Abstract
The engineering of synthetic gene networks has mostly relied on the assembly of few characterized regulatory elements using rational design principles. It is of outmost importance to analyze the scalability and limits of such a design workflow. To analyze the design capabilities of libraries of regulatory elements, we have developed the first automated design approach that combines such elements to search the genotype space associated to a given phenotypic behavior. Herein, we calculated the designability of dynamical functions obtained from circuits assembled with a given genetic library. By designing circuits working as amplitude filters, pulse counters and oscillators, we could infer new mechanisms for such behaviors. We also highlighted the hierarchical design and the optimization of the interface between devices. We dissected the functional diversity of a constrained library and we found that even such libraries can provide a rich variety of behaviors. We also found that intrinsic noise slightly reduces the designability of digital circuits, but it increases the designability of oscillators. Finally, we analyzed the robust design as a strategy to counteract the evolvability and noise in gene expression of the engineered circuits within a cellular background, obtaining mechanisms for robustness through non-linear negative feedback loops.
INTRODUCTION
Over the past decade, we have witnessed the expansion of synthetic biology (1), where the attempts for cell reprogramming to perform new tasks have fructified in the engineering of several synthetic regulatory circuits (2–20). Usually, the design of synthetic circuits has been inspired on the use of mathematical models (21,22) and empirical engineering rules inferred from natural examples (23,24), although requiring in many cases a genetic fine-tuning to achieve the desired behavior (25). It is expected that the widespread use of libraries of previously well-characterized genetic regulatory elements (26–29), together with the ability of engineering combinatorially those elements (30), will allow avoiding trial-and-error procedures, which are not efficient for optimizing and implementing complex systems. Those designed circuits may be later fine-tuned with directed evolution techniques, although there is no general methodology for the de novo network engineering. In fact, this bottom-up approach is commonly used in other areas of engineering where a set of off-the-shelf parts with precise specifications of their operating points can be used to engineer sophisticated systems, and has been already successful to engineer novel biological circuits (12,19).
Large efforts in generating not only genetic diversity, especially libraries of promoters (19,31–36), but also post-transcriptional regulatory elements (6,14,37–39) and synthetic transcription factors (40,41), encourage to use a combinatorial approach to design artificial circuits (see Supplementary Data for further details). In addition, the quantitative characterization of these regulatory elements allows inferring simple phenomenological mathematical models, which could be used to construct the model of a system that assembles different elements. In that way, several synthetic biology-oriented design tools have been developed to make available a library of mathematical models created from that genetic diversity, together with an interface to create gene circuits by wiring elements (42–47). Notably, such a genetic diversity is translated into a functional diversity when assembling circuits, and these circuits could be readily compiled into nucleic acid sequences. However, the design is reduced to examine one-by-one all possible combinations (e.g. simulating the dynamical behavior), resulting in a tedious design process. Thereby, the evolutionary algorithms and optimization techniques (48–52) allow us to automate this process to find the desired circuits and finally depict the functional diversity of a library of regulatory elements. Our novel approach allows assembling models of regulatory elements from a library and couples this with an automated design strategy.
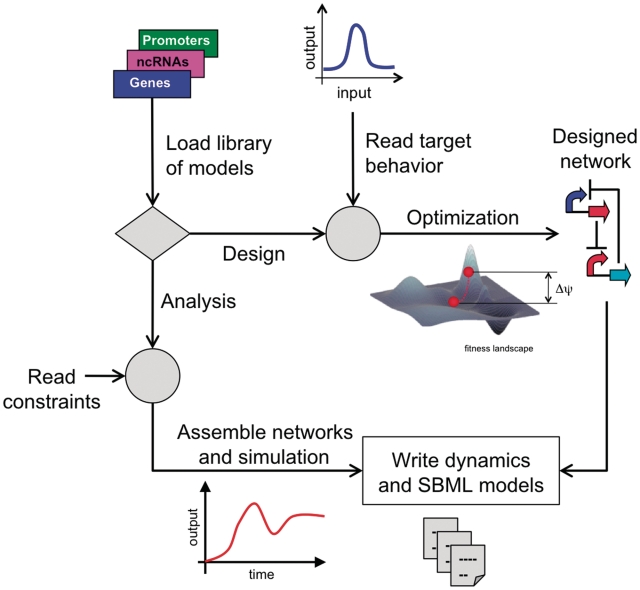
In this work, we tackle for the first time fundamental questions that naturally emerge from that approach. What functional circuits can we engineer with a given library of regulatory elements? What is the diversity of possible behaviors and what is the designability (defined as the fraction of assembled circuits that follow a given behavior) of each one? Is one behavior easier to design than others? Certainly, these features depend on the employed library. We also wonder what is the sensitivity of the results to the regulatory elements; in other words, how many functional circuits involve a given regulatory element? In addition, we look at the robustness of a circuit by locally perturbing its parameters and evaluating the resulting fitness. At fixed network topology, we further analyze the whole parameter space that provides the targeted functionality, which accounts for the robustness of all operative points and asymptotically tends to a value that we call asymptotic robustness. Indeed, this property accounts for the ability to design such a circuit given the limitation of the number of genetic elements, and it could be important to analyze the natural occurrence of certain genetic architectures. All in all, to solve these questions, we developed a computational framework to assemble, simulate and design circuits, and that allowed us to explore the functional diversity that came from the assembled circuits with certain behavior (Figure 1). The design of circuits was accomplished by a selection step according to a dynamical behavior-based fitness function that can also account for robustness. Initially, we applied the methodology to design several functional circuits with unlimited genetic diversity (given by the parameter space) and study their asymptotic robustness. Then, we designed complex circuits by plugging functional modules. Subsequently, we dissected the whole dynamical spectrum of a limited library of regulatory elements and analyzed the properties of the resulting circuits. We also analyzed the dependence of these results on the constituent library and how they could change when the stochasticity of the cell is taken into account. Later, we showed the application of our methodology to design a robust circuit. Finally, we discussed the reliability and implementability of the designed circuits.
Figure 1.
Scheme of the design platform adopted by harnessing a library of models of composable regulatory elements. We explore the functional networks that can be engineered either by exhaustive combinatorial assembly or by heuristic optimization.
MATERIALS AND METHODS
Mathematical model for gene regulatory circuits
For modeling genetic circuits, we used a coupled system of differential equations. We considered three different types of species: mRNA (it can also be non-coding), proteins (mainly transcription factors) and small molecules that interact with proteins to activate or inhibit their regulatory ability (e.g. isopropyl β-d-1-thiogalactopyranoside—IPTG—inhibits the activity of LacI). Likewise, the production of the ith mRNA (xi) from a regulated promoter follows
| (1) |
where the term f(yj,uj) is the transcription rate (yj and uj represent the concentrations of the j-th protein and its regulating chemical, respectively), C the gene copy number, δ the mRNA degradation coefficient and µ the growth rate of the cell (dilution term). C = 1 is assumed to be constant in this work. For the computational design of a circuit, we did not impose variability on µ but we assumed a constant value (e.g. µ = 0.02 min−1). For simplicity, we assumed that all genes in an operon (i.e. controlled by the same promoter) have the same mRNA expression. The term f(yj,uj) accounts for protein–DNA and protein–molecule interactions (22), and for constitutive promoters it is constant (see Supplementary Data for further details). Importantly, our approach is independent of the choice of this function, thus giving a big degree of freedom to the kinetic characterization from experimental data. Afterwards, the production of i-th protein (yi) is given by
| (2) |
where the term g(xi,xj) is the translation rate and β the protein degradation coefficient. The term g(xi,xj) accounts for post-transcriptional regulatory mechanisms, such as riboswitches, allowing a further genetic element, such as a trans-RNA, to control translation (6). In case of no post-transcriptional elements, the translation rate is proportional to the mRNA concentration. In addition, in this work we only considered first-order degradation kinetics. The description of the construction of the stochastic model is detailed in the Supplementary Data.
Library of models of regulatory elements
To construct a library, each genetic regulatory element was modeled by transfer functions that related the output to the input values. These functions can be fitted from experimental data. As DNA fragments, mathematical models can be assembled in a standard way to simulate the behavior of circuits. Here, we only allowed joining promoter and genes, or genes and genes (i.e. two consecutive promoters was not allowed; such a construction should be specified as a whole part). One useful format to store a mathematical model (molecular species, kinetic parameters and DNA sequence) is systems biology markup language (SBML) (53). Hence, we had a single SBML file for the model of each biological part; similarly as crystallographic data is stored in protein data bank (PDB) format. The models for promoter parts only account for the transcription rate, whereas for gene parts the model accounts for the translation rate and the degradation and dilution rates of mRNA and proteins. We selected Hill-function models because their overwhelming use in current characterization of transcription regulation works. In the future, when more advanced models may be used to fit characterization data, they could be readily used with our computational design procedure. A range of variation can be specified for some kinetic parameters; likewise, the corresponding value will be susceptible to be changed during the design process.
Exhaustive versus heuristic design
Multiple circuits can be constructed by harnessing the available regulatory elements. To computationally explore the functional diversity that offers such a library and the designability of certain behaviors, two different strategies can be adopted, and our approach provides an automated implementation of them. On the one hand, following the exhaustive design strategy, all possible circuits, up to a maximal number of elements, are constructed and simulated. Having the large collection of dynamics, a post-processing step is applied to find those circuits that behave according to the design specifications. This approach allows obtaining the whole functional diversity and designability. On the other hand, a heuristic design strategy provides a probabilistic sampling frame of the functional diversity. It allows iteratively assembling models of existing elements and evaluating the performance of the resulting circuit according to a dynamical behavior-based fitness function (54). For that, we used Monte Carlo Simulated Annealing (MCSA) as optimization scheme (55). A movement in the fitness landscape consists in a replacement, addition or deletion of a given regulatory element. To evaluate the fitness function, we first calculated the average distance (metric function) for all genes i considered as outputs, for a given target behavior k, between the current circuit dynamics (yik) and the target one (zik) according to
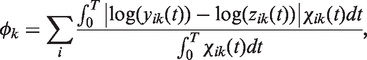 |
(3) |
where T is the final time (e.g. the time to reach the steady state). The metric is in logarithmic scale to properly balance the species concentrations, since they can vary in several orders of magnitude in biological systems. The function χ(t) is a weighting factor to only evaluate the circuit dynamics in a specified temporal domain (χ(t): [0,T] → [0,1]). Subsequently, the fitness function that aggregates all targets we used reads
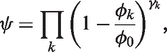 |
(4) |
where ϕ0 is a normalization constant to adjust the fitness value to the metric function (e.g. ϕ0 = 3), and γk gives the scalar weight in logarithmic scale for optimizing target k (e.g. γk = 10γl indicates that target k has 10 times more priority than l). If ϕk>ϕ0 for one k then we assumed ψ = 0. Importantly, this fitness function (ψ belongs to the interval [0,1]) penalizes those circuits that do not satisfy simultaneously all targets. Being Δψ the fitness update after a movement, this is accepted with probability max{1, exp(Δψ/TMCSA)}, where TMCSA is the MCSA temperature. TMCSA is continuously adjusted during the optimization process following an exponential cooling scheme.
RESULTS AND DISCUSSION
Circuit design and asymptotic robustness
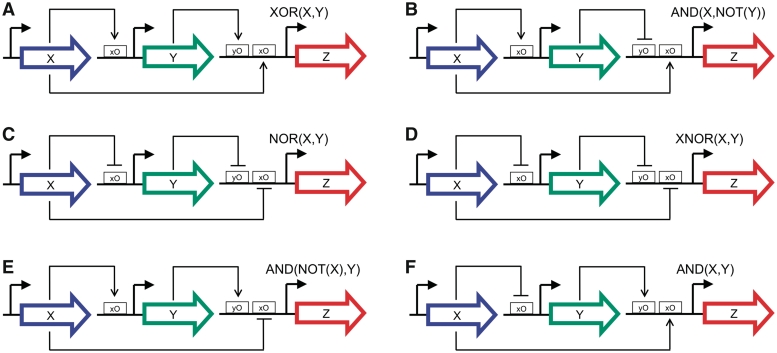
Initially, we constructed a library of artificial regulatory elements, including all types of logic combinatorial promoters of two entries. Additionally, the kinetic parameters characterizing those elements were specified as a range of variation. This feature allows that the genetic sequence of many biological parts could be easily modified to create a new part with diminished binding affinity or stability by a single mutation. Otherwise, it is much more difficult to find a suitable mutation that would increase the binding or stability. Therefore, by allowing this range in the parameter space, we would enlarge the search space while still maintaining the linking with the genotype, because the parts from an optimal solution could be readily engineered to follow a model agreeing with the designed parameters. The nominal values were taken from several experimental studies (11,12,27,36). Thus, the genetic diversity was almost unlimited, being the design space defined by topological and parameter modifications of the circuit. To explore this space we applied the heuristic design strategy to find the optimal assemblies of elements and parameterizations that gave functional circuits. First, we repeatedly applied the optimization method to design all possible circuits relying on a feed-forward loop (FFL) structure for one-stripe pattern formation (56–59). We found six different architectures for working as an amplitude filter (Figure 2), where five of them corresponded to incoherent FFLs (all except 2A), and in architectures 2B and 2E repression dominated over activation. Moreover, in Supplementary Figure S1, we show the circuits corresponding to inverse amplitude filters. Certainly, the two regulatory branches with opposite sign are responsible for such a behavior, and the combinatorial promoter of the downstream promoter is central to get a variety of functionally analogous circuits. Interestingly, some of those architectures have been found involved in developmental processes (60,61). In Supplementary Figure S2, we illustrate a possible implementation of a FFL-based circuit responding to intermediate concentrations of IPTG, together with its characteristic transfer function. Notably, we did not exhaustively construct all possible FFL circuits from the library for their scoring. Instead, we probabilistically sampled the fitness landscape and we always found a solution corresponding to one of the six FFL structures presented. Moreover, this approach can be applied to design functional circuits without accounting for the designability of the desired behavior, and then study the intrinsic properties of the circuit irrespective to the library, such as its asymptotic robustness.
Figure 2.
Schemes of several FFL-based gene circuits optimized to operate as amplitude filters (also called band detectors). The mathematical model for each circuit is provided in SBML format in the Supplementary File sbml.zip.
We then investigated the asymptotic robustness of those FFL circuits, which functioned as amplitude filters with a fold-change (F) of at least one order of magnitude at the detection point ( ). By constraining the sign of the regulations (fixed topology), we obtained a parameter space of ∼2×105 different combinations for each topology shown in Figure 2. Accordingly, the highest asymptotic robustness was reached by the architecture 2E with the 21.29%, followed by the architectures 2A with the 17.17% and 2F with the 17.66%, indicating that those circuits have a one-stripe pattern-prone structure. Interestingly, this could be because the input gene (X) has a non-monochromatic regulatory mode (i.e. both activator and repressor) in these topologies. On the contrary, the architecture 2C was highly sensitive to parameter variations with asymptotic robustness of only the 2.54%. The architectures 2B and 2D with the 8.60% and 7.69%, respectively, were in between. However, despite of its low asymptotic robustness, the structural core 2C is broadly found in many natural systems. For instance, in the Drosophila patterning circuitry, gene hb represses both genes kni and Kr and kni also represses Kr (60). In addition, the core 2B is the FFL motif most abundant within the regulatory map of bacteria and yeast (62). That the genes involved in the structures 2B and 2C have a monochromatic regulatory mode could explain the increasing presence of these circuits. Moreover, from a synthetic perspective, promoters type NOR and IMPLIES could be engineered by placing contiguously the corresponding operators in the promoter region (7,12,36).
). By constraining the sign of the regulations (fixed topology), we obtained a parameter space of ∼2×105 different combinations for each topology shown in Figure 2. Accordingly, the highest asymptotic robustness was reached by the architecture 2E with the 21.29%, followed by the architectures 2A with the 17.17% and 2F with the 17.66%, indicating that those circuits have a one-stripe pattern-prone structure. Interestingly, this could be because the input gene (X) has a non-monochromatic regulatory mode (i.e. both activator and repressor) in these topologies. On the contrary, the architecture 2C was highly sensitive to parameter variations with asymptotic robustness of only the 2.54%. The architectures 2B and 2D with the 8.60% and 7.69%, respectively, were in between. However, despite of its low asymptotic robustness, the structural core 2C is broadly found in many natural systems. For instance, in the Drosophila patterning circuitry, gene hb represses both genes kni and Kr and kni also represses Kr (60). In addition, the core 2B is the FFL motif most abundant within the regulatory map of bacteria and yeast (62). That the genes involved in the structures 2B and 2C have a monochromatic regulatory mode could explain the increasing presence of these circuits. Moreover, from a synthetic perspective, promoters type NOR and IMPLIES could be engineered by placing contiguously the corresponding operators in the promoter region (7,12,36).
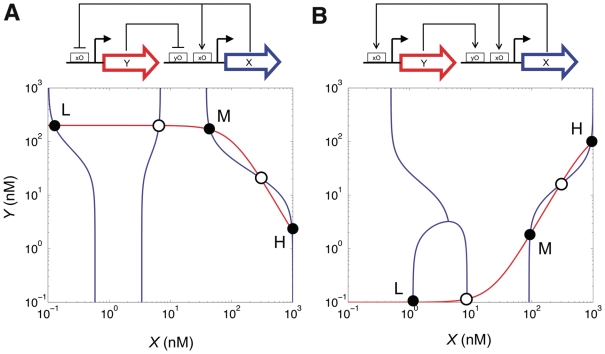
Subsequently, we used the optimization method to design a circuit able to count. This is an interesting example that could already unveil many of the issues we meet in more complex networks. Cells may take advantage of this sort of circuits to regulate fundamental processes, such us telomere length control (63), where a machinery to count molecules or events is required. Counters have different stable states and rely on memory-like architectures that allow retaining the initial state, unless a perturbation switches the system (3,14,18,64). Generally, the underlying mechanism of biological counters consists in overcome certain threshold after a specific number of consecutive pulse-like events. Herein, we attempted the design of a two-pulse counter, where we imposed that the system had to reach three different states. We applied the optimization method to design all possible two-gene circuits. We found that, within a delimited time domain, all possible circuits were functional and reached three states. However, those circuits based on an activator–repressor core had a meta-stable state, which falls into the basin of attraction of one of the two stable states after certain time (Supplementary Figure S3). In Figure 3, we show the phase diagrams for the circuits based on a monochromatic regulatory core and one self-activation (see in Supplementary Figure S4 the corresponding phase diagrams for the circuits based on an activator-repressor core). The addition of another self-activation on the buffer gene allows having a symmetric multi-stable device (64). We then computed the asymptotic robustness of these two circuits, by exploring exhaustively all possible parameterizations (for that we discretized the parameter space into ∼2 × 105 different combinations). The double repression core allowed tristability in the 0.2422% of the cases, whereas the double activation core in the 0.1787% (relatively low in both cases).
Figure 3.
Schemes of two two-gene circuits designed to reach tristability, showing the corresponding phase diagrams. Filled and open circles represent stable and unstable states respectively. The mathematical model for each circuit is provided in SBML format in the Supplementary File sbml.zip.
Next, we attempted the design of a two-pulse counter relying on just two states. As design specifications, we imposed pulses of 10 min within an interval of 50 min with amplitude of 100-fold. The designed circuit with a plausible implementation counting pulses of IPTG is shown in Supplementary Figure S5. However, the functioning of this system (i.e. number of pulses it is able to count) depends on the pulse length and interval. In addition, we attempted the automated design of a tunable genetic timer. These devices consist of memories that change the state of operation according to an external signal and the time to accomplish this transition (time to reach the steady state) can be modulated by another signal (19). The designed circuit with a plausible implementation is shown in Supplementary Figure S6, together with its characteristic transfer function. This circuit consisted in a coherent FFL coupled to a memory-like mechanism based on a self-activation, and it existed a threshold in the IPTG concentration from which the circuit responded to different levels of it.
Hierarchical design and modularity
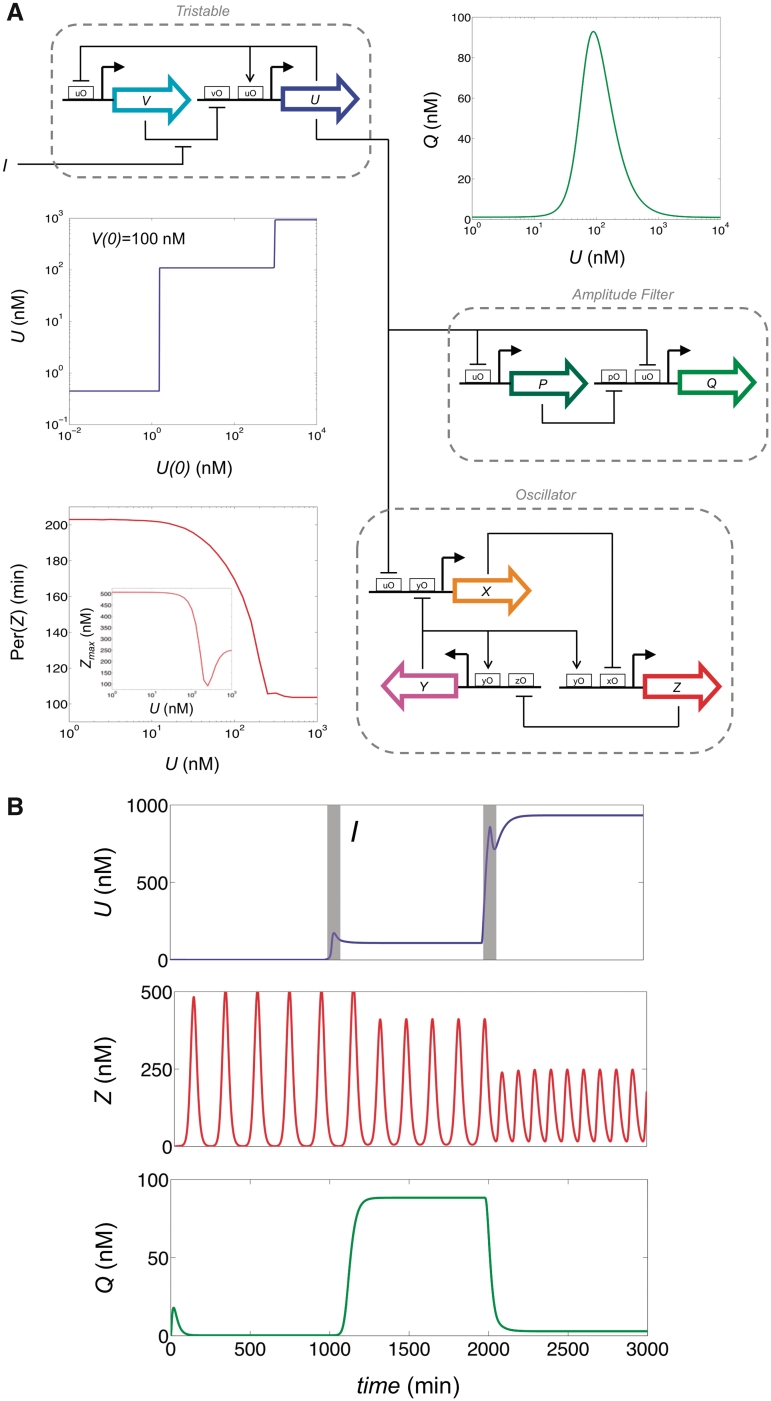
Once a functional genetic device is obtained, either from computational or rational design methods, and experimentally validated, it can be integrated in the library as a new element to be used in the construction of more complex systems. As a first approach, we included in our library of regulatory elements a circuit previously optimized to operate as a tristable. Remarkably, the incorporation into the design procedure of black-box modules enhances the optimization of the impedance matching, where the output of a device serves directly as the input of a downstream one, and could considerably enlarge the functional diversity of the library. Hence, following such a design approach, we were able to obtain complex functions with modular systems. In our particular case, we designed a system coupling a tristable, an amplitude filter, and a frequency-tunable oscillator (Figure 4). Initially, this tristable gave a low concentration. After a pulse of 20 min with amplitude of 1000-fold in the inducer, the device switched its state to reach the intermediate concentration level, and subsequently the oscillator changed its frequency and the amplitude filter, which operates as a detector of the intermediate state, reached its ON state. After a second pulse, the device switched to its high concentration point, inducing a new change in the frequency of the oscillator and giving the detector back to its OFF state. Interestingly, the frequency-tunable oscillator evolved to couple two different regulatory mechanisms, and the external signal switched from one mechanism to another, then changing the frequency of the oscillations. In addition, with the consideration of delayed reactions (e.g. due to translation and multimerization) we could obtain complex oscillations, which can drive to a route toward chaos (65). In fact, this mechanism has been previously applied to design genetic oscillators with a minimal number of elements (16). In Supplementary Figure S7, we show a genetic clock designed by optimization coupled to a tristable, which modulates its frequency and the shape of its oscillations according to the states of the switch-like circuit.
Figure 4.
(A) Scheme of a complex regulatory system comprising a frequency-tunable oscillator and a state detector, designed by using the tristable device as an element of the library. Moreover, we show the transfer functions of the different devices that form the system. (B) Dynamics of the output genes of the complex system. Pulses in the input (I) of 20 min and 1000-fold of amplitude were applied at t = 1000 and 2000 min.
However, one important issue in such an approach is the possibility of the loss of function of a device when plugging it to a downstream module. This effect, usually called retroactivity (66), emerges when a transcription factor plays two different roles in both modules, and is indeed a consequence of the limited protein amount. This result may have significant consequences on the dynamics of the system, even when the stochasticity of the cell is taken into account (67). Here, our modeling neglects this effect by assuming that the concentration of free protein is always much higher than the protein bound to DNA (22); also as an imposition to ensure modularity in the design and to be able to combine different elements from the library. Although for many systems this approach is valid (9), it could be found some examples where such a model is not too accurate. To solve this problem in practice, one strategy would be to impose as a design constraint that the output gene had no regulatory effects on the circuit. Likewise, this output could be used as the input in further downstream modules with increased guarantees of a proper functioning. Thereby, in our system of Figure 4, gene U could be split into two genes, one for working within the tristable device and another for setting the amplitude filter and the oscillator, although still it would exist a coupling between these two devices due to a common input.
Functional diversity and designability
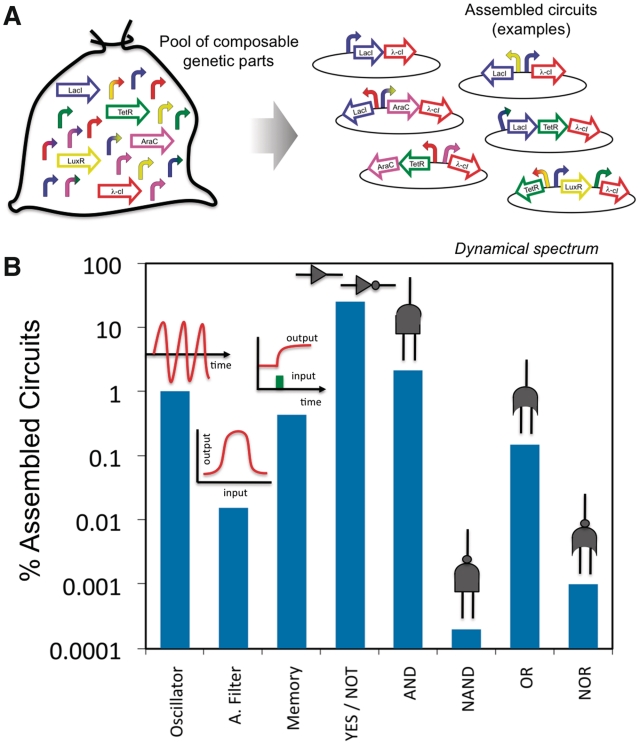
We further studied the designability of a given dynamical behavior. For that, we constructed a library of SBML models of well-characterized regulatory elements previously implemented in vivo. Likewise, the corresponding kinetic parameters were fitted from experimental data and kept fixed. Using this library, we constructed by in silico assembly all possible architectures up to three genes, giving 501 952 different circuits (see the different configurations in Figure 5A). We systematically imposed λ-cI as output gene in all circuits. Thereby, we computed the dynamics of all circuits to perform an analysis of the behaviors that could be obtained with such a library. For this work, we considered a library of 36 elements, involving 5 genes and 31 synthetic promoters. As genes, we contemplated the classical repressors LacI, TetR and λ-cI, and the activators AraC and LuxR. Moreover, we built a library with three constitutive promoters with different transcription rates, 16 single promoters involving 4 lacO, 4 tetO, 2 araO, 2 luxO (36), 2 λRO (11) and 2 λRMO (5) and 12 combinatorial promoters involving 4 lacO–tetO, 3 araO–lacO, 2 araO–tetO (36), 1 λRMO–lacO (12), 1 luxO–λRO (7) and 1 luxO–lacO (68). The models also accounted for the external molecules (IPTG, anhydrotetracycline –aTc–, L(+)-arabinose and acyl homoserine lactone—AHL—) that modify the regulatory ability of the transcription factors and represent the inputs of the circuits. These SBML models are provided as Supplementary Data and they contain the corresponding kinetic parameters. Then, for each external inducer we considered three different states (low, intermediate and high), giving 81 environmental conditions for all combinations, and four more conditions in which the inducers had a pulse-like dynamics.
Figure 5.
(A) Graphical representation of the exhaustive design strategy. Starting from a library of composable genetic regulatory elements (mathematical models provided in SBML format in the Supplementary File sbml.zip), we constructed all possible circuits up to three genes for simulation. (B) Dynamical spectrum of the library by exhaustive exploration (functional diversity). We represent the percentage of circuits that behave as oscillators, amplitude filters, memories and logic gates (designability). To differentiate between two states of a circuit, we imposed at least one order of magnitude in concentration.
By compiling all numerical results (details in Supplementary Data), we were able to dissect the dynamical spectrum of the library (i.e. its functional diversity), which included circuits operating as oscillators, amplitude filters, memories and different logic gates (Figure 5B). As expected, the majority of the circuits functioned as logic gates, and because the external signals (IPTG, aTc, arabinose and AHL) always activated transcription, the set of NAND and NOR gates was highly reduced. In addition, ∼1% of the assembled circuits was able to exhibit oscillations. Furthermore, we found amplitude filters in the 0.016% of the cases, and memories in the 0.436%. Certainly, this spectrum depends on the value of F specified to differentiate between two concentration levels (F gives their ratio). Herein, we imposed 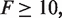 although we also performed a screening to see the effect of different values of F. Not surprisingly, as higher is F, the number of functional circuits decreases (Supplementary Table S1). In addition, we studied the effect of the initial condition on the output gene finding that the results were almost independent of this. Certainly, the initial condition only affects in memory-like circuits, but this effect was captured by imposing pulse-like dynamics on the input. Interestingly, the repertoire of designed circuits was essentially based on minimal cores that provided the required functional mechanism (Supplementary Figure S8). These cores illustrate the design principles in which the dynamical spectrum is based on. However, the use of a limited library and a partial set of input conditions, while allowing an exhaustive exploration, prevent obtaining a comprehensive analysis of the design principles. For instance, as we have shown above, a double activation core gives a memory-like mechanism but it was not found in the repertoire of circuits. In addition, all amplitude filters were based on the FFL architecture 2C, although further circuits, not necessarily FFLs, can be employed to read morphogen gradients (59). We did not obtain further topologies because the monochromatic regulatory mode and the lack of cooperation between transcription factors.
although we also performed a screening to see the effect of different values of F. Not surprisingly, as higher is F, the number of functional circuits decreases (Supplementary Table S1). In addition, we studied the effect of the initial condition on the output gene finding that the results were almost independent of this. Certainly, the initial condition only affects in memory-like circuits, but this effect was captured by imposing pulse-like dynamics on the input. Interestingly, the repertoire of designed circuits was essentially based on minimal cores that provided the required functional mechanism (Supplementary Figure S8). These cores illustrate the design principles in which the dynamical spectrum is based on. However, the use of a limited library and a partial set of input conditions, while allowing an exhaustive exploration, prevent obtaining a comprehensive analysis of the design principles. For instance, as we have shown above, a double activation core gives a memory-like mechanism but it was not found in the repertoire of circuits. In addition, all amplitude filters were based on the FFL architecture 2C, although further circuits, not necessarily FFLs, can be employed to read morphogen gradients (59). We did not obtain further topologies because the monochromatic regulatory mode and the lack of cooperation between transcription factors.
Scalability and sensitivity of designability
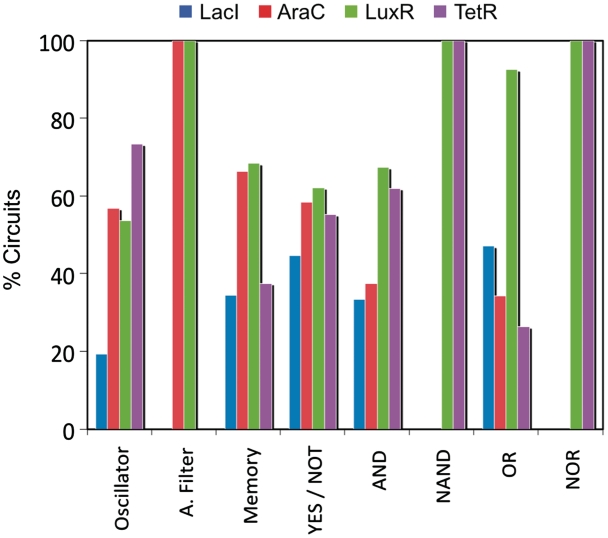
Furthermore, we investigated the dependence of the designability of a function on the existing elements of the library by calculating the degree of sensitivity of each regulator over the resulting dynamical spectrum (Figure 6). Accordingly, LacI appeared to be the most important regulator, indeed for this particular case of study, since it participated in the majority of the functional circuits. In the specific case of the amplitude filters, since their mechanism relied on two different repressions (Supplementary Figure S8), LacI and TetR participated in all circuits. Certainly, the addition of more regulatory elements in the library would enlarge the designability of the different behaviors, and the identification of the regulatory cores in Supplementary Figure S8 would lead to rationally decide on the elements of more interest. In addition, we studied whether the designability could be estimated by sampling a small subset of assembled circuits instead of an exhaustive exploration. This would provide further support to the heuristic exploration by means of optimization methods. Interestingly, we found similar results for the dynamical spectrum of the library when analyzing the dynamics of about 1000 circuits (corresponding to the 0.2% of the total circuits) (Supplementary Figure S9). This suggests that even a small fraction of assembled circuits is representative of the whole population of circuits. By exploiting this fact, we could analyze the functional diversity and designability of several libraries of models at a minimal computational cost or we could study how to enrich the library with new regulatory elements.
Figure 6.
Sensitivity analysis of the dynamical spectrum. We release one regulatory element of the library (in particular, one gene) to analyze its contribution to the dynamical spectrum (we represent the remaining number of functional circuits relative to the total).
We further studied the designability of the different behaviors when considering the stochasticity inherent to the cellular processes (we focused on intrinsic noise, details in Supplementary Data) (69). Since the stochastic simulation entails a higher computational cost, we considered a subset of circuits as before to perform this study, and because it is expected this will not strongly affect the results. For each condition of inputs, we considered the average value and standard deviation of the output (computed using the time dynamics after a transient period). In general, we found similar results as in the deterministic regime (Supplementary Figure S10). We could explain this by the fact that in most cases gene expression is high enough, which minimizes the effect of intrinsic noise, although in some cases a particular circuit topology could also help in such a noise reduction (59). However, we found an increase of almost a doubling in the number of oscillators. By examining the circuits, we realized that circuits based on an activation–repression mechanism with fast damped oscillations in the deterministic regime and that were identified as stable circuits were then selected as noise-induced oscillators. For the other behaviors, the designability results in the stochastic regime were slightly lower. The maximal reduction in designability was of ∼20% in the case of YES/NOT gates. In the circuits that were selected according to the deterministic solution but not to the stochastic one, there is an increase of noise in protein expression that prevents identifying different states of operation.
Afterwards, we wondered whether a unique circuit could exhibit different behaviors. Interestingly, we observed special circuits that displayed multifunctionality according to different input conditions (e.g. oscillators working as amplitude filters, memories or logic gates). Supplementary Table S2 shows the number of circuits with two functions and the corresponding statistical significance. For instance, the 0.3% of the total set of circuits functioning as oscillators and memories held the two functions by properly setting the environmental factors (statistical significance assessed by bootstrapping). In addition, we calculated the number of circuits with multifunctionality (Supplementary Table S3) showing a tendency log-normal in the distribution (slope=−1.463, R2 = 0.982, P < 0.1). This sort of circuits is appealing for cellular regulation and organization because the circuitry rewiring instrumental to change the function is accomplished by an on-the-fly reprogramming sentence (70). As well as a single gene can attain several functions [e.g. a protein with different enzymatic properties (71)], a multifunctional circuit can be exploited by the cell to exert a conditional control of different responses.
Robust design for a cell environment
One of the main problems in synthetic biology is the reliability of the engineered circuits when they are deployed in a continuous cellular background. Certainly, if there is not a selective pressure to maintain a circuit and it results in a cost for the cell, the genetic parts that form it will accumulate mutations (involving point mutations, insertions and deletions) and a quantity of them will entail the loss of function of the circuit (27,72). Thereby, we implemented a strategy based on robust design to counteract this undesirable effect by which the resulting circuit has the ability of maintaining the same behavior under perturbations in the kinetic parameters of the model (i.e. a sort of mutational robustness) (54)]. This also deals with the eventual parameter uncertainty of the inferred models. To this end, we expanded the metric function to account for robustness, scalarizing the corresponding multi-objective optimization problem, given by
| (5) |
where λ is the degree of robustness specified (i.e. for  no robustness is imposed), and
no robustness is imposed), and 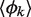 is the average of the metric function over a large number of perturbations. As a case of study, and since the promoter region is usually a preferential site to accumulate mutations (72), we just focused on perturbations in the binding affinities that randomly change the parameter values up to 2-fold (changes in all parameters simultaneously). In addition, to accelerate the convergence future work could just evaluate that function in a more reduced neighborhood with a first-order approximation (73).
is the average of the metric function over a large number of perturbations. As a case of study, and since the promoter region is usually a preferential site to accumulate mutations (72), we just focused on perturbations in the binding affinities that randomly change the parameter values up to 2-fold (changes in all parameters simultaneously). In addition, to accelerate the convergence future work could just evaluate that function in a more reduced neighborhood with a first-order approximation (73).
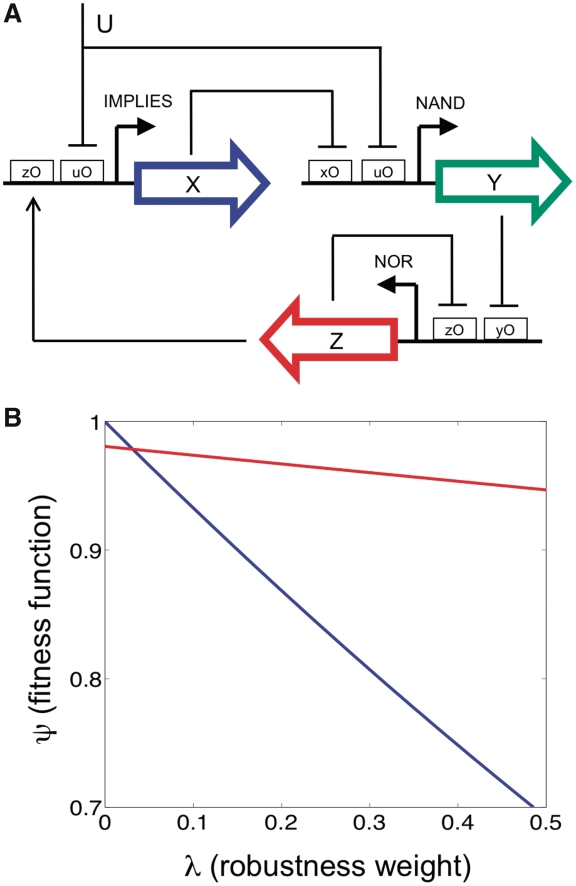
Accordingly, we attempted the design of a robust amplitude filter satisfying the (input, output) specifications of 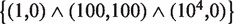 (Figure 7A). The architecture of the resulting circuit involves a FFL and for its implementation we would need a promoter type NAND, which could be engineered by using artificial operator sites that would be recognized by heterodimers from chimeric proteins (74). We then used this circuit as starting point (keeping fixed the topology) to further apply the optimization procedure with
(Figure 7A). The architecture of the resulting circuit involves a FFL and for its implementation we would need a promoter type NAND, which could be engineered by using artificial operator sites that would be recognized by heterodimers from chimeric proteins (74). We then used this circuit as starting point (keeping fixed the topology) to further apply the optimization procedure with  . As we could observe, the new circuit reaches a higher fitness at
. As we could observe, the new circuit reaches a higher fitness at  , which indicates that it exists a cost for robustness (Figure 7B). While being this cost low, this allows the circuit to maintain an operative mode even under genetic perturbations.
, which indicates that it exists a cost for robustness (Figure 7B). While being this cost low, this allows the circuit to maintain an operative mode even under genetic perturbations.
Figure 7.
(A) Scheme of a genetic circuit optimized to operate as a robust amplitude filter. U and Z are the input and output, respectively. (B) Fitness function versus the robustness weight for the robust circuit shown above (red) and the same circuit optimized with  (blue), illustrating the cost of robustness.
(blue), illustrating the cost of robustness.
In addition to the evolvability of living systems, gene circuits are subjected to stochastic processes that may prevent the expected behavior. The use of stochastic models could allow designing more robust circuits, or at least circuits that maintain their deterministic behavior. One way to explore the effect of noise in the design of circuits would be to in silico evolve a circuit to be robust against noise and compare it with a circuit not evolved with such a selective pressure. We applied our optimization method to design circuits that maintained a targeted dynamics in presence of noise (both intrinsic and extrinsic), obtaining different circuits with self-repressions and low translation rates (Supplementary Figure S11) (69). On the contrary, this approach could be applied to design circuits displaying high noise levels (75), since they may provide to the cell certain advantage under unpredictable environmental conditions (20).
CONCLUSION
In this work, we have tackled the problem of the designability of a given gene dynamics provided a library of composable regulatory elements, considering that the functional circuits come from combining different elements of the library. This measure of designability quantifies the entropy of a given dynamical behavior. For that, we have developed a computational methodology that allows exploring the diversity of behaviors that can be obtained by assembling circuits by means of two different design strategies: one based on heuristic optimization and other based on exhaustive simulation of circuits. We have taken advantage of current characterizations of regulatory elements into libraries of mathematical models (26–29), allowing to rapidly select the regulatory element of interest for our circuit. Although the emergence of unexpected behaviors is always an issue in synthetic biology, it is anticipated that the use of standardized parts allows reducing the endless tweaking process when engineering a synthetic gene circuit (12,19). Using a proper mathematical formulation, we were able to generate a large collection of genetic circuits by assembling those regulatory elements, and identify the functional subset according to certain specifications. Initially, we constructed an artificial library of models to design circuits by optimization toward a configuration satisfying the specifications. We designed filters and counters of gene expression, which allowed us to find new regulatory mechanisms able to provide such behaviors. Sometimes the behavior requires a very precise genotype, making improvable to get many cells with such behavior in a heterogeneous population. To investigate this, we have defined the concept of asymptotic robustness, which provides a measure of the maximum genotypic heterogeneity for a given of phenotypic behavior. In the long term, it is expected that synthetic biology projects will provide many examples of standardized circuits with a given dynamics, which could be incorporated into the available libraries. Then, one could extend our analysis to such cases. One issue here would involve the interfacing of such modules. To analyze this, we exploited one of our designed circuits as a single element of the library to obtain a complex system involving such a functional unit, illustrating a hierarchical design approach and allowing the design of plug-and-play devices with optimal impedance matching.
Given a library of regulatory elements, it is possible to construct many circuits with various dynamical behaviors. But some behaviors occur more often than others. To quantitatively analyze this, we computed the designability of a set of useful behaviors. There, we constructed a more reduced library of regulatory elements to assemble all possible circuits up to three genes and process their dynamics. Remarkably, the library involved promoters that had been previously characterized and even used for engineering various synthetic circuits in the bacterium Escherichia coli. Interestingly, we found that a limited library could encode a large number of behaviors. Certainly, our computational method allowed constructing and simulating the dynamics of this large set of circuits and assisted to dissect the spectrum of dynamical behaviors and study their designability. We found that a same genotype could have several functions depending on the external signals. Nevertheless, as the size of the circuits and the number of elements of the library increases, the exhaustive design strategy becomes unpractical, thus requiring heuristic methods. Since noise is an important factor that affects the dynamics of a circuit, we also included it in our analysis. The consideration of intrinsic noise slightly reduces the designability of digital circuits, but it increases the designability of oscillators. This is understandable from the fact that digital devices are steady-state based circuits, where noise could only spoil the behavior. On the other hand, oscillatory circuits are dynamical systems, where the noise could contribute to enhance the behavior. In addition, we expect that our results would be maintained when the library is enhanced by incorporating more accurate experimental measurements of the transcription regulation elements. That the new models could be more elaborated and accurate, they would not much change the fitness landscape and thus the designability of behaviors.
Herein, as opposite to other optimization-based design approaches (48–52) where it is difficult or even impossible to compile into a reliable DNA sequence the designed circuit, our automated approach scrutinizes in a combinatorial way the available genetic diversity to optimize a circuit with the desired function, and finally output a DNA sequence that encodes this designed circuit. However, the evolvability of genetic circuits in a continuous cellular background is a delicate issue. Importantly, the robust design approach entails that the expected behavior can be also observed throughout the cells of a population. Indeed, natural systems are robust to counteract mutations that, in most cases, entail unpredictable changes in the function of these elements (76). Moreover, as higher is the load of the circuit for the cell (i.e. quantity of resources required for function expression), faster will be the loss of function of the circuit because deleterious mutations will be selected (27,72). Hence, we tackled the design of robust circuits by accounting for parameter sensitivity into the objective function. We illustrated that such a circuit pays a cost by decreasing its fitness in order to gain robustness, which will lead to a higher evolutionary stability. Alternatively, or even in combination, we could design circuits that could, while preserving its central function, induce a benefit to the cell, by accounting for the interface between the circuit and the cellular interactome (77), and then assess the selection and reliability of the engineered circuit.
Interestingly, one possible extension to our work would be the development of a more complex, hierarchically distributed design platform (26). Herein, more diverse, characterized regulatory elements would be considered, involving transcriptional, riboregulatory, metabolic and signaling elements. These different regulatory elements would be combined to yield complex functional genetic circuits, involving different regulatory mechanisms. In addition to new elements, inherent effects such as the variation of cell growth rate due to different culture media, the delay in the biochemical reactions and the parameter uncertainty of the models are important questions that would be explored. Furthermore, the design of circuits could be combined with tools for the design of synthetic DNA sequences. This would exploit the interactions between nucleic acids and the reengineering of natural proteins. Promoters with targeted transcription rates or multiple operators (35,36), small RNAs with targeted secondary structures (6), or chimeric proteins acting as new transcription factors (74) are examples of what we could design computationally. These all elements would be modeled by transfer functions and these would be stored in a library (virtual or real). Importantly, it could be also specified a degree of evolvability, by which the value of the kinetic parameters characterizing that element would be susceptible to be changed after specific mutations in its sequence. Finally, the cellular chassis in which the circuit is going to be deployed could be also introduced as a generalized element by modeling the host elements that require the circuit for its expression (78). This would allow to provide a prediction of the response of the engineered cell under the conditions for which the circuit was designed, and consequently improve the design process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Generalitat Valenciana (BFPI-2007-160); HPC-Europa programme (RII3-CT-2003-506079); Spanish Ministry of Education and Science (TIN-2006-12860); Structural Funds ERDF; FP6-NEST 043340 (BioModularH2); FP7-ICT-043338 (BACTOCOM); FP7-ICT-265505 (CADMAD); ATIGE Genopole/UEVE (A3405); Fondation pour la Recherche Medicale.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Suarez and J. Forment for helping with programming and computational resources.
REFERENCES
- 1.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006.0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 3.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an autoregulatory genetic module. Proc. Natl Acad. Sci. USA. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 7.Basu S, Mehreja R, Thiberge S, Chen M, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 12.Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 13.Anderson J, Voigt C, Arkin A. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 15.Balagaddé FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 18.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cagatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Suel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 21.deJong H. Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 2002;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- 22.Bintu L, Buchler NE, Garcia H, Gerland U, Hwa T, Kondev J, Philips R. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wall ME, Hlavacek WS, Savageau MA. Design of gene circuits: lessons from bacteria. Nat. Rev. Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- 24.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 25.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 27.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 28.Voigt CA. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 31.Dubendorff JW, Studier FW. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 32.Edelman GM, Meech R, Owens GC, Jones FS. Synthetic promoter elements obtained by nucleotide sequence variation and selection for activity. Proc. Natl Acad. Sci. USA. 2000;97:3038–3043. doi: 10.1073/pnas.040569897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imburgio D, Rong M, Ma K, McAllister WT. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39:10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- 34.Mey M, Maertens J, Lequeux GJ, Soetaert WK, Vandamme E. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol. 2007;7:34. doi: 10.1186/1472-6750-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy KF, Balázsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl Acad. Sci. USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox RS, III, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Che AJ, Knight TF., Jr Engineering a family of synthetic splicing ribozymes. Nucleic Acids Res. 2010;38:2748–2755. doi: 10.1093/nar/gkq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krueger M, Scholz O, Wisshak S, Hillen W. Engineered Tet repressors with recognition specificity for the tetO-4C5G operator variant. Gene. 2007;404:93–100. doi: 10.1016/j.gene.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Rodrigo G, Carrera J, Jaramillo A. Asmparts: assembly of biological model parts. Syst. Synth. Biol. 2007;1:167–170. doi: 10.1007/s11693-008-9013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchisio MA, Stelling J. Computational design of synthetic gene circuits with composable parts. Bioinformatics. 2008;24:1903–1910. doi: 10.1093/bioinformatics/btn330. [DOI] [PubMed] [Google Scholar]
- 44.Cai Y, Hartnett B, Gustafsson C, Peccoud J. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics. 2007;23:2760–2767. doi: 10.1093/bioinformatics/btm446. [DOI] [PubMed] [Google Scholar]
- 45.Chandran D, Bergmann FT, Sauro HM. TinkerCell: modular CAD tool for synthetic biology. J. Biol. Eng. 2009;3:19. doi: 10.1186/1754-1611-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Densmore D, Hsiau TH, Kittleson JT, DeLoache W, Batten C, Anderson JC. Algorithms for automated DNA assembly. Nucleic Acids Res. 2010;38:2607–2616. doi: 10.1093/nar/gkq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooling MT, Rouilly V, Misirli G, Lawson J, Yu T, Hallinan J, Wipat A. Standard virtual biological parts: a repository of modular modeling components for synthetic biology. Bioinformatics. 2010;26:925–931. doi: 10.1093/bioinformatics/btq063. [DOI] [PubMed] [Google Scholar]
- 48.François P, Hakim V. Design of genetic networks with specified functions by evolution in silico. Proc. Natl Acad. Sci. USA. 2004;101:580–585. doi: 10.1073/pnas.0304532101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paladugu SR, Chickarmane V, Deckard A, Frumkin JP, McCormack M, Sauro HM. In silico evolution of functional modules in biochemical networks. IEE Proc. Syst. Biol. 2006;153:223–235. doi: 10.1049/ip-syb:20050096. [DOI] [PubMed] [Google Scholar]
- 50.Rodrigo G, Carrera J, Jaramillo A. Genetdes: automatic design of transcriptional networks. Bioinformatics. 2007;23:1857–1858. doi: 10.1093/bioinformatics/btm237. [DOI] [PubMed] [Google Scholar]
- 51.Tagkopoulos I, Liu Y, Tavazoie S. Predictive behavior within microbial genetic networks. Science. 2008;320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dasika MS, Maranas CD. OptCircuit: An optimization based method for computational design of genetic circuits. BMC Syst. Biol. 2008;2:24. doi: 10.1186/1752-0509-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 54.Rodrigo G, Carrera J, Elena SF. Network design meets in silico evolutionary biology. Biochimie. 2010;92:746–752. doi: 10.1016/j.biochi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 56.Entus R, Aufderheide B, Herbert M, Sauro MH. Design and implementation of three incoherent feed-forward motif based biological concentration sensors. Syst. Synth. Biol. 2007;1:119–128. doi: 10.1007/s11693-007-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaplan S, Bren A, Dekel E, Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol. Syst. Biol. 2008;4:203. doi: 10.1038/msb.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Kwon YK, Cho KH. The biphasic behavior of incoherent feed-forward loops in biomolecular regulatory networks. Bioessays. 2008;30:1204–1211. doi: 10.1002/bies.20839. [DOI] [PubMed] [Google Scholar]
- 59.Cotterell J, Sharpe J. An atlas of gene regulatory networks reveals multiple three-gene mechanisms for interpreting morphogen gradients. Mol. Syst. Biol. 2010;6:425. doi: 10.1038/msb.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- 61.Reeves GT, Muratov CB, Schupbach T, Shvartsman SY. Quantitative models of developmental pattern formation. Dev. Cell. 2006;11:289–300. doi: 10.1016/j.devcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 62.Mangan S, Alon U. Structure and function of the feedforward loop network motif. Proc. Natl Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1999;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 64.Guantes R, Poyatos JF. Multistable decision switches for flexible control of epigenetic differentiation. PLoS Comput. Biol. 2008;4:e1000235. doi: 10.1371/journal.pcbi.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mackey MC, Glass L. Oscillation and chaos in physiological control systems. Science. 1977;197:287–289. doi: 10.1126/science.267326. [DOI] [PubMed] [Google Scholar]
- 66.Del Vecchio D, Ninfa AJ, Sontag ED. Modular cell biology: retroactivity and insulation. Mol. Syst. Biol. 2008;4:161. doi: 10.1038/msb4100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim KH, Sauro HM. Measuring retroactivity from noise in gene regulatory networks. Biophys. J. 2011;100:1167–1177. doi: 10.1016/j.bpj.2010.12.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sayut DJ, Niu Y, Sun L. Construction and enhancement of a minimal genetic and logic gate. Appl. Environ. Microbiol. 2009;75:637–642. doi: 10.1128/AEM.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 70.Segal ME, Frieder O. On-the-fly program modification: systems for dynamic updating. IEEE Software. 1993;10:53–65. [Google Scholar]
- 71.Stark GR. Multifunctional proteins: one gene - more than one enzyme. Trends Biochem. Sci. 1977;2:64–66. [Google Scholar]
- 72.Sleight SC, Bartley BA, Lieviant JA, Sauro HM. Designing and engineering evolutionary robust genetic circuits. J. Biol. Eng. 2010;4:12. doi: 10.1186/1754-1611-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsutsui S, Ghosh A. Genetic algorithms with a robust solution searching scheme. IEEE Trans. Evol. Comput. 1997;1:201–208. [Google Scholar]
- 74.Hollis M, Valenzuela D, Pioli D, Wharton R, Ptashne M. A repressor heterodimer binds to a chimeric operator. Proc. Natl Acad. Sci. USA. 1988;85:5834–5838. doi: 10.1073/pnas.85.16.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu T, Ferry M, Weiss R, Hasty J. A molecular noise generator. Phys Biol. 2008;5:036006. doi: 10.1088/1478-3975/5/3/036006. [DOI] [PubMed] [Google Scholar]
- 76.Kitano H. Biological robustness. Nat. Rev. Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 77.Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.