Abstract
Endonucleases that generate double-strand breaks in DNA often possess two identical subunits related by rotational symmetry, arranged so that the active sites from each subunit act on opposite DNA strands. In contrast to many endonucleases, Type IIP restriction enzyme BcnI, which recognizes the pseudopalindromic sequence 5′-CCSGG-3′ (where S stands for C or G) and cuts both DNA strands after the second C, is a monomer and possesses a single catalytic center. We show here that to generate a double-strand break BcnI nicks one DNA strand, switches its orientation on DNA to match the polarity of the second strand and then cuts the phosphodiester bond on the second DNA strand. Surprisingly, we find that an enzyme flip required for the second DNA strand cleavage occurs without an excursion into bulk solution, as the same BcnI molecule acts processively on both DNA strands. We provide evidence that after cleavage of the first DNA strand, BcnI remains associated with the nicked intermediate and relocates to the opposite strand by a short range diffusive hopping on DNA.
INTRODUCTION
Type IIP restriction endonucleases (REases) interacting with symmetric (palindromic) DNA sites are often arranged as dimers comprised of identical subunits (1). In the REase–DNA complex the DNA and the protein dimer share a dyad symmetry, placing the catalytic center and sequence recognition elements of one subunit against the scissile phosphate and DNA bases in one half of the recognition site and the other subunit against the symmetry related half-site (1–3). This strategy, which is shared by many Type II enzymes, enables the REase to recognize two symmetrical DNA half sites of different polarity and cut phosphodiester bonds on opposite strands to generate a double-strand break.
In contrast to orthodox Type IIP REases, MutH nuclease involved in DNA repair is a monomer and nicks the unmethylated strand in the hemimethylated 5′-GATC-3′ site (4). Methylation of the A base in one DNA strand breaks the target site symmetry and directs MutH cleavage to the phosphodiester bond on the unmethylated DNA strand (5). Surprisingly, crystal structures of the Type IIP REases MvaI and BcnI, which generate a double-strand break respectively at the 5′-CC/WGG-3′ and 5′-CC/SGG-3′ sequences (W stands for A or T, S for C or G, ‘/’ designates the cleavage position), share striking similarity with the MutH repair enzyme (6–8). Moreover, similarly to MutH, MvaI and BcnI are monomers and contain a single active site. However, unlike MutH which nicks its recognition site, BcnI and MvaI make a double-strand break within their target sequences. This raises the question of how monomeric REases such as BcnI and MvaI accomplish cleavage of the double-stranded DNA.
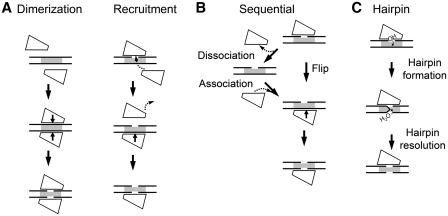
In principle, several alternative mechanisms may be proposed. (i) Dimerization/recruitment model, which assumes a transient interaction of two monomers on DNA (Figure 1A). The transient dimerization mechanism was first proposed for the FokI restriction enzyme that recognizes asymmetric DNA sequence and cuts phosphodiester bonds on both strands away of the target site (9,10). Later it was demonstrated for other REases (11). In theory, one cannot exclude the possibility that instead of making a transient dimer, the DNA-bound monomer may recruit from the solution another monomer, which binds in the opposite orientation and displaces the DNA-bound monomer (Figure 1A, ‘recruitment’ mechanism). (ii) Consecutive nicking model, which assumes that the monomeric enzyme cleaves DNA in two sequential nicking reactions (Figure 1B). As the two DNA strands run in opposite directions, the enzyme must switch its orientation on DNA between the two cleavage steps. If strand switching requires enzyme release into bulk solution after the first strand cleavage, only one DNA strand will be cleaved per binding event. Alternatively, if an enzyme is able to flip to the opposite strand without being physically separated from DNA, as shown for the methyl–CpG-binding domain (12), both DNA strands can be cut by the same enzyme molecule during a single binding event. A similar mechanism was recently demonstrated for BfiI REase that cleaves both DNA strands by rotating a single catalytic center (13). (iii) The reaction mechanism involving a hairpin intermediate (Figure 1C). This mechanism has been previously demonstrated for retroviral integrases, transposases and V(D)J recombination enzymes of DDE family (14–16). It assumes that the enzyme first cuts one DNA strand to leave a 3′-hydroxyl, which then attacks the scissile phosphate in the opposite DNA strand to generate a hairpin intermediate, which is subsequently hydrolyzed to produce a double-strand break.
Figure 1.
Possible mechanisms for the double-stranded DNA cleavage by monomeric Type IIP REases. Protein monomer is shown as a quadrangle shape, DNA is depicted by two parallel black lines, gaps indicate cleaved DNA strands and a grey rectangle marks the target site. (A) Dimerization/recruitment mechanism, (B) sequential cleavage, (C) reaction via hairpin intermediate.
In this study we employed single-turnover and steady-state kinetics to dissect the mechanism of double-stranded DNA cleavage by BcnI. In the available crystal structures [PDB ID: 2ODI (7) and 3IMB] the BcnI monomer is bound to its target site in two distinct orientations which bring the catalytic center in the vicinity of either the C- (5′-CCCGG-3′) or the G- (5′-CCGGG-3′) strand. Structural analysis excludes simultaneous binding of two monomers on the same recognition site due to steric conflict, making ‘dimerization’ mechanism unlikely (7). However, alternative reaction mechanisms, including recruitment of another monomer from the solution (Figure 1A, ‘recruitment’ mechanism), consecutive nicking (Figure 1B) or cleavage via a hairpin intermediate (Figure 1C) are still possible and were subjected to experimental analysis.
MATERIALS AND METHODS
Mutagenesis
The catalytically deficient D55A mutant of BcnI was obtained as described in (17). Sequencing of the entire gene of the mutant confirmed that only the designed mutation had been introduced.
Protein expression and purification
Wild-type (wt) and the D55A mutant were expressed in Escherichia coli and purified to >99% homogeneity as described earlier (7). Concentrations of both proteins were determined from A280 measurements using extinction coefficient of 21 430 M−1 cm−1.
DNA substrates
All oligodeoxynucleotides used in this study were purchased from Metabion (Martinsried, Germany). Oligoduplexes used in the DNA cleavage experiments are listed in Table 1. Substrate assembly and radiolabeling procedures are described in the Supplementary Data section. Double-stranded phage ΦX174 DNA was obtained from Fermentas (Vilnius, Lithuania).
Table 1.
Oligonucleotide substrates
| Duplex | Sequencea | Specification |
|---|---|---|
| HP | 5′-CTTCGCAGTACGCCpGGGCAATAACGCACGT\ | A hairpin substrate with a single BcnI recognition sequence.b |
| 3′-GAAGCGTCATGCGGCpCCGTTATTGCGTGAT/ | ||
| 30/30 | 5′-CTTCGCAGTACGCCpGGGCAATAACGCACGT-3′ | Same as HP, but lacks the hairpin loop. Was used as unlabeled trap DNA. |
| 3′-GAAGCGTCATGCGGCpCCGTTATTGCGTGCA-5′ | ||
| G-nick | 5′-CTTCGCAGTACGCCGGGCAATAACGCACGT-3′ | As 30/30, except for the nick in the scissile position of the G-strand (designated by a gap).c |
| 3′-GAAGCGTCATGCGGCpCCGTTATTGCGTGCA-5′ | ||
| C-nick | 5′-CTTCGCAGTACGCCpGGGCAATAACGCACGT-3′ | As 30/30, except for the nick in the scissile position of the C-strand (designated by a gap).c |
| 3′-GAAGCGTCATGCGGCCCGTTATTGCGTGCA-5′ | ||
| GG | /TAGCAGTACGCC*GGGCAACGCCpGGGCAATAACGCA-3′ | A hairpin substrate that carries two BcnI sites in direct repeat orientation. The internal 33P label is designated by *. |
| \TGCGTCATGCGGCpCCGTTGCGGCpCCGTTATTGCGT-5′ |
aThe BcnI recognition sequences are underlined; scissile positions are designated by ‘p’.
bThe HP substrate carried a radiolabel either at the 5′ or the 3′ terminus.
cThe G-nick and the C-nick substrates carried a radiolabel at the 5′ terminus of the 30 nt strand and a 5′-terminal phosphate at the position of the nick.
Reactions with oligonucleotide substrates
Single turnover reactions were carried out at 25°C in a Kin-Tek RQF-3 quench-flow device. For pre-mix reactions, BcnI was preincubated with the radiolabeled DNA in Buffer Y (33 mM Tris–acetate, pH 7.9 at 25°C, 66 mM potassium acetate and 0.1 mg/ml BSA) supplemented with 0.2 mM EDTA and the reaction started by mixing with an equal volume (16 µl) of 20 mM magnesium acetate solution in Buffer Y. For post-mix reactions starting with enzyme and DNA in separate solutions, 16 µl of BcnI in Buffer Y containing 20 mM magnesium acetate was mixed with an equal volume of DNA substrate in Buffer Y supplemented with 0.2 mM EDTA. In both cases, the final reactions contained 200–400 nM of BcnI, 2 nM of radiolabeled DNA, 10 mM magnesium acetate and <0.12 mM EDTA. In the alternative set of reactions, the magnesium acetate solution used for the pre-mix reactions was supplemented either with 4 µM of unlabeled DNA (30/30, G-nick or C-nick, Table 1) or 16 µM of the D55A mutant. In control experiments, wt BcnI was preincubated with the radiolabeled substrate in the presence of either 4 µM unlabeled DNA or 16 µM of D55A mutant prior to mixing with magnesium acetate.
The reactions were quenched by mixing with 2 M HCl. The recovered samples (∼100 µl) were mixed with 45 µl of Neutralization Solution (3.5 M Tris and 3% SDS) and 60 µl of denaturing loading dye (95% v/v formamide, 25 mM EDTA, 0.01% bromphenol blue). Reaction products were separated by high resolution denaturing PAGE (20% 19:1 acrylamide/bis–acrylamide with 8 M urea in TBE buffer thermostated at 60°C). Radiolabeled DNA was detected and quantified by phosphorimager.
Steady state reactions were initiated by manually adding magnesium acetate to the premixed BcnI and oligoduplex solution. The final reactions contained 50–200 nM of DNA, 1–2 nM BcnI and 10 mM magnesium acetate in Buffer Y. Samples (8 µl) were collected at timed intervals, quenched by mixing with 10 µl of denaturing loading dye and analyzed as described above.
Reactions with supercoiled DNA
The single turnover experiments on the supercoiled ΦX174 DNA were carried out in a quench-flow device as described above, but quenched with 6 M guanidinium chloride instead of HCl. The final reactions contained 2 nM ΦX174 DNA, 100–200 nM enzyme and 10 mM magnesium acetate. For DNA- and mutant-trap experiments the reactions also contained 2000 nM of the unlabeled oligoduplex 30/30 (Table 1) or 8 µM of the D55A mutant, respectively. DNA was recovered by ethanol precipitation and analyzed by electrophoresis through agarose and densitometric analysis of ethidium bromide-stained gels as described (18). Multiple-turnover cleavage reactions were performed at 25°C with 1–4 nM ΦX174 DNA and 0.1 nM wt BcnI. Aliquots (25 µl) were removed at timed intervals, quenched by adding 8 µl of non-denaturing loading dye solution (50% v/v glycerol, 75 mM EDTA, 0.01% bromphenol blue) and electrophoresed through agarose.
Data analysis
During BcnI reactions on radiolabeled oligoduplexes, a fraction of DNA (typically ∼10%) remained uncleaved even after prolonged incubation, presumably due to the incorrectly annealed substrate. We experimentally determined this fraction for every oligonucleotide duplex and eliminated it during data analysis. This correction was not applied to steady-state reactions. The preparation of supercoiled ΦX174 DNA used in this study contained 10% of the randomly nicked OC form. Assuming that randomly nicked DNA is equivalent to the intact substrate, we corrected the experimentally determined amounts of supercoiled (SC) and nicked (OC) DNA at each time point using the following equations: [SC]corrected = [SC]experimental/0.9, [OC]corrected = 100% − [SC]corrected − [FLL]experimental.
BcnI reaction on the 5′-labeled oligoduplex HP (Table 1) results in formation of radiolabeled 45 and 14 nt products. The 45-nt fragment results from nicking of the C-strand, and therefore corresponds to the C-nick reaction intermediate. The 14-nt product may result either from nicking of the G-strand or from a double-strand break, and therefore corresponds to the sum of the G-nick and the final product PP. The reaction on the 3′-labeled hairpin oligoduplex HP results in the 46- and 15-nt products, the former corresponding to the G-nick intermediate, and the latter to the sum of C-nick and PP. Thus, by performing BcnI reactions on both the 5′- and 3′-labeled hairpin substrate, we could follow the amount of three DNA forms: intact DNA SS, G-nick intermediate and C-nick intermediate. Amount of the final product PP was determined by subtracting concentrations of substrate, G-nick and C-nick from the total DNA concentration. If BcnI cleavage of the HP oligoduplex proceeded via a hairpin intermediate, the hairpin closure reaction on the G-nick intermediate would result in a novel ∼30-nt radiolabeled hairpin which was not detected in the gels.
Analysis of the two-site DNA (duplexes GG and GC) cleavage data and the equations used to quantify the single-turnover BcnI reactions on the HP, G-nick and C-nick substrates are provided in the Supplementary Data section. The steady-state reaction rates were determined by linear regression. Single-turnover reactions on ΦX174 DNA were analyzed as described in (19) to obtain the rate constant k1 for DNA nicking and the rate constant k2 for the second strand cleavage. All fitting procedures used KyPlot 2.0 software (20). Determined rate constants are presented as the optimal value ±1 standard error.
RESULTS
BcnI reactions on the supercoiled ΦX174 DNA
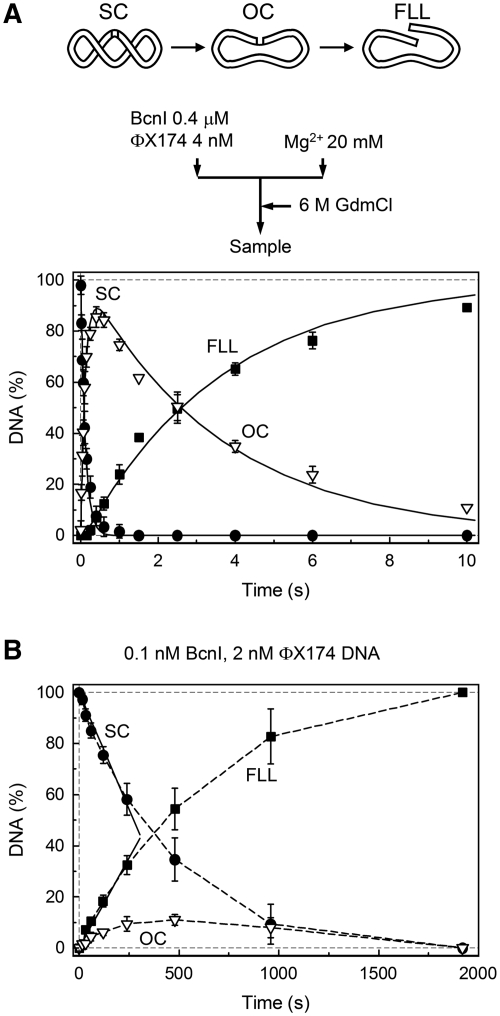
Initially we studied the BcnI cleavage of a supercoiled phage ΦX174 DNA that carries a single recognition site 5′-CCSGG-3′ (Figure 2). Cleavage reactions were first performed under single-turnover conditions, with the enzyme in molar excess over the substrate, to monitor possible reaction intermediates which should remain bound to the enzyme during the reaction course (18). Single turnovers of BcnI were performed in the pre-mix mode (Figure 2A), i.e. the supercoiled ΦX174 DNA was preincubated with a 100-fold excess of BcnI prior to initiation of the reaction with Mg2+ ions (see ‘Materials and Methods’ section for details). This reaction setup eliminates the possible effect of enzyme–DNA association rate on the subsequent reaction steps and therefore reports the actual cleavage rate of the enzyme-bound DNA. Under these conditions, BcnI rapidly converted the supercoiled DNA SC into the nicked intermediate OC (k1 ≈ 8 s−1) followed by a slow cleavage of the nicked intermediate to generate a linear product FLL (k2 ≈ 0.3 s−1) (Figure 2A). High yield of the nicked intermediate OC (up to 90%) and the ratio of rate constants (k1/k2 ≈ 25) indicate that BcnI reactions on the first and on the second DNA strands are not equivalent.
Figure 2.
BcnI reactions on the supercoiled ΦX174 DNA. The scheme above panel (A) illustrates the various DNA forms that can exist during BcnI reactions on supercoiled phage ΦX174 DNA bearing a single BcnI recognition site. (A) Single turnover reaction with 2 nM of DNA substrate and 200 nM of BcnI. The flow diagram above the graph schematically depicts the experiment performed in a quench-flow device (see ‘Materials and Methods’ section for details). Three DNA forms are shown: supercoiled DNA (SC, filled circles), open circular intermediate (OC, down triangles) and full length linear product (FLL, filled squares). All data points are presented as mean values from three independent experiments ±1 SD. Continuous lines are the best fit to the reaction equation SC − k1→OC − k2→FLL that gave k1 = 8.3 ± 0.2 s−1 (the rate constant for nicking of the first DNA strand) and k2 = 0.27 ± 0.1 s−1 (the rate constant for cleavage of the second DNA strand). (B) Steady-state experiment. All data points are mean values from seven independent experiments ±1 SD. The reaction contained 2 nM DNA and 0.1 nM BcnI. The initial SC cleavage and FLL formation rates (0.0036 ± 0.0003 nM s−1 and 0.0028 ± 0.0003 nM s−1, respectively), determined from linear fits (solid lines), indicate that BcnI converts 77% (ratio 0.0028/0.00365) of the initial substrate SC into the final reaction product FLL during a single binding event. The reaction kcat equals 0.036 ± 0.003 s−1 (0.0036 nM s−1/0.1 nM BcnI).
Unlike single-turnover reactions, steady-state experiments provide the total turnover rate, which theoretically may be limited by distinct reaction steps including DNA binding, cleavage and/or product dissociation. Analysis of the steady-state reaction products that are released by the enzyme and accumulate in solution may reveal the number of phosphodiester bonds cleaved by an enzyme during a single binding event. If BcnI dissociates into bulk solution after cutting one DNA strand, the nicked intermediate should accumulate in solution. Alternatively, if both DNA strands are cut during a single binding event, the linear DNA should predominate. BcnI steady-state reactions were performed at 10- to 40-fold excess of DNA (1–4 nM ΦX174) over BcnI (0.1 nM) (Figure 2B). The reaction rate was independent of the substrate concentration (data not shown), indicating that KM value was significantly <1 nM, the lowest substrate concentration used in experiments. Therefore, the determined initial reaction rate v0 is equal to vMAX and the ratio vMAX/[BcnI] ≈ 0.036 s−1 corresponds to kcat. Surprisingly, the major reaction product is a linear FLL form with both DNA strands cut at the BcnI recognition site; the nicked intermediate comprises only 10% of total DNA (or 23% of reaction products, Figure 2B), indicating that only a minor fraction of the enzyme is released into solution before cleavage of the second strand.
Steady-state reactions of ΦX174 DNA cleavage exclude an obligatory dissociation of the nicked OC intermediate in the BcnI reaction pathway (Figure 1B, ‘dissociation’), and also argues against a transient dimerization or ‘recruitment’ mechanisms (Figure 1A), as dimerization is unlikely under [BcnI] << [DNA] conditions (there would simply be no free enzyme required for dimerization in the presence of a saturating DNA concentration). On the other hand, steady state cleavage data are consistent with the sequential mechanism occurring without an excursion into solution (Figure 1B, ‘flip’) or an alternative mechanism involving a hairpin intermediate (Figure 1C). However, reactions on the ΦX174 DNA did not allow us to discriminate between these two mechanisms since the hairpin intermediate cannot be resolved by electrophoresis in agarose gel.
Single turnover reactions on the oligoduplex substrate
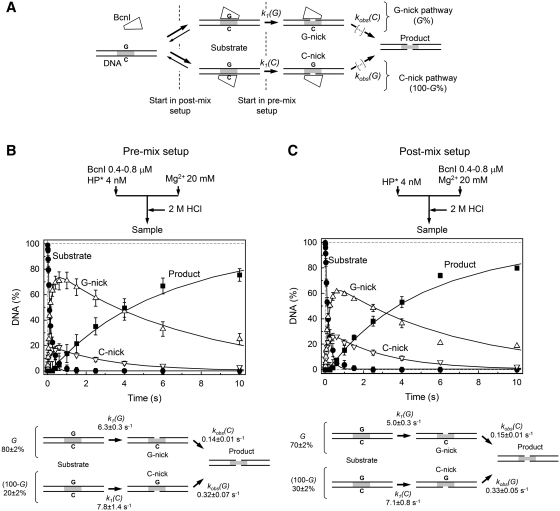
To overcome the limitations of the ΦX174 DNA substrate, we have analyzed BcnI reactions using the synthetic 30-bp oligoduplex HP (Table 1) assembled by self-annealing of the 60-nt hairpin-forming oligodeoxynucleotide. It contained a single cognate 5′-CCSGG-3′ site located at the center and flanked by 5-nt sequences identical to those surrounding the BcnI cleavage site in ΦX174 DNA. The 5′- or 3′-ends of the HP oligoduplex were radiolabeled and reaction products analyzed by denaturing PAGE to monitor and quantify all DNA forms generated during BcnI reaction (Figure 3A) as described in ‘Materials and Methods’ section. First, the HP oligoduplex cleavage was analyzed in the pre-mix setup (Figure 3B) by mixing large excess of BcnI (400–800 nM) over the radiolabeled oligoduplex (4 nM) in one syringe and initiating the reaction by Mg2+ solution contained in another syringe. Under these conditions the HP oligoduplex was converted into the mixture of G-nick and C-nick intermediates that were further transformed into the final reaction product with a double-strand break (Figure 3B). No other intermediates were detected in the BcnI reaction, ruling out DNA cleavage via a hairpin intermediate (Figure 1C). Previous studies revealed that BcnI is able to bind to its target site in two alternative orientations (17), which lead into two distinct routes for DNA cleavage depending whether the G- or C-strand is cleaved first (Figure 3A). In the pre-mix setup the majority of the oligoduplex was nicked at the G-strand (Figure 3B), while the C-strand intermediate formed a minor fraction. Concomitant formation of the G-nick and C-nick products during the HP substrate cleavage implies that the open circular intermediate OC formed during the ΦX174 DNA cleavage (Figure 2A) is also a mixture of DNA nicked in the G- and in the C-strand.
Figure 3.
BcnI reactions on oligoduplex DNA. (A) Scheme for DNA cleavage by BcnI. BcnI may bind to the target site in two alternative orientations placing the active site in the vicinity of either the G- (5′-CCGGG-3′) or the C-strand (5′-CCCGG-3′). The ensuing nicking reactions [rate constants k1(G) and k1(C)] result in the nicked intermediates G- and C-nick, respectively. The fraction of substrate cleaved via the G-nick intermediate is designated by G(%); the fraction of substrate that follows the C-nick pathway corresponds to 100–G%. To complete the reaction after cutting the first DNA strand, BcnI must switch its orientation on the recognition site. All reaction steps that lead to cleavage of the second DNA strand are described by the rate constants kobs(G) and kobs(C). (B and C) Cleavage of the hairpin oligoduplex HP (Table 1) in the pre-mix and post-mix reactions, respectively. The flow diagrams above the graphs schematically depict the two types of experiments performed in a quench-flow device (see ‘Materials and Methods’ section for details). The amounts of four DNA forms are shown: intact substrate (filled circles), G-nick intermediate (up triangles), C-nick intermediate (down triangles), and final product (filled squares). All data points are presented as mean values from three or more independent experiments ±1 SD. Solid lines are the fit of the scheme in panel A to experimental data, assuming that the enzyme binding steps do not contribute to the observed DNA cleavage rates. Determined rate constants are listed below each graph.
Reaction profiles obtained at 200 and 400 nM of BcnI were undistinguishable (combined data are provided in Figure 3B), suggesting that the observed reaction rates are limited by conversion of the enzyme-bound DNA rather than by enzyme–DNA association. Therefore, the explicit BcnI reaction scheme (Figure 3A) could be simplified (Figure 3B) by eliminating enzyme-binding steps and fitted to experimental data (see Supplementary Data for details) to yield rate constants for individual reaction steps and the fraction of DNA cleaved via the G-nick and the C-nick routes (G% and 100–G%) (Figure 3B). In the intact substrate BcnI rapidly cuts both G- and C-DNA strands [k1(G) ≈ 6 s−1, k1(C) ≈ 8 s−1], but cleavage rates of the same strands in the nicked intermediates are reduced by ∼20- and ∼60-fold, respectively [kobs(G) ≈ 0.3 s−1, kobs(C) ≈ 0.14 s−1]. These data are in agreement with the data for the supercoiled ΦX174 DNA cleavage (Figure 2A). However, while experiments on the ΦX174 DNA provide only the averaged rates for DNA nicking and linearization, reactions on the HP oligoduplex allow for a complete specification of all DNA cleavage steps in the alternative G-nick and C-nick reaction pathways.
Strand preference of BcnI
The fraction of DNA cleaved in the pre-mix mode via the G-nick and C-nick pathways reflects the initial BcnI distribution between the G- and C-strand bound states in the absence of divalent metal ions (Figure 3A). In the pre-mix mode ∼80% of DNA is cleaved via the G-nick intermediate (Figure 3B) independent of DNA sequences flanking the recognition site (data not shown). It suggests that under equilibrium binding conditions in the pre-mix setup the BcnI–DNA complex with the catalytic center close to the G-strand is more stable than the complex in the opposite DNA orientation. However, the relative contribution of the G- and C-nick complexes may change in the post-mix setup where enzyme is mixed with DNA in the presence of Mg2+ ions. Indeed, if an enzyme association rate to the G- and C-strands is the same and each encounter is equally productive, equal amounts of DNA should be cleaved via the G-nick and the C-nick intermediates. Post-mix reactions conducted at two different enzyme concentrations (200 and 400 nM) were undistinguishable (combined data are displayed in Figure 3C) and provided rates for individual DNA strand cleavage that were close to the values obtained in the pre-mix experiments (Figure 3B and C). Most importantly, the G-nick pathway still prevailed in the post-mix mode (G = 70%, Figure 3C), suggesting that either BcnI binds the G-strand faster than the C-strand, or the equilibration rate between the alternative BcnI–DNA complexes is comparable to DNA cleavage rate.
Single-turnover reactions on nicked intermediates
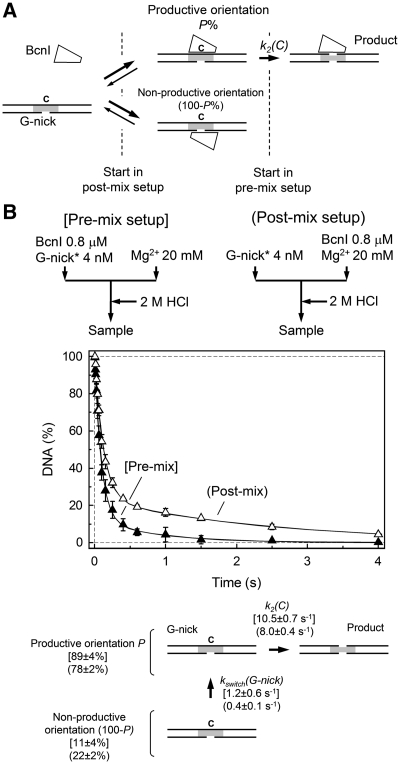
BcnI cuts the first DNA strand >20-fold faster than the second strand (Figure 3B and C). This difference could be due to the differences in the actual cleavage rates or distinct rate-limiting steps. According to the proposed BcnI reaction mechanism (Figure 3A), cleavage of the first DNA strand is limited by the reaction chemistry, but hydrolysis of the second DNA strand must be preceded by the switch in enzyme orientation (Figure 3A). If an enzyme flip is slower than DNA hydrolysis, it may limit the reaction rate of the second DNA strand cleavage. To determine BcnI cleavage rates of the nicked intermediates, we assembled the pre-nicked substrates G-nick and C-nick from three DNA strands (see ‘Materials and Methods’ section). Except for the hairpin loop and the nick, the G-nick and C-nick duplexes were otherwise identical to the HP oligoduplex and were phosphorylated at the 5′-terminus at the nick site to mimic the BcnI reaction intermediates (Table 1). BcnI may bind the nicked intermediate in either of two orientations, positioning the active site close to the continuous or to the nicked strand (Figure 4A). If the active site faces the intact DNA strand (C-strand in the G-nick intermediate, Figure 4A), the binding mode is productive, as an enzyme may directly proceed to hydrolysis of the second DNA strand. Alternatively, if the BcnI active site faces the nicked strand (G-strand in the G-nick duplex), the binding mode is non-productive and an enzyme must switch its orientation before it can cut the intact strand (Figure 4A). The non-productive and productive binding modes on the nicked substrate mimic different stages of the BcnI reaction (Figure 3A): the non-productive orientation corresponds to the BcnI–DNA complex immediately after cleavage of the first DNA strand, and the productive mode corresponds to the situation after the presumed switch in enzyme orientation.
Figure 4.
BcnI cleavage of the G-nick intermediate. (A) Scheme for BcnI reaction on the G-nick intermediate. At the initial moment of the reaction, BcnI may bind the nicked DNA in either of two alternative orientations. In the productive orientation, the catalytic center is positioned at the scissile phosphodiester bond in the intact C-strand to proceed with the cleavage [rate constant k2(C)]. In the alternative orientation, the catalytic center of BcnI is placed against the nicked DNA strand G, thus the enzyme has to dissociate and then re-bind in the opposite orientation before cleavage can occur. This rearrangement is described by the rate constant kswitch(G-nick). The fraction of substrate bound in the productive orientation is designated by P(%). (B) Cleavage of the G-nick intermediate in the pre-mix (filled triangles) and post-mix (open triangles) reactions. Flow diagrams schematically depict the two types of experiments performed in a quench-flow device. All data points are presented as mean values from three or more independent experiments ±1 SD. Solid lines represent the fit of the reaction scheme in panel A to experimental data, assuming that the enzyme binding steps are fast and do not affect the observed DNA cleavage rates. Determined rate constants for the pre-mix and post-mix reactions are provided below the graph in square brackets and parentheses, respectively.
Pre-mix and post-mix experiments on the G-nick intermediate, performed at different enzyme concentrations (200 and 400 nM) were undistinguishable (combined data are displayed in Figure 4B), suggesting that enzyme–DNA association is fast and does not contribute to the observed rate of DNA cleavage. Quantification of BcnI reactions on the G-nick substrate using the two-step reaction scheme (Supplementary Data) provided the rate constant k2(C) for the C-strand cleavage by the enzyme in the productive orientation, kswitch(G-nick) for the switch in enzyme orientation, and P (%), the fraction of enzyme bound in the productive orientation at the initial reaction moment (Figure 4B). In both pre-mix and post-mix experiments BcnI predominantly bound the G-nick oligoduplex in the productive orientation (fraction P close to 90 and 80% for the pre-mix and post-mix reactions, respectively) and rapidly cleaved the C-strand [k2(C) ≈ 10 s−1]. The remaining substrate was cleaved at a much lower rate, presumably due to the slow switch to the alternative enzyme orientation [kswitch(G-nick) ≤ 1 s−1]; this rate could not be reliably determined due to a small fraction of substrate bound in the non-productive orientation. Similar results were also obtained with the C-nick intermediate (Supplementary Figure S1): the predominant fraction of BcnI bound the C-nick substrate in the productive orientation (P ≈ 85%) and rapidly cleaved the intact G-strand [k2(G) ≈ 8 s−1].
Taken together, BcnI cuts the pre-formed G-nick and C-nick oligoduplexes much faster than the nicked intermediates generated in situ during the HP oligoduplex cleavage [cf. k2(G) ≈ 8 s−1 and kobs(G) ≈ 0.3 s−1; k2(C) ≈ 10 s−1 and kobs(C) ≈ 0.14 s−1; Figures 3, 4 and Supplementary Figure S1]. Thus, slow cleavage of the nicked intermediate in the case of the HP substrate (Figure 3B and C) must be limited by the BcnI–DNA complex rearrangement after the first DNA strand cleavage. Indeed, to cut the second DNA strand, BcnI must rotate 180° around the axis perpendicular to the DNA to match the opposite polarity of second scissile phosphate. However, the direct flip of BcnI between DNA strands is structurally restricted. Due to the Ω-like clamp structure in the DNA bound form, 180° rotation of BcnI should result in a steric clash between DNA and the protein. The structural problem for the enzyme rotation could be evaded if BcnI dissociates into bulk solution after cutting the first strand and then re-associates in the opposite orientation. However, such mechanism is inconsistent with the steady-state experiment (Figure 2B) which shows that nicked intermediate is not released into bulk solution during DNA cleavage.
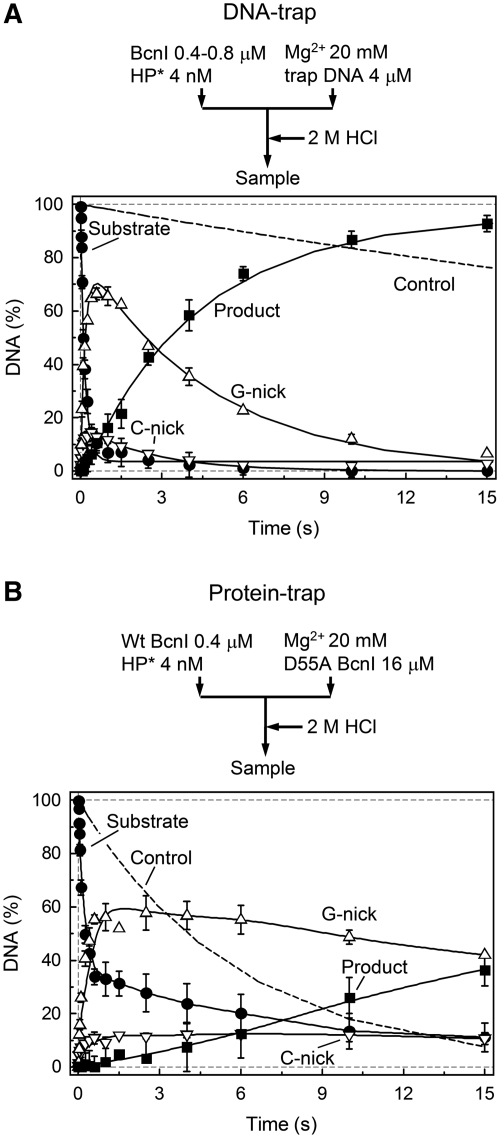
DNA-trap experiments
To better understand the mechanism of the BcnI switch between DNA strands, we have performed single turnover experiments in the presence of DNA or protein trap (Figure 5). In the DNA-trap setup, BcnI and radiolabeled DNA were pre-mixed in one syringe and DNA cleavage initiated by mixing with Mg2+ and 1000-fold excess of unlabeled DNA in the second syringe (Figure 5A). Any enzyme unbound to the radiolabeled DNA at the initial reaction moment or released into bulk solution during the reaction course should be trapped by unlabeled DNA. Therefore, if the DNA strand switch is accompanied by BcnI dissociation into bulk solution (i.e. diffusion away from the nicked DNA to such distance that re-binding to the original DNA molecule becomes unlikely), an enzyme will be trapped by the unlabeled DNA and cleavage of the second DNA strand will be inhibited.
Figure 5.
Trap experiments. In both panels, change in the amount of intact DNA (filled circles), G-nick intermediate (up triangles), C-nick intermediate (down triangles) and final reaction product (filled squares) is shown. All data points are presented as mean values from two to six independent experiments ±1 SD. (A) Pre-mix experiment with trap DNA. A sample of enzyme preincubated with radiolabeled DNA was mixed with magnesium acetate and excess of unlabeled DNA. Data was quantified as in Figure 3B (solid lines). The fitting procedure gave k1(G) = 5.7 ± 0.4 s−1, k1(C) = 10.5 ± 4.0 s−1, kobs(C) = 0.21 ± 0.02 s−1, kobs(G) = 0.39 ± 0.16 s−1; the majority of DNA was cleaved via the G-nick intermediate (G = 84 ± 2%). The dashed line shows cleavage of DNA substrate in the control experiment where enzyme was added to the mixture of radiolabeled substrate and trap DNA. (B) Pre-mix experiment with trap protein. A sample of enzyme, preincubated with radiolabeled DNA was mixed with magnesium acetate and excess of the inactive BcnI mutant D55A. The dashed line shows cleavage of DNA substrate in the control experiment where wt BcnI was preincubated with both substrate and excess of D55A mutant prior to initiation of the reaction with Mg2+ ions.
In fact, trap DNA had virtually no effect on the BcnI reaction (Figure 5A), as the cleavage pattern of the HP oligoduplex in the presence of DNA trap was nearly identical to the cleavage in the pre-mix setup (Figure 3B). Oligoduplex trap also had no effect on the cleavage pattern of the supercoiled ΦX174 DNA (Supplementary Figure S2A). This provides direct evidence that BcnI cleaves both DNA strands without dissociation into bulk solution and comes in a full support of the steady state experiments (Figure 2B and Supplementary Figure S3) which show that the majority of substrate (77% of ΦX174 DNA and 92% of HP oligoduplex) is converted into final reaction products during a single binding event. Furthermore, even unlabeled G- and C-nick DNA traps had no effect on the HP oligoduplex cleavage (data not shown), excluding the possibility that BcnI dissociates from the nicked DNA into bulk solution, but then re-associates with the nicked intermediate rather than an intact substrate molecule due to higher affinity for the nicked DNA. No preference of BcnI for the nicked intermediates over the HP oligoduplex was observed in the steady-state reactions performed with equimolar mixtures of nicked and intact DNA (Supplementary Figure S4).
Mutant-trap experiments
In the mutant-trap setup (Figure 5B), BcnI was first preincubated with the radiolabeled oligoduplex and then mixed with a large excess of the catalytically impaired D55A mutant (7) in the presence of Mg2+ (final concentrations: 2 nM radiolabeled DNA, 200 nM wt BcnI and 8 µM D55A mutant). In a control experiment, the D55A mutant effectively inhibited the single-turnover reaction of wt BcnI, presumably through binding of the radiolabeled substrate (Figure 5B, dashed line). Therefore, in the protein-trap experiment any radiolabeled DNA (intact substrate or nicked intermediate) liberated from the wt BcnI during the reaction should be trapped by the D55A mutant.
In the presence of the catalytically deficient mutant trap, BcnI rapidly converted the majority of oligoduplex into the G-nick and C-nick intermediates. However, contrary to the DNA-trap experiments, the reaction was nearly terminated after the first strand cleavage (Figure 5B). Similar cleavage pattern was also obtained during the BcnI reaction on the supercoiled ΦX174 DNA in the presence of the D55A mutant (Supplementary Figure S2B). These experiments suggest that after nicking of the first DNA strand, BcnI releases the target site which then becomes occupied by the D55A mutant. Mutant-trap experiments seem to be in conflict with the DNA-trap and steady-state reactions (Figure 5A, Figure 2B and Supplementary Figure S3), which demonstrate that BcnI cuts both DNA strands during a single binding event. These seemingly contradictory data, however, can be reconciled assuming that after cleavage of the first DNA-strand BcnI leaves the nicked recognition site, but instead of diffusing into bulk solution, it remains associated with a random sequence on the same DNA molecule. Since BcnI stays in the immediate proximity of the original substrate molecule, trap DNA has no effect on the BcnI reaction. On the other hand, during the random walk BcnI transiently leaves the recognition site which may become occupied by the catalytically inactive mutant in the mutant-trap experiment. The above model may also account for the different extent of substrate DNA cleavage in mutant- and DNA-trap experiments (Figure 5 and Supplementary Figure S2). Complete substrate cleavage in the DNA-trap experiment indicates that at the initial reaction moment all substrate is bound by an enzyme and the dissociation rate of the BcnI–DNA complex is much slower in comparison to the first DNA strand hydrolysis rate; however, a fraction of BcnI sites may remain free due to an equilibrium enzyme distribution between the target site and random DNA sequences. Excess of the trap DNA, introduced into the reaction together with Mg2+, does not interfere with the diffusional walk of BcnI on the substrate, and therefore the enzyme bound at the random sites will translocate to the target site and cut DNA. In contrast, in the mutant-trap setup, the inactive mutant, introduced into the reaction at a very high concentration, may locate the unoccupied target sites before the wt enzyme, and thereby inhibit their cleavage (the slowly cleaved fraction corresponds to ∼30% of the substrate, Figure 5B and Supplementary Figure S2B).
Processivity of BcnI
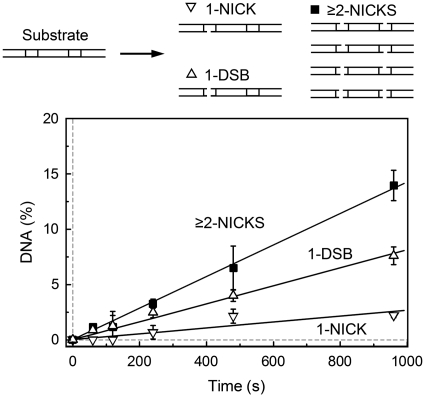
The proposed BcnI reaction mechanism involving two consecutive nicking reactions interrupted by a random walk is similar to the reactions of processive DNA enzymes that act on multiple recognition sites by diffusing along the DNA (21–26). If after the first strand cleavage BcnI relocates to a random sequence on the same DNA molecule, it may either switch its orientation and back-track to the same site to cut the second strand, or move to the next recognition site if it is present on the same DNA molecule. To test this possibility, we analyzed BcnI cleavage reactions on the two-site substrate GG that carries two BcnI sites in a direct repeat orientation separated by 10 bp (Table 1). In this case, to generate final reaction products cut at both strands at both sites, BcnI must cleave four phosphodiester bonds. Theoretically, 14 different reaction intermediates with one, two or three phosphodiester bonds cleaved may be formed during the reaction on the two-site oligoduplex (Supplementary Figure S5). However, all possible reaction products cannot be quantified by denaturing polyacrylamide gels analysis. Therefore, we determined only the total amount of DNA containing a single nick (‘1-NICK’) and the amount of DNA containing a single double-strand break (‘1-DSB’, equivalent to two nicks at the same target sequence). The reaction intermediates containing two or more nicks distributed between the two target sequences and the final reaction product—DNA with two double-strand breaks—were quantified as a single fraction ‘≥2-NICKS’ (see Supplementary Data for details).
To determine the number of phosphodiester bonds cleaved during a single binding event of BcnI, we analyzed two-site DNA cleavage reactions under the steady-state conditions (Figure 6). Only a minor fraction (∼11%) of all products released into solution contained a single nick; this finding is in agreement with steady-state experiments on the single site oligoduplex substrate, where G-nick and C-nick intermediates comprise a similar fraction (∼8%, Supplementary Figure S3). The ‘1-DSB’ DNA with a double-strand break in one BcnI site (and the intact second site) comprised ∼32% of all DNA cleavage products, and the ‘≥2-NICKS’ products with two or more nicks distributed between different sites comprised the remaining 57% of cleaved DNA. Similar results were obtained with the two-site substrate GC that carried two BcnI sites in the inverse orientation (data not shown). The abundance of the ‘≥2-NICKS’ products indicates that BcnI indeed is able to cut phosphodiester bonds in neighboring recognition sites during a single binding event. This finding is consistent with the hypothesis that the switch in BcnI orientation involves a diffusional walk of the enzyme on the DNA substrate.
Figure 6.
Steady-state reactions on the two-site DNA. The reactions contained 1 nM BcnI and 100 nM of the two-site hairpin oligoduplex GG (Table 1). Changes in the relative amount of various DNA forms (schematically depicted above the graph) are shown: products with a single nick (‘1-NICK’, down triangles), products with one double-strand break (‘1-DSB’, up triangles) and the sum of products with two or more nicks at both recognition sites (‘≥2-NICKS’, filled squares). All data points are presented as mean values from three independent experiments ±1 SD. Linear regression (solid lines) was used to quantify the formation rates of ‘1-NICK’ (0.0027 ± 0.0005 nM s−1, 11% of all products), ‘1-DSB’ (0.0082 ± 0.0008 nM s−1, 32% of all products) and ‘≥2-NICKS’ (0.014 ± 0.002 nM s−1, 57% of all products).
DISCUSSION
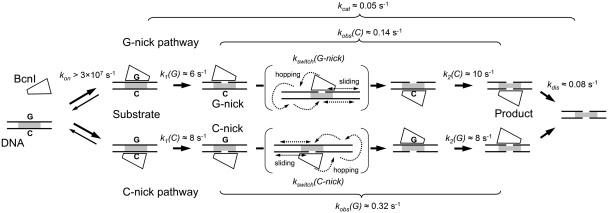
Kinetic analysis of BcnI reactions provided in this report is consistent with the sequential reaction mechanism involving (i) rapid enzyme–DNA association and hydrolysis of the first DNA strand, (ii) slow reorientation to the opposite strand followed by rapid cleavage of the second DNA strand, and (iii) slow product release (Figure 7).
Figure 7.
Mechanism of double-stranded DNA cleavage by BcnI. BcnI rapidly associates with substrate DNA (rate constant kon) placing the catalytic center in the vicinity of either the C- (5′-CCCGG-3′) or the G- (5′-CCGGG-3′) strand. Rapid hydrolysis of the first DNA strand [rate constants k1(G) and k1(C)] converts the substrate into a mixture of G- and C-nick intermediates. The second strand is cleaved at a much lower rate [rate constants kobs(C) and kobs(G)]. The latter process consists of two phases: (i) a relatively slow switch of BcnI on DNA [rate constants kswitch(G-nick) and kswitch(C-nick)] involving a diffusional walk on DNA; and (ii) a rapid hydrolysis reaction [k2(C) and k2(G)]. BcnI preferentially binds intact DNA in the orientation that places the catalytic center close to the G-strand, therefore the major fraction of DNA (70–80%) is cleaved via the G-nick intermediate. The reaction cycle is completed by the product release (rate constant kdis). The rate constants for cleavage of the first and the second DNA strands together with kdis define the turnover number of BcnI kcat. Occasional dissociation of BcnI from the nicked intermediate (rate constant ∼0.02 s−1) is not depicted.
DNA binding and strand preference of BcnI
Mixing of BcnI and DNA solutions results in fast complex formation. Under single turnover conditions (Figure 3C), the BcnI–DNA association rate is faster than the subsequent DNA hydrolysis step (∼6–8 s−1), indicating that conformational rearrangements that may occur during the specific complex formation do not limit the cleavage rate. The estimated lower limit of 3 × 107 M−1 s−1 (6 s−1/200 nM) for the BcnI–DNA association rate constant kon is close to the diffusion-limited rate of 1 × 108 M−1s−1 (27).
BcnI is a monomer in solution, which binds to the degenerate sequence 5′-CCSGG-3′ and cuts both DNA strands after the second C. In the BcnI–DNA complex the enzyme is distributed between two alternative binding orientations, which place the catalytic center in the vicinity of either the G- or the C-strands (7,17). Cleavage of the first DNA strand in these two distinct complexes results in a concomitant formation of the G-nick and the C-nick intermediates (Figure 3). Surprisingly, amounts of the two reaction intermediates are not equal. In the pre-mix setup (Figure 3B), ∼80% of DNA is converted into the G-nick intermediate, and the remaining 20% into the C-nick intermediate. Thus, the equilibrium binding distribution of BcnI shows ∼4-fold preference for the G-strand. Structural or thermodynamic mechanisms of this bias are not clear. However, we have recently demonstrated that this preference can be considerably altered by mutations of BcnI residues H219 and H77 that contact the central C:G base pair: H219Q substitution converts BcnI into the G-strand specific nicking enzyme, while the H77A replacement converts BcnI into the C-strand specific nicking endonuclease (17).
Surprisingly, the wt BcnI strand preference is lost on the pre-nicked oligoduplex substrates. In this case BcnI binds both G-nick and C-nick intermediates almost exclusively in the orientation that places the catalytic center close to the intact strand (C-strand in the G-nick DNA, G-strand in the C-nick DNA, Figure 4B and Supplementary Figure S1B). It is possible that altered DNA conformation and/or the extra negative charge of the 5′-terminal phosphate in the nicked strand interferes with BcnI binding and re-directs enzyme to the intact strand. Similar role of the terminal phosphate has been suggested for the BfiI restriction enzyme (28).
The rearrangement of BcnI and the second strand cleavage
BcnI cuts the second strand of the intact DNA substrate ∼20-fold slower than the first strand (Figure 3B), but in the pre-assembled nicked intermediate the second strand is cut as fast as the first strand (Figure 4B and Supplementary Figure S1B). Thus, slow cleavage of the nicked intermediate formed in situ must be due to a slow BcnI rearrangement that brings the single catalytic center from the cleaved DNA strand to the scissile phosphate in the opposite strand (Figure 7). Based on the trap and steady-state experiments (Figures 2B, 5 and Supplementary Figure S3), we propose that upon nicking of the first DNA strand BcnI relocates to a random DNA sequence on the same DNA molecule by hopping/sliding (27) and then either switches orientation to cut the opposite strand (Figure 7) or occasionally dissociates into bulk solution releasing nicked DNA. The diffusional walk mechanism is directly supported by the steady-state experiments on the two-site substrate, which show that over 50% of products generated by BcnI during a single binding event contain two or more nicks distributed among the two adjacent recognition sites (‘≥2-NICKS’ products, Figure 6).
Many DNA acting enzymes, including Type II REases, DNA methyltransferases and glycosylases (21,24,25,29–33), employ hopping and/or sliding to facilitate location of their target among the large excess of background DNA. This mechanism also contributes to enzyme processivity, e.g. the ability to act on two (or more) target sites during a single binding event (21–25,34,35). Enzyme processivity on a two-site substrate is defined as the number of reactions occurring at both recognition sites relative to the total number of reactions. This ratio may be close to 1 on adjacent DNA sites (25), but it decreases as the sites are brought further apart. Noteworthy, processivity of the orthodox REases on linear DNA substrates is limited to 0.5 even with adjacent sites, as after cleavage of the first site the two DNA fragments diffuse away from each other, and there is only 50% probability that an enzyme will remain associated with the DNA fragment containing the second recognition site (22,23,27). The mechanism of double-stranded DNA cleavage proposed here for BcnI is reminiscent of the processive reactions: to complete DNA cleavage, BcnI must locate the target sequence, cut the first DNA strand, diffuse to the opposite strand of the same target sequence and cut the second DNA strand. After nicking of the first strand, the left-hand and the right-hand DNA fragments are still held together by the uncleaved DNA strand. Not surprisingly, the processivity of BcnI, defined as the number of double-strand breaks per total number of reactions at the recognition site occurring during a single binding event is very high (∼0.8 on ΦX174 DNA and ∼0.9 on oligoduplex substrate, Figure 2B and Supplementary Figure S3). The lower processivity of BcnI on the macromolecular substrate might be due to much longer arms of nonspecific DNA surrounding the target site. Indeed, the diffusional walk of BcnI on the oligoduplex substrate is restricted to just ∼30 bp, but an enzyme may diffuse much further on the ∼5.4-kb ΦX174 DNA, decreasing the probability for reassociation of BcnI with the nicked target sequence.
The contribution of sliding and hopping in the process of BcnI relocation between the two DNA strands remains to be determined. However, the obligatory change in BcnI orientation, required for cleavage of the second DNA strand, implies that the diffusional walk occurring between the two hydrolysis reactions must involve at least one hop of BcnI [physical separation of enzyme from DNA without diffusion into bulk solution (36)] that would enable rotation of the enzyme around the axis perpendicular to the DNA and subsequent re-binding. Switching in enzyme orientation during the processive action on adjacent DNA sites, attributed to hopping, was reported for several DNA acting enzymes, including REase BbvCI and some DNA repair glycosylases (22,24,25).
Product release step
In the steady state reactions, after cutting the second DNA strand at its target site, BcnI must release the final reaction product—DNA cut at both strands—before switching to another DNA molecule. The reaction cycle of many REases is limited by product release (37–40). However, this may not be the case for BcnI, as the turnover number kcat for cleavage of the oligoduplex substrate (0.050 s−1, Supplementary Figure S3) is comparable to the single-turnover rate of the second DNA strand cleavage in the predominant G-nick pathway [kobs(C) ≈ 0.14 s−1, Figure 3C]. The kcat and the rate constants for the individual first-order steps in the G-nick pathway (Figure 7) are related by Equation (1):
| (1) |
Insertion of the k1(G), kcat and kobs(C) values into Equation (1) yields kdis value of 0.08 s−1 for the enzyme release rate from the DNA after the double-strand break. Similar values of kdis (0.08 s−1) and kobs(C) (0.14 s−1) imply that the multiple turnover rate of BcnI is partially limited by two steps: cleavage of the second DNA strand (which in turn is limited by the slow BcnI flip to the second strand) and release of the final product.
Assuming that the second strand of the oligoduplex is cleaved with the observed rate of 0.14–0.30 s−1 [rate constants kobs(C) and kobs(G), Figure 7], and ∼10% of DNA escapes the enzyme as the nicked intermediate (Supplementary Figure S3), the estimate value of the rate constant for the nicked product dissociation must be close to 0.02 s−1 [∼10-fold lower than the cleavage rates kobs(C) and kobs(G)]. This value is ∼4-fold lower than the rate constant kdis for the release of the final product cut at both strands (Figure 7). Therefore, it is tempting to speculate that the newly generated double-strand break facilitates the escape of BcnI into bulk solution.
CONCLUSION
We provide here experimental evidence which support an unusual mechanism of DNA cleavage by the monomeric restriction enzyme BcnI. After cutting phosphodiester bond at one DNA strand at the 5′-CCSGG-3′ site, BcnI undertakes a random walk on DNA to switch its orientation before proceeding to the second strand cleavage. While the key role of 1D diffusion in the target site location has been previously demonstrated for many enzymes acting on DNA, we show here that BcnI cleavage at the single recognition site 5′-CCSGG-3′ includes obligatory enzyme hopping/sliding. A similar mechanism (‘intrasite processivity’) for the double methylation of the 5′-GATC-3′ site was recently inferred for the E. coli Dam methyltransferase (26). It remains to be determined whether the mechanism proposed here for BcnI is valid for other monomeric REases (41,42). Preliminary data for the structurally related REase MvaI (5′-CC/WGG-3′) indicate that cleavage of both DNA strands occurs during a single binding event (G. Sasnauskas and V. Siksnys, unpublished data); a relatively low amount of nicked DNA observed in HinPI reactions (43,44) is also consistent with the above mechanism.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Lithuanian Science Council (grant MIP-81/2010 to G.S.); Lithuanian State Studies Foundation Student Research Fellowship (to G.K.). Funding for an open access charge: FP7 Grant ‘MoBiLi’ (245721).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. S. Klimašauskas for the opportunity to use the quench-flow equipment and Dr Č. Venclovas for helpful discussions. The authors also acknowledge Drs Bochtler and Monika Sokolowska for making the BcnI structure in the alternative binding mode (PDB ID 3IMB) available before publication.
REFERENCES
- 1.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly TJ, Jr, Smith HO. A restriction enzyme from Hemophilus influenzae. II. J. Mol. Biol. 1970;51:393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal AK. Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol. 1995;5:11–19. doi: 10.1016/0959-440x(95)80004-k. [DOI] [PubMed] [Google Scholar]
- 4.Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J. Biol. Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 5.Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Kaus-Drobek M, Czapinska H, Sokolowska M, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically. Nucleic Acids Res. 2007;35:2035–2046. doi: 10.1093/nar/gkm064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokolowska M, Kaus-Drobek M, Czapinska H, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007;369:722–734. doi: 10.1016/j.jmb.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Sokolowska M, Kaus-Drobek M, Czapinska H, Tamulaitis G, Siksnys V, Bochtler M. Restriction endonucleases that resemble a component of the bacterial DNA repair machinery. Cell. Mol. Life Sci. 2007;64:2351–2357. doi: 10.1007/s00018-007-7124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–1720. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soundararajan M, Chang Z, Morgan RD, Heslop P, Connolly BA. DNA binding and recognition by the IIs restriction endonuclease MboII. J. Biol. Chem. 2002;277:887–895. doi: 10.1074/jbc.M109100200. [DOI] [PubMed] [Google Scholar]
- 12.Inomata K, Ohki I, Tochio H, Fujiwara K, Hiroaki H, Shirakawa M. Kinetic and thermodynamic evidence for flipping of a methyl-CpG binding domain on methylated DNA. Biochemistry. 2008;47:3266–3271. doi: 10.1021/bi7019029. [DOI] [PubMed] [Google Scholar]
- 13.Sasnauskas G, Zakrys L, Zaremba M, Cosstick R, Gaynor JW, Halford SE, Siksnys V. A novel mechanism for the scission of double-stranded DNA: BfiI cuts both 3′-5′ and 5′-3′ strands by rotating a single active site. Nucleic Acids Res. 2010;38:2399–2410. doi: 10.1093/nar/gkp1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 15.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 17.Kostiuk G, Sasnauskas G, Tamulaitiene G, Siksnys V. Degenerate sequence recognition by the monomeric restriction enzyme: single mutation converts BcnI into a strand-specific nicking endonuclease. Nucleic Acids Res. 2011;39:3744–3753. doi: 10.1093/nar/gkq1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagunavicius A, Sasnauskas G, Halford SE, Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 19.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Conversion of the tetrameric restriction endonuclease Bse634I into a dimer: oligomeric structure-stability-function correlations. J. Mol. Biol. 2005;348:459–478. doi: 10.1016/j.jmb.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 20.Yoshioka K. KyPlot – a user-oriented tool for statistical data analysis and visualization. CompStat. 2002;17:425–437. [Google Scholar]
- 21.Stanford NP, Szczelkun MD, Marko JF, Halford SE. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl Acad. Sci. USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terry BJ, Jack WE, Modrich P. Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J. Biol. Chem. 1985;260:13130–13137. [PubMed] [Google Scholar]
- 24.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl Acad. Sci. USA. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedglin M, O’Brien PJ. Hopping enables a DNA repair glycosylase to search both strands and bypass a bound protein. ACS Chem. Biol. 2010;5:427–436. doi: 10.1021/cb1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coffin SR, Reich NO. Escherichia coli DNA adenine methyltransferase: intrasite processivity and substrate-induced dimerization and activation. Biochemistry. 2009;48:7399–7410. doi: 10.1021/bi9008006. [DOI] [PubMed] [Google Scholar]
- 27.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasnauskas G, Halford SE, Siksnys V. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl Acad. Sci. USA. 2003;100:6410–6415. doi: 10.1073/pnas.1131003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 30.Jack WE, Terry BJ, Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc. Natl Acad. Sci. USA. 1982;79:4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardone G, George J, Chirikjian JG. Differences in the kinetic properties of BamHI endonuclease and methylase with linear DNA substrates. J. Biol. Chem. 1986;261:12128–12133. [PubMed] [Google Scholar]
- 32.Bonnet I, Biebricher A, Porte PL, Loverdo C, Benichou O, Voituriez R, Escude C, Wende W, Pingoud A, Desbiolles P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehbrecht HJ, Pingoud A, Urbanke C, Maass G, Gualerzi C. Linear diffusion of restriction endonucleases on DNA. J. Biol. Chem. 1985;260:6160–6166. [PubMed] [Google Scholar]
- 34.Gowers DM, Halford SE. Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling. EMBO J. 2003;22:1410–1418. doi: 10.1093/emboj/cdg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berkhout B, van Wamel J. Accurate scanning of the BssHII endonuclease in search for its DNA cleavage site. J. Biol. Chem. 1996;271:1837–1840. doi: 10.1074/jbc.271.4.1837. [DOI] [PubMed] [Google Scholar]
- 36.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 37.Sasnauskas G, Jeltsch A, Pingoud A, Siksnys V. Plasmid DNA cleavage by MunI restriction enzyme: single-turnover and steady-state kinetic analysis. Biochemistry. 1999;38:4028–4036. doi: 10.1021/bi982456n. [DOI] [PubMed] [Google Scholar]
- 38.Sapranauskas R, Sasnauskas G, Lagunavicius A, Vilkaitis G, Lubys A, Siksnys V. Novel subtype of type IIs restriction enzymes. BfiI endonuclease exhibits similarities to the EDTA-resistant nuclease Nuc of Salmonella typhimurium. J. Biol. Chem. 2000;275:30878–30885. doi: 10.1074/jbc.M003350200. [DOI] [PubMed] [Google Scholar]
- 39.Nobbs TJ, Szczelkun MD, Wentzell LM, Halford SE. DNA excision by the Sfi I restriction endonuclease. J. Mol. Biol. 1998;281:419–432. doi: 10.1006/jmbi.1998.1966. [DOI] [PubMed] [Google Scholar]
- 40.Erskine SG, Baldwin GS, Halford SE. Rapid-reaction analysis of plasmid DNA cleavage by the EcoRV restriction endonuclease. Biochemistry. 1997;36:7567–7576. doi: 10.1021/bi970155s. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Horton JR, Maunus R, Wilson GG, Roberts RJ, Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–1901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasko T, Der A, Klement E, Slaska-Kiss K, Posfai E, Medzihradszky KF, Marshak DR, Roberts RJ, Kiss A. BspRI restriction endonuclease: cloning, expression in Escherichia coli and sequential cleavage mechanism. Nucleic Acids Res. 2010;38:7155–7166. doi: 10.1093/nar/gkq567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.