Abstract
In dividing fission yeast Schizosaccharomyces pombe cells, the balance between Wee1 kinase and Cdc25 phosphatase which control the cyclin-dependent kinase (CDK) at the G2–M transition determines the rod-shaped cell length. Under nitrogen source starvation or glucose limitation, however, cell size determination is considerably modulated, and cell size shortening occurs for wild-type cells. For several mutants of kinases or phosphatases, including CDK, target of rapamycin complex (TORC) 1 and 2, stress-responsive mitogen-activated protein kinase (MAPK) Sty1/Spc1, MAPK kinase Wis1, calcium- and calmodulin-dependent protein kinase kinase-like Ssp1, and type 2A and 2A-related phosphatases inhibitor Sds23, this cell shortening does not normally occur. In tor1 and ssp1 mutants, cell elongation is observed. Sds23 that binds to and inhibits 2A and 2A-related phosphatases is synergistic with Ssp1 in the cell size determination and survival under low glucose and nitrogen source. Tor2 (TORC1) is required for growth, whereas Tor1 (TORC2) is needed for determining division size according to different nutrient conditions. Surprisingly, in growth-diminished tor2 mutant or rapamycin-treated cells, the requirement of separase/Cut1-securin/Cut2 essential for chromosome segregation is greatly alleviated. By contrast, defects of tor1 with secruin/cut2 or overproduction of Cut1 are additive. While Tor1 and Tor2 are opposite in their apparent functions, both may actually coordinate cell division with growth in response to the changes in nutrients.
Keywords: Tor1, Tor2, Ssp1, protein phosphatase, separase, starvation
1. Introduction
In the book ‘The biology of the cell cycle’, Mitchison [1] wrote that one of the basic questions to ask about the cell cycle is what is the pattern of overall cell growth between one division and the next. This question still remains. It is surprising how little we know about the basic mechanism of cell growth in the relationship to the cell cycle. Mitchison pointed out the criteria that can be used for measuring cell growth; they are cellular volume, constituents including water, total dry mass, macromolecular dry mass and low molecular weight compounds pool, protein and other macromolecules such as RNA, DNA and carbohydrates. Precise measurements of these parameters have actually not advanced well technically since the time of writing the book, so that growth data are often missing or not satisfactory in many cell cycle studies. The fission yeast Schizosaccharomyces pombe, a eukaryotic micro-organism, is convenient for estimating the cell volume by simply measuring cell length as its cell shape in the vegetative phase is rod-like, so Mitchison pioneered the use of S. pombe as a eukaryotic model for understanding growth versus cell cycle. The growing phase (e.g. cell length increase) of S. pombe in the standard (rich) culture medium occurs after DNA replication, whereas the cell length is constant during the phases of mitosis and cell division [2,3].
Thuriaux et al. [4] and Nurse & Thuriaux [5] isolated S. pombe mutants that were thought to be altered in the control coordinating cell division with cell growth. More than 50 mutant strains—most severely altered in this control—were isolated, which showed the same growth rate as wild-type, but divided at a much shorter cell size. The great majority of the mutants were genetically mapped within the single wee1 locus (wee means little), and the remaining one mutant turned out to be an allele of cdc2, originally called wee2. At that time, Wee1 and Cdc2 were predicted to be involved in a control initiating mitosis when the cell attains a critical cell length. The wee1+ gene was postulated to code for a negative element or inhibitor, and cdc2+ to code for a positive element or activator in the mitotic control. We now know that these elements are indeed cell cycle-regulating protein kinases; Wee1 directly phosphorylates Cdc2 and inhibits the kinase activity. The original mutation wee2-1 (cdc2-w1) escapes the negative regulation by Wee1 resulting in premature division with regard to cell size. The success in identifying Wee1 as the negative regulator of mitotic entry came from the genetic screen for mutants displaying the strongest wee phenotype. In addition, the growth rate was shown to be normal in these mutants, separating the growth issue from the cell cycle control. In retrospect, there were a number of mutants that showed the semi-wee phenotypes, which were wisely not investigated at that time. After 30 years since the discovery of wee1 mutants, however, the time may be ripe to shed light on broad mutations that produce the less severe, ‘wee-like’ phenotypes, many of which may include the defects in growth and/or cell cycle control.
Cdc25, another important regulator for mitotic entry, was discovered by Fantes [6] through the analysis of interactions between wee and various cdc (cell division cycle) mutants. The block of mitotic entry or the prolonged G2 interphase caused by a defective cdc25 allele is suppressed when combined with the wee mutants. Suppression of the temperature-sensitive (ts) cdc25 phenotype by wee1 is almost complete. Other cdc2-w mutations (e.g. cdc2-3w) are sensitive to Wee1 function, but largely abolish Cdc25 requirement. Cdc25 turned out to be a protein phosphatase [7,8] that competes with Wee1 and is an activator of Cdc2 by dephosphorylating the tyrosine residue (Y15) of Cdc2. Not only cdc25, cdc13 (mitotic cyclin mutant) and most cdc2 ts alleles are blocked at the boundary of G2–M transition. Note that the loss of Cdc25 and Cdc2–Cdc13 blocks mitotic entry but not cell growth, leading to the formation of highly elongated cells arrested in the G2–M boundary but continuing growth. The loss of cyclin-dependent kinase (CDK) activation disrupts the cell cycle control and also affects the cell size determination as clearly exemplified by wee1 mutation. It is obvious, though often forgotten, that the cell size is strongly affected by cell cycle control, growth control or both. In wee1 mutant cells, growth is not inhibited, but prematurely committed mitosis and following cytokinesis take precedence over growth to produce small cells.
2. Extensive shortening of cell size occurs by division under nitrogen deficiency
Wild-type S. pombe cells respond to nutritional change by changing the cell size. When S. pombe is transferred from the complete synthetic Edinburgh Minimal Medium (designated EMM2) to EMM2 –N lacking the nitrogen source (NH4Cl), cells can divide approximately twofold, an approximately fourfold increase in number in the absence of the growth phase, producing short and round cells, which are arrested at the G1 phase (figure 1a,b; [9–12]). Note that the EMM2 medium has no amino acids, so that NH4Cl is the sole nitrogen source. These divisions in EMM2 –N are thought to occur and resulting quiescent cells are maintained through recycling the intracellular nitrogen sources. If cell populations are competent for meiosis in the EMM2 –N medium, then cells exit from the G1, conjugate with other mating type cells, and irreversibly commit meiotic divisions. If cell populations are heterothalic (mono-sexual), however, then solitary cells enter the G0 phase at around 12 h and remain viable in the quiescent phase for quite long times (greater than one month). The capability to mate with opposite mating type cells is lost upon entry into the G0 phase. The size of small G0 cells remains constant. The nitrogen-starvation-induced G0 cells thus lack both growth and division, but are metabolically active [12,13].
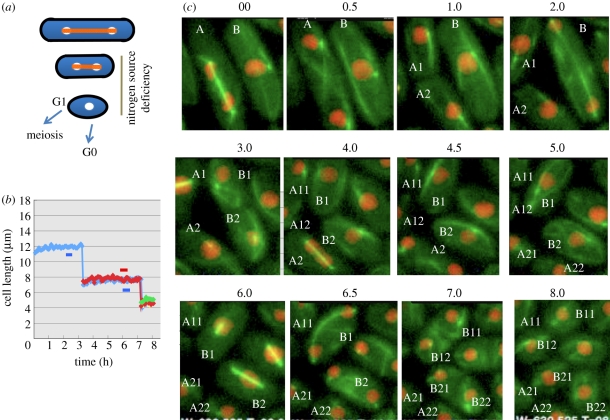
Figure 1.
Nitrogen source deficiency-induced cell size shortening of wild-type. (a) Schizosaccharomyces pombe wild-type cells under the absence of nitrogen source (NH4Cl) divide twice and arrest at a temporal G1 phase followed by meiosis or the entry into quiescent G0 phase dependent on the presence or absence of mating. The orange bar represents the spindle. (b) Cell length is shortened dramatically during two divisions after transfer (time 0) from the complete EMM2 to the NH4Cl-deficient EMM2 medium. (c) Light micrographs of time course changes of the two cells in the complete medium (time 00 h) shifted to the nitrogen-deficient medium for 8.0 h. The first division produced daughter cells A1, A2 and B1, B2 cells. The second division produced A11, A12, A21, A22 and B11, B12, B21, B22 cells. Red, histone H2A; green, tubulin and the Sid4 SPB protein (see text).
During nitrogen source deficiency (designated N-starvation hereafter)-induced divisions, the reduction of cell size occurs from the average 12 µm long rod of vegetative cells in EMM2 to 5 µm diameter round-shaped G0 phase cells in EMM2 –N (figure 1b). The short bars in figure 1b represent the timing of mitosis prior to the first and second cell division. Cell length shortening from mother (blue) to daughter (blue, red) and grand-daughter (red, green) is clearly shown. Note that the growth phase (cell length increase) is missing in these divisions. The calculated ellipsoidal cell volume and the protein content measured for these small G0 cells are 1/3 and 1/6, respectively, of those of vegetative cells [14]. These small G0 cell features are not called the wee phenotype as they are seen in N-starvation, and, besides, cells are arrested while the wee1 mutant cells grow and divide. G0 cells are round-shaped, suggesting that the cytoskeletal architecture is altered from that of wee1 in the presence of nitrogen source. Having stated these differences, there may exist some parallel: accelerated mitoses occur twice in the EMM2 –N medium prior to the arrest. Hence, during nutrient deficiency, wild-type cells try to make a final division before quiescence, which occurs prematurely at a time when the cell size is still small. The wee phenotype therefore may represent the commitment of mitosis under starvation conditions. In other word, N-starvation can produce wee-like cells.
Microarray and high-resolution analysis of transcripts indicate that one half of approximately 5000 whole genome genes had significantly changed levels by simply removing NH4Cl from the medium [14,15]. Mitosis under N-starvation was analysed by movies (figure 1c). Two example of living cells (A and B) were observed by colour-tagged histone H2A-RFP (red, chromatin), alpha-tubulin-GFP (green, microtubule; [16]) and Sid4-GFP protein (green) localized at the spindle pole body (SPB; [17]). They divided at a time interval of approximately 3 h and were arrested (e.g. B11, B12, B21 and B22). The mode of cell division was normal except for the absence of growth, and the decline of cell length by division. The anaphase spindle was short owing to the shortened cell length, or occasionally elongated as the curved form.
How is Wee1 kinase involved during the nitrogen-deficiency-induced ‘premature’ division? Paradoxically, Wee1 kinase is required for the meiotic entry under N-starvation, and phosphorylates the Y15 residue of Cdc2 (CDK1) kinase [18]. The activity of Wee1 is thus not lost. Hence, while not identified, other kinases and phosphatases may be involved in cell size shortening under N-starvation. The state of Cdc2 kinase in the G0 phase is presumably inactive with abundant Rum1, an inhibitor of Cdc2 [14,19], and by Cdc2 Y15 phosphorylation [20].
3. Sty1 and Ssp1 in addition to Cdc2 are required for shortening under N-starvation
To identify mutants that are defective in cell size shortening under N-deficiency, 600 ts strains were searched for any that remained rod-shaped while the cells divided [13]. Only six classes of mutants remained rod-shaped while they divided before the arrest at 26°C, the permissive temperature. Genetic analyses indicated that these have mutations in protein kinase genes that are implicated in cell cycle control and stress-responsive signalling [13]: mitogen-activated protein kinase (MAPK) sty1/spc1-989, MAPK kinase (MAPKK) wis1-558,-982, CDK cdc2-974, mitotic cyclin cdc13-563, calcium- and calmodulin-dependent protein kinase kinase (CaMKK)-like ssp1-412. In databases, p38-like MAPK Sty1 and MAPKK Wis1 are classified as involved in cell cycle, cell growth, gene expression, translation and cellular response to stress, whereas Ssp1 is involved in cell cycle, cell growth, cell morphology, actin cytoskeleton and response to stress. Under N-starvation, stress-responsive Wis1 and Sty1 and calcium-, calmodulin-dependent Ssp1 kinases are thus coordinated with the cell cycle regulator Cdc2–Cdc13 to appropriately change the cell size and shape.
Examples of cells in the nitrogen-deficient medium are shown in figure 2a in comparison with the wild-type control. Interestingly, mutants sty1 and wis1 retained high viability immediately after the two rounds of divisions, but greatly lost the viability during the transition from the transient G1 to the quiescent G0 phase [13]. Resulting long rod-shaped sty1 and wis1 mutant cells displayed large nuclei (figure 2b). Other mutants (cdc2, cdc13 and ssp1) also revealed rod-shaped cells, but retained high viability in the G0 phase at the permissive temperature, suggesting that being rod-shaped per se during the divisions under N-starvation does not cause the loss of viability.
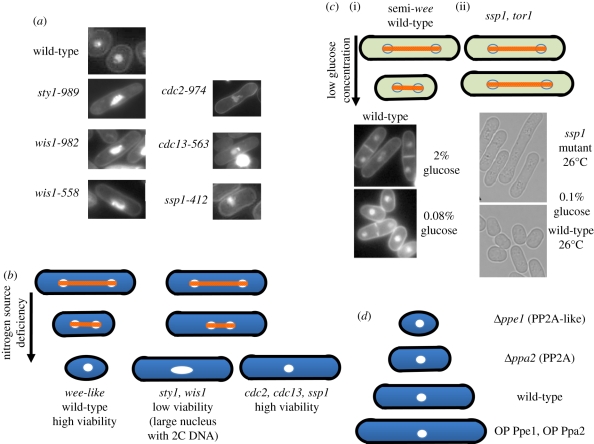
Figure 2.
Genes required for cell size shortening under limited nitrogen or glucose. (a) In the absence of nitrogen source, mutant cells sty1-989, wis1-982, -558, cdc2-974, cdc13-563 and ssp1-412 remained rod-shaped at 26°C, the permissive temperature. (b) Schematic of mutants that fail to shorten cell size upon the transfer to the culture deficient of the nitrogen source. (c) Length of dividing wild-type cells is shortened in EMM2 medium containing low (0.1%) glucose instead of standard 2% glucose concentration. Mutant ssp1-412 is elongated in 0.1% glucose at 26°C rather than the shortening in wild-type. A similar result is obtained for tor1 mutant cells at the semi-permissive temperature. (a,b) Based on Sajiki et al. [13]; (c) based on Hanyu et al. [21]. (d) The phosphatase deletion mutant Δppe1 or Δppa2 results in the production of small, round or short semi-wee cells, respectively. By contrast, overproduction of Ppe1 or Ppa2 causes the semi-cdc25 elongation phenotype [22,23].
Nevertheless, these different classes of protein kinases are vital for shortening divisions under N-starvation. It is thus plausible to speculate that Ssp1, MAPK (Sty1) and MAPKK (Wis1) may directly or indirectly regulate the activation of Cdc2 to cause accelerated mitosis under N-starvation. In the published movies of sty1-989 and wis1-982 mutant cells, the growth phase clearly exists during the division cycles in EMM2 –N medium, while cells keep their rod shape [13]. Sty1 and Wis1 are thus essential for the cessation of growth in N-starvation. It is truly surprising how sty1- and wis1-deficient cells manage to commit divisions while keeping an apparent growth phase without an outsource of nitrogen. One possible reason for such aberrant growth might be due to the use of an abundant carbon source (glucose) for cell size increase, while a nitrogen source may be available by recycling. Indeed, the rate of glucose consumption is high in sty1 mutant cells (L. Uehara & A. Mori 2011, unpublished data). Based on the reason why MAPK and MAPKK mutants post-division lose their viability after entry into the G0 phase, we suspect that the remodelling of nuclear chromatin required for the survival during long-term G0 quiescence may not occur, because the nuclei in these mutants were abnormally expanded [13].
There have been a number of reports showing the close relationships between S. pombe CDK (Cdc2–Cdc13) and stress-responsive pathway (SRP) through various genes, such as protein kinase A (PKA), response regulator Mcs4, Polo-like kinase Plo1 and target of rapamycin (TOR) kinase [24–29]. Plo1 may be important in cell size shortening, as it regulates cytokinesis [30–32]. Petersen & Nurse [27] reported that TOR kinase, which is modulated by nutritional conditions and inhibited by rapamycin, controls the entry into mitosis through stress-responsive Sty1. However, overall understanding of the relationships among CDK, SRP and TOR is still meagre. It is crucially important to set up a clear-cut experimental system to relate growth with the cell cycle. In this regard, the growth-absent, size-shortening division under N-starvation is an excellent model to understand the suppression of growth by nutritional stress. This system may resemble the early embryonic egg cleavage.
4. Under low glucose, the semi-wee is induced in wild-type but not in two kinase mutants
Glucose is a source of energy for cells as well as the source of cell structure, and the cellular mode of its utilization may be centrally important for understanding cell growth, division and quiescence. We investigated the cell division–quiescence behaviour of S. pombe under diverse glucose concentrations from excess, regular diet and starvation to fasting (111–0 mM; [33]). The division mode (observed under a microscopic perfusion system that constantly supplied the medium) was surprisingly normal except for the shortening of cell length (20–30%) when glucose concentrations were highly diluted (5.6 mM, 1/20 concentration of the standard culture medium that contains 111 mM (2%) glucose). This semi-wee length-shortening property is observed in a range of low glucose levels equivalent to human blood sugar concentrations (figure 2c(i)). Normal human blood glucose content is around 4 mM (0.08%) before breakfast. Schizosaccharomyces pombe may be a good model, with regard to understanding the cellular uptake and utilization mechanism of normal blood glucose, which is defective in certain patients with type II diabetes.
When glucose concentration is further reduced to a level of starvation, the nature of division becomes stochastic in addition to cell shortening, accompanied by a curious epigenetic inheritance of division timing. A sharp transition from division to quiescence takes place in a narrow glucose concentration range (from 2.2 to 1.7 mM). Under severe glucose starvation (1.1 mM), cells are mostly quiescent and only a small population of cells divide. Under fasting condition (0 mM), division is immediately arrested, and fasting cells have a short chronological lifespan (16 h) if the shift was abrupt. If, however, the shift to fasting is slow, then the resulting lifespan greatly increases. Various biomarker compounds specific for different glucose concentrations have been identified. Glucose concentrations thus control the cell size, the doubling time, the uniformity of cell division pattern and even epigenetic behaviour among different cell lineages.
It is surprising to find that the doubling time in 111 and 4.4 mM glucose is the same, but the semi-wee phenotype, 20–30% reduction in cell size, partly explains the non-prolonged doubling time in low glucose. Taken together, S. pombe has a very wide range of optimal glucose concentrations for the rate of division with regard to the doubling time. Schizosaccharomyces pombe under low glucose may thus sacrifice the growth phase in order to keep the same rate of increase in cell number. Here, the linked regulation between growth and division clearly exists. Note that glucose limitation or starvation, or even fasting does not affect cell shape, whereas nitrogen starvation causes the deviation of cell shape (to round) from rod.
A subsequent question is what kind of gene function converts the information about limited glucose to cell size determination. Hanyu et al. [21] reported that, under low glucose concentrations, Ssp1 kinase described above plays an important role in the size control; mutant ssp1 cells remain long rod in approximately 0.1 per cent glucose (2% in the regular medium) at 26°C, the permissive temperature, displaying the semi-cdc25 phenotype (figure 2c(ii)). At the semi-permissive temperature under low glucose, the phenotype becomes severer as mutant cells fail to divide. Furthermore, the measurements of remaining glucose concentrations in the medium showed that the rate of glucose consumption is considerably slower in ssp1 mutant cells than that of wild-type under limited glucose, suggesting that the utilization of glucose is impaired in ssp1 mutant cells. Note that ssp1 cells fail to reduce cell size under both nitrogen and glucose limitation, so that it may have a broad role for the utilization of nutrients, such as in incorporation or transport of nutrients. Indeed, Ssp1 is the cell cortex protein. Ssp1 kinase may function in parallel with Gsk3 kinase and oppose PP2A and PP2A-related phosphatases [21].
Another protein kinase identified is Tor1, the catalytic subunit of TORC2 kinase, the mutant of which fails to reduce the cell length under limited glucose (figure 2c; [20]). A new tor1 substitution mutant tor1-L2045D was constructed using the information of the ts tor2 mutation site that resides in the highly conserved phosphatidyl inositol kinase domain of the catalytic subunit [34]. Only the substitution mutant tor1-L2045D displayed the ts phenotypes among the five different substitutions made at the same site. This tor1-L2045D mutant (tor1-D hereafter) grows normally at 26°C but fails to grow at 36°C or under low glucose concentration at the semi-restrictive temperature, displaying semi-cdc25 phenotype at 36°C or in low glucose at a semi-permissive temperature (the deletion mutation Δtor1 is more severe than the tor1-D mutation). Taken together, two nutrient-sensitive protein kinases Ssp1 and Tor1 are responsible for the cell size reduction in response to limited glucose. Screening a large number of ts and deletion mutants grown under the low glucose identified these mutants (details described elsewhere).
5. CaMKK-like Ssp1 is related to SIT4-like Ppe1 phosphatase, cortex actin and AMPK-like Ssp2 kinase
The ssp1+ gene was originally identified as one of the extragenic ts suppressors for the cold-sensitive (cs) deletion phenotypes of Ppe1 [35]. Ppe1 is a member of the evolutionarily conserved type 2A-related phosphatase family, similar to budding yeast SIT4 and mammalian PP6 [36–39]. Ssp1 kinase is involved in salt–stress responses as it is rapidly recruited to the plasma membrane during high salt-induced osmotic pressure [32,40]. While the mutant phenotype resembles that of sty1 stress-responsive MAPK mutants, Ssp1 and Sty1 do not seem to act through the same pathway. Ssp1 controls the state of cortical actin [32,35,40]: ssp1 mutant cells grow in a monopolar fashion and arrest at the G2–M boundary, but the relationship between Cdc2 and Ssp1 in the cell cycle is unclear. Cortical actin distribution in growing ssp1 mutant cells is also monopolar. Ssp1 is hence required to promote the bipolar, rather than monopolar, cell elongation. Overproduction of Ssp1 kinase caused the dispersion of actin, resulting in round cell shape [35]. By contrast, the Δppe1 deletion mutant causes the dispersal of actin, resulting in small round cells. Judging from the mutant phenotypes, Ssp1 kinase and Ppe1 phosphatase may be opposing. They might act on the same substrate important for responding to nutrient limitation. One candidate is Ssp2 (one of the two AMPKs, see below).
Ssp2 kinase was also identified as an extragenic suppressor for Δppe1 [35]. Ssp2 is an S. pombe homologue of mammalian AMP-dependent protein kinase (AMPK) and budding yeast Snf1, containing three distinct subunits [21]. AMPK is thought to be a central player for carbohydrate catabolic processing. Schizosaccharomyces pombe actually has two AMPK-like catalytic subunits, Ppk9 and Ssp2, and 1 β and 1 γ subunit homologues (Amk2/Spcc1919.03c and Cbs2, respectively). The regulatory subunit Cbs2 is essential to maintain the viability of N-starved G0 cells. The cell cycle phenotypes of AMPK catalytic subunit mutants remain to be investigated.
6. Roles of PP2A Ppa2 and PP2A-related Ppe1 phosphatases for mitotic entry and cell size control
It has been known that PP2A and PP2A-related catalytic subunits, Ppa2 and Ppe1, respectively, affect the size of dividing S. pombe cells [22,23]. The cs phenotype of the Δppe1 mutant is rescued by overproduction of the catalytic subunits of PP2A. Both Δppe1 and Δppa2 mutants are small, but differ in shape, round and rod, respectively (figure 2d). Schizosaccharomyces pombe has the second PP2A catalytic subunit Ppa1. The double mutant Δppa1 Δppa2 is lethal, producing the small rod cells indistinguishable from wee1 [41]. If okadaic acid is added to the culture of single mutant Δppa2, then basically the same result was obtained. By contrast, overproduction of Ppa2 results in the semi-cdc25-like elongation, producing long rod cells. Taken together, PP2A and PP2A-related phosphatases may play similar roles in the cell size control. Judging from cell shape of the mutants, Δppe1 might be more related to growth defect, whereas Δppa2 is defective in mitotic entry. Kinoshita et al. [22] showed that Ppa2 interacts genetically with the cell cycle regulators Cdc25 tyrosine phosphatase and Wee1 kinase in S. pombe: the Δppa2 mutant is lethal when combined with wee1-50, but partially suppresses the phenotype of cdc25-22, suggesting that Ppa2 and Wee1 may function in parallel.
In higher eukaryotes, PP2A is sharply downregulated during mitosis by greatwall kinase through its target proteins, alpha-endosulphine and Arpp19, which are inhibitors of PP2A [42,43]. Greatwall kinase phosphorylates and activates the inhibitors of the PP2A-B55delta holoenzyme. In S. pombe, homologues of greatwall and Arpp19/endosulphine are Ppk18 and Mug134, respectively, judging from the database search, whereas in Saccharomyces cerevisiae, RIM15 kinase and IGO1/2 represent the counterparts, respectively [44]. RIM15 is known to phosphorylate IGO1, which is required for initiation of G0 phase. It is of considerable interest whether these budding yeast and fission yeast counterparts of greatwall and alpha-endosulphine (and/or Arpp19) also negatively regulate PP2A homologues during mitosis. In S. pombe, the B55delta counterpart subunit is probably Pab1. The Δpab1 deletion phenotype suggests that Pab1, the regulatory subunit of PP2A, may control the polar actin distribution [45], possibly linked to cell shape control. Our recent results indicate that Δppe1, Δekc1 and Δpab2 lose their viability in nitrogen-starved G0 cells (K. Sajiki 2011, unpublished data), suggesting that these phosphatases may also be involved in cell size and shape change under nitrogen starvation.
7. Sds23 is related to diverse functions by binding to and inhibiting PP2A and PP2A-related phosphatases
Hanyu et al. [21] reported that Sds23 is a key to linking phosphatases with the utilization of low glucose and related kinase Ssp1. Sds23 was identified as one of the three high-copy suppressors for cs dis2-11 that is defective in PP1 phosphatase [46]. The remaining two other suppressors are Sds21, the second PP1 catalytic subunit [47,48], and Sds22, the positive regulatory subunit of PP1 to promote metaphase–anaphase progression [49–51]. The mammalian homologue of Sds22 is implicated in cancer [52]. Sds23 is known to be related to diverse functions through its ability as a high-copy suppressor for mutants of PP1, APC/cyclosome subunits, Ssp1 and others (figure 3a; [21,46]; Y. Hanyu & M. Yanagida 2011, unpublished data). Sds23 is also involved in inducing sexual development as Moc1 is identical to Sds23 [53]. Conversely, high-copy plasmids carrying the PP1 dis2+, APC/C cut9+ and ssp1+ gene rescue the deletion of Sds23, so that the suppression is reciprocal (figure 3a).
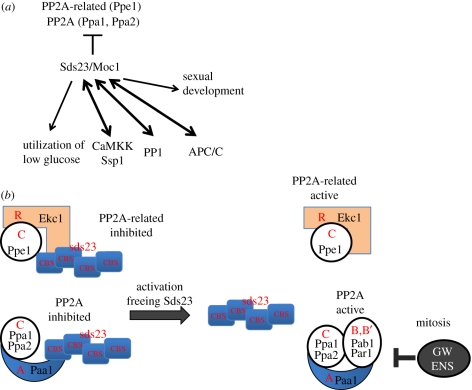
Figure 3.
Diverse roles of Sds23. (a) High-copy plasmid carrying the sds23+ gene suppresses the mutations of ssp1, protein phosphatase PP1 and APC/cyclosome [21,46,51]. Sds23/Moc1 is required for the utilization of low glucose and sexual development. (b) Sds23 stably associates with PP2A-related Ppe1–Ekc1 and PP2A phosphatases (see text). C, R: catalytic and regulatory subunits of PP2A-related phosphatase. C, A, B: catalytic, regulatory A and B subunits of PP2A. The phosphatase free from Sds23 seems to be active [21]. GW and ENS represent greatwall kinase and alpha-endosulphine, respectively (see text).
A critical finding to understand these diverse functions of Sds23 is that Sds23 binds to PP2A and PP2A-related phosphatases, and inhibits in vitro the PP2A-related phosphatase activity of Ppe1 [21]. Analyses of mass spectroscopy and two-hybrid interactions demonstrate that Sds23 is bound directly to the regulatory subunits Ekc1 (SAP-like) and Paa1 (the subunit A-like, figure 3b; [21]). As the B and B′ subunits (Pab1 and Par1, respectively) of PP2A are scarce or missing in the immunoprecipitates, Sds23 might associate with an intermediate assembly form of PP2A holoenzymes. Taken together, high-copy suppression of many mutations by Sds23 appears to be due to the collective negative modulation of two major phosphatases, PP2A and PP2A-related phosphatases, through direct inhibition of the regulatory subunits Paa1 and Ekc1. The inhibitory role of Sds23 is reminiscent of the PP2A inhibitor alpha-endosulphine (IGO1/2, Mus134) that is the target of greatwall kinase. However, Sds23 acts in interphase or throughout the cell cycle. Homologues of Sds23 are found in all fungi and in cellular slime mould, but not in higher eukaryotes so far [21]. It is similar to, but distinct from, the γ subunit of AMPK: both Sds23 and AMPK γ subunit contain the two cystathionine-β-synthase domains.
Sds23 is required to use low glucose: the deletion mutant Δsds23 fails to proliferate in 0.1 per cent glucose and slowly consumes glucose [21]. In sds23-deficient cell extracts, the phosphatase activity greatly increases, and is diminished by the addition of okadaic acid, an inhibitor of PP2A and related phosphatases. The high phosphatase activity of PP2A and PP2A-related may hence be inhibitory to use low glucose. Ssp1 and the phosphatases may be opposing, and Sds23 and Ssp1 synergistically cooperate to use low glucose. The downregulation of PP2A and PP2A-related phosphatases appears to be required for using the low concentration of glucose in the culture medium.
8. Reverse cell size phenotypes of tor1 and tor2 mutants
The target of rapamycin complex (TORC) 1 and 2 exist in eukaryotes (figure 4a; [54]). A variety of cell functions involved in cell growth in response to nutritional cues are controlled by TORCs: TORCs are thought to be the central regulators of growth upon nutritional alterations. Rapamycin is an antiproliferative drug that prolongs the life of model animals, and might be useful in the treatment of certain cancers. Rapamycin bound to FKBP12 (a peptidyl–prolyl cis–trans isomerase) inhibits TORC1 [55]. The mammalian TOR is the sole catalytic subunit, whereas S. cerevisiae and S. pombe contain two highly similar catalytic subunits, Tor1 and Tor2 (figure 4a).
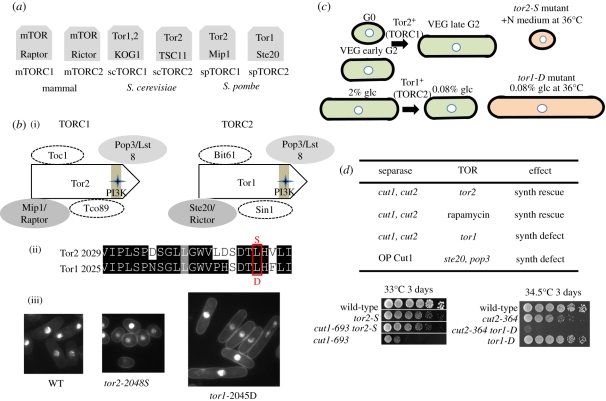
Figure 4.
Functional relationship of TORCs with securin and separase. (a) Two TORCs exist in mammals, Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mammalian TOR is the sole catalytic subunit in mammals, while budding and fission yeasts have two distinct catalytic subunits. (b)(i) The subunit constituents of TORC1 and 2 in S. pombe. The crosses indicate the mutation sites of tor1-D and tor2-S in the PI3K domain of Tor1 and Tor2. (b)(ii) The mutated residues are indicated in red colour in the conserved amino acid sequences in the PI3K domain. (b)(iii) DAPI-stained micrographs of wild-type, tor2-S and tor1-D cultured at 36°C for 6 h. (c) Schematic of the role of Tor2 and Tor1 in cell size determination (see text). glc, glucose. (d) Synthetic rescue or defect observed in the pair of mutations in securin/cut2, separase/cut1 or overproduction (OP) of Cut1 and mutations of tor1, tor2, regulatory subunits (ste20 and pop3) or the addition of rapamycin. See text. Two examples of the spot test at the semi-permissive temperature, showing the (left) synthetic rescue and (right) defect, are shown (see text).
Saccharomyces cerevisiae TORC1 and TORC2 mediate the control of many cellular events [56–61]. The C-terminal domain of catalytic subunits contains a lipid kinase motif, which places them in the phosphatidyl inositol-kinase-related kinase (PIKK) family [62]. Tor1 and Tor2 form two functionally distinct TOR complexes [58]. TORC1, which is responsible for many of the known TOR functions [63], contains either Tor1 or Tor2 (figure 4a). TORC2, which is not sensitive to rapamycin, helps regulating actin cytoskeleton polarization [60], and its complex function is less understood.
Similar to S. cerevisiae, S. pombe has two TOR kinase genes, tor1+ and tor2+ [34,64–68]. However, the nomenclature of TOR kinase in S. pombe is unfortunate; S. pombe Tor2 is similar to S. cerevisiae Tor1, whereas S. pombe Tor1 is similar to S. cerevisiae Tor2. Accordingly, TORC1 and TORC2 in S. pombe contain distinct catalytic subunits, Tor2 and Tor1, respectively (figure 4a,b). Mass spectrometric analysis with immunoprecipitation experiments indicated that S. pombe TORC1 contains only Tor2 [20,34].
To couple extracellular nutrient signals with cell growth, S. pombe TORC1 and TORC2 are reported to be controlled by the small GTPases, Rheb1 [69] and Ryh1 [70], respectively. Wild-type S. pombe is insensitive to rapamycin [71]. However, S. pombe becomes sensitive to rapamycin under conditions of starvation [72] or in tor2 mutants [34].
Each of S. pombe TORC1 and TORC2 contains four evolutionarily conserved regulatory subunits (figure 4b; [34]). Mip1 and Ste20 are homologues of mammalian Raptor and Rictor, respectively, while Pop3/Wat1 is a homologue of Lst8 that associates with both TORC1 and TORC2. The mutation site of a ts and rapamycin-sensitive tor2-S is the substitution from L2048 to S (figure 4b(ii); [34]). A new ts mutant tor1-D was constructed by introducing the D (aspartate) residue at the conserved 2045L as described above. This tor1-D mutant is useful for critical comparison between tor1 and tor2 phenotypes as the mutation sites are basically identical (the inset of sequences; [20]). The cellular phenotypes of tor1-D and tor2-S at 36°C in the complete medium are virtually opposite (figure 4b(iii); [20]). The short, ellipsoidal-shaped phenotype was produced by tor2-S, whereas the elongated cells were observed for the tor1-D mutant, suggesting that their mode of involvement in the cell size control, and possibly also in their relation to the cell cycle, may be opposing.
9. Nutrient-dependent cell size control by Tor1 and Tor2 kinases
The opposite phenotypes of tor1-D and tor2-S in cell size may be explained by the different roles of Tor1 and Tor2 in responding to nutritional signals (figure 4c). As the currently held view on TORC1 [73], Tor2 facilitates the intracellular use of nitrogen source and supports growth (the increase of cell volume). In tor2-S mutants even in the ample presence of environmental nitrogen source, protein and nucleic acid synthesis become defective, resulting in the inhibition of growth, but one or two rounds of the size-shortening divisions occur, reminiscent of the wild-type cell behaviour in N-starvation. If tor2-S cells that arrested in the G0 phase at 26°C are shifted to 36°C in the presence of nitrogen source, then cells do not exit from the G0 phase [34]. These results indicate that the small cell size of tor2-S at 36°C is due to the residual rounds of cell division in the absence of growth. As this phenotype at 36°C suggests, Tor2 is surely required for growth, but it is uncertain how it relates to cell cycle control.
While Tor2 is essential, Tor1 is dispensable. The phenotype of tor1-D is becomes clearly defective under low glucose (less than 0.1%): tor1-D mutant is defective in shortening cell size under low glucose, like ssp1 mutants (figure 4c). The failure to reduce the cell length under glucose limitation leads to semi-cdc25-like cell elongation for tor1-D [20]. Tor1 and Ssp1 may thus sense low glucose and reduce the cell length through regulating mitosis and cytokinesis. In this regard, Tor1 is like a cell cycle-controlling gene. Indeed, the genetic interaction of the deletion Δtor1 with cdc25 mutation was reported [74,75].
The timing of cell cycle events in tor1-D after the release from the G0 phase was monitored. DNA replication occurred with the same timing as the wild-type, but the entry into mitosis and subsequent cytokinesis is delayed, resulting in the increase of cell size [20]. The delay is due to the delay in Cdc2 mitotic activation, as Y15 phosphorylation of Cdc2 remains in elongated cells. Interestingly, actin was abundant at the single cell tip in the interphase of tor1-D mutants. Bipolar cell elongation seems to be absent in tor1-D mutant cells. The intensity of actin at the equator was also high, whereas myosin makes a normal-looking contractile ring. Interphase monopolar growth of tor1-D mutants that accompanied abundant actin localization at one single tip might cause the delay in Cdc2 activation. Proper localization of actin may be required for signalling the transition of nutrient state or utilization, which may be the upstream event for regulating the G2–M transition.
10. Requirement of separase/Cut1-securin/Cut2 is alleviated in tor2-diminished cells
Surprisingly, the complex of securin/Cut2–separase/, essential for chromosome segregation, strongly interacts with both TORC1 and 2. Alleles of cut1 were synthetically rescued by tor2-S mutation (one example of the spot test is shown in figure 4d; [20]). The addition of rapamycin to cut1 mutants also strongly suppressed the ts phenotype. As the main target of rapamycin is Tor2 in S. pombe, these results suggest that the downregulation of Tor2 lessens the necessity for Cut1. Consistently, the synthetic rescue by tor2-S mutation and rapamycin was also found for the mutant alleles of cut2. As the levels of Cut1 and Cut2 are not restored at all in cells suppressed by tor2-S or rapamycin, the necessity of Cut1–Cut2 is greatly alleviated, or the mode of chromosome segregation under the diminished Tor2 situation may be drastically altered, regarding the requirement of Cut1–Cut2. It remains to be determined which of the Cut1 functions that include proteolytic and non-proteolytic activities in S. pombe [64,65,76] are actually suppressed.
The additive defects were observed between cut2 and tor1 (the spot test result is shown in figure 4d). Between cut1 and tor1, the additive effect also exists. These results suggest that the Cut2–Cut1 complex shares the essential function with Tor1. The synthetic defect was also found between the overproduction of Cut1 and the tor1-D mutant. The previous study [77] showed that overproduction of Cut1 causes the synthetic defect with ste20 and pop3 mutations that are defective in the regulatory subunits of TORCs. These apparently opposite results between Tor1 and Tor2 in the interaction with Cut1–Cut2 mutations again suggest that the roles of Tor1 and Tor2 are opposing (discussed below).
Other unexpected finding is that the ts phenotype of the cut2 mutant is partly suppressed by Δfkh1, the deletion of FKBP12-like Fkh1, in the absence of rapamycin [20]. Fkh1 encodes a peptidylproline cis–trans isomerase enzyme, which accelerates the folding of proteins. While Fkh1 is necessary for tor2-S and tor1-D mutants to be sensitive to rapamycin, suppression of the ts phenotype of cut2 by the deletion Δfkh1 occurs in the absence of rapamycin. Fkh1 appears to be needed for the ts phenotype of the cut2 mutant. Since Fkh1 affects the protein conformation, the result might fit with a notion that Cut2 is a chaperone-inhibitor of Cut1 [54]: Fkh1 may cause instability of mutant Cut2 protein.
11. Discussion
The aim of this review is to discuss the perspective regarding facts and hypotheses on size control during the cell division cycle under limited nutrients and their implication in the mode of mitosis. First, it should be emphasized that cell size control is the meeting point for cell division cycle and growth control. When considering growth control, it is not surprising that glucose and nitrogen source are determinant factors for the cell size. The cell length of S. pombe at the time of division is pre-determined, depending on different concentrations of nitrogen source and glucose in the culture medium. The wee or semi-wee cellular phenomenon occurs in wild-type cells under limited nutrients. Second, protein phosphorylation and dephosphorylation are closely implicated in the cell size control. Several kinases and phosphatases are found to control cell size under the nutritional limitations. Discussions are mostly restricted to the cases of S. pombe rather than budding yeast and mammalian systems. The reason is that critical examination of mammalian cell size control during the somatic cell division cycle has been scarce and that the role of SWE1 (Wee1 homologue) and MIH1 (Cdc25 homologue) in cell size control at the time of S. cerevisiae division is unclear.
As is the case of S. pombe Cdc25 and Wee1, which are opposing phosphatase and kinase, but actually coordinate the timing of Cdc2 activation for mitotic entry through the change of their activities, TORC1 (Tor2) and TORC2 (Tor1) may be opposing, but actually coordinate growth, mitosis and cell size control in response to nutritional cues. Premature mitosis is observed in the tor2-S mutant, whereas cells are elongated in the tor1-D mutant, reminiscent of wee1 and cdc25 mutants. TORC1 and TORC2 are both kinases, so that they are unlikely to target the same substrate to control the cell size by their opposing functions. It is possible that TORC1 and TORC2 are the upstream regulators for Cdc2 activation, and affect directly or indirectly Wee1 and Cdc25 depending on the levels of nutrients. Note that S. pombe has the second Wee1-like kinase, Mik1, which may also be involved in the nutritional control for Cdc2 activation. The hypothesis that TORC1 and TORC2 control Cdc2 activation at the G2–M boundary in opposing manners remains to be tested. Nutrient-sensitive Ssp1 kinase (and possibly also Ssp2 AMPK) and PP2A (Ppa2), PP2A-related (Ppe1) phosphatases are also opposing. The Ssp1–Ppa2/Ppe1 signalling may also be considered as the upstream regulators for Cdc2 activation. They are implicated in calcium signalling in addition to nutrient uptake. However, their relationship to TOR kinases, dependent or independent, remains unclear. The Ssp1–Ppa2/Ppe1 signalling may target the same substrate(s) to control the cell size under different nutrients.
We argue that Sds23 is a key molecule to understand the role of Ppa2 and Ppe1 phosphatases for the nutritional utilizations because Sds23 restrains the phosphatases by stable binding. The loss of Sds23 leads to the hyperactivation of the phosphatases, causing the failure to consume the low concentration of glucose for cell proliferation [21] and resulting in loss of viability of the G0 cells under nitrogen starvation [14]. The effect of Δsds23 deletion on the glucose consumption is exceptionally strong among thousands of mutant strains examined. Measurement indicates that the consumption rate of glucose in Δsds23 cells is extremely slow (L. Uehara & A. Mori 2011, unpublished data). The biomarker compound cytidine diphosphate-choline, the level of which greatly increases under glucose fasting in S. pombe [33], considerably increases in the extracts of Δsds23 deletion mutants (T. Pluskal 2011, unpublished data), strongly suggesting that the intracellular nutrient state of Δsds23 is close to glucose starvation, whereas glucose is abundant in the culture medium. The uptake of low glucose is clearly defective in Δsds23. In higher glucose concentration (2%), Δsds23 cells can proliferate, suggesting that the glucose transport becomes very inefficient. We consider that this kind of analysis might be useful for understanding the human disease type II diabetes.
The cell length of Δsds23 is long and rod-shaped [46], resembling the overproduction phenotype of Ppa2 and Ppe1 phosphatases. Sds23 is highly phosphorylated. Judging from the phosphorylated residues determined by mass spectrometry, candidate kinases are PKA, protein kinase C and MAPK. It is important to determine whether phosphorylation controls the binding to phosphatase. We consider that Sds23 acts in parallel with PP1 and APC/cyclosome, and negatively regulates PP2A and PP2A-related phosphatases in interphase or throughout the cell cycle. The role of Sds23 or the downregulation of PP2A and PP2A-related phosphatases becomes essential when the levels of glucose and nitrogen source are low. The transcript level of Sds23 sharply increases by oxidative (H2O2) stress, and transiently increases by cadmium, heat and sorbitol treatments [78].
The role of PP2A in cell size control and its interactions with Cdc25 and Wee1 was documented many years ago in S. pombe [22,23,41], but their negative role for glucose consumption has only recently been realized through the study on Sds23. Furthermore, the discovery of mitotic inhibition of PP2A by phosphorylation of alpha-endosulphine and Arpp19 by greatwall kinase [42,43] strongly suggested the importance of PP2A downregulation during mitotic progression. It is conceivable that the downregulation of PP2A and PP2A-related phosphatases during mitosis enhances the utilization of glucose and the maximal production of energy source.
The role of CaMKK-like Ssp1 kinase is similar to that of Sds23. Ssp1 is essential to properly respond to the starvation of nitrogen source and also glucose [13,14,21], suggesting that Ssp1 may broadly contribute to survival under limited nutrients. However, its molecular function, particularly in the relation to calcium and calmodulin, has been little studied. It is scarcely understood how actin localization, and glucose and nitrogen source utilization are integrated with Ssp1 kinase. The short, conserved stretch sequence present between the kinase- and calmodulin-binding domains is essential for maintaining rod-like cell shape [21]. Mass spectrometric analysis shows that Ssp1 is bound to 14-3-3 homologues, Rad24 and Rad25, suggesting that Ssp1 shuttles between the nucleus and cytoplasm. Ssp1 is highly phosphorylated: seven phosphopeptides have been identified [21]. Overproduction of Ssp1 disperses cortex actin, resulting in the production of round cells. By contrast, under nitrogen starvation, ssp1 mutants keep the rod cell shape and fail to reduce cell size. The role of Ssp1 based on the interaction with Sty1 and Wis1 is of considerable interest. Our knowledge is still scarce for the relationship between Ssp1 and Sty1 in the regulation of cell size under nitrogen starvation.
To critically compare the phenotypes between tor1 and tor2 mutants, the ts mutant tor1-D that has the substitution at the same PI3K site as that of the previously isolated tor2-S mutant is useful [20]. Their phenotypic comparison indicates that Tor1 and Tor2 are opposite in many aspects including the cell size and also in the mode of interactions with securin and separase mutations. Rapamycin is highly inhibitory to tor2-S, but only slightly to tor1-D. As mutant TORC1 and TORC2 kinases are purified and their TOR kinase activities are found to be greatly diminished [20], different phenotypes may be due to the substrate specificities of TORCs kinases.
We interpreted that the opposite cell size phenotypes of tor1-D and tor2-S are due to their distinct responses to nutritional conditions. One important role of Tor1 is to reduce the cell size for division upon decrease of glucose concentration. Effects of other nutrients on the cell size control by Tor1 remain investigated. By contrast, the role of Tor2 is to support growth by increasing the cell size until cells reach the critical size for division. Hence, Tor1 and Tor2 may be considered to coordinate growth with the determination of the timing of cell division depending on nutritional conditions, although their apparent phenotypes and deduced functions look to be opposing. To substantiate such a hypothesis, more mechanistic studies are necessary in future, particularly, in relation to Cdc2 activation as discussed above.
The final issue is how to interpret the synthetic rescue of cut1 and cut2 mutations by tor2-S or rapamycin. The suppression by rapamycin requires Fkh1, a FKBP homologue. This unexpected rescue is strong so that the functional loss of Cut1–Cut2 complex is clearly alleviated if Tor2 (TORC1) is diminished. Chromosome segregation looks quite normal in the suppressed mutant cells, but the low level of separase and securin does not increase at all, as if only a small amount of Cut1–Cut2 is needed in tor2 or rapamycin-treated cells [20]. The complex of Cut1–Cut2 is absolutely needed for proper chromosome segregation in wild-type cells cultured in the regular medium, but the necessity is known to be greatly lessened in the cut1 and cut2 mutant cells in medium containing high concentrations of salt or sorbitol [64]. However, effect of salt differs from that of rapamycin, as the high salt increases the level of Cut1–Cut2 in a Sty1-dependent manner. It is well known in mammalian cells that immunological response (transplant rejection) is suppressed after rapamycin treatment. Additionally, the progression of certain cancers is prevented, and the lifespan is extended in mice. Although the mechanism is scarcely understood, diminished Tor2 (TORC1) greatly lessens the necessity of both Cut1 and Cut2, suggesting that the mode of chromosome segregation might be dramatically altered by the nutritional change via Tor2 (TORC1).
Our results suggest that the segregation role of Cut1–Cut2 is opposite to TORC1 (Tor2), but in parallel with TORC2 (Tor1). There is a popular concept that growth opposes cell division. Conversely, in S. pombe, the period of mitosis restrains growth (cell elongation). TORC1 may restrain the Cut2–Cut1 function in order to prevent premature mitosis, while TORC2 is in parallel with Cut2–Cut1 as its role is to determine the critical timing of division under different nutritional conditions. The close interaction of the TORC complexes with Cut2–Cut1 suggests that nutritional control may be mediated at the metaphase–anaphase transition. This is a surprising possibility, and calls for further study. The nutritional control might be exerted on the metaphase–anaphase progression through the functions of Cut1–Cut2, which might require the balanced regulation of TORC1 function. Cut2 is phosphorylated ([64]; Y. Hanyu & M. Yanagida 2011, unpublished data). Further study is definitively needed for understanding these interactions of Cut1–Cut2 with TORC1 and TORC2.
In summary, we identified Sty1, Wis1, Ssp1, Tor1 and Tor2 kinases, and Ppa2 and Ppe1 protein phosphatases to be important for regulating the cell division cycle control under different nutritional conditions. Their contributions become strikingly apparent in mutant cells under nutritional limitations. These kinases and phosphatases seem to affect Cdc2 kinase activation, though the mechanisms are little understood. Our results revealed the surprising dependency of mitotic metaphase to anaphase transition on the nutrient-sensing TORCs. The block of chromosome segregation by diminished Cut1–Cut2 by mutations is restored by diminished TORC1 by mutation or rapamycin. The mode of chromosome segregation may have to be controlled by TORCs in order to respond to nutritional changes.
Acknowledgements
This review is dedicated to John Murdoch Mitchison, the great founder of the S. pombe growth and cell cycle control field. The authors thank all the members of the G0 cell unit of OIST, particularly to Ayaka Mori, Lisa Uehara and Nobuyasu Ikai, a former member of the laboratory in Kyoto University. The generous support of OIST is gratefully acknowledged. The present study was partly conducted by the CREST research project of the Japan Science and Technology (JST Corporation) grant while M.Y. was in Kyoto University.
References
- 1.Mitchison J. 1971. The biology of the cell cycle. London, UK: Cambridge University Press. [Google Scholar]
- 2.Mitchison J. M. 1957. The growth of single cells. I. Schizosaccharomyces pombe. Exp. Cell Res. 13, 244–262 10.1016/0014-4827(57)90005-8 (doi:10.1016/0014-4827(57)90005-8) [DOI] [PubMed] [Google Scholar]
- 3.Nurse P., Thuriaux P., Nasmyth K. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146, 167–178 10.1007/BF00268085 (doi:10.1007/BF00268085) [DOI] [PubMed] [Google Scholar]
- 4.Thuriaux P., Nurse P., Carter B. 1978. Mutants altered in the control co-ordinating cell division with cell growth in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 161, 215–220 10.1007/BF00274190 (doi:10.1007/BF00274190) [DOI] [PubMed] [Google Scholar]
- 5.Nurse P., Thuriaux P. 1980. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics 96, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fantes P. 1979. Epistatic gene interactions in the control of division in fission yeast. Nature 279, 428–430 10.1038/279428a0 (doi:10.1038/279428a0) [DOI] [PubMed] [Google Scholar]
- 7.Dunphy W. G., Kumagai A. 1991. The cdc25 protein contains an intrinsic phosphatase activity. Cell 67, 189–196 10.1016/0092-8674(91)90582-J (doi:10.1016/0092-8674(91)90582-J) [DOI] [PubMed] [Google Scholar]
- 8.Gould K. L., Moreno S., Tonks N. K., Nurse P. 1990. Complementation of the mitotic activator, p80cdc25, by a human protein-tyrosine phosphatase. Science 250, 1573–1576 10.1126/science.1703321 (doi:10.1126/science.1703321) [DOI] [PubMed] [Google Scholar]
- 9.Costello G., Rodgers L., Beach D. 1986. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr. Genet. 11, 119–125 10.1007/BF00378203 (doi:10.1007/BF00378203) [DOI] [Google Scholar]
- 10.Nurse P., Bissett Y. 1981. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292, 558–560 10.1038/292558a0 (doi:10.1038/292558a0) [DOI] [PubMed] [Google Scholar]
- 11.Su S. S., Tanaka Y., Samejima I., Tanaka K., Yanagida M. 1996. A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J. Cell Sci. 109, 1347–1357 [DOI] [PubMed] [Google Scholar]
- 12.Yanagida M. 2009. Cellular quiescence: are controlling genes conserved? Trends Cell Biol. 19, 705–715 10.1016/j.tcb.2009.09.006 (doi:10.1016/j.tcb.2009.09.006) [DOI] [PubMed] [Google Scholar]
- 13.Sajiki K., et al. 2009. Genetic control of cellular quiescence in S. pombe. J. Cell Sci. 122, 1418–1429 10.1242/jcs.046466 (doi:10.1242/jcs.046466) [DOI] [PubMed] [Google Scholar]
- 14.Shimanuki M., et al. 2007. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells 12, 677–692 10.1111/j.1365-2443.2007.01079.x (doi:10.1111/j.1365-2443.2007.01079.x) [DOI] [PubMed] [Google Scholar]
- 15.Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C. J., Rogers J., Bahler J. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243 10.1038/nature07002 (doi:10.1038/nature07002) [DOI] [PubMed] [Google Scholar]
- 16.Tatebe H., Goshima G., Takeda K., Nakagawa T., Kinoshita K., Yanagida M. 2001. Fission yeast living mitosis visualized by GFP-tagged gene products. Micron 32, 67–74 10.1016/S0968-4328(00)00023-8 (doi:10.1016/S0968-4328(00)00023-8) [DOI] [PubMed] [Google Scholar]
- 17.Morrell J. L., et al. 2004. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 14, 579–584 10.1016/j.cub.2004.03.036 (doi:10.1016/j.cub.2004.03.036) [DOI] [PubMed] [Google Scholar]
- 18.Wu L., Russell P. 1997. Roles of Wee1 and Nim1 protein kinases in regulating the switch from mitotic division to sexual development in Schizosaccharomyces pombe. Mol. Cell Biol. 17, 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labib K., Moreno S. 1996. rum1: a CDK inhibitor regulating G1 progression in fission yeast. Trends Cell Biol. 6, 62–66 10.1016/0962-8924(96)81016-6 (doi:10.1016/0962-8924(96)81016-6) [DOI] [PubMed] [Google Scholar]
- 20.Ikai N., Nakazawa N., Hayashi T., Yanagida M. In press The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in S. pombe. Open Biol. (doi:10.1098/rsob.110007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanyu Y., et al. 2009. Schizosaccharomyces pombe cell division cycle under limited glucose requires Ssp1 kinase, the putative CaMKK, and Sds23, a PP2A-related phosphatase inhibitor. Genes Cells 14, 539–554 10.1111/j.1365-2443.2009.01290.x (doi:10.1111/j.1365-2443.2009.01290.x) [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita N., Yamano H., Niwa H., Yoshida T., Yanagida M. 1993. Negative regulation of mitosis by the fission yeast protein phosphatase ppa2. Genes Dev. 7, 1059–1071 10.1101/gad.7.6.1059 (doi:10.1101/gad.7.6.1059) [DOI] [PubMed] [Google Scholar]
- 23.Shimanuki M., Kinoshita N., Ohkura H., Yoshida T., Toda T., Yanagida M. 1993. Isolation and characterization of the fission yeast protein phosphatase gene ppe1+ involved in cell shape control and mitosis. Mol. Biol. Cell 4, 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George V. T., Brooks G., Humphrey T. C. 2007. Regulation of cell cycle and stress responses to hydrostatic pressure in fission yeast. Mol. Biol. Cell 18, 4168–4179 10.1091/mbc.E06-12-1141 (doi:10.1091/mbc.E06-12-1141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishimoto N., Yamashita I. 2000. Cyclic AMP regulates cell size of Schizosaccharomyces pombe through Cdc25 mitotic inducer. Yeast 16, 523–529 (doi:10.1002/(SICI)1097-0061(200004)16:6<523::AID-YEA546>3.0.CO;2-5) [DOI] [PubMed] [Google Scholar]
- 26.Petersen J., Hagan I. M. 2005. Polo kinase links the stress pathway to cell cycle control and tip growth in fission yeast. Nature 435, 507–512 10.1038/nature03590 (doi:10.1038/nature03590) [DOI] [PubMed] [Google Scholar]
- 27.Petersen J., Nurse P. 2007. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat. Cell Biol. 9, 1263–1272 10.1038/ncb1646 (doi:10.1038/ncb1646) [DOI] [PubMed] [Google Scholar]
- 28.Shieh J. C., Wilkinson M. G., Buck V., Morgan B. A., Makino K., Millar J. B. 1997. The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev. 11, 1008–1022 10.1101/gad.11.8.1008 (doi:10.1101/gad.11.8.1008) [DOI] [PubMed] [Google Scholar]
- 29.Shiozaki K., Shiozaki M., Russell P. 1997. Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol. Biol. Cell 8, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson A. E., Gould K. L. 2011. Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J. 30, 341–354 10.1038/emboj.2010.317 (doi:10.1038/emboj.2010.317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng S. S., Papadopoulou K., McInerny C. J. 2006. Regulation of gene expression and cell division by Polo-like kinases. Curr. Genet. 50, 73–80 10.1007/s00294-006-0077-y (doi:10.1007/s00294-006-0077-y) [DOI] [PubMed] [Google Scholar]
- 32.Robertson A. M., Hagan I. M. 2008. Stress-regulated kinase pathways in the recovery of tip growth and microtubule dynamics following osmotic stress in S. pombe. J. Cell Sci. 121, 4055–4068 10.1242/jcs.034488 (doi:10.1242/jcs.034488) [DOI] [PubMed] [Google Scholar]
- 33.Pluskal T., Hayashi T., Saitoh S., Fujisawa A., Yanagida M. 2011. Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. FEBS J. 278, 1299–1315 10.1111/j.1742-4658.2011.08050.x (doi:10.1111/j.1742-4658.2011.08050.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., Yanagida M. 2007. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 12, 1357–1370 10.1111/j.1365-2443.2007.01141.x (doi:10.1111/j.1365-2443.2007.01141.x) [DOI] [PubMed] [Google Scholar]
- 35.Matsusaka T., Hirata D., Yanagida M., Toda T. 1995. A novel protein kinase gene ssp1+ is required for alteration of growth polarity and actin localization in fission yeast. EMBO J. 14, 3325–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afshar K., Werner M. E., Tse Y. C., Glotzer M., Gonczy P. 2010. Regulation of cortical contractility and spindle positioning by the protein phosphatase 6 PPH-6 in one-cell stage C. elegans embryos. Development 137, 237–247 10.1242/dev.042754 (doi:10.1242/dev.042754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastians H., Ponstingl H. 1996. The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J. Cell Sci. 109, 2865–2874 [DOI] [PubMed] [Google Scholar]
- 38.Cherkasova V., Qiu H., Hinnebusch A. G. 2010. Snf1 promotes phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell Biol. 30, 2862–2873 10.1128/MCB.00183-10 (doi:10.1128/MCB.00183-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton A., Immanuel D., Arndt K. T. 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell Biol. 11, 2133–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rupes I., Jia Z., Young P. G. 1999. Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast. Mol. Biol. Cell 10, 1495–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinoshita N., Ohkura H., Yanagida M. 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63, 405–415 10.1016/0092-8674(90)90173-C (doi:10.1016/0092-8674(90)90173-C) [DOI] [PubMed] [Google Scholar]
- 42.Gharbi-Ayachi A., Labbe J. C., Burgess A., Vigneron S., Strub J. M., Brioudes E., Van-Dorsselaer A., Castro A., Lorca T. 2011. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330, 1673–1677 10.1126/science.1197048 (doi:10.1126/science.1197048) [DOI] [PubMed] [Google Scholar]
- 43.Mochida S., Maslen S. L., Skehel M., Hunt T. 2011. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330, 1670–1673 10.1126/science.1195689 (doi:10.1126/science.1195689) [DOI] [PubMed] [Google Scholar]
- 44.Talarek N., et al. 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′–3′ mRNA decay pathway. Mol. Cell 38, 345–355 10.1016/j.molcel.2010.02.039 (doi:10.1016/j.molcel.2010.02.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinoshita K., Nemoto T., Nabeshima K., Kondoh H., Niwa H., Yanagida M. 1996. The regulatory subunits of fission yeast protein phosphatase 2A (PP2A) affect cell morphogenesis, cell wall synthesis and cytokinesis. Genes Cells 1, 29–45 10.1046/j.1365-2443.1996.02002.x (doi:10.1046/j.1365-2443.1996.02002.x) [DOI] [PubMed] [Google Scholar]
- 46.Ishii K., Kumada K., Toda T., Yanagida M. 1996. Requirement for PP1 phosphatase and 20S cyclosome/APC for the onset of anaphase is lessened by the dosage increase of a novel gene sds23+. EMBO J. 15, 6629–6640 [PMC free article] [PubMed] [Google Scholar]
- 47.Dawson J. F., Holmes C. F. 1999. Identification of sds21 in fission yeast in an inhibitor-resistant high molecular mass protein phosphatase-1 complex. Biochem. Cell Biol. 77, 551–558 10.1139/o99-062 (doi:10.1139/o99-062) [DOI] [PubMed] [Google Scholar]
- 48.Ohkura H., Kinoshita N., Miyatani S., Toda T., Yanagida M. 1989. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell 57, 997–1007 10.1016/0092-8674(89)90338-3 (doi:10.1016/0092-8674(89)90338-3) [DOI] [PubMed] [Google Scholar]
- 49.Posch M., Khoudoli G. A., Swift S., King E. M., Deluca J. G., Swedlow J. R. 2010. Sds22 regulates aurora B activity and microtubule–kinetochore interactions at mitosis. J. Cell Biol. 191, 61–74 10.1083/jcb.200912046 (doi:10.1083/jcb.200912046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stone E. M., Yamano H., Kinoshita N., Yanagida M. 1993. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr. Biol. 3, 13–26 10.1016/0960-9822(93)90140-J (doi:10.1016/0960-9822(93)90140-J) [DOI] [PubMed] [Google Scholar]
- 51.Ohkura H., Yanagida M. 1991. S. pombe gene sds22 + essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell 64, 149–157 10.1016/0092-8674(91)90216-L (doi:10.1016/0092-8674(91)90216-L) [DOI] [PubMed] [Google Scholar]
- 52.Jiang Y., Scott K. L., Kwak S. J., Chen R., Mardon G. 2011. Sds22/PP1 links epithelial integrity and tumor suppression via regulation of myosin II and JNK signaling. Oncogene 30, 3248–3260 10.1038/onc.2011.46 (doi:10.1038/onc.2011.46) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldar M. M., Nishie T., Ishikura Y., Fukuda T., Takegawa K., Kawamukai M. 2005. Functional conservation between fission yeast moc1/sds23 and its two orthologs, budding yeast SDS23 and SDS24, and phenotypic differences in their disruptants. Biosci. Biotechnol. Biochem. 69, 1422–1426 10.1271/bbb.69.1422 (doi:10.1271/bbb.69.1422) [DOI] [PubMed] [Google Scholar]
- 54.Nagao K., Yanagida M. 2006. Securin can have a separase cleavage site by substitution mutations in the domain required for stabilization and inhibition of separase. Genes Cells 11, 247–260 10.1111/j.1365-2443.2006.00941.x (doi:10.1111/j.1365-2443.2006.00941.x) [DOI] [PubMed] [Google Scholar]
- 55.Zheng X. F., Florentino D., Chen J., Crabtree G. R., Schreiber S. L. 1995. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82, 121–130 10.1016/0092-8674(95)90058-6 (doi:10.1016/0092-8674(95)90058-6) [DOI] [PubMed] [Google Scholar]
- 56.Beck T., Hall M. N. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692 10.1038/45287 (doi:10.1038/45287) [DOI] [PubMed] [Google Scholar]
- 57.Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271–3279 10.1101/gad.13.24.3271 (doi:10.1101/gad.13.24.3271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 10.1016/S1097-2765(02)00636-6 (doi:10.1016/S1097-2765(02)00636-6) [DOI] [PubMed] [Google Scholar]
- 59.Powers T., Walter P. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt A., Kunz J., Hall M. N. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl Acad. Sci. USA 93, 13 780–13 785 10.1073/pnas.93.24.13780 (doi:10.1073/pnas.93.24.13780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., Ohsumi Y. 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell Biol. 25, 7239–7248 10.1128/MCB.25.16.7239-7248.2005 (doi:10.1128/MCB.25.16.7239-7248.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lempiainen H., Halazonetis T. D. 2009. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28, 3067–3073 10.1038/emboj.2009.281 (doi:10.1038/emboj.2009.281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin D. E., Hall M. N. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17, 158–166 10.1016/j.ceb.2005.02.008 (doi:10.1016/j.ceb.2005.02.008) [DOI] [PubMed] [Google Scholar]
- 64.Kawasaki Y., Nagao K., Nakamura T., Yanagida M. 2006. Fission yeast MAP kinase is required for the increased securin–separase interaction that rescues separase mutants under stresses. Cell Cycle 5, 1831–1839 10.4161/cc.5.16.3010 (doi:10.4161/cc.5.16.3010) [DOI] [PubMed] [Google Scholar]
- 65.Nakamura T., Nagao K., Nakaseko Y., Yanagida M. 2002. Cut1/separase C-terminus affects spindle pole body positioning in interphase of fission yeast: pointed nuclear formation. Genes Cells 7, 1113–1124 10.1046/j.1365-2443.2002.00586.x (doi:10.1046/j.1365-2443.2002.00586.x) [DOI] [PubMed] [Google Scholar]
- 66.Alvarez B., Moreno S. 2006. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119, 4475–4485 10.1242/jcs.03241 (doi:10.1242/jcs.03241) [DOI] [PubMed] [Google Scholar]
- 67.Matsuo T., Otsubo Y., Urano J., Tamanoi F., Yamamoto M. 2007. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell Biol. 27, 3154–3164 10.1128/MCB.01039-06 (doi:10.1128/MCB.01039-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uritani M., Hidaka H., Hotta Y., Ueno M., Ushimaru T., Toda T. 2006. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells 11, 1367–1379 10.1111/j.1365-2443.2006.01025.x (doi:10.1111/j.1365-2443.2006.01025.x) [DOI] [PubMed] [Google Scholar]
- 69.Urano J., Sato T., Matsuo T., Otsubo Y., Yamamoto M., Tamanoi F. 2007. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl Acad. Sci. USA 104, 3514–3519 10.1073/pnas.0608510104 (doi:10.1073/pnas.0608510104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tatebe H., Morigasaki S., Murayama S., Zeng C. T., Shiozaki K. 2010. Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr. Biol. 20, 1975–1982 10.1016/j.cub.2010.10.026 (doi:10.1016/j.cub.2010.10.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weisman R., Choder M., Koltin Y. 1997. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J. Bacteriol. 179, 6325–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weisman R., Finkelstein S., Choder M. 2001. Rapamycin blocks sexual development in fission yeast through inhibition of the cellular function of an FKBP12 homolog. J. Biol. Chem. 276, 24 736–24 742 10.1074/jbc.M102090200 (doi:10.1074/jbc.M102090200) [DOI] [PubMed] [Google Scholar]
- 73.Wullschleger S., Loewith R., Hall M. N. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484 10.1016/j.cell.2006.01.016 (doi:10.1016/j.cell.2006.01.016) [DOI] [PubMed] [Google Scholar]
- 74.Ikeda K., Morigasaki S., Tatebe H., Tamanoi F., Shiozaki K. 2008. Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 7, 358–364 10.4161/cc.7.3.5245 (doi:10.4161/cc.7.3.5245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schonbrun M., Laor D., Lopez-Maury L., Bahler J., Kupiec M., Weisman R. 2009. TOR complex 2 controls gene silencing, telomere length maintenance, and survival under DNA-damaging conditions. Mol. Cell Biol. 29, 4584–4594 10.1128/MCB.01879-08 (doi:10.1128/MCB.01879-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomonaga T., et al. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14, 2757–2770 10.1101/gad.832000 (doi:10.1101/gad.832000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuasa T., et al. 2004. An interactive gene network for securin-separase, condensin, cohesin, Dis1/Mtc1 and histones constructed by mass transformation. Genes Cells 9, 1069–1082 10.1111/j.1365-2443.2004.00790.x (doi:10.1111/j.1365-2443.2004.00790.x) [DOI] [PubMed] [Google Scholar]
- 78.Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229 10.1091/mbc.E02-08-0499 (doi:10.1091/mbc.E02-08-0499) [DOI] [PMC free article] [PubMed] [Google Scholar]