multifunctional protein that integrates light signals to the circadian clock as a core repressive component of the oscillator.
Abstract
Arabidopsis thaliana EARLY FLOWERING3 (ELF3) is essential for the generation of circadian rhythms. ELF3 has been proposed to restrict light signals to the oscillator through phytochrome photoreceptors, but that has not been explicitly shown. Furthermore, the genetic action of ELF3 within the clock had remained elusive. Here, we report a functional characterization of ELF3 through the analysis of the elf3-12 allele, which encodes an amino acid replacement in a conserved domain. Circadian oscillations persisted, and unlike elf3 null alleles, elf3-12 resulted in a short circadian period only under ambient light. The period shortening effect of elf3-12 was enhanced by the overexpression of phytochromes phyA and phyB. We found that elf3-12 was only modestly perturbed in resetting of the oscillator and in gating light-regulated gene expression. Furthermore, elf3-12 essentially displayed wild-type development. We identified targets of ELF3 transcriptional repression in the oscillator, highlighting the action at the morning gene PSEUDO-RESPONSE REGULATOR9. Taken together, we identified two separable roles for ELF3, one affecting the circadian network and the other affecting light input to the oscillator. This is consistent with a dual function of ELF3 as both an integrator of phytochrome signals and a repressor component of the core oscillator.
INTRODUCTION
The plant circadian clock coordinates physiological and metabolic processes by anticipating daily changes in the environment. Numerous cellular and physiological processes, including transcript accumulation, hormone signaling, photosynthesis, growth, and plant–pathogen interactions, are circadian regulated (Davis and Millar, 2001; Hanano et al., 2006; Covington et al., 2008; Michael et al., 2008; Roden and Ingle, 2009; Graf et al., 2010; Wang et al., 2011). Accordingly, plants with an internal clock matching the diurnal cycle have been shown to display enhanced growth, thus highlighting the contribution of the clock to fitness (Dodd et al., 2005). The synchronization of internal to external time is achieved by the perception of diurnal light and temperature changes (Harmer, 2009), and these environmental cues are termed zeitgebers (time givers). The zeitgebers, of which light is the major signal, confer daily clock resetting mainly at dawn, a process referred to as entrainment. It has been proposed that the phytochrome (phy) photoreceptor family is important in this process (Somers et al., 1998).
In Arabidopsis thaliana, a series of interconnected transcription-translation feedback loops constitute a regulatory network of the circadian clock. Several of these loops have been defined. The Myb-like transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) peak in the morning and antagonize the expression of the pseudo-response regulator (PRR) TIMING OF CAB EXPRESSION1 (TOC1), which thus peaks at night. These three genes are considered the core loop of the oscillator (Alabadí et al., 2001; Locke et al., 2006; Zeilinger et al., 2006; Ding et al., 2007a). Additional loops interlock with this central loop. During the day, CCA1 and LHY promote the expression of the TOC1-related PRR9 and PRR7 (Locke et al., 2006; Zeilinger et al., 2006), whereas PRR9, PRR7, and PRR5 act sequentially to repress CCA1/LHY expression (Nakamichi et al., 2010; Pokhilko et al., 2010). GIGANTEA (GI) is incorporated with the unknown component Y in a feedback loop with TOC1 (Locke et al., 2006). Additionally, the GARP transcription factor LUX ARRHYTHMO (LUX)/PHYTOCLOCK (Hazen et al., 2005; Onai and Ishiura, 2005) connects evening gene expression to the PRR9/PRR7 morning loop, where LUX acts as a repressor specifically targeting PRR9 (Helfer et al., 2011). This action is similar to that of another evening gene, EARLY FLOWERING4 (ELF4), which is a genetic repressor of PRR9, PRR7, and GI (Kolmos and Davis, 2007; McWatters et al., 2007; Kolmos et al., 2009).
ELF3 was the first plant clock gene described to possess the zeitnehmer (time taker) function. The zeitnehmer concept is defined as a circadian-controlled input pathway to the clock, a feature that is also a clock-gating mechanism (McWatters et al., 2000). The light- and circadian-regulated expression of ELF3, and rapid dampening in constant darkness (DD), supported a light-gating function of ELF3 (Liu et al., 2001). Studies performed with a null elf3 allele revealed a gating defect in phase response assays, where the acute activation of CHLOROPHYLL A/B BINDING2 (CAB2) expression by light was monitored over the 24-h cycle. In the wild type, light-induced acute CAB2 expression occurs during the subjective day, whereas in the elf3 mutant, light also induced CAB2 during the subjective night (McWatters et al., 2000; Covington et al., 2001). Under diurnal cycles, one cycle could be driven in elf3 (Hicks et al., 2001; Nozue et al., 2007), but under constant conditions, the clock in elf3 mutants displayed arrhythmia (Hicks et al., 2001; Thines and Harmon, 2010). Accordingly, expression profiling revealed that elf3 had lost normal circadian regulation of clock transcript accumulation. elf3 was found to have reduced expression of the morning genes CCA1 and LHY and a high level of the evening genes GI and TOC1 under constant light (LL) (Schaffer et al., 1998; Fowler et al., 1999; Alabadí et al., 2001; Kikis et al., 2005). Thus, ELF3 represses light input to the clock and acts during the night phase of the circadian cycle. It has also been established that elf3 is defective in entrainment by temperature cycles, and this demonstrates that ELF3 has a general role in clock processes (Thines and Harmon, 2010). Notably, previous studies described a strong hypomorphic allele elf3-7, which has some limited oscillator function, especially after temperature entrainment (McWatters et al., 2000; Reed et al., 2000). The residual ELF3 protein in elf3-7 might represent a minimum amount necessary for marginal clock function (McWatters et al., 2000; Reed et al., 2000; Hicks et al., 2001). Taken together, these genetic studies revealed that ELF3 is critical for clock function and light input to the clock, but the arrhythmic nature of published elf3 alleles has precluded an understanding of the processes that connect the two.
In diurnal organisms such as Arabidopsis, light input to the circadian clock follows Aschoff's Rule: Increasing light intensities result in a shortening of the free-running period (Aschoff, 1979; Millar et al., 1995b). Light input to the clock is under the action of phyA and phyB (Devlin and Kay, 2000), and many responses to red light are mediated by phyB (Anderson et al., 1997). Notably, phyB mutants display a long circadian period predominantly under continuous red (Rc) light; similarly, phyA displays long period under Rc and constant blue (Bc) light (Somers et al., 1998). A connection of phyB-mediated light signaling to ELF3 was established by phyB-ELF3 protein–protein interaction, which may provide a mechanism for ELF3-mediated gating of light to the oscillator (Liu et al., 2001). By contrast, in the light-mediated inhibition of hypocotyl growth, elf3 and phyB mutations have additive defects, indicating that ELF3 and phyB genes can act partly independently for growth control (Reed et al., 2000). The role of phytochrome action relative to ELF3 has thus remained elusive.
ELF3 contributes to the timing of photoperiodic development. The elf3 mutant has an early and photoperiod-insensitive flowering time, and it has been concluded that ELF3 is upstream of GI and CONSTANS (CO) in the photoperiod pathway and that this genetic function includes a negative regulation of the flowering activator CO (Zagotta et al., 1996; Suárez-López et al., 2001). In addition, ELF3 is involved in regulation of GI stability via the ubiquitination pathway (Yu et al., 2008). Furthermore, it was suggested that ELF3 has a broad role in flowering time regulation based on the observation that elf3 early flowering can be independent of CO (Kim et al., 2005). Furthermore, under diurnal growth conditions, elf3 fails to suppress hypocotyl elongation growth around dusk, and this phenotype is not as pronounced under LL conditions (Dowson-Day and Millar, 1999; Nozue et al., 2007). Taken together, the loss of ELF3 function results in multiple photoperiodic phenotypes of development.
ELF3 is an evening-expressed gene that encodes a nuclear-localized protein (Liu et al., 2001). It has no sequence similarity to proteins of known function. The Arabidopsis genome contains a single ELF3 paralog, ESSENCE OF ELF3 CONSENSUS (EEC), which has no reported role in the circadian clock (Hall et al., 2003). Putative orthologs of ELF3 have been isolated from plants, and four conserved domains have been proposed. However, the information from the protein sequence phylogeny has not been connected to ELF3 function (Liu et al., 2001). Different domains of the ELF3 protein mediate physical interaction with phyB, COP1, and GI (Liu et al., 2001; Yu et al., 2008). Additionally, general amino acid features throughout the ELF3 protein suggest its involvement in transcriptional regulation (Hicks et al., 2001). These observations indicate that ELF3 is a multifunctional protein. From studies in rice (Oryza sativa), it was found that ELF3-related genes are not necessarily clock regulated, and this was interpreted as meaning that ELF3 function is not fully conserved (Murakami et al., 2007). Collectively, much remains to be learned about ELF3 structure as it relates to function.
In this article, we explore the ELF3-encoded sequence as related to light input to the circadian clock. This started with the isolation of a new elf3 allele (elf3-12) from a forward genetic screen. The defined phenotype of elf3-12 is distinct from previously characterized elf3 alleles, and the site of the mutation thus delineates a functional domain. We used elf3-12 to characterize the action of ELF3 within the circadian system and on developmental processes. We found that the expression of a subset of clock components is affected in elf3-12. Additionally, elf3-12 displayed a light-dependent short circadian period. Elucidating the role of light input in this elf3 allele, the overexpression of photoreceptors PHYA and PHYB was found to enhance the shortening effect of elf3-12 on light-dependent phenotypes. Moreover, we found that the elf3-12 circadian clock was impaired both in regulating light-induced gene expression and in resetting of the oscillator. We conclude that elf3-12 is attenuated in its capacity for repressive light input signaling to the clock while maintaining its capacity to sustain oscillator function.
RESULTS
The elf3-12 Mutation Demarks a Functional Domain of ELF3
From a forward genetic screen of seedlings with altered CAB2:LUCIFERASE (LUC; 2CA/C) rhythms in DD (Kevei et al., 2006), one mutant was isolated, originally termed G12. The acrophase of the luminescence peak in this mutant was ~3 h early compared with the wild-type C24 (Figure 1A). Positional mapping of the G12 mutation revealed tight genetic linkage to the ELF3 locus. Sequencing of ELF3 in G12 revealed a GGT-to-GAT transition mutation encoding the G326D change. The G12 mutant was thus named elf3-12. Interestingly, the affected Gly residue (Gly-326) was fully conserved among ELF3 sequences from a variety of plants (Figure 1B; see Supplemental Figure 1 online). The alignments of the middle part of ELF3 shown in Figure 1B and Supplemental Figure 1 online correspond to Block II, as defined by Liu et al. (2001). By our identification of the elf3-12 mutant, we thus propose Block II to be functionally important.
Figure 1.
The elf3-12 Point Mutation Causes a Short Period Length under Light.
Seedlings harboring LUC were entrained in 12L:12D and transferred to constant conditions at ZT0. Error bars represent se. Time refers to time under constant conditions.
(A) Free-running profile of CAB2:LUC (2CA/C) expression in elf3-12 and the wild type in DD. This was the phenotype that led to isolation of the elf3-12 mutation. The phase advance in elf3-12 (~3.5 h in sidereal time) is indicated. c.p.s., counts per second.
(B) Alignment of Block II in ELF3-like sequences. The residues are shaded according to the degree of conservation. The arrow indicates the position of Arabidopsis G326. The DNA mutation in elf3-12 leads to an MboI site as indicated. Ah, Arachis hypogaea; Af, Aquilegia formosa; At, Arabidopsis; Ee, Euphorbia esula; Gh, Gossypium hirsutum; Lg, Lemna gibba; Lp, L. paucicostata; Mc, Mesembryanthemum chrystallinum; Os, rice; Pp, Physcomitrella patens; Ta, Triticum aestivum.
(C) Free-running profile of CAB2:LUC expression in elf3-1, elf3-7, elf3-12, and the wild type under Rc.
(D) Period estimates of rhythms shown in (C). See also Supplemental Table 1 online.
(E) Allelic strength of elf3-12 CAB2:LUC crossed to elf3-1 (F1). Free-running profiles under LL.
(F) Period versus RAE estimates of rhythms shown in (E). See also Supplemental Table 1 online.
To determine the properties of the circadian clock in elf3-12, we analyzed the free-running rhythm of elf3-12 under LL conditions. Seedlings harboring the CAB2:LUC+ reporter were entrained for 1 week under 12L:12D cycles and then transferred to Rc. Compared with the wild type, elf3-12 displayed an ~2-h shorter circadian period (Figures 1C and 1D; see Supplemental Table 1 online) in agreement with the early phase we observed for this reporter in DD (Figure 1A). Interestingly, the elf3-12 rhythm was robustly rhythmic, as it displayed low relative amplitude of error (RAE) values (Figure 1D), which defines the successful capacity of the FFT-NLLS curve fitting to assign a trigonometric function to the data, and this is thus an estimate of overt rhythmicity. The robust rhythms (low RAE values) of elf3-12 was in sharp contrast with the arrhythmic phenotypes (high RAE values; Figure 1D) of elf3-1 (null) and elf3-7 (strong hypomorph).
To confirm that the elf3-12 short-period phenotype was caused by the missense mutation within the ELF3 locus, we generated trans-heterozygous elf3 plants from elf3-1 crossed with elf3-12. The F1 plants from the elf3-1 × elf3-12 cross displayed the same short period under LL of CAB2:LUC+ expression as homozygous elf3-12 (Figures 1E and 1F; see Supplemental Table 1 online). Additionally, we found that the elf3-12 mutation was recessive, as the control F1 cross of elf3-12 to the wild type resulted in a free-running periodicity similar to the wild type crossed to itself (the progeny that would contain two fully functional ELF3 alleles).
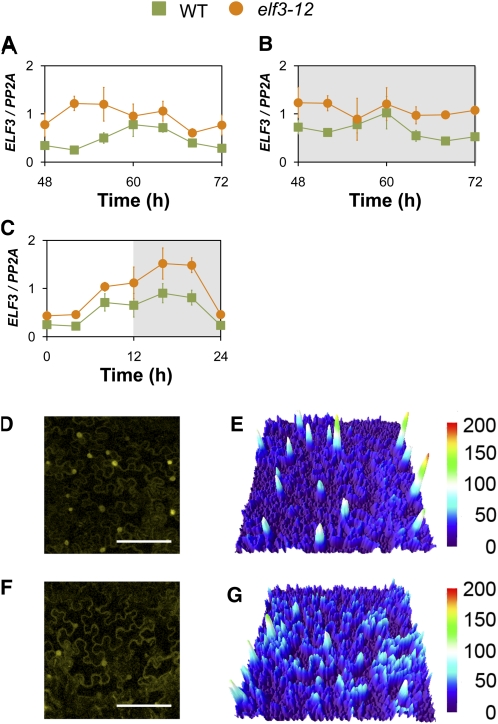
ELF3 has been proposed to repress circadian function (Thines and Harmon, 2010); thus, a reduction of ELF3 transcript levels in a reduced-function, hypomorphic allele could plausibly explain a short-period phenotype. We sought to exclude the possibility that the phenotypes in elf3-12 could be explained by a simple reduction of expression level. For this, we first measured ELF3 transcript accumulation and then assessed the capacity of the ELF3-12 transcript to generate protein. ELF3 transcript accumulation in elf3-12 and the wild type was measured under diurnal light-dark (LD) cycles, LL, and in DD. In all cases, we found that the mean ELF3-12 transcript levels were elevated relative to the wild type (Figures 2A to 2C). This elevation does not explain the short-period phenotype seen in elf3-12, as ELF3 elevation leads to a deceleration of clock periodicity (Covington et al., 2001). We therefore examined the capacity of ELF3-12 to generate cellular-accumulated protein, as assessed by an imaging assay. For that, ELF3-YFP and ELF3-12-YFP fusions were generated under the control of the native ELF3 promoter, and these constructs were used for expression in tobacco (Nicotiana benthamiana). Leaf material was imaged at dusk, the time of maximal ELF3 accumulation (Liu et al., 2001). We found that both ELF3-YFP and ELF3-12-YFP accumulated in the nuclei (Figures 2D and 2F, respectively). Additionally, fluorescence signal was found in the cytoplasm for both fusion proteins (Figures 2D and 2F, respectively). Interestingly, this cytoplasmic-nuclear distribution was different for ELF3-12-YFP. Notably, the ELF3-12-YFP nuclear pool was decreased, and its cytoplasmic pool was increased (Figures 2E and 2G). Taken together, it was the cellular distribution of ELF3 protein, and not its levels, that correlated to the elf3-12 phenotype.
Figure 2.
ELF3 Transcript and ELF3-YFP Accumulation in elf3-12.
(A) to (C) Transcript accumulation of ELF3 in elf3-12 and wild-type (WT) Arabidopsis under LL (A), DD (B), and LD (C). Time (h) for LL and DD is from the last lights on period. Expression levels are normalized for PROTEIN PHOSPHATASE 2a subunit A3 (PP2A).
(D) and (F) Confocal microscopy images of N. benthamiana leaf epidermal cells expressing ELF3:ELF3-YFP (D) and ELF3:ELF3-12-YFP (F), imaged at dusk. The images show the average projections as a 10-μm stack. White bar indicates 150 μm.
(E) Heat map of YFP intensity from ELF3:ELF3-YFP in (D).
(G) Heat map of YFP intensity from ELF3:ELF3-12:YFP in (F). x and y axes of both heat maps correspond to image coordinates from (D) and (F). z axis is YFP intensity.
As ELF3 interacts with phyB (Liu et al., 2001), we then tested if this interaction could be affected in ELF3-12. We found that the elf3-12 point mutation had no effect on the phyB-ELF3 interaction in a yeast two-hybrid assay (see Supplemental Figure 2 online). As the location of the elf3-12 mutation is outside of the phytochrome-interacting region (Liu et al., 2001), this result was expected. We concluded that both ELF3 transcript regulation and ELF3 nuclear accumulation are altered in elf3-12 in a way consistent with the ELF3-12 protein maintaining circadian activity.
The elf3-12 Mutant Displays Wild-Type Gross Morphology
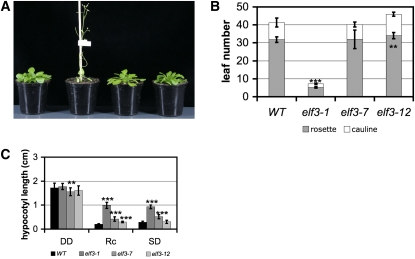
The circadian clock is key in the control of photoperiodic signaling, such as in the timing of the floral transition, an output pathway of the circadian system (de Montaigu et al., 2010). As elf3 loss-of-function leads to photoperiod-independent early flowering (Zagotta et al., 1996), we examined the flowering time of elf3-12 by determining the number of rosette leaves at the time of bolting and cauline leaves on the main inflorescence. Under both short- (ShD) and long-day (LgD) conditions, we found that the flowering time of elf3-12 was similar to that of the wild type and not as strikingly early as is seen in the null allele elf3-1 (Figures 3A and 3B; see Supplemental Figure 3A and Supplemental Table 1 online). Under LgD conditions, elf3-12 plants flowered marginally later than the wild type, and an analysis of the ratio of days to bolting and days to anthesis excluded the possibility that this is caused by a plastochron effect (see Supplemental Figure 3B online). The elf3-12 mutant is thus not early flowering. Taken together, the elf3-12 allele encodes a protein that remains functional for normal flowering-time control.
Figure 3.
The elf3-12 Mutant Has Wild-Type Gross Morphology.
(A) Flowering time of 35-d-old plants of C24 wild type, elf3-1, elf3-7, and elf3-12 (from left to right) under ShD.
(B) Leaf number at time of bolting under ShD, depicted as average rosette leaf number and average cauline leaf number on the main inflorescence (n = C24, 30; elf3-1, 22; elf3-7, 21; elf3-12, 20). Error bars correspond to sd. WT, wild type.
(C) Hypocotyl length of 7-d-old seedlings in DD and under ShD and Rc, respectively. Average hypocotyl length (±sd; cm). Mean values that are significantly different from the wild type according to the t test are indicated by *, **, or *** for P values < 0.05, 0.01, or 0.001, respectively. See also Supplemental Table 1 online.
[See online article for color version of this figure.]
Hypocotyl elongation growth is a developmental process that is diurnally regulated (Nozue et al., 2007). In the absence of ELF3 function, hypocotyls are elongated under Rc and also under diurnal conditions (Zagotta et al., 1996; Reed et al., 2000). Similar to the lack of a dramatic flowering-time phenotype (above), we observed a hypocotyl length for elf3-12 under ShD that resembled that of the wild type (Figure 3C). Under Rc, elf3-12 displayed a marginally elongated hypocotyl (~0.1 cm longer than the wild type; P < 0.001), whereas elf3-1 and elf3-7 displayed long hypocotyls (~0.2 to 0.8 cm longer; Figure 3C; see Supplemental Table 1 online), consistent with the literature (Reed et al., 2000). In DD, all elf3 alleles tested were similar to the wild type (Figure 3C; see Supplemental Table 1 online) (Liu et al., 2001). Additionally, we measured the hypocotyls of these elf3 alleles grown under Rc, while applying entraining temperature cycles to set circadian phase (see Supplemental Figure 4B online). We found that only elf3-1 displayed a long hypocotyl under this regime. Taken together, we observed that elf3-12 lacked the null phenotype for major developmental responses.
PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5 are circadian-controlled transcription factors important for phase-specific hypocotyl elongation at the end of the night (Nozue et al., 2007). To investigate further the subtle hypocotyl phenotype of elf3-12, we assayed the expression of PIF4 and PIF5 under Rc and ShD (see Supplemental Figure 4A online). Under Rc, we found no detectable PIF4 and PIF5 expression differences between elf3-12 and the wild type. In stark contrast, as expected, the expression levels of these genes were higher in elf3-1 and elf3-7. We observed similar expression differences for the elf3 genotypes under ShD, where elf3-12 displayed merely a tendency toward an elevated PIF4 level at the end of the night, and elf3-1 and elf3-7 had markedly elevated PIF4 and PIF5 expression throughout the night period (see Supplemental Figure 4A online). Thus, the ELF3-12 protein clearly contains capacity to repress PIF4 and PIF5 transcript accumulation.
Changes in the Clock Network in elf3-12
Altered expression levels of the oscillator genes CCA1 and LHY accompany elf3 loss of function (Schaffer et al., 1998; Kikis et al., 2005). Unlike elf3-1 and elf3-7 alleles, circadian oscillations persisted in elf3-12; thus, this allele allowed us to determine the placement of ELF3 within the clock network. For this, we assayed transcript accumulation of different oscillator genes during a 24-h window. Under diurnal conditions, the phase of clock gene expression in elf3-12 was similar to the wild type (see Supplemental Figure 5 online). We note that elf3-12 had small alterations in amplitude, such as slightly reduced peaks for CCA1 and LHY (see Supplemental Figures 5C and 5D online), and a subtle derepression of TOC1 and LUX levels in the night (see Supplemental Figures 5E and 5F online). These mild phenotypes were in stark contrast with the strong reduction of CCA1 and LHY expression in elf3-1 and elf3-7. In addition, elf3-1 displayed derepressed and low-amplitude levels of PRR9 and PRR7 (see Supplemental Figures 5A and 5B online), whereas the amplitude of TOC1 was increased and the profiles of GI and LUX were caused by the changes of light and dark and are termed diurnally driven rhythms (see Supplemental Figures 5E to 5G online). The high levels of PRR9 and PRR7 were indicated in earlier reports (Thines and Harmon, 2010), and the evening gene phenotypes of elf3-1 were in accordance with earlier studies (Fowler et al., 1999; Alabadí et al., 2001).
As elf3-12 displayed a new elf3 circadian phenotype (short period, not arrhythmic; Figure 1C), we characterized its transcript profile of clock genes under both LL and in DD. In agreement with the short period of the elf3-12 LUC rhythms, we found that all gene expressions assayed under LL displayed an early phase (Figure 4). This was in contrast with arrhythmic elf3-1 and elf3-7, which had constant low levels of CCA1 and LHY (Figures 4C and 4D), high levels of morning loop genes PRR9 and PRR7 (Figures 4A and 4B), and high levels of evening genes TOC1, LUX, and GI (Figures 4E to 4G) (Alabadí et al., 2001; Kikis et al., 2005). In DD, elf3-12 displayed low CCA1 and LHY expression. For CCA1, the trough level was similar to the wild type, whereas for LHY it was lower. The amplitude was, if present at all, strongly reduced (Figures 4J and 4K). By contrast, the mean level of PRR9 and PRR7 expression in elf3-12 was higher than in the wild type (Figures 4H and 4I), and the level of PRR7 was comparable to that of elf3-1 (Figure 4I). The expression level of PRR9 in elf3-12 was higher than in the wild type, but stayed below the level in elf3-1 and elf3-7 (Figure 4H). The mean levels of GI and LUX were only slightly higher than in the wild type, whereas elf3-12 displayed no detectable phenotype for TOC1 expression in DD (Figures 4L to 4N). It was striking that in DD, elf3-12 expression rhythms were more similar to elf3 loss-of-function rhythms than they were under LL. Collectively, it seems that elf3-12 has a light-dependent effect on circadian period length, which does not directly correlate with clock amplitude.
Figure 4.
The Transcript Accumulation Pattern of Clock Genes in Various elf3 Alleles.
Transcript profiling of clock gene expression in elf3-1, elf3-7, elf3-12, and the wild type (WT) under LL and in DD. Time (h) is from the last lights on period. All gene expressions represent the mean of expression of three replicates of the same biological sample and are normalized for PROTEIN PHOSPHATASE 2a subunit A3 (PP2A). Error bars represent sd of the technical replicates. LL: (A) PRR9, (B) PRR7, (C) CCA1, (D) LHY, (E) TOC1, (F) LUX, and (G) GI. DD: (H) PRR9, (I) PRR7, (J) CCA1, (K) LHY, (L) TOC1, (M) LUX, and (N) GI.
The Short Circadian Period of elf3-12 Is Light Dependent
As the transcript accumulation phenotypes of elf3-12 were distinct under LL (Figure 4), we examined the circadian period of LUC reporter genes in elf3-12 under LL compared with in DD. Under LL, elf3-12 LHY:LUC displayed an ~2.5-h-shorter period and a marginally reduced amplitude compared with the wild type (Figure 5A). This phenotype was markedly different from the null elf3-1, which effectively displayed no LHY expression (Figure 5A; Kikis et al., 2005). Interestingly, we did not find a periodicity phenotype for elf3-12 in DD (albeit it had an early phase; Figure 5D; see Supplemental Table 1 online). Thus, the reduced period length of LHY:LUC in elf3-12 only under LL indicated an attenuated function of ELF3 under light. We next used a LUC reporter gene fused to the COLD AND CIRCADIAN REGULATED2 (CCR2) promoter (also known as GRP7; Schöning et al., 2007), which displays robust circadian activity independent of light (Covington et al., 2001). Consistent with earlier reports, we found that elf3-1 CCR2:LUC had no detectable circadian rhythms both under LL and in DD (Figures 5B to 5D; Reed et al., 2000; Thines and Harmon, 2010). By contrast, the elf3-12 allele displayed a significantly shortened circadian period of CCR2:LUC expression under LL but not in DD (Figures 5B to 5D). Taken together, elf3-12 displayed a light-dependent short-period phenotype that was unlike the arrhythmia of elf3-1.
Figure 5.
Mutant Clock Period Properties Are Light Dependent in elf3-12.
(A) Free-running profile of LHY:LUC expression in elf3-1, elf3-12, and the wild type (Wt) under LL. Note that in elf3-1 the LUC levels are arrhythmic and >1000-fold lower than in the wild type. Error bars correspond to se. cps, counts per second.
(B) Free-running activity of CCR2:LUC in elf3-1, elf3-12, and the wild type under LL.
(C) Free-running activity of CCR2:LUC in elf3-1, elf3-12, and the wild type in DD.
(D) Circadian period length of LHY:LUC and CCR2:LUC rhythms (as shown in [A] to [C]). Under LL, the period of elf3-12 is short, but in DD, no period phenotype is found. Error bars correspond to se. Error significance is as defined in Figure 3. See also Supplemental Table 1 online.
Entrainment Properties in the elf3-12 Oscillator
Since the elf3-12 circadian phenotype was light dependent, we analyzed if this mutant was defective in entrainment to LD cycles. Wild-type clocks will entrain only to every second cycle (demultiply) if the zeitgeber cycle is close enough to half of the endogenous period length (or every fourth if one-quarter period, etc.), whereas mutants with dysfunctional clocks will display driven rhythms, where the LD cycle becomes dominant in rhythm generation (Nozue et al., 2007; Thines and Harmon, 2010). We performed a demultiplication assay of elf3-12 to test whether the period of the oscillator was functionally shorter from the zeitgeber (T) cycle to significantly alter its entrainment properties. After growth under a 12L:12D regime (T = 24) for 1 week, we transferred seedlings harboring CCR2:LUC to 6L:6D (T = 12) and monitored luminescence for ~72 h (see Supplemental Figure 6 online). The strong elf3 mutants failed to maintain the driven 24-h rhythms after transfer into 6L:6D (T = 12). Both elf3-1 and elf3-7 maintained their driven phase for the first one-and-a-half run of the 12-h T-cycle, but thereafter elf3-1 became arrhythmic, whereas elf3-7 displayed driven 6-h rhythms. By contrast, elf3-12 displayed standard 24 h oscillations under 6L:6D, as under 12L:12D, which is similar to the wild type (see Supplemental Figure 6 online). Thus, the elf3-12 oscillator can resist extreme T-cycles in a manner similar to the wild type, whereas strong elf3 alleles become sensitive to the effects of this demultiplication and display a driven rhythm dependent on the nature of the LD cycle.
The elf3-12 Mutant Has Gating and Phase-Resetting Defects
One hallmark of the elf3 loss-of-function phenotype is a defect in clock repression of light-regulated gene expression. This phenotype of elf3 in so-called gating assays helped classify this mutant as defective in light input to the clock (McWatters et al., 2000). To characterize elf3-12 with respect to circadian control of light input to the oscillator, we performed a gating experiment. We monitored the acute induction of CAB2:LUC expression in dark-adapted seedlings following 5-min light pulses every 3 h of the circadian cycle. As previously reported, the elf3-1 mutant displayed increased acute responses to light during the subjective night (see Supplemental Figure 7 online; McWatters et al., 2000). By contrast, elf3-12 displayed a gating defect in the late subjective night, ~ZT18 to 24 (Figure 6A). We noted that the early peak time (phase) of elf3-12 in DD was ~ZT26, whereas the wild-type peak was at ~ZT30, which was ~3 h later. This phase of elf3-12 at ~ZT26 was later than the observed increased acute responses following the light pulses, at ~ZT18 to 24. We therefore consider that the gating defect in elf3-12 does not readily follow its short-period phenotype. Thus, compared with the gating pattern in the wild type, elf3-12 responded with increasing acute responses during those late hours (~2- to 3-fold higher than the wild-type response) (Figure 6A). Thus, elf3-12 has an unclosed gate only during late night.
Figure 6.
Both Clock Gating and Resetting Are Altered in elf3-12.
(A) Gating of acute CAB2:LUC expression in dark-adapted seedlings of elf3-12. Connected points are control seedlings kept in DD. Error bars represent sd. Time is time since last lights on (ZT).
(B) PRC of CCR2:LUC expression in elf3-12 and the wild type. Phase shift of the first acro-peak of CCR2:LUC induced by hour-long red light pulses plotted against the circadian time (CT) at which pulses were applied. Phase advances are plotted as positive values and delays as negative values. Error bars represent the pooled se of phase values for pulsed and nonpulsed populations.
The phase response curve (PRC) is a measure of the clock's time-dependent sensitivity to resetting cues (zeitgebers; Pittendrigh, 1981). The degree of clock resetting was reported to be inversely correlated with the level of ELF3 expression, suggesting that ELF3 might be a buffering mechanism to limit phase resetting in the middle of the night (Covington et al., 2001). We sought to confirm this idea by analyzing the clock-regulated resetting response of CCR2:LUC expression to red-light pulses in elf3-12. We found that the clock in elf3-12 plants was hypersensitive to the resetting cue during half of the circadian cycle (Figure 6B). During the initial subjective evening and early night (CT12 to CT18), light pulses delayed the phase similarly in elf3-12 and the wild type. By contrast, elf3-12 had increased phase advances (up to 4 h) during the late subjective night and the following early subjective day (CT18 to CT27). Thus, from the shape of this PRC, we conclude that elf3-12 can reset its oscillator, but it is hypersensitive to phase advancement.
The Effect of elf3-12 on PHYA Signaling
The photoreceptors phyA and phyB play a major role in light input signaling to the circadian clock (Anderson et al., 1997; Devlin and Kay, 2000), and it was previously shown that ELF3 physically interacts with phyB (Liu et al., 2001). As elf3-12 is light defective to the clock, we sought to assess the effect of its light-dependent short period on phy-mediated signaling to the circadian system. For this, we chose established PHY overexpression lines (PHYA-ox and PHYB-ox), available in the C24 ecotype (Boylan and Quail, 1991; Wagner et al., 1991; Anderson et al., 1997). The use of PHY-ox lines in part reduces complexities from partial redundancy of phy loss-of-function mutants. We integrated each of the PHY-ox transgenes into elf3-12 harboring different LUC reporters. Subsequently, we performed assays under free-running conditions under different light regimes: Rc, constant far-red (FRc), and in DD. We note here that the overt rhythmicity in elf3-12 allowed the evaluation of ELF3 function in PHYA and PHYB signaling. This benefit is noted as previous analyses were hampered by the complete elf3-1 arrhythmicity (and even the partial elf3-7 arrhythmicity) that masked the circadian effects of PHYA-ox and PHYB-ox under Rc and in DD (see Supplemental Figures 8 to 11 online).
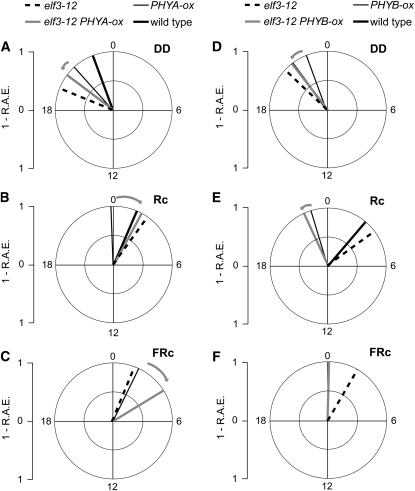
The PHYA-ox line was reported to generate a short circadian period (Anderson et al., 1997). We tested the effect of the elf3-12 mutation on PHYA-ox signaling to the clock using the LHY:LUC reporter (Figure 7; and Supplemental Figure 8 online). As expected, we found a short-period phenotype for PHYA-ox under all light regimes tested. Similarly, we found short-period phenotypes for elf3-12 under all light regimes, and these did not significantly differ from the PHYA-ox period (see Supplemental Table 2 online). Interestingly, under Rc, we found the period of the elf3-12 PHYA-ox double mutant to be ~2 h shorter than both elf3-12 and PHYA-ox (Figure 7B; see Supplemental Table 2 online), indicating an additive effect of elf3-12 on circadian period. In DD, we did not detect an effect of elf3-12 on PHYA-ox period length (Figure 7A; see Supplemental Table 2 online). In addition, when we determined the circadian phase, we found that elf3-12 PHYA-ox and elf3-12 had the same phase under Rc but not in DD (Figures 8D and 8B, respectively; see Supplemental Table 2 online). This suggested that elf3-12 was epistatic for phase to PHYA-ox, but only in the presence of light. Both under light and in DD, elf3-12 reduced the circadian amplitude seen in PHYA-ox by ~50% under Rc, and this was even more pronounced in DD (see Supplemental Figure 8 online). Taken together, elf3-12 shortened circadian periodicity and delayed circadian phase of PHYA-ox in a light-dependent manner.
Figure 7.
phyA and phyB Effects on Period in the Context of the elf3-12 Allele.
Period estimates of LHY:LUC expression in elf3-12, PHYA-ox, elf3-12 PHYA-ox, and the wild type in DD (A) and under Rc (B) and FRc (C) and in elf3-12, PHYB-ox, elf3-12 PHYB-ox, and the wild type in DD (D) and under Rc (E) and FRc (F). Error bars indicate sd; n = 24. Statistical assessment of these data can be found in Supplemental Tables 2 and 3 online. Error significance is as defined in Figure 3.
Figure 8.
The Effects of phyA and phyB on Phase in the Context of the elf3-12 Allele.
Averages of circadian phase estimates of LHY:LUC expression in elf3-12, PHYA-ox, elf3-12 PHYA-ox, and the wild type in DD (A) and under Rc (B) and FRc (C) and in elf3-12, PHYB-ox, elf3-12 PHYB-ox, and the wild type in DD (D) and under Rc (E) and FRc (F). The gray arrow denotes the change of phase. Statistical assessment of these data can be found in Supplemental Tables 2 and 3 online.
The Effect of elf3-12 on PHYB Signaling
We next analyzed the effect of the elf3-12 mutation with relation to phyB signaling to the circadian clock, which was motivated by the known physical association of ELF3 with phyB (Liu et al., 2001). It was previously shown that PHYB-ox had a dose-dependent reduced period length and early phase under free-running conditions (Anderson et al., 1997; Somers et al., 1998; Hall et al., 2002). Similar to PHYA-ox, we found that PHYB-ox LHY:LUC displayed high amplitude and short-period rhythms under Rc and in DD (Figure 7; see Supplemental Figure 9 and Supplemental Table 3 online). The phase of PHYB-ox was ~2 to 3 h advanced compared with the wild type under Rc, whereas PHYB-ox displayed no phase phenotype in DD (Figure 8; see Supplemental Table 3 online). Analysis of the elf3-12 mutation in the PHYB-ox background revealed an additive period-shortening effect of elf3-12 on PHYB-ox under Rc (~1 h, Figure 7E; see Supplemental Table 3 online) and under a combination of blue and red light (~1.5 h; see Supplemental Table 3 online). This additive effect was not observed in DD (Figure 7; see Supplemental Table 3 online). Moreover, we found that circadian phase in elf3-12 PHYB-ox was similar to that of PHYB-ox and significantly different, by 1 to 3 h, from elf3-12 under all light regimes tested and in DD (Figure 8; see Supplemental Table 3 online). This was in contrast with the effects of elf3-12 on PHYA-ox, where the phase was found to be similar to the elf3-12 phase under Rc (Figure 8B). Thus, PHYB-ox appears to suppress the effect of elf3-12 on circadian phase. Additionally, elf3-12 reduced the amplitude of PHYB-ox ~50% under Rc and Bc and in DD (see Supplemental Figure 9 online). We conclude that for phyB-mediated signaling to the circadian clock, the elf3-12 mutation shortens the circadian period under Rc and reduces the amplitude but has no effect on circadian phase.
ELF3 Signaling under FRc
The effect of FRc input to the circadian clock is not well understood. However, recent findings indicated a role for phyA in controlling clock-regulated gene expression under FRc (Wenden et al., 2011). Accordingly, we sought to investigate the role of ELF3 in FR-mediated input to the clock. First, we analyzed elf3-1 and elf3-7 for circadian rhythms under FRc. Since LHY:LUC dampened rapidly for the wild type under FRc (see Supplemental Figure 8C online), we used instead the robust output reporter CCR2:LUC to assess the effect of various elf3 mutations on circadian periodicity under FRc.
Surprisingly, we found circadian rhythmicity for both elf3-1 and elf3-7 under FRc. A clear amplitude persisted for at least two circadian cycles, and we scored ~40% of individual lines as rhythmic (see Supplemental Figures 10 and 11 and Supplemental Tables 4 and 5 online). We note that this elf3 precision phenotype was different from rhythms in DD, where elf3-7 rhythms had a higher precision than elf3-1 (~60 and ~30% of rhythmic plants, respectively; see Supplemental Tables 4 and 5 online). Thus, functional elements of circadian activity could be detected in strong elf3 alleles under FRc.
As phyA is the main far-red photoreceptor (Dehesh et al., 1993), we tested genetic interactions of PHYA-ox to elf3 under FRc. We found that elf3-1 PHYA-ox rhythms were more precise under FRc than in DD (~60 and ~40% of rhythmic plants, respectively; see Supplemental Table 4 and Supplemental Figure 10 online). Interestingly, the elf3-7 PHYA-ox double mutant was overtly rhythmic both under FRc and in DD (~90% rhythmic plants; see Supplemental Figures 10B and 10D and Supplemental Table 5 online). To determine further the role of ELF3 under FRc, we next analyzed elf3-12 LHY:LUC under FRc. Surprisingly, we found robust, low-amplitude rhythms for elf3-12 LHY:LUC expression under FRc, which for the wild type dampened rapidly (see Supplemental Figure 8 and Supplemental Table 2 online). Similar to the phenotype of elf3-12 under Rc (Figure 7), we found a period-shortening effect of elf3-12 on PHYA-ox LHY:LUC under FRc (Figure 7C). However, the circadian phase of elf3-12 PHYA-ox was delayed ~2 h when compared with elf3-12 (note that under Rc elf3-12 was epistatic to PHYA-ox; Figure 8D; see Supplemental Table 2 online). We found that PHYB-ox LHY:LUC was not rhythmic under FRc. However, elf3-12 PHYB-ox was rhythmic but with reduced amplitude compared with elf3-12 (see Supplemental Figure 9 online). Collectively, the detected rhythmicity in elf3-1, and especially elf3-7, under FRc indicates that ELF3 has a more limited role in controlling FRc signaling to the oscillator than under other wavelengths. Under FRc, elf3-12 and PHYA-ox also have an additive effect on period shortening. During circadian phase setting under FRc, elf3-12 and PHYA-ox displayed a synergistic interaction.
DISCUSSION
The elf3-12 Site Demarcates a Domain Controlling Circadian Period
ELF3 function was shown to be crucial for generation of circadian rhythms and the perception of light and temperature zeitgebers, as the loss of ELF3 led to clock arrest under free-running conditions, seen as arrhythmicity of all outputs (Fowler et al., 1999; Alabadí et al., 2001; Covington et al., 2001; Hicks et al., 2001; Kikis et al., 2005; Nozue et al., 2007; Thines and Harmon, 2010). However, residual clock function could be detected in elf3-7, which presumably generates a limited pool of ELF3 protein (McWatters et al., 2000; Reed et al., 2000; Hicks et al., 2001). In this study, we identified elf3-12 as a new elf3 allele with a light-dependent short-period phenotype. The elf3-12 defect was distinct from known mutants elf3-1 (null) and elf3-7 (strong hypomorph), as robust rhythms were clearly detectable (Figure 1). The site of the elf3-12 mutation implies that Block II modulates ELF3 circadian function (Figure 1B).
The elf3-12 Mutant Retains ELF3 Output Function on Growth
The determination of ELF3's role in clock output pathways, such as flowering time and hypocotyl elongation growth, has been complicated due to crosstalk with other signaling pathways, such as light, hormone, and temperature signaling (Covington et al., 2001; Kim et al., 2005; Yu et al., 2008; Strasser et al., 2009; Yoshida et al., 2009). The elf3-12 allele was used to resolve this issue. We found that there was essentially no phenotypic effect of the elf3-12 mutation on flowering time (Figures 3A and 3B). ELF3 has previously been shown to interact physically with GI in the nucleus to regulate GI protein stability, and ELF3 presence thus shapes the GI accumulation pattern (Yu et al., 2008). Furthermore, in a yeast two-hybrid assay, the N-terminal half of ELF3 (residues 1 to 440), including the Gly-326 that is mutated in elf3-12, could interact with GI protein (Yu et al., 2008). As elf3-12 lacks a flowering-time defect (Figures 3A and 3B), and as GI is downstream of ELF3 in the flowering-time network (Yu et al., 2008), it is plausible that the encoded ELF3-12 protein is capable of fulfilling its upstream role in GI-mediated signaling. Regarding early seedling development, we detected only a mild hypocotyl phenotype in elf3-12 under Rc (Figure 3C). It was reported that elf3 mutants fail to suppress hypocotyl elongation growth around dusk under diurnal cycles, a phenotype that is less pronounced under LL (Dowson-Day and Millar, 1999; Nozue et al., 2007). Additionally, it was recently found that temperature cycles fail to entrain elf3-1 mutants (Thines and Harmon, 2010), whereas elf3-7 mutants still have sufficient ELF3 protein to entrain to temperature cycles (McWatters et al., 2000). In agreement with these reports, and considering the light-dependent circadian phenotypes of elf3-12 (this study), the hypocotyl phenotype of elf3-12 (and also elf3-7) more resembled the wild type when the Rc regime was supplemented with temperature cycles (see Supplemental Figure 4B online).
The absence of strong developmental defects in elf3-12 implied that the early flowering and long hypocotyl phenotype of elf3-1 was due to ablated circadian function (Dowson-Day and Millar, 1999; McWatters et al., 2000; Reed et al., 2000; Nozue et al., 2007) and not only to defects in light input to the oscillator. Multiple light perception pathways lead to the circadian oscillator, as evidenced by genetic enhancement of elf3-1 by the time for coffee (tic) mutant, which is compromised for light input at dawn (Hall et al., 2003; Ding et al., 2007b). As ELF3 is a night-acting factor, how dawn signals enter the oscillator in a TIC-dependent manner remains to be resolved.
ELF3 as a Repressor of PRR9 and PRR7 in the Morning Loop of the Circadian Clock
The zeitgeber cycle can drive a rhythm in many clock mutants and therefore mask an underlying clock oscillation (McWatters et al., 2000; Yamashino et al., 2008). The elf3-1 loss-of-function mutant under free-running conditions has been shown to display major defects in the expression of the central oscillator genes CCA1, LHY, TOC1, and GI (Schaffer et al., 1998; Fowler et al., 1999; Alabadí et al., 2001; Kikis et al., 2005). Our analyses with the hypomorphic elf3-12 allele revealed an early phase for all clock transcripts examined (Figures 2 and 4), in agreement with its short period under LL (Figures 1 and 5). We note here that ELF3 is autoregulatory, as the elf3-12 mutant displayed elevated ELF3 transcript levels (Figures 2A to 2C). This implies that ELF3 represses its own transcription, a process that could be indirect. For other core clock genes, the amplitude of expression was basically unaffected in elf3-12 under LL, whereas we found that elf3-1 and elf3-7 had constant wild-type peak levels of morning genes PRR9 and PRR7 and also peak levels of the evening genes, including LUX (Figure 4). This is in agreement with earlier reports (Fowler et al., 1999; Alabadí et al., 2001; Kikis et al., 2005). The amplitude phenotype of elf3-12 relative to elf3-1 and elf3-7 was interesting. Compared with under LL, the elf3-12 amplitude was slightly different in DD, with reduced levels of CCA1 and LHY, essentially resembling elf3-1 and elf3-7. Recently, it was shown that PRR9 and PRR7 work as direct repressors of CCA1 and LHY expression (Nakamichi et al., 2010; Pokhilko et al., 2010). Therefore, the low expression levels of CCA1 and LHY in elf3-12 in DD could be due to the higher expression levels of PRR9 and PRR7. Recently, it was also determined that both LUX and ELF3 bind to the PRR9 promoter (Dixon et al., 2011; Helfer et al., 2011), but how the elevated level of PRR9 connects to both LUX and ELF3 activity is unclear and will need further investigation. Taking all these gene expression data into account, and the finding that ELF3 can associate with the PRR9 promoter (Dixon et al., 2011), we propose that ELF3 directly represses PRR9 and PRR7 and that this leads to indirect activation of CCA1 and LHY by ELF3 (Figure 9).
Figure 9.
Model for ELF3 Action in the Circadian Clock.
ELF3 is a repressor of circadian period by repressing PRR9 and PRR7 expression. ELF3 indirectly promotes the expression of LHY and CCA1 through this repression of PRR9 and PRR7. Under ambient light, phy-mediated input signals drive the oscillator by reducing the repressive function of ELF3. We propose that the ELF3 central domain (Block II) promotes ELF3 action by increasing its nuclear pool (nuclear anchor). Hence, the effects of phy-mediated repression and nuclear anchoring on ELF3 are additive.
With regard to circadian-regulated LUC rhythms, we found that elf3-12 was clearly different from elf3-1 and elf3-7. Most notably, the clock in elf3-12 was overtly rhythmic and had light-dependent short-period rhythms for expression of both clock components (LHY) and clock outputs (CAB2 and CCR2; Figures 1 and 5). This mutant phenotype was opposed to the long-period phenotype of ELF3-ox (Covington et al., 2001), supporting the hypothesis that ELF3 is a repressor of circadian period (Covington et al., 2001; Thines and Harmon, 2010).
The elf3-12 Mutant Has Gating and Phase-Resetting Defects
Since the elf3-12 period phenotype was light dependent, we reasoned that elf3-12 might be defective in integrating light signals to the clock. In a frequency demultiplication assay, we found that elf3-1 mutants failed to entrain to T-cycles shorter than 24 h, consistent with a previous study (Thines and Harmon, 2010). By contrast, elf3-12 also demultiplied, as the wild type (see Supplemental Figure 6 online), confirming that the oscillator in elf3-12 is functional.
ELF3 has a crucial function in gating of light signals to the circadian clock and is therefore important for proper clock resetting, which mainly takes place at dawn (Millar and Kay, 1996; McWatters et al., 2000). We found that elf3-12 had only a subtle gating phenotype, at the end of the night (Figure 6A). Furthermore, by constructing a PRC, we found resetting defects around the late subjective night, when phase advances in elf3-12 were enhanced, whereas most of the delays had normal amplitude (Figure 6B). This phenotype was different from ELF3-ox, which responded with reduced advances and also reduced delays to red light resetting pulses (Covington et al., 2001). Both gating and phase-resetting phenotypes indicate that elf3-12 hypersensitivity to red light signaling is caused by an attenuated function of the ELF3-12 protein.
ELF3 Has Limited Control of FR Input to the Circadian Clock
We found a more limited function of ELF3 in FRc signaling to the oscillator, as an overt rhythm was detected in both elf3-1 and elf3-7. Importantly, the elf3-7 mutation did not reduce the rhythmicity in the PHYA-ox background (see Supplemental Figure 10 online), indicating that the assumed residual ELF3 function in elf3-7 was sufficient for proper PHYA-ox rhythms under FRc. Regarding elf3-12 under FRc, elf3-12 conferred a delay in the phase of PHYA-ox. This was different from the elf3-12 PHYA-ox double mutant phenotype under Bc and Rc (Figures 7 and 8; see Supplemental Figure 8 and Supplemental Table 2 online). We interpret this observation as genetic enhancement of elf3-12 by PHYA-ox under FRc. This is evidence of overlap between the respective genetic pathways controlling FRc input to the clock, as also indicated by the epistasis of elf3-12 to PHYA-ox under other light qualities. In summary, ELF3 has a role in the clock under FRc, but, surprisingly, a weak oscillation persists even after genetic depletion of ELF3, supporting FR-specific roles of additional evening loop genes (TOC1, GI, and ELF4), as suggested by Wenden et al. (2011) in a recent study.
Enhanced Repression of elf3-12 by PHY-ox
Under LL, increasing light intensities result in a shortening of the circadian period (Aschoff's Rule; Aschoff, 1979; Millar et al., 1995b; Somers et al., 1998; Devlin and Kay, 2000). Light input to the clock is dominated by the action of phyA and phyB (Anderson et al., 1997; Somers et al., 1998; Devlin and Kay, 2000). Accordingly, due to increased light input to the circadian clock, the PHY-ox lines used in this study displayed short-period rhythms (Wagner et al., 1991; Anderson et al., 1997; Figures 7 and 8; see Supplemental Tables 2 and 3 online). On the contrary, ELF3-ox was found to lead to long period under LL (Bc and Rc; Covington et al., 2001). Hence, the phys and ELF3 have opposite roles in regulating circadian periodicity. Consistent with an attenuated ELF3 function in elf3-12, we found light-dependent short-period circadian rhythms in this allele (Figure 5). Since the effect of PHY-ox was additive to elf3-12, thus leading to enhanced short period under LL (Figure 7; see Supplemental Tables 2 and 3 online), and the binding of phyB to ELF3 was intact in elf3-12 (see Supplemental Figure 2 online), we hypothesize that the physical phyB-ELF3 interaction leads to repression of ELF3-mediated lengthening of the period of the circadian clock.
ELF3 has been shown to be a nuclear protein (Liu et al., 2001; Yu et al., 2008), and it was reported that ELF3 works as a transcriptional repressor associating to the PRR9 promoter (Dixon et al., 2011). Hence, the decrease of the ELF3 nuclear pool that we observed in ELF3:ELF3-12-YFP (Figures 2F and 2G) could explain the attenuated ELF3 function (light-dependent short periodicity; Figures 5 and 6). Therefore, we hypothesize that the ELF3 central domain (Block II) activates ELF3 by increasing its nuclear pool (Figure 9) and that elf3-12 is defective in this activation step. Thus, we propose that in elf3-12, the lack of activation is additive to a light-dependent increased repression by phyB. This model could account for both the attenuated ELF3 function and the additive effect of PHY-ox on the elf3-12 period phenotype that we only observed under LL (Figure 9).
Signaling mediated by the phys is known to modulate the phase of the oscillator under different light qualities (Salomé et al., 2002, 2006; Hanano et al., 2006). Our circadian analysis revealed light-dependent epistasis of elf3-12 to PHYA-ox regarding the phase property of the oscillator (Figure 8; see Supplemental Table 2 online). This was a new finding, as earlier, using elf3-1, it was concluded that ELF3 and PHYA work in independent genetic pathways, including developmental and growth-related traits (Reed et al., 2000; Liu et al., 2001). By contrast, we found that PHYB-ox suppressed elf3-12 circadian phase both under Rc and in DD (Figure 8; see Supplemental Table 3 online), supporting an increased repression effect of the phyB-ELF3 interaction in the elf3-12 mutant background (above). We did not find an additive effect of PHYA-ox on the phase of elf3-12, indicating that period and phase effects of phys on ELF3 have different mechanisms.
Taken together, our circadian characterization of the hypomorphic elf3-12 allele led us to delineate a functional domain required to inhibit phy action on ELF3 in the nucleus. We found that the elf3-12 mutant had a light-dependent short-period phenotype, leading to an alteration in ELF3 cellular distribution, a defect in gating of light signaling, and enhanced resetting responses by light. Our findings are consistent with ELF3 being a multifunctional protein that integrates light signals to the clock and does so as a core, repressive oscillator component.
METHODS
Plant Materials and Growth Conditions
The elf3-12 allele is in the Arabidopsis thaliana C24 ecotype. The mutants and overexpression lines used in this study were as follows: elf3-1 (Hall et al., 2003), elf3-7 (McWatters et al., 2000), elf3-12 (this study), PHYA-ox (AOX; Boylan and Quail, 1991; Anderson et al., 1997), and PHYB-ox (BOX; Wagner et al., 1991; Anderson et al., 1997). All lines were backcrossed an additional three times to the C24 wild type to remove the CAB2:LUC (2CA/C) reporter gene and homogenize the accession background. Homozygous plants were subsequently identified in the BC3-F2 population using specific markers (see Supplemental Table 6 online). The LUC reporter lines used were CAB2:LUC+ (Hall et al., 2001), LHY:LUC+ (Kevei et al., 2006), and CCR2:LUC+ (Doyle et al., 2002; Kevei et al., 2006), which were integrated into the mutant genomes by crossing.
For LUC assays, seeds were surface sterilized and plated on Murashige and Skoog medium (4.4 g/L, pH 5.7) containing 3% Suc. Following ~3 d of stratification at 4°C, seedlings were entrained under 12L:12D cycles (~10 μmol m−2 s−1) with a constant temperature of 22°C. For flowering-time measurements, plants were grown on soil containing a 3:1 mixture of substrate and vermiculite in a growth chamber with 8 h light/16 h dark and 22°C/18°C temperature cycles, as described (Domagalska et al., 2007). Relative humidity was ~50%, and the light intensity was ~200 μmol m−2 s−1. The flowering time was scored at the time of bolting (i.e. shoot 0.5 cm above rosette) as the total leaf number. For hypocotyl assays, seedlings were grown from germination on Murashige and Skoog medium (2.2 g/L, pH 5.7) without Suc, as described (Davis et al., 2001). Hypocotyl length was determined for seedlings grown under ShD (8L:16D), in DD, or Rc for 7 d (light intensity: ShD, 120 μmol m−2 s−1; Rc, 15 μmol m−2 s−1). For expression analysis of PIF4 and PIF5 and the respective hypocotyl measurements, seedlings were grown under Rc with 12-h 20°C/12-h 18°C cycles. Seedlings were scanned and hypocotyl elongation was measured as described (Davis et al., 2001). These experiments were performed at least three times with similar results.
Mapping
The elf3-12 mutant was isolated and mapped from a forward genetic screen of ethyl methanesulfonate–mutagenized populations harboring the CAB2:LUC (2CA/C) reporter gene (Millar et al., 1995a; Kevei et al., 2006). Rough mapping of its phase advance phenotype (Figure 1A) revealed linkage to the ELF3 gene (At2g25930) and subsequent sequencing of ELF3 identified the transition mutation shown in Figure 1B.
Sequence Alignment
ELF3-like sequences were identified from sequences in the database of plant transcript assemblies (Childs et al., 2007) at The Institute of Genomic Research (http://blast.jcvi.org/euk-blast/plantta_blast.cgi) and Phytozome (www.phytozome.net). Sequences were aligned with Block II of ELF3, and its only Arabidopsis sequence relative EEC (At3g21320), using the multiple alignment tool ClustalX (Thompson et al., 1997). The alignment was formatted as previously described (Kolmos et al., 2008).
Expression Analysis
RNA extraction was performed on ~7-d-old seedlings at the time points indicated. Growth conditions, quantitative RT-PCR, and primer sequences were previously described (Kolmos et al., 2009). Additional primers are as follows: ELF3, forward, 5′-GATGCCCACCATAATGAACC-3′, and reverse, 5′-TTGCTCGCGGATAAGACTTT-3′; PIF4, forward, 5′-GATCATCTCCGACCGGTTTGCTAGATACAT-3′, and reverse, 5′-CGGTGGTCTTCGTCGGCACAGACGACGGTT-3′; PIF5, forward, 5′-CGGAGCAGCTCGCTAGGTACATGGGCAGGA-3′, and reverse, 5′-ACCCATATGAAGACTGTCGGTGGTCGCCGG-3′. Primer efficiencies were determined for an annealing temperature of 58°C. The quantitative RT-PCR was analyzed with the Bio-Rad software package version 2.0 according to the manufacturer's recommendations. Samples were normalized to the expression of PROTEIN PHOSPHATASE 2a subunit A3 (PP2A; At1g13320): forward fwd, 5′-TATCGGATGACGATTCTTCGTGCAG-3′; reverse, 5′-GCTTGGTCGACTATCGGAATGAGAG-3′. The relative gene expression is shown as the average of three technical replicates, and error bars represent sd among the technical replicates. The results presented are representative of at least three biological replicates.
Expression of ELF3-YFP
The Gateway cassette of the pDESTR4R3 vector (Invitrogen) was cloned into the binary vector pPZP211 (Hajdukiewicz et al., 1994) to obtain the modified pPZP211R4R3 vector. The ELF3 promoter (3 kb upstream of the 5′ untranslated region), the ELF3 genomic coding region (including 3′ untranslated region), and the YFP cDNA were amplified by PCR and cloned into the vectors pDONR221:P4-P1R, pDONR201, and pDONR221:P2R-P3 (Invitrogen), respectively (see Supplemental Table 7 online for primer sequences). The three constructs were then recombined into the pPZP211R4R3 by Multi-Gateway LR reaction (Invitrogen). The final plasmid was introduced into Agrobacterium tumefaciens GV3101. Agroinfiltration of Nicotiana benthamiana was performed as described (Voinnet et al., 2003). After 3 d, infiltrated leaves were excised at ~ZT16 and imaged using a Leica TCS SP2 AOBS confocal laser scanning microscope. Identical microscope settings were used to image both wild-type and ELF3-12 YFP fusion proteins. Average intensity projections of 10-μM stacks formed by 10 1-μm sections were generated using Leica confocal software. Heat maps of the YFP intensity were generated using the Interactive 3D Surface Plot plug-in in ImageJ (available at http://rsb.info.nih.gov/ij). Infiltration followed by microscopy observation were performed three times with similar results. Each time, leaf excisions from at least two different leaves were imaged.
Yeast Two-Hybrid Experiments
ELF3 and PHYB full-length cDNAs were amplified with Pfu-Ultra II Fusion HS (Stratagene) and recombined into the pDONR201 vector (Invitrogen) (see Supplemental Table 8 online for primer sequences). Quikchange mutagenesis (Stratagene) was subsequently used to introduce the analogous point mutation present in the elf3-12 allele into the pDONR201 vector containing ELF3 full-length cDNA to obtain pDONR ELF3-12 (see Supplemental Table 8 online for primer sequences). Then, the corresponding amplicons were shuttled from the pDONR201 into pDEST22 (Gal4 AD) and pDEST32 (Gal4 DB) (Invitrogen) to obtain BD-ELF3, BD-ELF3-12, and AD-PHYB. The recombinant pDEST22 (Gal4 AD) and pDEST32 (Gal4 DB) vectors were cotransformed into Saccharomyces cerevisiae AH109 strain by Li-Ac Small Scale Transformation (Clontech). Yeast two-hybrid experiments were performed using minimal synthetic defined base (Clontech) supplemented with the appropriate amino acid dropout (−Leu/−Trp/−His/−Ade; Clontech) to select for positive interactions. For plating, a 10-fold dilution series was started from a 3-mm colony of double-transformed yeast that was resuspended in 200 μL sterile TE (10 mM Tris and 1 mM EDTA, pH ).
Analysis of LUC Expression Profiles
LUC expression profiles were monitored and processed as described by Hanano et al. (2008) and Boikoglou et al. (2011). The gating experiment was performed as described by McWatters et al. (2000). Briefly, plants were entrained to LD 12/12, and on day 6, plants were transferred to DD. Then, replicate samples were exposed to a 20-min white light pulse every 2 h. Acute induction of CAB2:LUC was calculated by subtracting the luminescence of each seedling before the light pulse to that of the luminescence after the pulse. In these experiments, 24 seedlings were analyzed per genotype. The PRC was constructed similarly to Covington et al. (2001), with the modification that phase was based on the first acro-peak following the resetting pulse (below). More than 24 seedlings were analyzed per genotype per time point. Briefly, for this PRC, plants were grown for 7 d under white 12L:12D and then transferred to DD for one full day before 1-h red light pulses (40 μmol m−2 s−1) were applied every 3 h. The time of the first peak after each pulse was calculated for the pulsed and nonpulsed populations. The circadian period of each population after the pulse was calculated and used to transform phase values to circadian phase (CT). Phase advances were plotted as positive and phase delays as negative values with error bars that represent pooled se (y axis). The time of the pulse was corrected to circadian time (CT; x axis). The LUC assays were performed at least twice with similar results.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers defined in Supplemental Figure 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Full-Length ELF3 Sequences.
Supplemental Figure 2. Yeast Two-Hybrid Interaction between ELF3-12 and phyB.
Supplemental Figure 3. Flowering Time under LgD.
Supplemental Figure 4. PIF4 and PIF5 Expression and Hypocotyl Elongation under Rc and ShD.
Supplemental Figure 5. Transcript Profiling of Clock Genes in elf3 Mutants under LD.
Supplemental Figure 6. Frequency Demultiplication is Normal in elf3-12.
Supplemental Figure 7. Confirmation of the Gating Phenotype of elf3-1.
Supplemental Figure 8. PHYA-ox Enhances the Short Circadian Period of elf3-12 under Light, but elf3-12 Controls the Phase.
Supplemental Figure 9. PHYB-ox Enhances the Short Circadian Period of elf3-12 under Light, and elf3-12 Does Not Control the Phase.
Supplemental Figure 10. elf3-1 PHYA-ox and elf3-7 PHYA-ox Rhythms.
Supplemental Figure 11. elf3-1 PHYB-ox and elf3-7 PHYB-ox Rhythms.
Supplemental Table 1. Summary of Period Estimates, Hypocotyl Length, and Flowering Time
Supplemental Table 2. Circadian Period and Phase Estimates of LHY:LUC in elf3-12 PHYA-ox.
Supplemental Table 3. Circadian Period and Phase Estimates of LHY:LUC in elf3-12 PHYB-ox.
Supplemental Table 4. Circadian Rhythmicity of CCR2:LUC in elf3-1 and PHYA-ox.
Supplemental Table 5. Circadian Rhythmicity of CCR2:LUC in Various elf3-7 Genotypes.
Supplemental Table 6. dCAPS Genotypic Markers.
Supplemental Table 7. ELF3-YFP Primers.
Supplemental Table 8. Yeast Two-Hybrid Primers.
Acknowledgments
We thank A.M. Davis for technical assistance and R. Saini for the expanded ELF3 alignment. We also thank L. Kozma-Bognar for comments on the manuscript and A. Viczian for assistance to the yeast two-hybrid experiments. S.J.D. received funding from the Max-Planck-Gesellschaft and Deutsche Forschungsgemeinschaft (Grant DA 1061/4-1; SPP 1530/1). A.J.M. received funding from the Biotechnology and Biological Sciences Research Council (Award G10325).
AUTHOR CONTRIBUTIONS
S.J.D., F.N., and A.J.M conceived the project. E.K., E.H., N.B., R.T., and P.G. designed the experiments and carried out the work. E.K., E.H., and N.B. wrote the article under guidance from A.J.M., F.N., and S.J.D.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Anderson S.L., Somers D.E., Millar A.J., Hanson K., Chory J., Kay S.A. (1997). Attenuation of phytochrome A and B signaling pathways by the Arabidopsis circadian clock. Plant Cell 9: 1727–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff J. (1979). Circadian rhythms: Influences of internal and external factors on the period measured in constant conditions. Z. Tierpsychol. 49: 225–249 [DOI] [PubMed] [Google Scholar]
- Boikoglou E., Ma Z., von Korff M., Davis A.M., Nagy F., Davis S.J. (2011). Environmental memory from a circadian oscillator: The Arabidopsis thaliana clock differentially integrates perception of photic versus thermal entrainment. Genetics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan M.T., Quail P.H. (1991). Phytochrome a overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88: 10806–10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K.L., Hamilton J.P., Zhu W., Ly E., Cheung F., Wu H., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P. (2007). The TIGR plant transcript assemblies database. Nucleic Acids Res. 35(Database issue): D846–D851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Panda S., Liu X.L., Strayer C.A., Wagner D.R., Kay S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Bhoo S.H., Durski A.M., Walker J.M., Vierstra R.D. (2001). The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol. 126: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Millar A.J. (2001). Watching the hands of the Arabidopsis biological clock. Genome Biol. 2: REVIEWS1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K., Franci C., Parks B.M., Seeley K.A., Short T.W., Tepperman J.M., Quail P.H. (1993). Arabidopsis HY8 locus encodes phytochrome A. Plant Cell 5: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montaigu A., Tóth R., Coupland G. (2010). Plant development goes like clockwork. Trends Genet. 26: 296–306 [DOI] [PubMed] [Google Scholar]
- Devlin P.F., Kay S.A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Doyle M.R., Amasino R.M., Davis S.J. (2007a). A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Millar A.J., Davis A.M., Davis S.J. (2007b). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19: 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Domagalska M.A., Schomburg F.M., Amasino R.M., Vierstra R.D., Nagy F., Davis S.J. (2007). Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Dowson-Day M.J., Millar A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Schlereth A., Stitt M., Smith A.M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hall A., Bastow R.M., Davis S.J., Hanano S., McWatters H.G., Hibberd V., Doyle M.R., Sung S., Halliday K.J., Amasino R.M., Millar A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Kozma-Bognár L., Bastow R.M., Nagy F., Millar A.J. (2002). Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 32: 529–537 [DOI] [PubMed] [Google Scholar]
- Hall A., Kozma-Bognár L., Tóth R., Nagy F., Millar A.J. (2001). Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol. 127: 1808–1818 [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Domagalska M.A., Nagy F., Davis S.J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Hanano S., Stracke R., Jakoby M., Merkle T., Domagalska M.A., Weisshaar B., Davis S.J. (2008). A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genomics 9: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K.A., Albertson T.M., Wagner D.R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei E., et al. (2006). Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 140: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Hicks K.A., Somers D.E. (2005). Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 139: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Davis S.J. (2007). ELF4 as a central gene in the circadian clock. Plant Signal. Behav. 2: 370–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Nowak M., Werner M., Fischer K., Schwarz G., Mathews S., Schoof H., Nagy F., Bujnicki J.M., Davis S.J. (2009). Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 3: 350–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Schoof H., Plümer M., Davis S.J. (2008). Structural insights into the function of the core-circadian factor TIMING OF CAB2 EXPRESSION 1 (TOC1). J. Circadian Rhythms 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wagner D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J.C.W., Kozma-Bognar L., Gould P.D., Feher B., Kevei E., Nagy F., Turner M.S., Hall A., Millar A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters H.G., Bastow R.M., Hall A., Millar A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- McWatters H.G., Kolmos E., Hall A., Doyle M.R., Amasino R.M., Gyula P., Nagy F., Millar A.J., Davis S.J. (2007). ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 144: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.J., Carré I.A., Strayer C.A., Chua N.H., Kay S.A. (1995a). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar A.J., Kay S.A. (1996). Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 93: 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.J., Straume M., Chory J., Chua N.H., Kay S.A. (1995b). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Murakami M., Tago Y., Yamashino T., Mizuno T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48: 110–121 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Onai K., Ishiura M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972 [DOI] [PubMed] [Google Scholar]
- Pittendrigh C.S. (1981). Circadian systems: Entrainment. In Biological Rhythms. Handbook of Behavioral Neurobiology, Vol. 4, Aschoff J., (New York: Plenum Press; ), pp. 95–124 [Google Scholar]
- Pokhilko A., Hodge S.K., Stratford K., Knox K., Edwards K.D., Thomson A.W., Mizuno T., Millar A.J. (2010). Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol. Syst. Biol. 6: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Bastow R.M., Solomon K.S., Dowson-Day M.J., Elumalai R.P., Millar A.J. (2000). Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 122: 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden L.C., Ingle R.A. (2009). Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21: 2546–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., Michael T.P., Kearns E.V., Fett-Neto A.G., Sharrock R.A., McClung C.R. (2002). The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 129: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., To J.P., Kieber J.J., McClung C.R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18: 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schöning J.C., Streitner C., Page D.R., Hennig S., Uchida K., Wolf E., Furuya M., Staiger D. (2007). Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Strasser B., Alvarez M.J., Califano A., Cerdán P.D. (2009). A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J. 58: 629–640 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Thines B., Harmon F.G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wagner D., Tepperman J.M., Quail P.H. (1991). Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell 3: 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Barnaby J.Y., Tada Y., Li H., Tör M., Caldelari D., Lee D.U., Fu X.D., Dong X. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenden B., Kozma-Bognar L., Edwards K.D., Hall A.J., Locke J.C., Millar A.J. (2011). Light inputs shape the Arabidopsis circadian system. Plant J. 66: 480–491 [DOI] [PubMed] [Google Scholar]
- Yamashino T., Ito S., Niwa Y., Kunihiro A., Nakamichi N., Mizuno T. (2008). Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 49: 1839–1850 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Fekih R., Fujiwara S., Oda A., Miyata K., Tomozoe Y., Nakagawa M., Niinuma K., Hayashi K., Ezura H., Coupland G., Mizoguchi T. (2009). Possible role of early flowering 3 (ELF3) in clock-dependent floral regulation by short vegetative phase (SVP) in Arabidopsis thaliana. New Phytol. 182: 838–850 [DOI] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta M.T., Hicks K.A., Jacobs C.I., Young J.C., Hangarter R.P., Meeks-Wagner D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zeilinger M.N., Farre E.M., Taylor S.R., Kay S.A., Doyle F.J. (2006). A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]