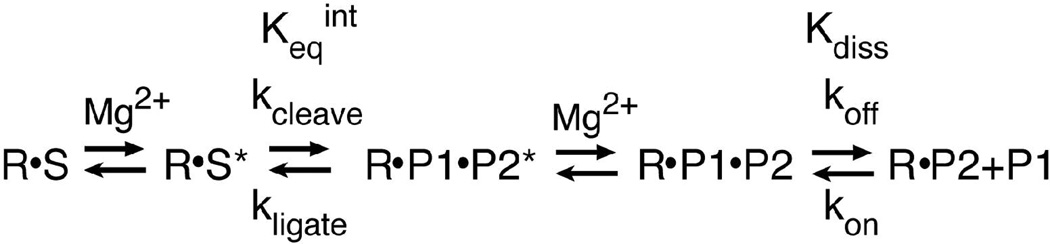Abstract
The hammerhead ribozyme from Schistosoma mansoni is the best characterized of the natural hammerhead ribozymes. Biophysical, biochemical, and structural studies have shown that the formation of the loop-loop tertiary interaction between stems I and II alters the global folding, cleavage kinetics, and conformation of the catalytic core of this hammerhead, leading to a ribozyme that is readily cleaved under physiological conditions. This study investigates the ligation kinetics and the internal equilibrium between cleavage and ligation for the Schistosoma hammerhead. Single turnover kinetic studies on a construct where the ribozyme cleaves and ligates substrate(s) in trans showed up to 23% ligation when starting from fully cleaved products. This was achieved by a ~2,000-fold increase in the rate of ligation compared to a minimal hammerhead without the loop-loop tertiary interaction, yielding an internal equilibrium that ranges from 2–3 at physiological Mg2+ ion concentrations (0.1 –1 mM). Thus, the natural Schistosoma hammerhead ribozyme is almost as efficient at ligation as it is at cleavage. The results here are consistent with a model where formation of the loop-loop tertiary interaction leads to a higher population of catalytically active molecules, and where formation of this tertiary interaction has a much larger effect on the ligation than the cleavage activity of the Schistosoma hammerhead ribozyme.
Ribozymes are the catalytic molecules that take part in a variety of biochemical reactions including peptidyl transfer (1, 2), self-splicing (3–5), tRNA maturation (6, 7), regulation of translation in Gram-positive bacteria (8), and processing of replication intermediates in plant pathogenic RNAs such as viroids and satellite RNAs of viruses (9–11). These plant pathogenic RNAs replicate via a rolling circle mechanism where hammerhead and hairpin ribozymes self-cleave the long multi-genomic RNA strand into single genomes (11–15). These linear single genomes must be subsequently re-circularized to continue the replication cycle. The hammerhead ribozyme has been proposed to also function as the RNA ligase where it reacts in cis on a linear RNA to give the circular genome (11, 16–18). Other possibilities for the in vivo ligation reaction are that the ribozyme requires the assistance of a host protein (19) or that a host protein performs the ligation reaction (20–22).
The hammerhead ribozyme belongs to the group of small catalytic RNAs that includes the hairpin, VS1, and HDV ribozymes (23). These ribozymes catalyze the same cleavage reaction where nucleophilic attack of the 2’-OH at the cleavage site phosphate leads to products containing a 2’, 3’-cyclic phosphate and a 5’-OH (11, 24). The minimal hammerhead ribozyme consists of a three helix junction that folds into a Y-shaped structure surrounding a 15-nucleotide catalytic core where 11 of these nucleotides are conserved in all hammerhead sequences (15, 24, 25). This minimal hammerhead has been extensively studied by biochemical, biophysical and structural techniques (26) and efficient cleavage activity in vitro is achieved only at relatively high levels of Mg2+ ions (cleavage rate constants of ~1.0 min−1 at 10 mM Mg2+) (27). Although this minimal hammerhead exhibits ligation activity in vitro, cleavage is highly preferred, with the ratio of fraction cleaved to uncleaved of ~130 (27). More recent studies have shown that when the minimal sequence of the hammerhead ribozyme is extended to include naturally occurring loop-loop tertiary interactions between stems I and II (Figure 1), catalysis takes place under physiological conditions both in vivo and in vitro (28, 29). This loop-loop tertiary interaction stabilizes the catalytically active conformation (30), which increases observed cleavage rate constants by up to 50-fold, allowing efficient cleavage even at low concentrations of metal ions (cleavage rate constants of ~0.2 – 1.4 min−1 in 100 µM Mg2+ (28, 31, 32)).
Figure 1.
Sequence and secondary structure of the trans Schistosoma hammerhead ribozyme constructs. The full construct is shown for Schist26, whereas only stem III and the corresponding P1 are shown for Schist24 through Schist20 since the rest of the construct is identical to Schist26. Construct names correspond to the number of bases in the substrate strand. The double-headed arrow indicates the tertiary loop-loop interaction between stems I and II. Canonical base pairs are shown as solid lines and the Hoogsteen base pair is shown with a ●. The conserved nucleotides that make up the catalytic core are shown in outline and numbered according to standard convention. Bold nucleotides indicate the ribozyme strand and the single headed arrow points to the scissile bond. The cleavage experiments employ a two-stranded construct consisting of the ribozyme and the full-length substrate. The ligation experiments employ a three-stranded construct consisting of the ribozyme, the 5’-cleavage product, P1, which has a 2', 3' cyclic phosphate at C17, and the 3’-cleavage product, P2, which has a 5’-OH on C18. For the UUCG control, the sequence in the stem II loop is replaced with a UUCG sequence (boxed).
The hammerhead ribozyme from the Schistosoma mansoni satellite DNA (33, 34) is the best characterized of the natural hammerhead systems with data on the cleavage kinetics, global folding by FRET, local and global dynamics by EPR, and cross-linking studies (31, 35–39). These studies have shown that the formation of the loop-loop tertiary interaction alters the structure, populations of active conformations, global folding and cleavage kinetics of the natural hammerhead, leading to a ribozyme that is readily cleaved under physiological conditions. In addition, Scott and coworkers have solved the X-ray structure of a construct of the Schistosoma mansoni hammerhead ribozyme, which contains the natural loop-loop tertiary interaction (40). This structure shows large conformational changes in the catalytic core compared to the structure of the minimal hammerhead ribozyme and represents an improved model for understanding the catalytic mechanism of the hammerhead ribozyme (30, 40).
The cleavage reaction for several natural hammerhead ribozymes has been studied (28, 29, 32), but the ligation kinetics have only been characterized for the sTRSV hammerhead (32). Here we report on the ligation kinetics and internal equilibrium of the best characterized hammerhead, the Schistosoma ribozyme. This Schistosoma construct is an efficient ligase and yields up to 23% ligated product starting from the fully cleaved substrate, which is the highest level of ligation reported for any unmodified hammerhead. This Schistosoma ribozyme shows an increased rate of ligation ~2,000-fold compared to the minimal hammerhead and has a Keqint between 2–3 at physiological Mg2+ concentrations. Thus the loop-loop tertiary interaction leads to a ribozyme that is both an efficient nuclease and ligase in vitro.
MATERIALS AND METHODS
RNA Synthesis and Sample Preparation
A set of Schistosoma mansoni hammerhead (33, 34) ribozyme constructs were employed that differ primarily in the length of stem III (Figure 1). The cleavage constructs consist of two RNA strands, a ribozyme strand and a substrate strand, whereas the ligation construct consists of the ribozyme strand and the two cleavage products, P1 and P2. RNAs were either chemically synthesized (W. M. Keck Facility, Yale University and Dharmacon Inc.) or generated by in vitro transcription using T7 RNA polymerase (41). Synthetic DNA templates for in vitro transcription were purchased from Integrated DNA Technologies. Transcription products were purified by 20% (w/v) denaturing PAGE and full-length RNAs were excised from the gel, eluted and ethanol precipitated. The RNAs were dissolved in excess EDTA (10 mM), heated to 65 °C for 5 min to remove any Mg2+ ions, and cooled at room temperature for 10 min. All RNAs were exchanged into reaction buffer containing 50 mM Tris pH 7.0, 100 mM NaCl, and 0.1 mM EDTA by repeated dilution/concentration in Microcon YM-3 centrifugal filter devices (Millipore) to remove the excess EDTA.
The substrate strands were 5’ 32P end-labeled using T4 polynucleotide kinase and purified by 20% (w/v) denaturing PAGE. To obtain 32P-labeled P1, which has the 2’3’-cyclic phosphate required for ligation reaction, full-length 32P-labeled substrate was annealed to 10-fold excess ribozyme by heating to 70 °C and then cooling to 25 °C over 10 min. Cleavage of the substrate strand was initiated by addition of MgCl2 to a final concentration of 20 mM Mg2+ and the reaction was allowed to cleave at 25 °C for 20 min. The reaction was then heated to 70 °C, slowly cooled and allowed to cleave at 25 °C for 20 min. This cycle was repeated 2 more times to achieve nearly 100% cleavage and then 32P-P1 was gel purified, as described above. The unlabeled P1 that was employed as the chase in dissociation reactions and P2 were chemically synthesized and both RNAs have a terminal OH at their 3’ and 5’ ends.
The buffers were purified over chelating resin (iminodiacetic acid, Sigma) to remove any trace of Mg2+ or other divalent metal ions. The stock solutions of buffer, MgCl2 and NaCl were analyzed for contaminating metals by inductively coupled plasma optical emission spectroscopy.
Ligation and Cleavage Experiments
Single turnover ligation experiments were performed by first combining 0.5 µM ribozyme strand, 1 µM P2 and trace 32P-P1 in 110 µL of reaction buffer, heating to 70 °C for 2 min and cooling to 25 °C over 10 min. The ligation reactions were then initiated by the addition of MgCl2 to the indicated final concentrations. At times ranging from 8 s to 20 min, samples were removed and quenched in 80% formamide, 0.02% bromophenol blue and excess EDTA and immediately frozen on dry ice. The reaction products were separated by 20% (w/v) denaturing PAGE. 32P radioactivity of both ligated and unligated bands was quantified by phosphorimager analysis using a Typhoon 9400 Variable Mode Imager and ImageQuant 5.2 software (Amersham Biosciences). The observed ligation rate constants were determined with KaleidaGraph 3.5 (Synergy Software) where the fraction of 32P-labeled ligated full-length substrate versus time was fit to the equation Ft = Fo+ F∞(1–e−kt), where Ft is the fraction of the full-length substrate at time t, Fo is the initial fraction of full-length substrate, F∞ is the maximum fraction of full-length substrate and k is the observed rate constant.
For experiments where the observed rates were too fast to measure manually, a KinTek Chemical Quench-Flow Model RQF-3 instrument was used. These experiments were performed at room temperature by first annealing the ribozyme strand, 2-fold excess P2 and trace 32P-P1 as described above. Ligation was initiated by rapidly mixing (2 ms dead time) ~15 µL of annealed hammerhead with ~15 µL 2X Mg2+ in reaction buffer, followed by a rapid quench with 90 µL gel loading buffer (80% formamide, excess EDTA, and 0.02% bromophenol blue). The radioactively labeled reaction products were resolved by 20% denaturing PAGE, and quantified by phosphorimager analysis as above.
Single turnover cleavage experiments were performed under the same conditions and analyzed by the same procedure as the ligation experiments except that trace full-length 32P-labeled substrate was annealed to 0.5 µM ribozyme strand.
Measurement of P1 Dissociation Rate Constants
Pulse-chase experiments were used to measure P1 dissociation rate constants (27) where 1 µM ribozyme strand, 2 µM P2, and trace 32P-P1 were annealed as described above. Schist21 and Schist 23 P1s were annealed to the Schist23 ribozyme strand whereas Schist24 and Schist26 P1s were annealed to the Schist26 ribozyme strand. P2 was identical for all experiments. The annealed hammerheads were incubated in reaction buffer containing 1 mM Mg2+ for 30 min at room temperature. 50 µM of unlabeled P1 was then added to initiate the chase reaction. Aliquots were removed at various times, added to 2 µL native gel loading buffer (50% sucrose, 0.02% each bromophenol blue and xylene cyanol) and then immediately loaded on a running 12% native gel containing 1 mM Mg2+. The fraction of 32P-P1 complexed to the ribozyme was quantified by phosphorimager analysis and the P1 dissociation rate constant was determined by fitting the fraction of complexed 32P-P1 versus time as described above.
RESULTS AND DISCUSSION
Ligation and Internal Equilibrium in the Schistosoma Hammerhead Ribozyme
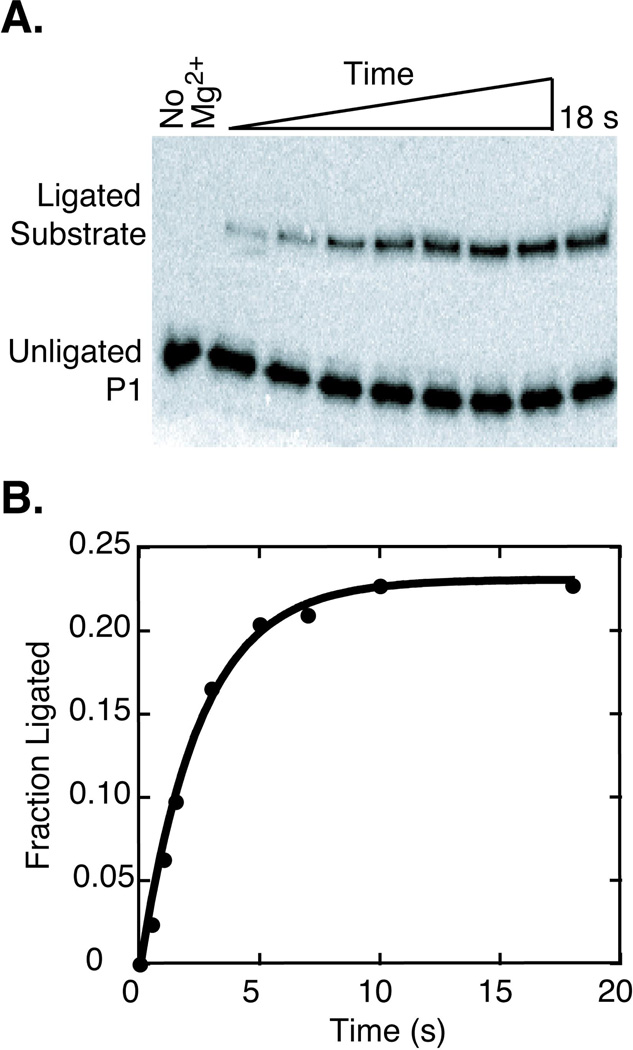
The hammerhead ribozyme is involved in rolling circle replication of circular RNA genomes, where it cleaves replicated mulitmeric genomes into monomeric genomes (9, 11, 13–15). The hammerhead is also thought to re-circularize these RNAs through the reverse ligation reaction (11, 16–18) (Figure 2). We previously reported the observed cleavage rate constant (kobs, cleave) for a trans construct of the Schistosoma hammerhead ribozyme as a function of Mg2+ concentration and pH (31). The ligation activity and the internal equilibrium of this hammerhead are studied here. For these studies the previously characterized trans Schistosoma construct (Schist23, Figure 1) was extended by 3 base pairs in Stem III (Schist26, Figure 1) to reduce dissociation of P1 from the ribozyme strand during the lifetime of the experiment (see below). Figure 3 shows a ligation experiment on Schist26 in 10 mM Mg2+ at 25 °C under single turnover conditions. This ligation reaction plateaus at ~23% ligated substrate, which is a higher level of ligation than previously reported for other hammerhead constructs (27, 32, 39). This indicates the ligation reaction of the Schistosoma hammerhead is quite efficient in vitro, where a significant percentage of a cleaved substrate can re-ligate. The kinetic data in Figure 3B are fit well by a single exponential, yielding an observed rate constant for ligation, kobs, ligate, of ~26 min−1. Ligation experiments were performed on Schist26 at various Mg2+ concentrations, yielding kobs, ligate ranging from 1.5 to 74 min−1 in 0.5 to 50 mM Mg2+ (Table 1). Cleavage experiments were also performed on Schist26 starting from the full-length substrate. In the Schist26 construct, kobs represents the approach to equilibrium and thus the sum of kcleave and kligate. Therefore, kobs,cleave should equal kobs, ligate for the same reaction conditions, and as seen in Table 1, within error these observed rate constants are within a factor of two for all Mg2+ concentrations.
Figure 2.
Reaction scheme of the trans hammerhead ribozyme cleavage and ligation reaction. R is the ribozyme strand, S is the substrate strand, and P1 and P2 are the 5' and 3' cleavage products, respectively. kcleave is the cleavage rate constant, kligate is the ligation rate constant, koff is the P1 dissociation rate constant, kon is the P1 association rate constant. Keqint is the equilibrium constant for the cleavage-ligation step of the reaction, and Kdiss is the equilibrium constant for dissociation of P1. The asterisks indicate the catalytically active forms of the cleaved and ligated complexes.
Figure 3.
Single turnover ligation kinetics of the Schist26 hammerhead ribozyme. A) A 20% denaturing polyacrylamide gel used to separate 32P-labeled ligated full-length substrate from 32P-P1 in the ligation experiment for Schist26 in 10 mM Mg2+. The different lanes correspond to samples removed from the ligation reaction at various times after addition of Mg2+. B) Plot of the fraction of ligated substrate vs. time where the solid line shows the fit to a single exponential with a kobs, ligate of ~26 min−1.
Table 1.
Mg2+ dependence of the rate constants and internal equilibrium constants for the internal equilibrium of the Schistosoma hammerhead ribozymea
| Mg2+ (mM) |
Schist26 kobs, cleave (min−1) |
Schist26 kobs, ligate (min−1) |
Schist21 kcleave (min−1) |
kligate (min−1)b | Keqint(c) | Keqint(d) |
|---|---|---|---|---|---|---|
| 0.1 | 0.2 | n.d. | 0.13 | 0.07 ± 05 | 1.9 ± 0.7 | ≥1.3 |
| 0.5 | 3 | 1.5 | 2.1 | 0.9 ± 0.7 | 2.3 ± 0.8 | 1.3 – 47 |
| 1 | 5.9 | 2.7 | 4.4 | 1.5 ± 1.5 | 2.9 ± 1.0 | 1.3 – 16 |
| 5 | 30 | 16 | 9.4 | 20 ± 6 | 0.5 ± 0.4 | 0.5 – 3.3 |
| 10 | 40 | 26 | 23 | 17 ± 9 | 1.4 ± 0.6 | 0.5 – 3.2 |
| 15 | 46 | 45 | 27 | 19 ± 11 | 1.4 ± 0.6 | 0.5 – 3.0 |
| 25 | 74 | 60 | 33 | 41 ± 16 | 0.8 ± 0.4 | 0.5 – 3.0 |
| 50 | 120 | 74 | 42 | 78 ± 25 | 0.5 ± 0.4 | 0.5 – 3.4 |
kobs, cleave, kobs, ligate and kcleave all have errors of less than ± 20% based on replicate experiments. The errors for kligate and Keqint (calculated from kcleave/kligate) were determined from standard error propagation assuming 20% errors for kobs, cleave and kcleave.
kligate was calculated by subtracting kcleave of Schist 21 from kobs, cleave of Schist26. kobs, cleave was used for this calculation instead of kobs, ligate to eliminate the additional complication that ligation experiments start with 3 strands instead of 2 strands.
Calculated from kcleave/kligate.
Range of values calculated from the fraction of cleaved substrate in the cleavage and ligation experiments of Schist26, assuming 100% active RNA (see text).
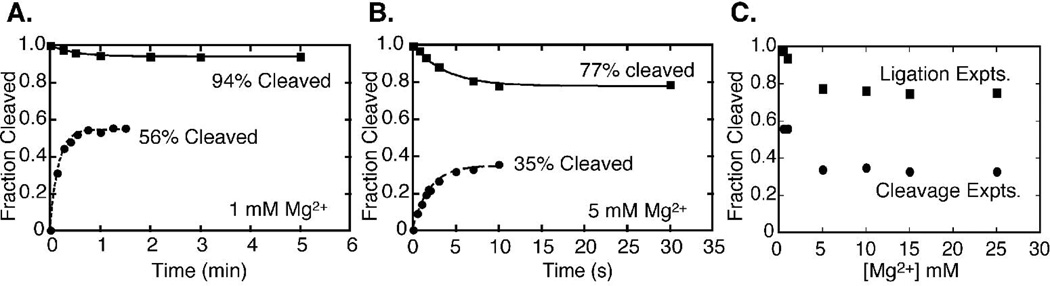
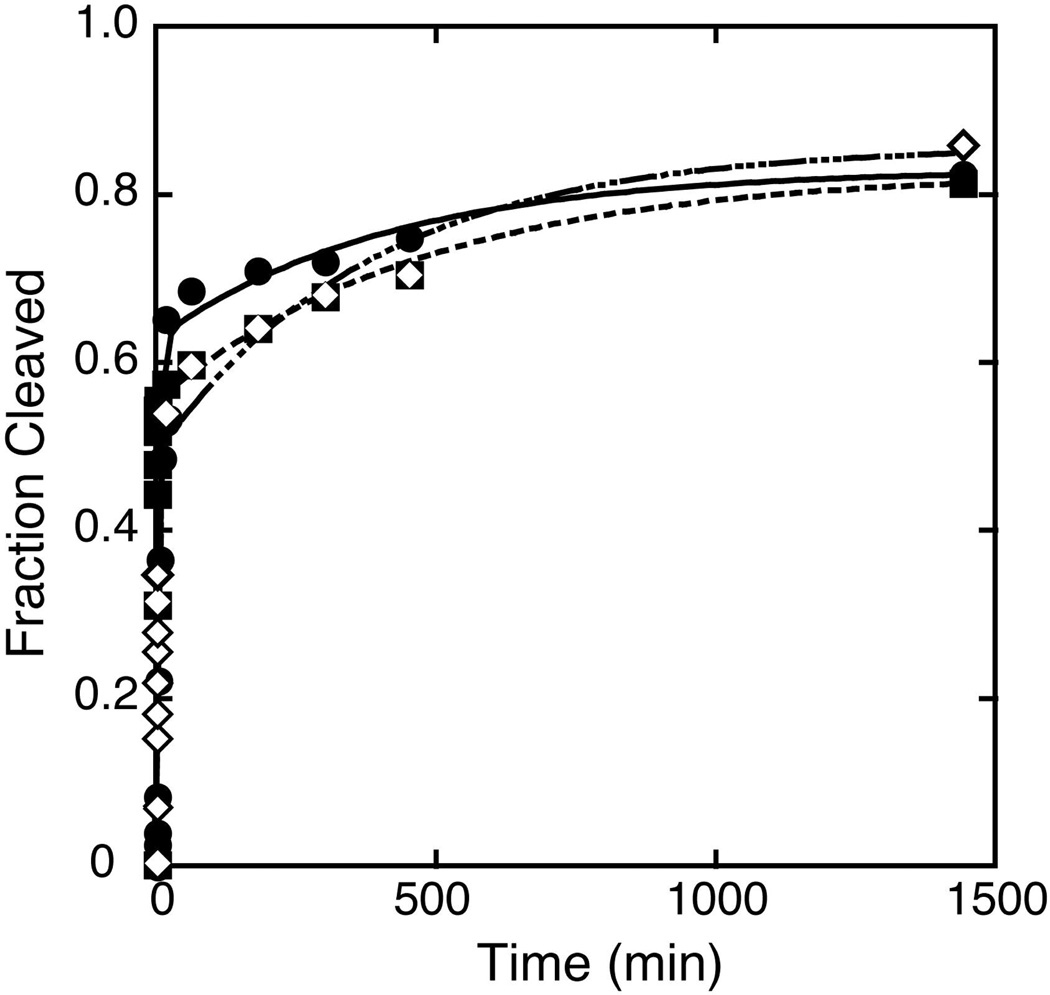
Several methods were next used to estimate the internal equilibrium (Keqint) between cleavage and ligation in vitro for the Schistosoma hammerhead construct. In one approach, limits on Keqint were calculated from the final fraction of cleaved substrate in the ligation or cleavage experiments. Figures 4A and B show the fraction cleaved substrate versus time starting from the fully cleaved and fully ligated substrates in 1 mM and 5 mM Mg2+, respectively. If all the substrates and ribozymes are fully active then at equilibrium (i.e. when fraction cleaved plateaus) the fraction of cleaved substrate should be identical when initiated from either side of the reaction. This is not the case for Schist26, where the fractions of cleaved substrate at the plateaus differ by ~40% between the cleavage and ligation experiments at every Mg2+ concentration (Figure 4C). These data indicate that, as is the case with many RNAs (42–45), Schist26 has a significant, long-lived inactive population, and the conversion between these inactive and active species is slow compared to the cleavage and ligation rates (see below).
Figure 4.
Fraction of cleaved substrate in cleavage and ligation experiments. Plots of the fraction of cleaved substrate vs. time from the cleavage (circles) and ligation (squares) experiments in A) 1 mM Mg2+ and B) 5 mM Mg2+. C) Maximum fraction of cleaved substrate observed in the cleavage experiments used to determine kobs, cleave (circles) and in the ligation experiments used to determine kobs, ligate (squares) at varying Mg2+ concentrations.
Despite the complications of an inactive population, the plateaus of the kinetics experiments provide upper and lower limits for Keqint (= fraction cleaved/fraction ligated). For example, at 1 mM Mg2+ (Figure 4A), 56% of the substrates were cleaved which means at least 56% of the species are active and cleavable and these data yield a lower limit on Keqint of 1.3. Similarly, the plateau for the ligation reaction was 94% cleaved substrate, which leads to an upper limit on Keqint of 16. Using the same rationale for the data at 5 mM Mg2+ (Figure 4B) gives a range of 0.5 ≤ Keqint ≤ 3.3. The ranges for Keqint determined by this method at the various Mg2+ concentrations are given in Table 1. There are larger ranges for Keqint at the lower Mg2+ concentrations, which arise from the low level of ligated product in the ligation experiments under these conditions (Figure 4C).
A second approach was employed that calculates Keqint from the ratio of the cleavage and ligation rate constants, kcleave/kligate. If the rate constant for conversion of inactive to active molecules is much slower than kcleave or kligate, then the measured rates are not affected by presence of such long-lived, inactive populations. A cleavage-only construct, Schist21, (Figure 1) was designed with a shorter P1 so that the rate of dissociation of P1 is much faster than the ligation rate (see below). Thus kobs is assumed to be equal to kcleave for this construct and was measured as a function of Mg2+ concentration. The measured kcleave is then subtracted from kobs,cleave for Schist26 giving kligate which is then used to calculate Keqint (= kcleave/kligate) (Table 1). This method yields Keqint values between 0.5 and 3 over the entire range of Mg2+ concentrations, which fall within the limits determined from the cleavage plateaus. Table 1 shows that although there are very large changes in the cleavage and ligation rates as a function of Mg2+, there are relatively small changes in Keqint. For these data the ratio of the rate constants is a better method for estimating Keqint than using the plateaus of the cleavage and ligation reactions. This is because the calculation of the fraction of cleaved or ligated substrate at the plateaus is directly affected by the long-lived, inactive population, whereas kcleave or kligate are not since, as shown below, the conversion from inactive to active species is much slower than the rates of cleavage or ligation.
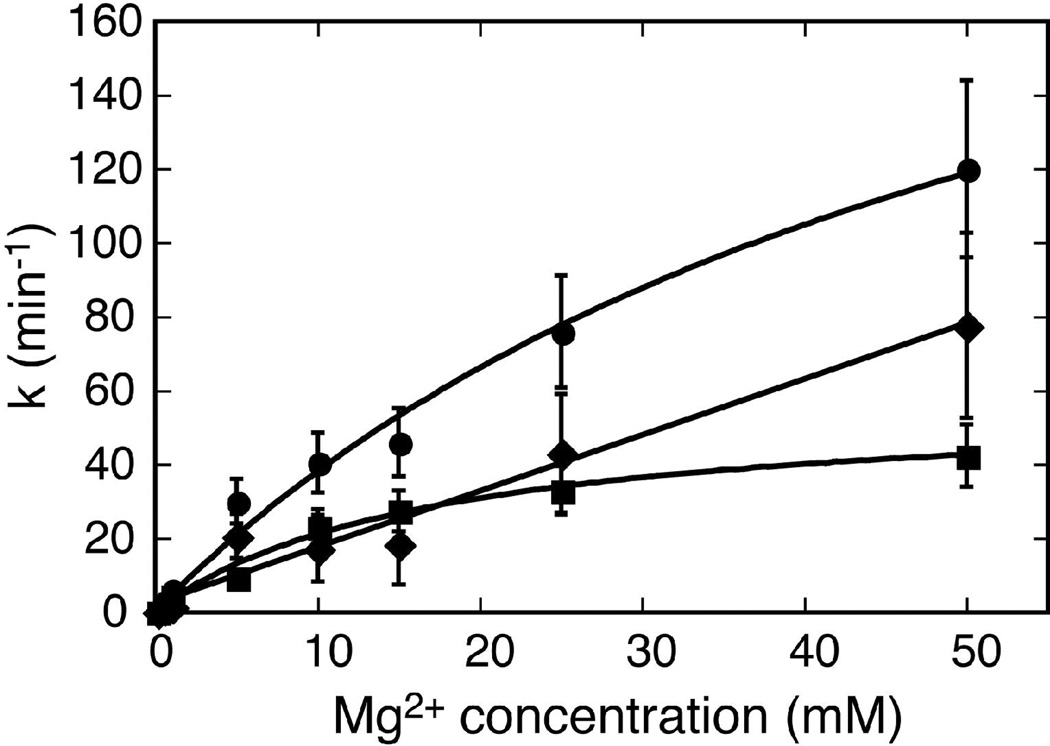
Analysis of the Mg2+-dependence of kobs, cleave for Schist26 and kcleave for Schist21 yields fits for [Mg2+]1/2 of ~55 and ~17 mM, respectively, where [Mg2+]1/2 is the concentration of Mg2+ required to reach half the maximum observed cleavage rate (Figure 5). kligate does not saturate at the concentrations of Mg2+ tested here indicating a higher value of [Mg2+]1/2 for the ligation than the cleavage reaction. There was no evidence for cooperative binding of Mg2+ in either reaction. These results indicate that at concentrations of Mg2+ >30 mM the rate-limiting step in the cleavage reaction is independent of Mg2+. However, the rate-limiting step in the ligation reaction shows a Mg2+ dependence even for the highest Mg2+ concentrations used here.
Figure 5.
Mg2+ dependence of the kinetics for the Schist26 and Schist21 constructs. A plot of kobs, cleave for Schist26 (circles), kcleave for Schist21 (squares), and calculated kligate (diamonds) vs. Mg2+ concentration. For kobs, cleave and kcleave the lines are fits of the Mg2+ dependence of the rate constant, k, to a two-state model for Mg2+ binding using the equation k = kmax [Mg2+]/([Mg2+] + [Mg2+]1/2), where kmax is the rate constant at saturating Mg2+, and [Mg2+]1/2 is the Mg2+ concentration required to achieve half the maximal cleavage rate constant. This analysis assumes a Hill coefficient of 1 since there was no evidence for cooperative binding of Mg2+ for any of the kinetic data. These fits yield [Mg2+]1/2 of 55 ± 10 mM for kobs, cleave for Schist26 and 17 ± 3 mM for kcleave for Schist21. The kligate does not saturate at the Mg2+ concentrations employed here making it impossible to determine a [Mg2+]1/2. Thus for kligate the solid line is a simple linear fit of kligate with [Mg2+]. The error bars are based on a 20% error between replicate experiments for the measured rate constants (see Table 1).
One potential problem in employing the cleavage only (Schist21) construct is that the measured cleavage rates could be affected by having only four base pairs in stem III, so that this helix may not be stably formed. To address this, the cleavage rates were also measured for the Schist20 and Schist22 substrates in 0.1, 0.5 and 1.0 mM Mg2+. The kobs for cleavage of Schist 21 and Schist22 are the same within error at each Mg2+ concentration (data not shown). These results demonstrate that only four base pairs in stem III are needed for full cleavage activity of the Schistosoma hammerhead ribozyme. This agrees with previous studies by Hertel et al. on the minimal hammerhead HH16 where 4 base pairs in stem III were also required for full cleavage activity (46). In contrast, Schist20 (which only forms three base pairs in stem III) had kobs values that were ~20-fold smaller than that of Schist21 and Schist22. Combined with the studies of the dissociation kinetics of P1 in the various constructs (see below), these results provide strong evidence that kobs = kcleave for the Schist21 construct.
Dissociation Kinetics of P1 from the Ribozyme Complex
The Schist21 and Schist26 constructs were designed so that the dissociation of P1 was either much faster or much slower than kobs, respectively. To test this assumption, pulse-chase experiments (27) were performed with a variety of substrates that contain between 4 (Schist21) and 9 (Schist26) base pairs in stem III (see Figure 1). Schist26 has the longest P1 and is expected to have the smallest dissociation rate constant, koff, P1. The pulse-chase experiment on Schist26 showed that P1 dissociates very slowly, with less than 10% dissociated after 24 hrs which corresponds to koff, P1 < 7 × 10−5 min−1 (data not shown). This koff, P1 is over 1,000 times smaller than the smallest kobs, cleave measured for this construct (0.2 min−1 in 0.1 mM Mg2+), meaning that the cleavage/ligation reaction is much faster than P1 dissociation. Estimates for koff can also be obtained from the Kdiss for RNA duplex formation (which is calculated using nearest neighbor rules (47)) and the association rate constant, kon, because koff = Kdiss kon. The kon has been shown to be relatively independent of the length and the sequence of the RNA for short duplexes (48), and a kon of 1.3 × 108 M−1 min−1 was used here (27). The nearest neighbor calculations for the 9 base pair stem III of Schist26 gives a Kdiss of 4 × 10−14 M which then yields a koff, P1 of 5.2 × 10−6 min−1. Thus, the measured and predicted rate constants for the P1 of Schist26 clearly demonstrate that product dissociation of P1 will not affect the measured cleavage/ligation data for this construct.
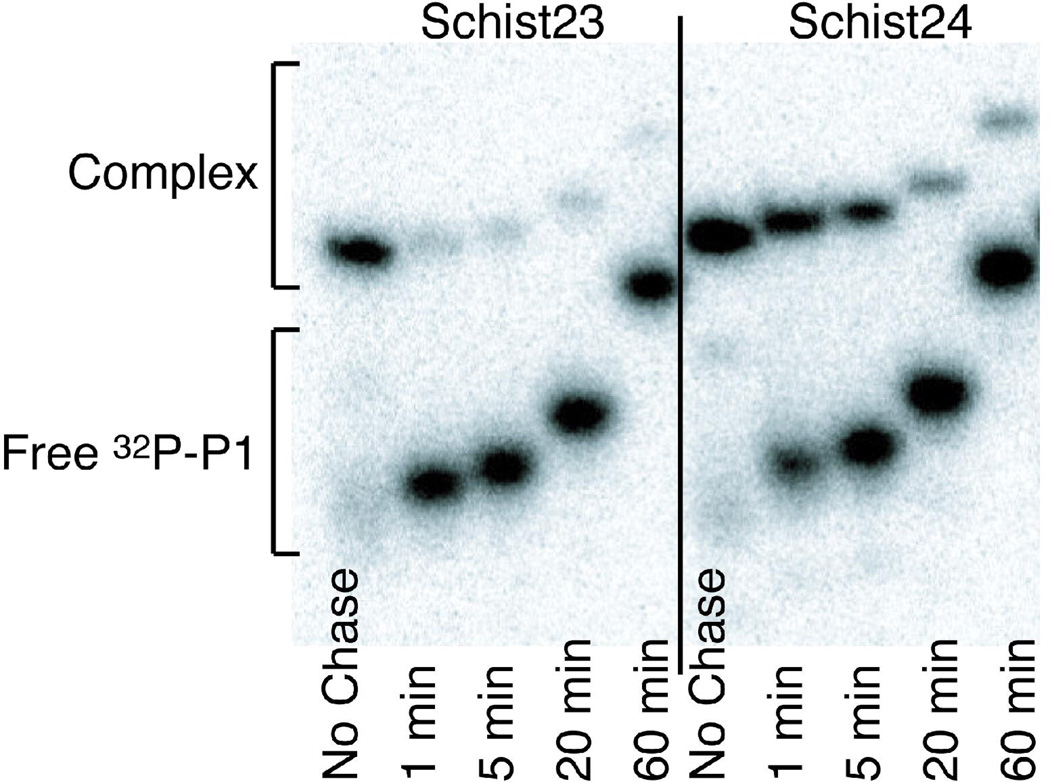
Pulse-chase experiments were also performed on Schist24 and Schist23 (Figure 6) and yielded koff, P1 values of 0.5 and >2 min−1 respectively. These are similar to the values predicted using nearest neighbor calculations, which give koff, P1 values of 0.1 min−1 for Schist24 and 3.5 min−1 for Schist23 and demonstrate the validity of this method for predicting the dissociation rates for P1. The good agreement between measured and the predicted values for koff, P1 for the Schist23 and Schist24 constructs means that this natural hammerhead ribozyme does not hold on to the 5’ product (P1) with higher affinity than expected from simple duplex formation in stem III. This is in contrast to the 4-way junction hairpin ribozyme that binds its 3’ cleavage product with ~50-fold higher affinity relative to the minimal hairpin (49). The P1 of Schist21 is predicted to dissociate very rapidly with koff, P1 of 4.5 × 104 min−1 which is 1,000 times faster than the fastest kobs measured for this construct (42 min−1 in 50 mM Mg2+). This is consistent with ligation experiments that showed no measurable ligation for Schist21 (data not shown). These results show that the P1 for Schist21 dissociates before the ligation reaction can occur, confirming that the kobs for Schist21 is only measuring the cleavage and not the ligation kinetics.
Figure 6.
Native polyacrylamide gels of 32P-labeled P1 for pulse-chase experiments used to measure rate constants for P1 dissociation from the ribozyme complex in 1 mM Mg2+. The times indicate when the sample was loaded on the gel after the unlabeled chase was added to the reaction. The samples were loaded on a running gel, so the bands are shifted up in the gel for the longer time points. The left half of the gel is data for the P1 of Schist23 dissociating from the Schist23 ribozyme and the right half of the gel is for the P1 of Schist24 dissociating from the Schist26 ribozyme (see Methods).
Inactive Populations
RNAs are well known to form alternate/multiple conformations, and this property can complicate the interpretation of kinetic or biochemical experiments (42, 45). In the case of this Schistosoma hammerhead, starting from either the fully cleaved or fully ligated substrates does not lead to the same populations of cleaved product (Figure 4), indicating a significant population of inactive species. Furthermore, it cannot be assumed that the inactive population is the same coming from either direction especially since the ligation reaction starts with three RNA strands and the cleavage reaction with only two strands. To test whether the inactive species in the cleavage construct could convert over time to active species, cleavage experiments were followed for 24 hrs on Schist26 in 0.1, 1, and 10 mM Mg2+. Figure 7 shows the cleavage data from the 24-hr experiments combined with data from the manual and quench flow experiments under the same conditions. After 24 hrs ~80% of the substrate is cleaved at all three Mg2+ concentrations. The pulse-chase experiments on the Schist26 ribozyme showed that a small fraction (<10%) of this 80% is potentially due to P1 dissociation. Thus using 70% cleaved gives a Keqint of ~2.3, which is similar to the Keqint calculated from the kinetic data.
Figure 7.
Single turnover cleavage kinetics followed for 24 hrs to monitor conversion of inactive to active species for Schist26. A plot of the fraction of cleaved substrate vs. time is shown for cleavage experiments in 0.1 mM (circles), 1 mM (squares), and 10 mM (open diamonds) Mg2+. The plots are a combination of the cleavage data from the 24-hr experiments and from the manual and quench flow experiments. The data are fit to a double exponential, where one exponential corresponds to the cleavage/ligation kinetics and the second exponential corresponds to kinetics for the conversion of inactive to active ribozymes. The fit to the second exponential yields kinactive→active of ~0.006 min−1.
The kinetic data from the 24 hr experiments (Figure 7) can be used to determine an apparent rate constant for the conversion of inactive to active ribozyme and yields a kinactive→active of 0.006 ± 0.003 min−1. Such a slow conversion to active ribozymes indicates the inactive, long-lived population does not affect the observed rate constants for the cleavage/ligation experiments discussed above.
The kobs for the cleavage and ligation experiments represent the approach to equilibrium and thus the sum of kcleave and kligate. One standard approach to separate these rate constants is to measure the initial rates of cleavage (or ligation) before any significant population of product has formed. Thus in the cleavage experiment the initial rate of product formation is kcleave[R•S]. Unfortunately it was not possible to accurately determine [R•S] here since this would require knowing the concentration the active ribozyme-substrate species. For example if only 50% of the ribozyme-substrate complexes are active, the kcleave determined by this initial rate method would be a factor of two smaller than the true kcleave. Under some conditions a factor of two error in the rate constant would not significantly effect the analysis but it has a large impact here because kcleave and kligate have similar values (Table 1).
Comparison of the Schistosoma Hammerhead with Other Ribozymes
To determine the effect of the loop-loop tertiary interaction on the cleavage and ligation rate constants, a Schistosoma control was designed that disrupted the loop-loop tertiary interaction between stems I and II by altering the loop sequence of stem II to UUCG. This control has a kobs, cleave of ~0.03 min−1 in 10 mM Mg2+ which is much smaller than the ~1 min−1 for the minimal hammerhead under the same conditions (27). It has previously been observed that increasing the length of stem II to greater than 4 base pairs reduces the rate of cleavage in the minimal hammerhead ~5-fold (50). Thus the longer stem II for the UUCG control appears to inhibit cleavage compared to the minimal hammerhead. Thus, we chose to compare the rate constants determined here for the Schistosoma to the rate constants observed in the well-characterized minimal construct, HH16 (27). The loop-loop tertiary interaction of the Schistosoma hammerhead ribozyme leads to a ~20-fold increase in the cleavage rate constant and a ~2000-fold increase in the ligation rate constant compared to HH16.
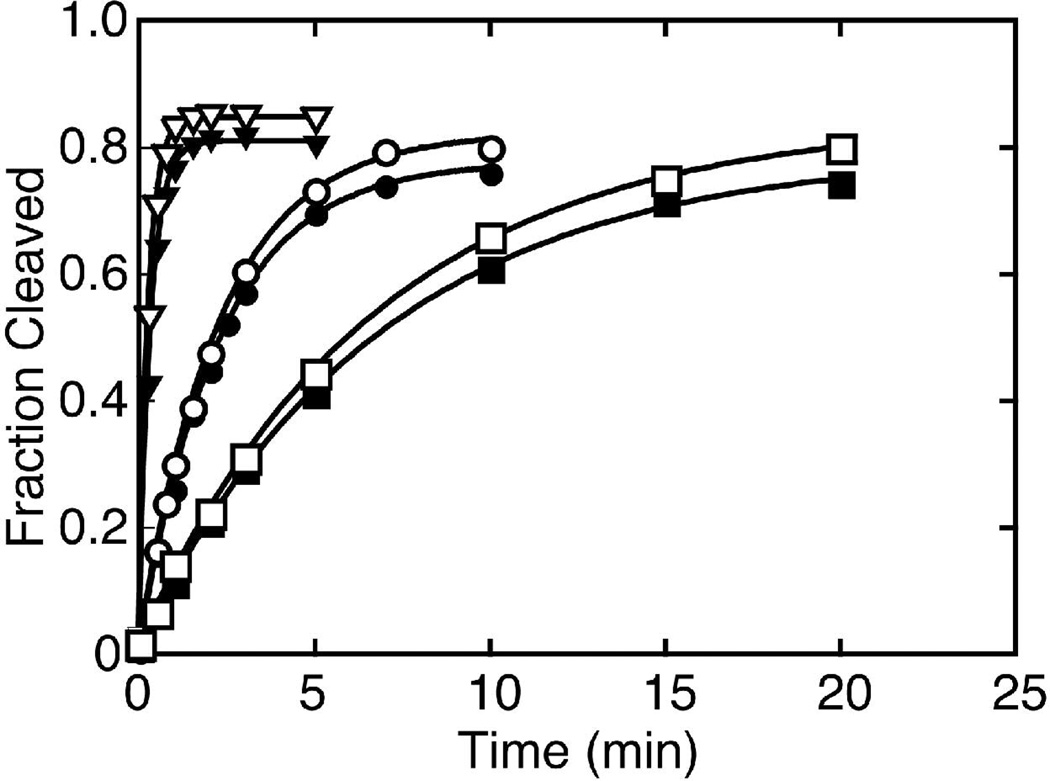
We also studied the kinetics of hammerhead constructs derived from RNA transcripts of satellite DNAs of two cave cricket species (51), Dolichopoda baccettii and Dolichopoda schiavazzii (Figure S1A). Cleavage experiments were performed under identical conditions as used with the Schistosoma constructs, and both cricket constructs cleaved ~80% of their respective substrates with kobs, cleave of ~0.15 min−1 in 0.5 mM Mg2+, ~0.4 min−1 in 1 mM Mg2+, and ~3 min−1 in 5 mM Mg2+ (Figure 8). These values are 10– to 20-fold smaller than the kobs, cleave values determined for Schist26 under the same Mg2+ concentrations, and are closer to cleavage rates for the minimal hammerhead (27). To determine the effect of the loop-loop tertiary interaction in these cave cricket constructs, the loop sequences in stem II were change to UUCG (Figure S1A) as in the Schistosoma control construct. When disrupting the loop-loop tertiary interaction, both cave cricket hammerheads show kobs, cleave in 10 mM Mg2+ ~ 0.03 min−1 (data not shown). This is the same rate constant observed for the Schistosoma UUCG control and 30-fold smaller than 1 min−1 for HH16, again most likely due to the longer stem II. Thus, although these cave cricket constructs are slower than the Schistosoma constructs, the loop-loop tertiary interactions increase the cleavage rate compared to the UUCG controls. Ligation experiments were performed at 1 mM Mg2+ on the cave cricket hammerhead ribozymes, but no ligated product was detected (the detection limit is >0.2% ligated, data not shown). Native gels on the ligation constructs indicated that the 3’ product may not have been annealing to the ribozyme strand. Thus, the sequence in stem I was redesigned to reduce the possibility of self-structure in the 3’ product, P2, but again no ligated product was detected. These results provide a lower limit on the internal equilibrium for these natural hammerhead ribozymes, where using the observed plateau in the cleavage experiments gives a Keqint ≥ 4.
Figure 8.
Single turnover cleavage kinetics for the cave cricket hammerheads. A plot of the fraction of cleaved substrate vs. time for Dolichopoda baccettii (solid symbols) and D. schiavazzii (open symbols) cricket hammerhead constructs at 0.5 mM (squares), 1 mM (circles) and 5 mM (triangles) Mg2+.
Hammerhead ribozymes derived from the positive and negative strands of the ASBVd (14) were also studied here and both cleaved to a high percentage; > 80% cleaved in 5 mM Mg2+, with kobs, cleave values for ASBVd(+) and ASBVd(−) 6- and 25-fold smaller, respectively, than those measured for the Schistosoma hammerhead (Figure S1B for constructs, data not shown). Controls that disrupted the loop-loop tertiary interactions were made by base-pairing the internal loop in stem II to create a continuous helix (Figure S1B). In 5 mM Mg2+ the controls for ASBVd(+) and ASBVd(−) show a ~5-fold and ~15-fold decrease, respectively, in kobs, cleave compared to their wild-type constructs, showing that the loop-loop tertiary interaction still has an effect on the observed cleavage rate. As observed for the cave cricket hammerheads, no ligation activity was detected in 1 mM Mg2+. One possibility for the lack of ligation in the ASBVd and cave cricket constructs studied here is that there may be a high percentage of inactive species in the ligation constructs for these ribozymes. Another possibility is that some hammerhead systems may require additional sequences and/or a host factor for full activity. For example, an avocado chloroplast RNA-binding protein has been shown to accelerate self-cleavage of the ASBVd(+) hammerhead in vitro (19). Furthermore, not all hammerhead ribozymes may be required to perform both cleavage and ligation activities in vivo (20). Although monomeric (−) circular RNA has been found in ASBVd-infected avocados (52), it is not known whether the self-cleaved RNA monomers in the cave crickets or Schistosomes are circularized in vivo (51). The role of hammerhead ribozymes in salamanders, cave crickets and Schistosomes has yet to be determined (53).
This Schistosoma hammerhead construct shows significantly higher fraction of ligated substrate (~20%) than other hammerhead ribozyme constructs reported to date with a Keqint of ~3 in 1 mM Mg2+ and ~1.5 in 10 mM Mg2+. HH16 has a Keqint of 130 in 10 mM Mg2+ (27). However, when a minimal hammerhead was chemically cross-linked to help bring stems I and II together the fraction of ligated substrate increased dramatically to a Keqint of ~1 (54). This change in Keqint was achieved by a ~100 fold increase in the ligation rate with little change in the cleavage rate, whereas the Schistosoma hammerhead shows increases in both the ligation and cleavage rates. Thus the cross-link between stems I and II only partially mimics the effects of the loop-loop tertiary interaction between these stems in the natural hammerhead systems. The sTRSV natural hammerhead also shows a higher fraction of ligated substrate than the minimal hammerhead, with Keqint ~16 in 1 mM Mg2+ and 7.3 in 10 mM Mg2+ (32). Previous studies on a Schistosoma construct similar to the ones used here reaches a maximum of 6% ligated substrate (with a reported Keqint of 17 at 1 mM Mg2+) (39). Thus all of the natural hammerheads have lower values of Keqint than the minimal construct. This smaller internal equilibrium constant is consistent with the proposed dual roles of cleavage and ligation for some hammerhead ribozymes in vivo.
The hammerhead ribozyme is classified as a small nucleolytic ribozyme along with the hairpin, VS and HDV ribozymes (55). All of these ribozymes are thought to be involved in the processing of replication intermediates, but they show quite different internal equilibrium constants. The minimal hairpin ribozyme with 2-way helical junction favors ligation with a Keqint of ~ 0.16 and the natural 4-way helical junction hairpin ribozyme from sTRSV favors ligation even more with a Keqint ~ 0.033 (49). This difference in Keqint between the 2-way and 4-way junction was achieved by an increase in the ligation rate with no change in the cleavage rate. Single molecule studies independently determined a Keqint of 0.029 for the 4-way hairpin (56). In contrast to these other ribozymes, the HDV ribozyme is thought to be a dedicated nuclease since no ligation activity has been detected under physiological conditions (57). The minimal construct of the VS ribozyme also lacks detectable ligation activity; however, extending the natural sequence revealed a region of complementarity 5’ and 3’ of the minimal self-cleaving domain that promotes the ligation reaction in vitro (58).
CONCLUSION
The well-characterized natural Schistosoma hammerhead ribozyme has been shown here to be efficient at both cleavage and ligation of RNA substrates in vitro. Formation of the loop-loop tertiary interaction in this natural hammerhead has a large affect on catalytic activity leading to ~20-fold and ~2,000-fold increases in the cleavage and ligation rates, respectively, compared to the minimal hammerhead. Thus, under physiological conditions in vitro this ribozyme has similar rates constants for cleavage and ligation and shows a slight preference for cleaved versus ligated product, with a Keqint of 2 – 3 at 0.1 − 1 mM Mg2+. The Mg2+ dependence of the cleavage kinetics show saturation of kcleave above ~30 mM indicating that there is a change in the rate-limiting step for the cleavage reaction as a function of Mg2+ concentration. kligate shows no evidence of saturation even at 50 mM Mg2+ so that the rate-limiting step for ligation is Mg2+ dependent over a wider range of Mg2+ concentrations (0.1 to 50 mM). These results illustrate the importance of measuring both the ligation and cleavage kinetics in the hammerhead ribozymes.
Peracchi et al. proposed a model where the hammerhead ribozyme exists in multiple conformations in solution but only a subset of these conformations are catalytically active (59). Nelson & Uhlenbeck extended this model on the basis of the crystal structure of the Schistosoma hammerhead where formation of the loop-loop tertiary interaction leads to a higher population of catalytically active molecules, resulting in higher catalytic activity for the ribozyme (30). The kinetic data here support this model where the large increase in apparent kligate for the natural versus minimal hammerhead does not result from a change in kcat, but instead arises from a higher population of catalytically active molecules in solution. Ensemble FRET has been used to study global folding of a natural hammerhead ribozyme that contains the loop-loop tertiary interaction (35). These studies employed non-cleavable substrates and so primarily give information about how this tertiary interaction affects folding prior to the cleavage reaction. Analogous studies on a cleaved substrate-ribozyme complex should also provide useful information on how the loop-loop tertiary interaction affects the folding of the cleaved hammerhead (J. Boots and A. Pardi, unpublished results). Single molecule FRET studies could prove even more informative, where it may be possible to determine both the populations and lifetimes of the catalytically active species in the hammerhead systems, analogous to recent single molecules studies on the hairpin ribozyme (60). Additionally, it would be interesting to see if the crystal structure of a cleaved natural hammerhead will have a similar conformation to that of the uncleaved hammerhead, since for the Schistosoma hammerhead the ligation reaction is nearly as efficient as the cleavage reaction.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Olke Uhlenbeck, Dr. Chris Downey and Michael Latham for valuable discussions.
Abbreviations
- VS
Neurospora Varkud satellite RNA
- HDV
Hepatitis Delta Virus
- FRET
fluorescence resonance energy transfer
- EPR
electron paramagnetic resonance
- (−)sTRSV
negative strand of the satellite RNA of tobacco ringspot virus
- PAGE
polyacrylamide gel electrophoresis
- EDTA
ethylenediaminetetraacetic acid
- Tris
tris(hydroxymethyl)aminomethane hydrochloride
- ASBVd(+)
positive strand of Avocado sunblotch viroid
- ASBVd(−)
negative strand of Avocado sunblotch viroid
Footnotes
This work was supported in part by grants from: NIH (AI 30726) and the W. M. Keck Foundation initiative in RNA science at the University of Colorado, Boulder
SUPPORTING INFORMATION AVAILABLE
The constructs used in the kinetics experiments for the cave cricket and ASBVd hammerhead ribozymes are presented in Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 2.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HAS, Kossoy E, Kessler M, Yonath A. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 3.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA - Auto-excision and auto-cyclization of the ribosomal-RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 4.Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 5.Vanderveen R, Arnberg AC, Vanderhorst G, Bonen L, Tabak HF, Grivell LA. Excised Group-II introns in Yeast mitochondria are lariats and can be formed by self-splicing invitro. Cell. 1986;44:225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]
- 6.Guerriertakada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of Ribonuclease-P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 7.Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 8.Winkler W, Nahvi A, Roth A, Collins J, Breaker R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:263–264. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 9.Symons RH. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores R, Delgado S, Gas ME, Carbonell A, Molina D, Gago S, De la Pena M. Viroids: the minimal non-coding RNAs with autonomous replication. FEBS Lett. 2004;567:42–48. doi: 10.1016/j.febslet.2004.03.118. [DOI] [PubMed] [Google Scholar]
- 11.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant-virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 12.Branch AD, Robertson HD. A replication cycle for viroids and other small infectious RNAs. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 13.Flores R, Hernandez C, de la Pena M, Vera A, Daros JA. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 2001;341:540–552. doi: 10.1016/s0076-6879(01)41175-x. [DOI] [PubMed] [Google Scholar]
- 14.Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of Avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active-sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 16.Buzayan JM, Hampel A, Bruening G. Nucleotide-sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus-RNA. Nucleic Acids Res. 1986;14:9729–9743. doi: 10.1093/nar/14.24.9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cote F, Perreault JP. Peach latent mosaic viroid is locked by a 2',5'-phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol. 1997;273:533–543. doi: 10.1006/jmbi.1997.1355. [DOI] [PubMed] [Google Scholar]
- 18.Cote F, Levesque D, Perreault JP. Natural 2',5'-phosphodiester bonds found at the ligation sites of peach latent mosaic viroid. J. Virol. 2001;75:19–25. doi: 10.1128/JVI.75.1.19-25.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daros JA, Flores R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002;21:749–759. doi: 10.1093/emboj/21.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branch AD, Robertson HD, Greer C, Gegenheimer P, Peebles C, Abelson J. Cell-free circularization of viroid progeny RNA by an RNA ligase from wheat-germ. Science. 1982;217:1147–1149. doi: 10.1126/science.217.4565.1147. [DOI] [PubMed] [Google Scholar]
- 21.Baumstark T, Schroder ARW, Riesner D. Viroid processing: Switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 1997;16:599–610. doi: 10.1093/emboj/16.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsagris M, Tabler M, Muhlbach HP, Sanger HL. Linear oligomeric potato spindle tuber viroid (PSTV) RNAs are accurately processed invitro to the monomeric circular viroid proper when incubated with a nuclear extract from healthy potato cells. EMBO J. 1987;6:2173–2183. doi: 10.1002/j.1460-2075.1987.tb02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 24.Uhlenbeck OC. A Small Catalytic Oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 25.Forster AC, Symons RH. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active-site. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- 26.Blount KE, Uhlenbeck OC. The structure-function dilemma of the hammer head ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 27.Hertel KJ, Herschlag D, Uhlenbeck OC. A kinetic and thermodynamic framework for the hammerhead ribozyme reaction. Biochemistry. 1994;33:3374–3385. doi: 10.1021/bi00177a031. [DOI] [PubMed] [Google Scholar]
- 28.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 29.De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson JA, Uhlenbeck OC. When to believe what you see. Mol. Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 32.Nelson JA, Shepotinovskaya I, Uhlenbeck OC. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry. 2005;44:14577–14585. doi: 10.1021/bi051130t. [DOI] [PubMed] [Google Scholar]
- 33.Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chartrand P, Harvey SC, Ferbeyre G, Usman N, Cedergren R. An oligodeoxyribonucleotide that supports catalytic activity in the hammerhead ribozyme Domain. Nucleic Acids Res. 1995;23:4092–4096. doi: 10.1093/nar/23.20.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DMJ. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim NK, Murali A, DeRose VJ. Separate metal requirements for loop interactions and catalysis in the extended hammerhead ribozyme. J. Am. Chem. Soc. 2005;127:14134–14135. doi: 10.1021/ja0541027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heckman JE, Lambert D, Burke JM. Photocrosslinking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- 38.Lambert D, Heckman JE, Burke JM. Three conserved guanosines approach the reaction site in native and minimal hammerhead ribozymes. Biochemistry. 2006;45:7140–7147. doi: 10.1021/bi052457x. [DOI] [PubMed] [Google Scholar]
- 39.Osborne EM, Schaak JE, DeRose VJ. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedor MJ, Uhlenbeck OC. Substrate sequence effects on hammerhead RNA catalytic efficiency. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 45.Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 46.Hertel KJ, Peracchi A, Uhlenbeck OC, Herschlag D. Use of intrinsic binding energy for catalysis by an RNA enzyme. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8497–8502. doi: 10.1073/pnas.94.16.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serra MJ, Turner DH. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 48.Bloomfield VA, Crothers DM, Tinoco I. Double helix formation by oligonucleotides without loops. In: Steifel J, editor. Nucleic Acids: Structures, Properties, and Functions. Sausalito: University Science Books; 2000. pp. 271–291. [Google Scholar]
- 49.Fedor MJ. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 50.ClouetdOrval B, Uhlenbeck OC. Hammerhead ribozymes with a faster cleavage rate. Biochemistry. 1997;36:9087–9092. doi: 10.1021/bi9710941. [DOI] [PubMed] [Google Scholar]
- 51.Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 2000;28:4037–4043. doi: 10.1093/nar/28.20.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daros JA, Marcos JF, Hernandez C, Flores R. Replication of Avocado sunblotch viroid - Evidence for a symmetrical pathway with 2 rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ugarkovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stage-Zimmermann TK, Uhlenbeck OC. A covalent crosslink converts the hammerhead ribozyme from a ribonuclease to an RNA ligase. Nat. Struct. Biol. 2001;8:863–867. doi: 10.1038/nsb1001-863. [DOI] [PubMed] [Google Scholar]
- 55.Lilley DMJ. Origins of RNA catalysis in the hairpin ribozyme. ChemBioChem. 2001;2:729–733. doi: 10.1002/1439-7633(20011001)2:10<729::AID-CBIC729>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 56.Nahas M, Wilson TJ, Hohng SC, Jarvie K, Lilley DMJ, Ha T. Observation of internal cleavage and ligation reactions of a ribozyme. Nat. Struct. Mol. Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 57.Taylor JM. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 2003;11:185–190. doi: 10.1016/s0966-842x(03)00045-3. [DOI] [PubMed] [Google Scholar]
- 58.Jones FD, Ryder SP, Strobel SA. An efficient ligation reaction promoted by a Varkud Satellite ribozyme with extended 5' and 3'-termini. Nucleic Acids Res. 2001;29:5115–5120. doi: 10.1093/nar/29.24.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peracchi A, Karpeisky A, Maloney L, Beigelman L, Herschlag D. A core folding model for catalysis by the hammerhead ribozyme accounts for its extraordinary sensitivity to abasic mutations. Biochemistry. 1998;37:14765–14775. doi: 10.1021/bi980867y. [DOI] [PubMed] [Google Scholar]
- 60.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.