Abstract
Mutant transcripts containing expanded CUG repeats in the untranslated region are a pathogenic factor in myotonic dystrophy type 1 (DM1). The mutant RNA sequesters the muscleblind-like 1 (MBNL1) splicing factor and causes misregulation of the alternative splicing of multiple genes that are linked to clinical symptoms of the disease. In this study, we show that either long untranslated CAG repeat RNA or short synthetic CAG repeats induce splicing aberrations typical of DM1. Alternative splicing defects are also caused by translated CAG repeats in normal cells transfected with a mutant ATXN3 gene construct and in cells derived from spinocerebellar ataxia type 3 and Huntington's disease patients. Splicing misregulation is unlikely to be caused by traces of antisense transcripts with CUG repeats, and the possible trigger of this misregulation may be sequestration of the MBNL1 protein with nuclear RNA inclusions containing expanded CAG repeat transcripts. We propose that alternative splicing misregulation by mutant CAG repeats may contribute to the pathological features of polyglutamine disorders.
INTRODUCTION
Human triplet repeat expansion diseases (TREDs) are caused by abnormally elongated trinucleotide repeats located in either coding or non-coding regions of specific genes. Although they have a similar type of underlying mutation, TREDs represent a clinically heterogeneous group of disorders with some shared symptoms (1,2). This observation implies that the phenotype for each disease is determined not only by the different genes harboring the mutations and their expression patterns but also by the mutated sequences themselves or their structural features (3–6). A subgroup of TREDs, polyglutamine (poly-Q) disorders, is caused by expansions of CAG repeats in translated sequences encoding poly-Q tracts in multiple functionally unrelated proteins (7). Huntington's disease (HD) and several types of spinocerebellar ataxia (SCA) belong to this group.
Diseases caused by expanded repeats located in untranslated regions (UTR) are another subgroup of TREDs. In myotonic dystrophy type 1 (DM1) and type 2 (DM2), a crucial role in pathogenesis is played by mutant DMPK and ZNF9 transcripts containing expanded CUG or CCUG repeats located in the 3′-UTR or in an intron, respectively (8,9). Both types of repeats form stable hairpin structures (10–12) that are toxic to cells because they trigger alternative splicing aberrations in multiple genes (13,14), including the insulin receptor (INSR), chloride channel 1 (CLCN1), sarcoplasmic/endoplasmic reticulum Ca+2 ATPase 1 and 2 (SERCA1 and SERCA2), cardiac troponin T (cTNT) and Tau (MAPT). Aberrant expression of these genes is linked to clinical symptoms of DM1 and DM2, such as insulin resistance, myotonia, muscle wasting, cardiac abnormalities and cognitive deficits (14–17). The splicing aberrations mainly result from an imbalance in the functional levels of two antagonistic alternative splicing regulators, muscleblind-like 1 (MBNL1) and CUGBP1 (18,19). MBNL1 is sequestered by mutant transcripts and accumulates in the nucleus in the form of inclusions in both DM1 and DM2 (20). CUGBP1 undergoes hyperphosphorylation, and its cellular level is elevated only in DM1 tissues (20–26).
The pathogenesis of poly-Q diseases is commonly attributed to expanded poly-Q tracts in proteins (27). Numerous cellular processes including transcriptional regulation, mitochondrial functions and axonal transport have been shown to be affected by proteins containing this type of mutation (27). Poly-Q expansions contribute to neurodegenerative diseases by both gain-of-function and partial loss-of-function mechanisms (28). The protein sequence context of the poly-Q tract was also shown to influence disease specificity (29). Mutant proteins or protein fragments containing poly-Q tracts form both nuclear and cytoplasmic inclusions (30,31), however, the toxic role of these deposits has been questioned in a number of studies (32–34). A contribution of the mutant transcript to the pathogenesis of poly-Q diseases has been postulated by several authors (35–37) due in part to the similar molecular architecture of CAG and CUG repeat RNAs (11,38–40) and the ability of MBNL1 protein to bind to the repeats (41). In addition, in the Drosophila eye model of spinocerebellar ataxia type 3 (SCA3), long CAG repeats were shown to generate some of the degenerative effects induced by CUG repeats (35).
In this study, we addressed the question of whether transcripts with expanded CAG repeats may cause alternative splicing aberrations similar to those triggered by CUG repeats. We performed comparative splicing analysis of a set of genes with misregulated splicing in DM1 cells, and we found that expression of long CUG and CAG repeats in normal cells triggers similar splicing changes. This effect was found for untranslated or translated repeats of exogenous and endogenous mutant transcripts. In cells expressing transcripts with expanded CAG repeats, we detected intranuclear RNA foci composed of the sense transcripts that co-localize with the MBNL1 protein. We conclude that, given the similarities between the mutant CUG and CAG repeat transcripts in triggering aberrant alternative splicing, the RNA-containing expanded CAG repeats may contribute to the pathogenesis of HD and SCA3 as well as other poly-Q diseases.
MATERIALS AND METHODS
Plasmids and synthetic oligonucleotides
DMPK fragments containing (CTG)74 and (CTG)200 were PCR amplified from DM1 patient genomic DNA. Shorter CTG and CAG fragments were generated by chemical synthesis and in vitro expansion events (42). All inserts were cloned as NotI fragments into the pEGFP-N3 vector (Clontech) in the 3′-UTR region of the enhanced green fluorescent protein (EGFP) cDNA. The EGFP-mutATXN3 construct (which drives expression of the EGFP-mutant ataxin 3 fusion protein) was also prepared in the pEGFP-N3 vector. The stop codon of EGFP was removed, and the human ATXN3 sequence containing 69 CAG repeats was inserted in frame as a BsrgI/NotI fragment. The insert sequences and number of repeats were confirmed by sequencing. All RNA oligonucleotides used in this study were synthesized by Metabion (Martinsried, Germany). The following synthetic RNAs were used: (CUG)7, (CAG)7, (CUG)20, (CAG)20, (AUG)17, (UUA)17, (UGG)17 and pre-miR-136 (43). The short interfering RNA (siRNA) duplexes used in this study were the same as described previously: ATXN3-specific siG16 (44), siMBNL1 (45) and siLuc (sense 5′-CGU ACG CGG AAU ACU UCG AUU-3′; antisense 5′-UCG AAG UAU UCC GCG UAC GUU-3′).
Cell culture and transfection
HeLa cells were maintained in RPMI 1640 medium (Bioshop) supplemented with 10% fetal bovine serum (HyClone) and antibiotics (Sigma). The human neuroblastoma SK-N-MC cell line (DSMZ–Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany) was maintained in DMEM (Sigma) supplemented with 10% fetal bovine serum, antibiotics and additional amino acids (Sigma). Fibroblasts from normal individuals (GM07492, GM00024, GM0321 and GM01650) as well as DM1 (GM03987), SCA3 (GM06153, GM06151) and HD patients (GM03864, GM04208 and GM04281) were obtained from the Coriell Cell Repositories and were grown in minimal essential medium (MEM, Gibco BRL Life Technologies) supplemented with 8% fetal bovine serum, antibiotics and MEM-vitamins. Cell transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Generating stable cell lines
To establish stable clones, HeLa and SK-N-MC cells were transfected using Lipofectamine 2000 with DraIII-linearized pEGFP-N3-derived plasmids carrying the neomycin resistance marker. Having completed 48 h after transfection, cells were transferred to medium containing 0.6 mg/ml G418 (Sigma) to select for stable integration of the plasmid. G418-resistant colonies were isolated from each transfection. Colonies were expanded and screened for exogene expression by EGFP-specific real-time reverse transcription polymerase chain reaction (RT–PCR). Stable clones were maintained in G418-containing medium.
MBNL1 over-expression
HeLa cell lines with the long CAG repeats that were chosen for pEGFP-MBNL1 transfection (74CAG K3, 200CAG K3 and 200CAG K4) were seeded on to a 6-well plate and 24 h after the cells were transfected using Lipofectamine 2000 with 0.5 μg pEGFP-C1MBNL1 plasmid that bears the sequence of MBNL1 protein 41 kDa isoform. Completing 48 h after transfection, cells were harvested and RNA was isolated. The cDNA synthesized using this template was further analyzed.
Fluorescence in situ hybridization and immunofluorescence
The detection of CUG and CAG transcripts in DM1, HD and SCA3 fibroblasts as well as in transfected HeLa cells was carried out by Fluorescence in situ hybridization (FISH) using Cy3-labeled DNA/LNA probes (CTG)6-CT or (CAG)6-CA, which were modified at positions 2, 5, 8, 13, 16 and 19 with LNA. For FISH and immunofluorescence (IF), the cells were fixed in 4% paraformaldehyde and were permeabilized with 2% pre-chilled acetone in phosphate buffered saline (PBS) at pH 7.4 for 5 min. For ribonuclease (RNase) treatment, cells were permeablized and then incubated with 0.01 μg/ml RNase A (Sigma) for 1.5 h at 37°C. Pre-hybridization was performed in 30% formamide and 2× SSC for 10 min, followed by hybridization in buffer containing 30% formamide, 2× SSC, 0.02% BSA, 66 µg/ml yeast tRNA, 10% dextran sulfate, 2 mM vanadyl ribonucleoside complex and 2 ng/µl DNA/LNA probe. Post-hybridization washing was done in 30% formamide and 2× SSC at 45°C for 30 min followed by 1× SSC at 37°C for the next 30 min. Slides were either mounted in Vectashield medium (Vector Laboratories, Inc. Burlingame, CA, USA) containing 4′,6-diamidino-2-phenylindole (DAPI) or were submitted to IF. For RNA FISH in combination with IF, the slides were treated as described above with no DAPI staining. The slides were incubated in blocking buffer (3% non-fat milk, 5% normal serum and 0.3% Triton X-100 in PBS) for 1 h at room temperature, and then incubated with rabbit anti-MBNL1 primary antibody (A2764) (a gift from Dr Charles Thornton) overnight at 4°C. After the reaction, the slides were washed in PBS and then incubated with a secondary anti-rabbit antibody conjugated to Cy2 (Jackson ImmunoResearch Laboratories, Inc.). Following PBS washes, the nuclei were stained with DAPI in Vectashield mounting medium. All FISH/IF images were acquired using a Zeiss Axiovert 200 M microscope equipped with a cooled AxioCam HRc camera and processed with LSM 510 software.
For DNase treatment cells were fixed and incubated O/N with 70% EtOH. The next day, cells were pre-incubated for 10 min in cytoskeletal buffer with sucrose (CSK) (10 mM HEPES, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100, 10 mM vanadyl riboncleoside, pH 6.8) [according to Ho et al. (36)] followed by incubation for 1.5 h at 37°C with 0.8 U/μl RNase-free DNase I (Epicentre) in the CSK buffer. After the reaction, DNA was removed by two additional washes with the ice-cold cytoskeletal buffer. DNase control slides were incubated under the same reaction conditions but DNase was omitted in the reaction buffer.
RT–PCR and real-time PCR
RT–PCR analysis was performed using standard protocols. Briefly, 1 µg of total RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocols with use of random hexamers. All reverse transcription reactions, after 10-fold dilution, were subjected to an optimal number of PCR cycles that were within the linear range of DNA amplification. PCR was conducted to analyze the splice products of several genes specified in Supplementary Table S1. RT–PCR products were resolved in 3% agarose gel containing 0.5 µg/ml ethidium bromide. The fraction of exon inclusion was calculated by dividing the intensity of the PCR band corresponding to the exon inclusion splice product by the total intensity of the RT–PCR products.
Real-time PCR was performed using a Rotor-Gene 3000 (Corbett Research) in the presence of SYBR green. Optimization of the real-time PCR was performed according to the manufacturer's instructions (Applied-Biosystems, SYBR Green PCR Master Mix reagent protocol). Appropriate primers were used for the detection and quantification of EGFP expression (forward 5′-CCGAGATAGGGTTGAGTGTTG; reverse 5′-CTTTCCCCGTCAAGCTCTAA) and GAPDH expression (forward 5′-GAAGGTGAAGGTCGGAGTC; reverse 5′-GAAGATGGTGATGGGATTTC).
Multiplex ligation-dependent probes amplification
The Multiplex ligation-dependent probes amplification (MLPA) assay was designed to assess the relative dosage of sense and antisense transcripts of triplet repeat-containing EGFP genes. The assay mixture included two antisense-specific probes and two sense-specific probes. The probe design strategy was as described previously (46). Target sequences of all designed probes were located within the EGFP sequence in close proximity to each other and to the triplet repeat tracts (Supplementary data). Probes (sequences are shown in Supplementary Table S3 and Supplementary Figure S3) were generated by chemical synthesis (Sigma). The MLPA reaction was performed as described previously (47), following the general directions provided by MRC-Holland (www.mlpa.com). Briefly, 5 μl of cDNA (prepared as described above and diluted 2×) or 5 µl of pEGFP-N3 (7.5 ng/µl) was incubated at 98°C for 5 min. After cooling the samples to room temperature, 1.5 μl of probe mix (containing 1 fmol of each probe) and 1.5 μl of SALSA hybridization buffer were added, and the solution was denatured at 95°C for 2 min and hybridized at 60°C for 16 h. Hybridized probes were ligated at 54°C for 15 min by the addition of 32 μl ligation mixture. Following heat inactivation, 10 μl of ligation reaction was mixed with 30 μl of PCR buffer, heated to 60°C, mixed with 10 μl PCR mixture (SALSA polymerase, dNTPs and universal primers, one of which was labeled with fluorescein) and subjected to 30 cycles of PCR amplification. All reagents except the synthesized oligonucleotide probes were obtained from MRC-Holland. Amplification products were diluted 1:40 in HiDi formamide (ABI) containing 1/36 volume of LIZ600 size standard (ABI) and then separated by size on an ABI 3100 Genetic Analyzer (ABI). Electropherograms were analyzed by GeneScan (ABI), and peak height data were exported to an Excel table. Excel programs were generated to transform the peak height data to normalized values. Briefly, peak heights (signals) for each probe were divided by the average signal from two sense-specific probes (run-to-run signal normalization), and these values were divided by corresponding values calculated from the reference vector plasmid DNA sample (relative dosage estimation). To facilitate comparisons between samples, calculated relative values were adjusted to obtain an average signal from sense-specific probes that was equal to 1.
Western blotting
Cells were lysed with 1x PB homogenate buffer (60 mM Tris–HCl, 2% SDS, 10% sucrose, 2 mM PMSF). A quantity of 20 µg of protein lysate were separated by SDS–PAGE and then transferred onto nitrocellulose membranes (Sigma). After blocking with 5% skim milk, membranes were incubated with primary antibodies. The following primary antibodies were used: anti-MBNL (EnCor; diluted 1:5000), anti-CUG binding protein 1 (CUG-BP1) (Abcam; diluted 1:500), anti-ataxin3 (Millipore, diluted 1:1000) and anti-GAPDH (Chemicon; diluted 1:20 000). After incubation with the primary antibody, membranes were washed in PBS containing 0.3% Tween-20 and then incubated with a secondary antibody conjugated to biotin. After several washes, proteins were visualized using the Sigma Fast BCIP/NBT kit (Sigma).
Filter binding assay
The filter binding assay was performed as described previously (41). Briefly, various concentrations of MBNL1 protein ranging from 0.1 nM to 200 nM were incubated with 500 fM of 5′-end-labeled CUG and CAG RNA fragments consisting of 7 or 20 triplet repeats in a reaction buffer (100 mM Tris–HCl pH 8.0, 50 mM NaCl, 50 mM KCl, 1 mM MgCl2). As controls, 5′-end-labeled (AUG)17, (UUA)17 and (UGG)17 RNA fragments were used. After 30 min of incubation at 37°C, samples were loaded onto filters (nitrocellulose and nylon membranes) and sandwiches were assembled on the slot-blot apparatus. Signal from both filters was visualized by phosphoimaging (Typhoon; Molecular Dynamics). The dissociation constant values (KD) were calculated based on three independent experiments using the one-site binding model (GraphPad Prism 5.00).
Statistics
Statistical significance was assessed by a two-tailed t-test using Prism V. 4.0 GraphPad Software (San Diego, CA, USA).
RESULTS
Cellular models expressing different lengths of untranslated CAG and CUG repeats
We sought to compare CUG and CAG repeats of different lengths in their ability to trigger aberrant alternative splicing characteristic for DM1. For this purpose, we used HeLa and SK-N-MC cell lines, which express the MBNL1 and CUGBP1 splicing factors at similar levels (Supplementary Figure S1). We generated stable clone lines expressing CAG and CUG repeats located in the 3′-UTR of pEGFP-N3 (Figure 1A). The recombinant exogene encoded 5, 30, 74 and 200 repeats of the CAG or CUG motif, and these lengths represent tracts that occur in normal (5, 30) and mutant (74, 200) alleles of genes implicated in DM1 and poly-Q diseases (28). Clones that maintained stable expression of the exogene were used for subsequent analyses. The level of exogene expression determined by quantitative RT–PCR was similar in all selected clonal lines.
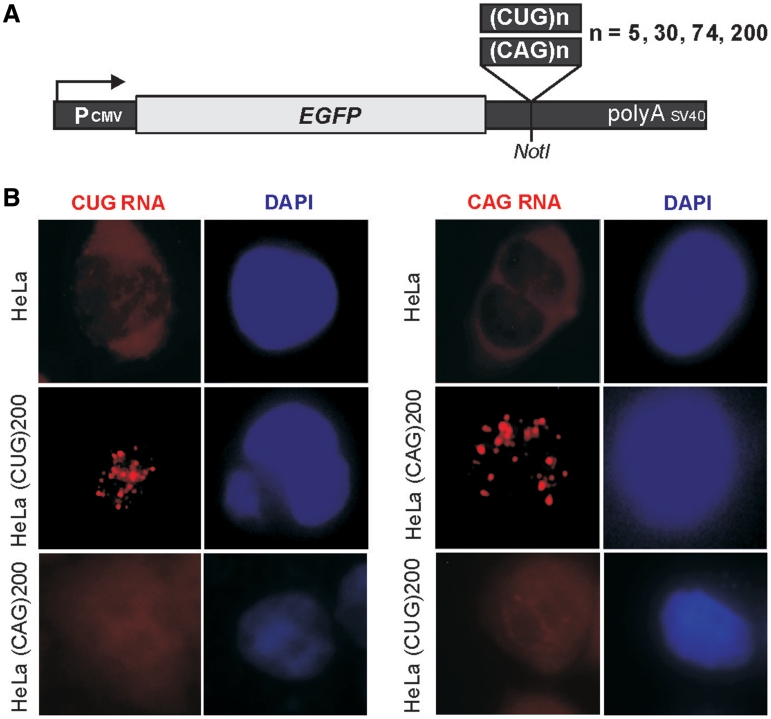
Figure 1.
Expression of untranslated CUG and CAG repeats and cellular localization of expanded repeat transcripts. (A) Diagram of the pEGFP-N3-derived constructs used to generate stable HeLa and SK-N-MC cell lines expressing 5, 30, 74 and 200 of the CUG or CAG repeats in the 3′-UTR of mRNA encoding EGFP. (B) Representative RNA-FISH images of HeLa cells cultured in the presence of plasmid encoding (CUG)200 and (CAG)200 repeat RNAs. Nuclear retention of the mutant transcripts is visible. Nuclei were stained with DAPI (blue); RNA foci are red; magnification ×60.
To resolve whether expression of CAG and CUG repeats is associated with nuclear retention of the repeat RNA, we performed RNA in situ hybridization. We found that only mutant variants of either the CUG or the CAG repeats (74 and 200 triplets) gave rise to numerous nuclear foci in HeLa cells (Figure 1B). The abundance of the intranuclear foci was greater in cells with the CUG repeats than in those with the CAG triplets. No foci were detected in cells expressing 5 or 30 CUG or CAG repeats.
Transcripts containing long untranslated CUG and CAG repeats trigger similar splicing aberrations in model cells
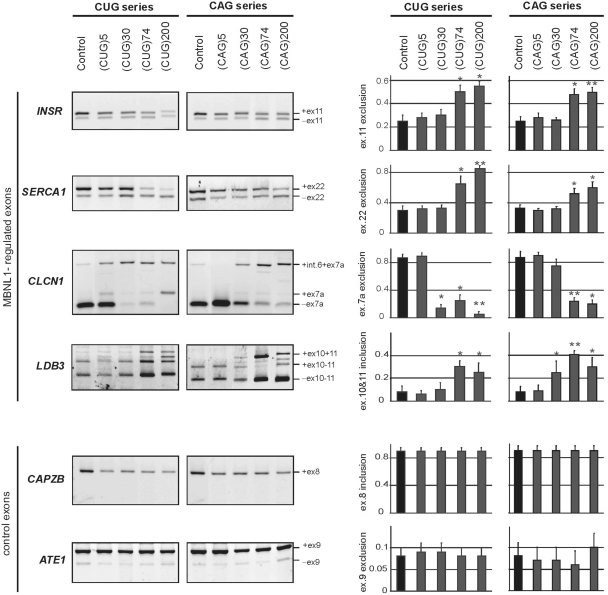
For alternative splicing analyses, we chose a set of endogenous transcripts instead of the exogenous minigenes typically analyzed for alternative splicing aberrations in DM1 (36). The pool of genes and splicing events that are known to undergo aberrant splicing in DM1 was limited to those that are expressed in HeLa and SK-N-MC cells and have a ratio of splice variants suitable for measurement. For MBNL1-sensitive genes, these criteria were fulfilled by the INSR (IR), CLCN1, SERCA1 (ATP2A1) and LDB3 (ZASP) genes in HeLa cells and by the CLCN1, SERCA1 and MAPT genes in SK-N-MC cells. For analysis of MBNL1-independent splicing events, we selected the CUGBP-regulated CAPZB gene, which shows an abnormal splicing pattern in DM1 patients as well as the ATE1 and FHL1 genes that are known to be alternatively spliced but do not exhibit any splicing abnormalities in DM (48).
The alternatively spliced INSR mRNAs give rise to receptors differing in their signaling properties. In normal adult skeletal muscle, the INSR mRNA containing exon 11, encodes an insulin receptor with greater signaling activity than that of the INSR mRNA lacking this exon. In DM1 skeletal muscle, the fetal isoform of INSR mRNA without exon 11 is retained and results in compromised signaling (15). We observed a higher degree of exon 11 exclusion (−ex.11) in HeLa cells expressing 74 or 200 CUG repeats (mean 67 ± 9%) than in the cells with 5 or 30 CUG repeats (mean 28 ± 5%). A very similar effect was observed for CAG repeats of the same length (Figure 2). The CLCN1 mRNA expressed in skeletal muscle of normal adults excludes exon 7a while, in DM1 tissue, the fetal isoform containing exon 7a is retained (CLCN1 +ex 7a), which results in transcript degradation by nonsense mediated decay (NMD) (49). We found that the CLCN1 isoform without exon 7a was the predominant isoform in non-transfected HeLa cells and in cells expressing either 5 CUG or 5 or 30 CAG repeats while the isoforms containing exon 7a were more prevalent in HeLa cells expressing 30, 74 or 200 CUG repeats or 74 or 200 CAG repeats (Figure 2). The same pattern of alternative splicing isoform expression was observed in SK-N-MC cells (Supplementary Figure S2). In the case of the SERCA1 mRNA, exon 22 is excluded in only a small fraction of the mRNA in normal adult skeletal muscle tissue, but it is excluded from most of the mRNA in tissue of DM1 patients (16,50). In our model system, the control HeLa cells lacking the exogenous repeats and those expressing 5 or 30 CUG or CAG repeats showed a higher proportion (mean 72 ± 5%) of longer SERCA1 mRNA containing exon 22. This proportion decreased significantly with increasing CUG or CAG repeat length (Figure 2). In SK-N-MC cells expressing CUG or CAG repeats, the same trend in splicing aberrations was observed (Supplementary Figure S2). The alternative splicing pattern was also analyzed for the LDB3 mRNA in HeLa cells and for MAPT mRNA in SK-N-MC cells. Consistently, similar changes in the distribution of alternative isoforms were observed in cells expressing CUG and CAG repeats (Figure 2 and Supplementary Figure S2). Most of the analyzed clonal lines expressing long CAG and GUG repeats have shown splicing misregulation. But not all of them, i.e. 3 out of 8 CAG HeLa lines and 3 out of 10 CUG HeLa lines did not show mis-regulated splicing. Among the genes selected as the alternative splicing controls, no substantial differences were observed for the CAPZB and ATE1 mRNAs in the presence of CUG or CAG repeats expressed in both cellular models.
Figure 2.
CUG and CAG repeats trigger similar pattern of the aberrant alternative splicing in HeLa cells. Total RNA isolated from cells of clonal HeLa lines expressing untranslated CUG or CAG repeat RNA of designated lengths was used to assess the levels of splicing variant transcripts from the INSR, SERCA1, CLCN1, LDB3, CAPZB, and ATE1 genes by RT–PCR. DM1-specific splicing aberrations were observed for the longest (74 and 200) both the CUG or CAG repeats and only for genes that are known to be regulated by MBNL1. The CAPZB, that is known to be regulated by CUGBP1 and ATE1 control gene that is not disrupted in DM1 patients, did not show significant changes in the proportion of transcript variants. The experiments were carried out in triplicate and quantitative results are shown as bar diagrams. The fraction of exon inclusion or exclusion (±SD) was calculated by dividing the signal of RT–PCR band corresponding to the inclusion/exclusion splice product by the total RT–PCR representing all splice products. *P = (0.05; 0.001), **P < 0.001 compared with the control.
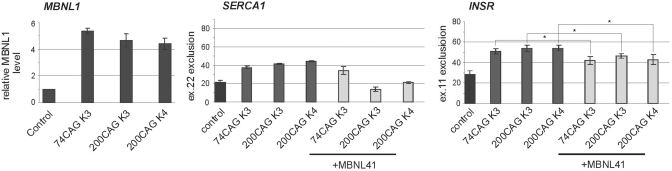
Partial reversion of splicing defects in cell lines expressing long tracts of CAG repeats after MBNL1 overexpression may suggest that observed changes are associated with MBNL1 sequestration. We transfected three Hela cell lines expressing long CAG tracts (74CAG K3, 200CAG K3 and 200CAG K4) with pEGFP-C1MBNL1 plasmid which is an expression vector bearing a sequence of 41 kDa isoform MBNL1 protein. Completing 48 h after transfection, the RNA was isolated. The analysis of SERCA1 and INSR endogenes showed that approximately 5-fold overexpression of MBNL1 produced a 24% increase in SERCA1 exon 22 inclusion and a 16% increase in INSR exon 11 inclusion in cells expressing 200 CAG repeats (Figure 3).
Figure 3.
Overexpression of MBNL1 partially reverses splicing abnormalities caused by long CAG repeats in HeLa cells. Control—untransfected cells; +MBNL41 lanes—cell lines bearing long CAG tracts transfected with pEGFP-C1MBNL1 plasmid. The level of transcript expressed from pEGFP-C1MBNL1 was determined by RT–PCR analysis. The experiments were carried out in duplicate and quantitative results are shown as bar diagrams. MBNL1 overexpression was estimated in comparison to GAPDH expression level. The fraction of exon inclusion or exclusion (±SD) was calculated by dividing the signal of RT–PCR band corresponding to the inclusion/exclusion splice product by the total RT–PCR signal representing all splice products. *P = (0.05; 0.001), compared with the cells that were not transfected with pEGFP-C1MBNL1 plasmid.
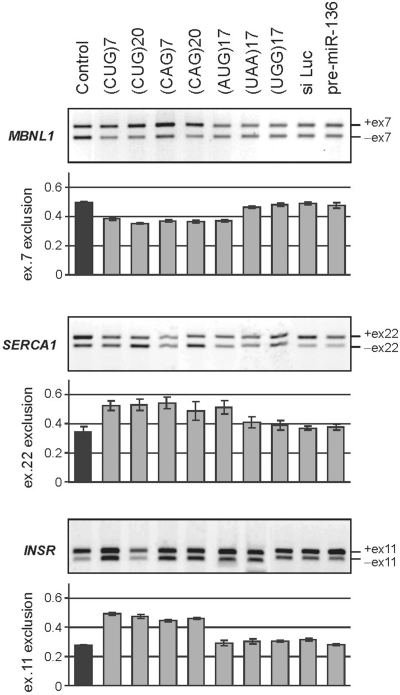
Synthetic short CUG and CAG repeat RNAs trigger splicing aberrations
In the experiments presented above, the shorter CUG and CAG repeat tracts were unable to induce the splicing aberrations that were triggered by the longer repeats. To determine whether a higher concentration of shorter repeats would induce aberrant splicing in cells, we transfected HeLa cells with 5, 10 and 40 nM of synthetic oligoribonucleotides (ORNs) composed of 7 or 20 CAG or CUG repeats. In addition, we used (AUG)17, (UUA)17 and (UGG)17 as well as control non-repeat ORNs [i.e. an siRNA duplex against the luciferase transcript (siLuc) and the precursor of microRNA-136]. Treatment with the highest 40 nM concentration of (CUG)7 resulted in considerable exclusion of exons from INSR and SERCA1 mRNA (the proportion of shorter isoforms increased from 27 ± 3% to 45 ± 6% and from 31 ± 2% to 52 ± 5%, respectively) and inclusion of exon 7 in MBNL1 mRNA (49 ± 2% to 65 ± 3%) (Figure 4). A similar effect was observed when 40 nM of (CAG)7 was used. Delivery of longer ORNs [i.e. (CUG)20 and (CAG)20] at the same concentration to HeLa cells, triggered similar effects on splicing aberrations. When four control ORNs were introduced into the cells (UUA and UGG triplet repeats, siLuc and pre-miR-136), splicing misregulation of the tested transcripts was not observed; however, (AUG)17 triggered changes in the exon inclusion of MBNL1 and SERCA1 similar to those observed after delivery of (CUG)20 and (CAG)20 ORNs. Alternative splicing of the control ATE1 and FHL1 genes was not affected by any of the utilized ORNs (data not shown). Furthermore, neither (CUG)7 nor (CAG)7 was capable of triggering measurable alternative splicing aberrations at concentrations of 5 and 10 nM.
Figure 4.
Short exogenous CUG and CAG repeat RNAs trigger DM1-specific splicing abnormalities in HeLa cells. RT–PCR products of the alternative splicing of MBNL1, SERCA1 and INSR genes after 48-h treatment of HeLa cells with designated synthetic oligoribonucleotides delivered in 40 nM concentration. Bar graphs show the percentage of the appropriate mRNA isoforms relative to the total amount of splice products. Splicing aberrations were observed for the CUG and CAG ORNs; (UAG)17 sequence also triggers some splicing misbalance. No changes were detected for control repeats (UAA)17, (UGG)17, siLuc, or pre-miR-136.
Poly-Q disease transcripts misregulate splicing in transfected cells and patient-derived cells
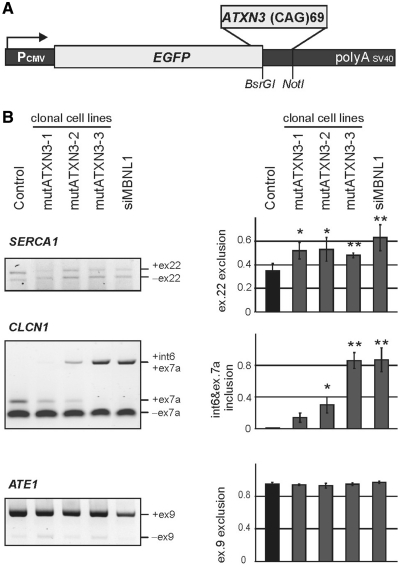
Next, we asked whether cells expressing translated CAG repeats would recapitulate splicing changes triggered by untranslated CAG repeats that were found in HeLa and SK-N-MC clonal lines. Therefore, we established stable SK-N-MC clones expressing an EGFP-mutant Ataxin 3 fusion protein (mutATXN3) (Supplementary Figure S4). SK-N-MC clones expressing an EGFP wild-type Ataxin 3 fusion protein (wtATXN3) (Supplementary Figure S5) were established as an additional control. In three out of six mutATXN3 clones, we found similar splicing alterations when compared with those induced by long untranslated CAG repeats (Figure 5 and Supplementary Figure S2). As shown in Figure 5, out of the three genes that were analyzed, two (CLCN1 and SERCA1) showed splicing misregulation. No changes were detected for control ATE1, which is consistent with the results obtained in HeLa and SK-N-MC clonal lines expressing long untranslated CAG repeats (Figure 5 and Supplementary Figure S2). The scale of alternative splicing differed from clone to clone.
Figure 5.
Splicing changes in cells expressing mutated Ataxin3 fragment with 69 glutamine residues. (A) Diagram of the pEGFP-mutATXN3 construct used to generate SK-N-MC cell lines expressing 69 CAG repeats in protein coding region. The stop codon of the EGFP protein was removed and a cDNA sequence of mutant human ATXN3 containing 69 CAG repeats was inserted in frame at the 3′-terminus to induce expression of EGFP-mutant Ataxin 3 fusion protein containing 69 Q-residues. (B) RT–PCR products of the alternative splicing of SERCA1, CLCN1 and control ATE1 gene. Quantification of the results is presented in the graphs as the percentage of splice products that include or exclude the indicated exons. The experiments repeated three times were carried out in three transgenic SK-N-MC lines (mutATXN3 1–3). *P = (0.05; 0.001), **P < 0.001 compared with the untreated cells (control). SK-N-MC cells with MBNL1 siRNA knocked down were used as a positive control (siMBNL1).
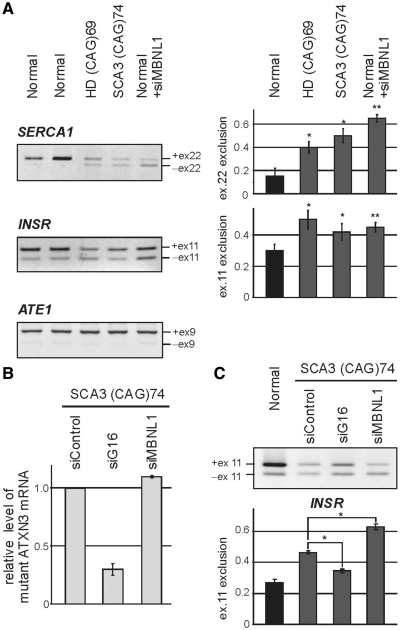
We next addressed the question of whether splicing misregulation occurs in cells derived from HD and SCA3 patients. Three HD cell lines expressing 44, 46 and 69 CAG repeats, two SCA3 lines with 69 and 74 CAG repeats and three control fibroblast cell lines were used to examine the expression of the INSR, SERCA1 and ATE1 mRNA splicing isoforms. Significant changes in the exclusion of INSR exon 11 (52 ± 5% in HD and 38 ± 4% in SCA3 versus 70 ± 2% in normal fibroblasts) and SERCA1 exon 22 (41 ± 6% in HD and 49 ± 4% in SCA3 versus 87 ± 5% in normal fibroblasts) were observed only in cells bearing the longest repeat tracts (i.e. 69 and 74 CAG) (Figure 6A). To examine whether the observed splicing changes were triggered by the expanded CAG RNA repeats, we knocked down the mutant ATXN3 transcript in SCA3 cells and anticipated a reversion of the splicing abnormalities (Figure 6B). Indeed, the 3-fold reduction of mutant ATXN3 levels through siRNA treatment resulted in partial but significant restoration of exon 11 splicing in the INSR transcript (Figure 6C).
Figure 6.
SERCA1 and INSR undergo alternative splicing changes in human HD and SCA3 fibroblasts. (A) RT–PCR analysis of endogenous SERCA1, INSR and ATE1 mRNA isoforms in fibroblasts derived from HD (69 CAG repeats in HTT) and SCA3 patients (74 CAG repeats in ATXN3) and healthy individuals (Normal). For comparison, normal cells after siRNA-mediated down-regulation of MBNL1 expression were used (+siMBNL1). A modest, but clearly visible disturbance of the balance of the SERCA1 and INSR splicing isoforms was detected in HD and SCA3 cell lines. MBNL1 knock-down resulted in analogical but stronger changes between splicing isoforms. In contrast, ATE1 mRNA expression remained unchanged. The percentages of exon 22 and exon 11 exclusion relative to the total transcripts indicate mean values ± SD of three independent experiments *P = (0.05; 0.001), **P < 0.001 compared with results obtained for normal fibroblasts). (B) The level of mutant ATXN3 transcript (74 CAG repeats) in SCA3 fibroblasts treated with three siRNAs: control, ATXN3-specific (siG16) and MBNL1-specific (siMBNL1). The relative expression level was determined by RT–PCR analysis of CAG repeat region of ATXN3 compared to GAPDH amplification product. (C) Treatment of SCA3 cells with siG16 resulted in partial reversion of INSR splicing misregulation, while MBNL1 silencing led to significant increasing of mRNA isoform without exon 11. Graph shows average values (±SD) for three independent transfection experiments [*P = (0.05; 0.001)].
Antisense CUG repeat transcripts are unlikely to trigger alternative splicing deregulation in cells expressing expanded CAG repeats
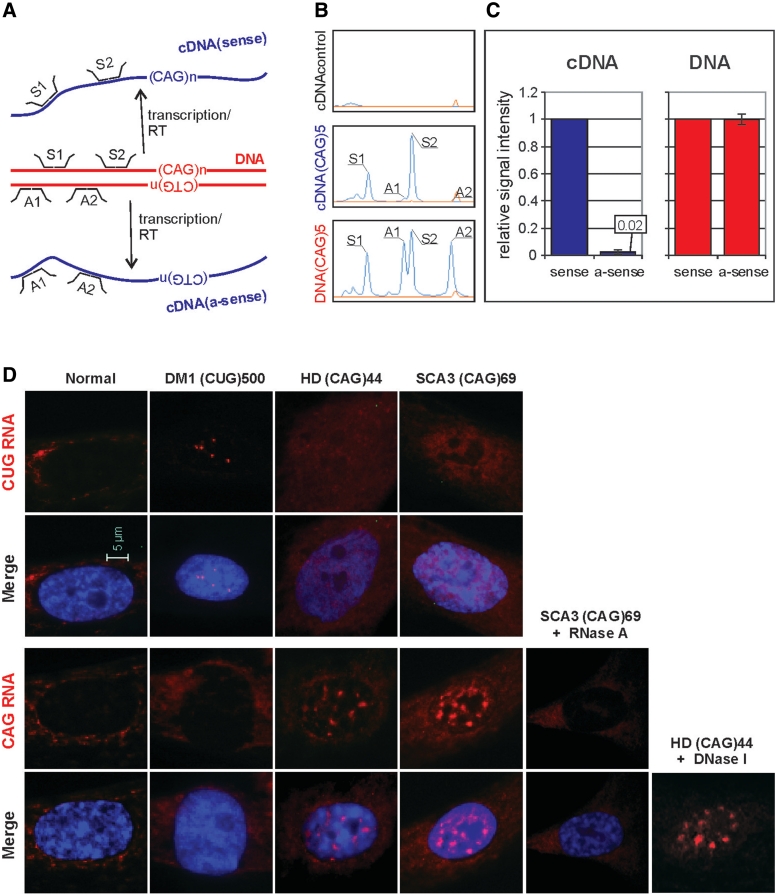
To determine whether splicing misregulation, which we detected in various cell lines in the presence of CAG expansions, was a result of antisense CUG repeat transcript co-expression, we designed and generated a strand-specific MLPA assay. In this multiplex assay, we used two probes specific for sense and two specific for antisense transcripts (Figure 7A). The MLPA was carried out for six cell lines. Five of the cell lines were derived from SK-N-MC cells transfected with EGFP-N3-derived plasmids encoding the following: (CAG)5, (CAG)74, (CUG)5, (CUG)74 and mutATXN3. Untransfected cells were used as a negative control and pEGFP-N3 DNA was used as a reference for the strand-specific MLPA assay. As shown in Figure 7B for (CAG)5-transfected cells, the signal from antisense-specific probes is very low when compared with the signal from sense-specific probes. Similar effects were observed for all other cell lines transfected with EGFP-N3. Average signals from antisense-specific probes account for 0.02 fraction of the signal from sense-specific probes (Figure 7C). These results indicate that the level of antisense transcript is very low (∼2%) in comparison to sense transcript in all analyzed model cell lines, regardless of the repeat length, type and sequence.
Figure 7.
Identification of sense and antisense repeat transcripts in cells expressing long CUG and CAG repeats. (A) Schematic representation of strand-specific MLPA assay design with probes located in the EGFP sequence close to the position of the changeable repeat tract. Signal from the probes hybridizing to plasmid DNA represents equal dosage from sense and antisense strands. The reverse transcription (RT) was performed with DNase treated total RNA. The signal from probe hybridizing to cDNA representing sense and antisense (a-sense) transcripts is proportional to the cellular level of these transcripts. (B) Representative MLPA electrophoregrams show results obtained for: cDNA from untransfected cells where no signals from sense and antisense probes are detected (top panel), cDNA from cells transfected with pEGFP-derived plasmid encoding (CAG)5 where very low, barely detectable signals from antisense-specific probes (A1 and A2) are visible (middle panel) and pEGFP-N3 plasmid DNA, where comparable signals from both the sense- and the antisense-specific probes are shown (lower panel). The signal from sense (S1 and S2) and antisense (A1 and A2) probes are indicated. (C) Bar plots show fraction of signal from the antisense-specific probes normalized to average signal from the sense-specific probe obtained from five cDNA samples of SK-N-MC cells transfected with the pEGFP-N3 encoding: (CAG)5, (CAG)74, (CUG)5 and (CUG)74 and mutATXN3 (left-hand side) and from two samples of pEGFP-N3 plasmid DNA (right-hand side). (D) Representative images of RNA-FISH in cultured human fibroblasts using Cy3-labeled DNA/LNA probes (CAG)6-CA (upper rows) and (CTG)6-CA (lower rows). HD cells expressing 44 CAG repeats and SCA3 cells expressing 69 CAG repeats from, respectively, the HTT and the ATNX3 genes show nuclear retention of sense transcripts harboring the CAG repeat expansions. Such CAG RNA inclusions were not detected in the nuclei of normal and DM1 fibroblasts when FISH was carried out with the same CTG probe. The presence of CAG repeat RNA in the nuclear inclusions was confirmed by RNase A (no FISH signal) and by DNaseI treatment (presence of FISH signal). The antisense transcripts through the repeat region in mutant HTT and ATXN3 genes were not detected with CAG probe in the nuclear RNA inclusions of HD and SCA3 cells. In this assay, DM1 cells were included as a positive control to detect ribonuclear CUG repeat inclusions. CAG and CUG nuclear RNA foci (red); DAPI nuclear stain (blue).
We also used in situ RNA hybridization with both the CAG and the CTG probes to test for the presence of nuclear RNA foci containing sense or antisense transcripts. For the FISH assay, we selected human HD and SCA3 fibroblasts expressing mutant HTT and ATXN3 mRNAs containing 44 and 69 CAG repeats, respectively. As a positive control for the FISH, we chose DM1 cells in which nuclear deposition of expanded CUG repeats (CUGexp) from mutant DMPK transcripts has been previously shown (21,51). The FISH analysis using the CTG probe demonstrated that numerous nuclear RNA inclusions containing the CAG repeats were detected in the HD and SCA3 cell lines (Figure 7D, lower panels). We did not observe CAG repeat RNA in the nuclear inclusions in the FISH experiments performed after RNase A treatment, whereas, the inclusions remained after DNase treatment. The use of CAG probes in the FISH assay revealed a lack of CUG RNA inclusions in the same cell lines, which may indicate that the HTT and ATXN3 antisense transcripts spanning the repeat region were not present in amounts detectable by FISH (Figure 7D, upper panels). For comparison, we detected typical CUG repeat RNA nuclear foci in DM1 cells in the same FISH assay.
MBNL1 co-localizes with mutant repeats in DM1, HD and SCA3 cells and shows similar affinity to CAG and CUG repeat RNAs
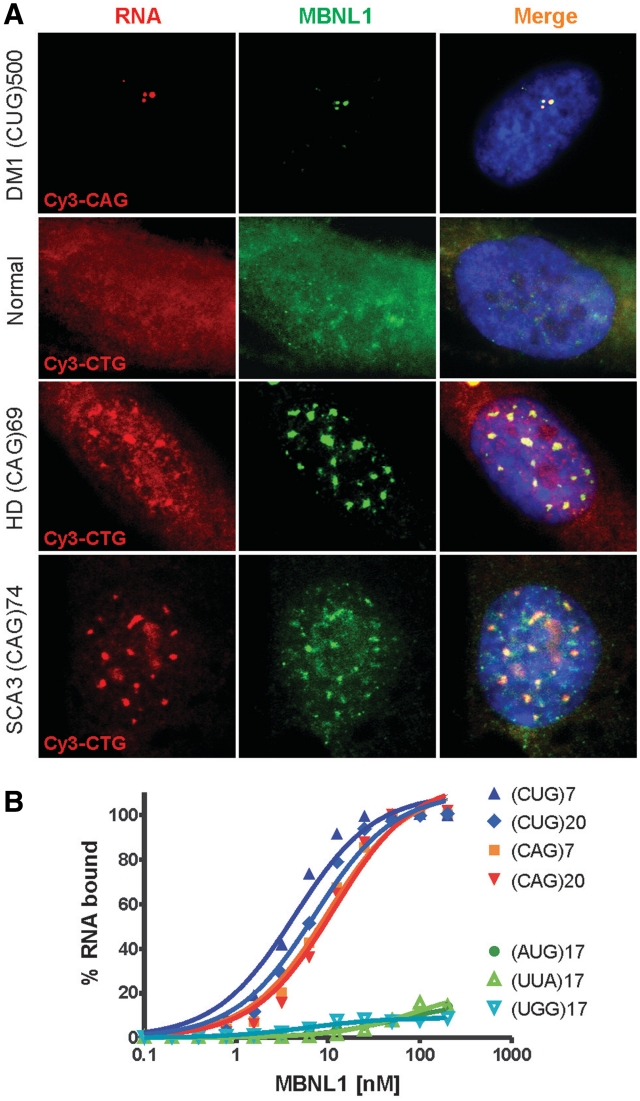
In DM1, the presence of CUG repeat RNA in the intranuclear foci is associated with sequestration of MBNL1 protein, which triggers aberrant splicing of specific genes (17,20,22). To investigate whether similar correlations occur between CAG repeat RNA foci formation and the splicing misregulation in HD and SCA3 cells, we performed combined FISH/immunostaining analysis to visualize localization of both CAG repeat RNA and MBNL1. As expected, the expression of mutant DMPK mRNA in DM1 fibroblast resulted in co-localization of punctate CUG repeat foci with MBNL1 (20). The MBNL1 splicing factor also co-localizes with mutant HTT and ATXN3 transcripts containing expanded CAG repeats presented in nuclear foci (Figure 8A). MBNL1 protein was present throughout the control fibroblasts lacking the CAG and CUG RNA expansions. This result indicates that the expression of either CUG or CAG expanded repeats may trigger sequestration of the MBNL1 protein.
Figure 8.
MBNL1 protein binds CAG and CUG repeats in vitro and co-localizes with repeat region of mutant HTT and ATXN3 transcripts in nuclear inclusions. (A) Combined FISH/IF assay using Cy3-labeled CAG probe in DM1 cells and CTG probe in normal, HD and SCA3 cells as well as rabbit polyclonal MBNL1 primary antibody (A2764) detected with secondary Cy2-labeled anti-rabbit antibody. Merged images show the colocalization of the MBNL1 protein with nuclear CAG repeat RNA inclusions in HD and SCA3 cells. Such colocalization is also demonstrated in DM1 cells with CUG RNA foci. In normal fibroblasts, MBNL1 is detected throughout the cell. DAPI staining (blue), MBNL1 (green), CAG and CUG nuclear RNA (red). (B) Results of filter binding assay to determine binding affinity of MBNL1 protein to 5′-end labeled transcripts of (CUG)7, (CUG)20, (CAG)7, (CAG)20, (AUG)17, (UUA)17 and (UGG)17. Affinity of MBNL1 to CUG and CAG RNA is similar with only slight preference for the CUG repeats.
Next, we determined the ability of MBNL1 splicing factor to bind CAG and CUG repeat RNAs in vitro. Using a filter binding assay (41), we compared the binding affinity of recombinant MBNL1 protein to the numerous synthetic oligoribonucleotides composed of different lengths of triplet repeats. The results shown in Figure 8B demonstrate that MBNL1 has similar affinity for CUG and CAG repeats with only a slight preference for the CUG repeats (KD(CUG)20 = 7.2 ± 1 nM versus KD(CAG)20 = 11.6 ± 1.8 nM). For shorter (CUG)7 RNA, the calculated KD was slightly lower (KD(CUG)7 = 4.3 ± 0.7 nM). The control triplet repeat RNAs, (AUG)17, (UUA)17 and (UGG)17, did not bind MBNL1 under our assay conditions. These results suggest that the observed co-localization of expanded CAG repeat RNAs with MBNL1 in HD and SCA3 cells is caused by direct RNA/protein interaction in nuclear foci and by depletion of the functional splicing factor in the nucleoplasm that leads to splicing misregulation of MBNL1-dependent transcripts.
DISCUSSION
Recent research has suggested a potential role for mutant RNA-containing CAG repeat expansions in the pathogenesis of poly-Q disorders (35,36). In this study, we performed comparative analysis of splicing misregulation in cells expressing exogenous and endogenous CAG or CUG repeat RNA to determine whether the expanded CAG repeat transcripts recapitulate some of the toxic features of CUG repeat RNA. We have shown that the expression of transcripts containing long CAG repeats results in RNA foci formation, MBNL1 co-localization and splicing aberrations similar to those detected for CUG repeat transcripts. We demonstrated abnormal alternative splicing using cellular model systems designed to express untranslated CAG and CUG repeats of various lengths. In those cells, we did not observe elevated expression of CUGBP1 protein (Supplementary Figure S6). The splicing defects were also induced by expression of exogenous mutant ATXN3 transcripts containing 69 CAG repeats in the translated sequence, and they were observed in cell lines derived from patients with SCA3 and HD. Given the similarity of the splicing defects caused by untranslated and translated CAG repeats, one could speculate that the presence of expanded CAG repeat RNA itself is sufficient to induce some pathogenic features of poly-Q expansion diseases.
Recently, involvement of antisense triplet repeat RNAs has been implicated in the pathogenesis and progression of SCA8, fragile X-associated tremor/ataxia syndrome (FXTAS) and Huntington's disease-like 2 (HDL2) (52,53). In this study, we also considered the possibility that bidirectional transcription through the repeat region resulting in high relative levels of the antisense versus sense transcripts could have influence on splicing misregulation which we attributed to CAG sense RNA. The presence of antisense CUG-containing transcripts was tested using a sensitive MLPA assay, and its potential participation in RNA foci formation was analyzed by RNA FISH. With those assays, we determined that (i) the average level of antisense CUG repeat transcripts was very low and accounted for about 2% of the CAG repeat transcripts, (ii) no antisense CUG repeat RNA foci formation was detected in cells expressing expanded CAG repeat RNA and (iii) distinct numerous CAG repeat RNA nuclear inclusions were visible in cells expressing mutant sense CAG transcripts. These results clearly indicate that the presence of minor but MLPA-detectable fractions of antisense transcripts did not localize within RNA foci. In DM1 cells, the presence of intranuclear foci mediates pathogenesis by affecting the accessibility of functional MBNL1 splicing factor, and, therefore, lack of such inclusions in tested cell lines strongly implies that misregulated alternative splicing was the result of factors other than antisense CUG transcripts.
Earlier studies have shown that DM-specific aberrant splicing patterns were reproduced in cell cultures in which endogenous MBNL1 was reduced by siRNAs (45) and in Mbnl1ΔE3/ΔE3 mice that lack most MBNL1 protein isoforms (54). Consistent with loss of the splicing factor, muscleblind knockout mice and transgenic DM1 HSALR mice expressing long CUG RNA repeats (LR) from the human skeletal actin promoter (HSA) demonstrated a similar phenotype, which clearly implies involvement of MBNL1 in DM pathogenesis (55). Our results reveal the occurrence of a DM1-characteristic pattern of splicing misregulation in the presence of CAG repeat RNA, and we propose this effect to be MBNL1-dependent and CUGBP1-independent. This proposition is consistent with the lack of CUGBP1 up-regulation and MBNL1 co-localization with mutant transcripts within nuclear foci. Similar splicing defects triggered by CUG and CAG repeats could be explained by the results of our in vitro analysis of the ability of MBNL1 to bind to these repeats as well as by previous reports (18,41,56). These results show that CUG and CAG repeats differ only slightly in their binding affinity for MBNL1. It is important to note, however, that even small changes in the cellular level of MBNL1 and toxic CUG repeat transcripts can create an imbalance in alternative splicing without changing the ability to form nuclear foci as observed in the DM1 HSALR mouse model (51) (X. Lin and C. Thornton, personal communication). This finding could explain our observation of splicing misregulation in the absence of foci formation using high concentrations of short synthetic RNAs (CUG)7, (CAG)7, (CUG)20 and (CAG)20 in HeLa cells. Therefore, a high concentration of the ORNs compensated for their reduced length and triggered the same alternative splicing aberrations as mutant repeat RNA. A similar observation was described earlier in a mouse model in which high expression of the DMPK transcript with short CUG repeats induced splicing abnormalities characteristic of DM1 (57). This unexpected effect of short repeat RNAs on splicing abnormalities should be taken into consideration when designing an antisense strategy against expanded CUG or CAG repeat transcripts. The chemistry of the oligomers could be an important factor in eliminating their undesirable effect on splicing. Two different types of antisense oligomers composed of CAG repeats, morpholinos (58) and 2′-O-metyl-phosphorotioates (59), were recently used to disturb the expanded CUG repeat/MBNL1 interaction in vivo and yielded significantly different splicing correction effects in the same mouse model of DM1.
Evidence for the contribution of CAG repeat RNA to the pathogenesis of poly-Q disorders has been provided by two earlier reports (35,36). Ho et al. (36) found that CAG repeat RNA mimics the effects of CUG mutant repeats on foci formation and MBNL1 co-localization. In their system, co-transfection of COSM6 cells with plasmids expressing long but interrupted CAG tracts and cTNT or INSR minigenes did not result in splicing alterations of the two tested pre-mRNAs. The splicing changes were detected only for long CUG transcripts. We believe that major sources of discrepancy between our results and those of Ho et al. (36) were cells used for transfections and different responsivness of endogenes and minigenes to mis-splicing triggers. In addition, differences in expression level between transgenes harboring different repeats could account for observed variations in alternative splicing abnormalities.
The contribution of expanded CAG repeat RNA to poly-Q toxicity was previously reported by Li et al. (35). In their Drosophila model, SCA3 expression of untranslated RNAs containing CAG repeats of pathogenic length was deleterious, indicating that SCA3 neurodegeneration may be manifested in the absence of poly-Q protein. Altering pure CAG repeat sequences to an interrupted tract within the poly-Q encoding region significantly mitigated the toxicity of the untranslated repeat RNA. Due to the fact that flies expressing pathogenic SCA3 poly-Q proteins encoded by interrupted repeat transgenes developed degenerative but less severe phenotypes than those of SCA3 poly-Q pure CAG, Li et al. (35) suggested that the RNA contributes to a shift of the toxicity curve rather than being solely responsible for toxicity. The results of up-regulation of ‘muscleblind’ activity on poly-Q-triggered toxicity in SCA3 Drosophila were strikingly different from what has been so far found for CUG repeat mutation. Surprisingly, poly-Q-triggered degeneration was enhanced after overexpression of either MblA or human MBNL1 suggesting that the protein may act on the RNA to add to poly-Q toxicity.
The splicing misregulation of the Clcn1 and Mbnl1 genes was recently described in the mouse model of Kennedy's disease AR113Q in which the AR gene expresses 113 CAG repeats from the mouse Ar promoter (60). The authors argued that the mis-splicing was triggered by mutant protein. However, the possibility that the poly-Q protein may exert effects on RNA splicing through the disruption of the organization of nuclear bodies seems rather unlikely in our case. We do not observe a general deregulation of the alternative splicing process and only specific mRNAs are affected. Our study shows that the expression of mutant transcripts alone is sufficient to trigger the mis-splicing events and that splicing deregulation is CAG repeat-dependent.
Lastly, as the formation of intranuclear inclusion bodies in HD cells has been suggested to be a beneficial response to the expression of toxic poly-Q (61), a similar protective effect might be attributed to nuclear deposition of mutant RNA. It is not known whether RNA foci formed by CAG repeats represent a fraction of transcripts designated for nuclear degradation or whether they become transiently blocked in the nucleus as soluble RNA–MBNL1 complexes that eventually reach the cytoplasm becoming available for toxic protein synthesis. The results of this and previous studies indicate that dwelling on toxic RNA gain-of-function mechanisms in mutant CAG-related disease might help explain some unknowns and uncertainties in our search for a cure of complex poly-Q disorders.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ministry of Science and Higher Education (grants PBZ-MNiI-2/1/2005, N301-112-32/3910, N302-260938, N N302 299536, N N302 278937); Operational Programme ‘Innovative economy' (POIG.01.03.01-30-098/08). Funding for open access charge: POIG.01.03.01-30-098/08.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Tom Cooper for his support, helpful suggestions and stimulating discussions. We thank Dr Charles A. Thornton for rabbit MBNL1 antibody A2764. The authors would like to thank also Maciej Figiel and Pawel Switonski for their comments on the article.
REFERENCES
- 1.Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 2.de Leon MB, Cisneros B. Myotonic dystrophy 1 in the nervous system: from the clinic to molecular mechanisms. J. Neurosci. Res. 2008;86:18–26. doi: 10.1002/jnr.21377. [DOI] [PubMed] [Google Scholar]
- 3.Underwood BR, Rubinsztein DC. Spinocerebellar ataxias caused by polyglutamine expansions: a review of therapeutic strategies. Cerebellum. 2008;7:215–221. doi: 10.1007/s12311-008-0026-z. [DOI] [PubMed] [Google Scholar]
- 4.Galvao R, Mendes-Soares L, Camara J, Jaco I, Carmo-Fonseca M. Triplet repeats, RNA secondary structure and toxic gain-of-function models for pathogenesis. Brain Res. Bull. 2001;56:191–201. doi: 10.1016/s0361-9230(01)00651-7. [DOI] [PubMed] [Google Scholar]
- 5.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merienne K, Trottier Y. SCA8 CAG/CTG expansions, a tale of two TOXICities: a unique or common case? PLoS Genet. 2009;5:e1000593. doi: 10.1371/journal.pgen.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 9.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum. Mol. Genet. 2006;15 doi: 10.1093/hmg/ddl181. (Spec No. 2), R162–R169. [DOI] [PubMed] [Google Scholar]
- 10.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 11.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tapscott SJ, Thornton CA. Biomedicine. Reconstructing myotonic dystrophy. Science. 2001;293:816–817. doi: 10.1126/science.1063517. [DOI] [PubMed] [Google Scholar]
- 13.Osborne RJ, Lin X, Welle S, Sobczak K, O'Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 15.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 16.Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, Dirksen RT, Takahashi MP, Dulhunty AF, Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 17.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 18.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. Embo J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler TM, Krym MC, Thornton CA. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007;17:242–247. doi: 10.1016/j.nmd.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 23.Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circ. Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 24.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J. Biol. Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cummings CJ, Zoghbi HY. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 29.Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 30.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 31.Lunkes A, Mandel JL. Polyglutamines, nuclear inclusions and neurodegeneration. Nat. Med. 1997;3:1201–1202. doi: 10.1038/nm1197-1201. [DOI] [PubMed] [Google Scholar]
- 32.Satoh M, Shimada A, Kawamura N, Chiba Y, Yoshikawa K, Ishii S, Furukawa A, Kumagai N, Hosokawa M. Neuronal toxicity of expanded polyglutamine depends on intracellular distribution in addition to the expression level. Neuropathology. 2008;28:485–496. doi: 10.1111/j.1440-1789.2008.00908.x. [DOI] [PubMed] [Google Scholar]
- 33.King MA, Goemans CG, Hafiz F, Prehn JH, Wyttenbach A, Tolkovsky AM. Cytoplasmic inclusions of Htt exon1 containing an expanded polyglutamine tract suppress execution of apoptosis in sympathetic neurons. J. Neurosci. 2008;28:14401–14415. doi: 10.1523/JNEUROSCI.4751-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 35.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 37.Birman S. Neurodegeneration: RNA turns number one suspect in polyglutamine diseases. Curr. Biol. 2008;18:R659–R661. doi: 10.1016/j.cub.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 38.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the ‘stretched U-U wobble': implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ordway JM, Detloff PJ. In vitro synthesis and cloning of long CAG repeats. Biotechniques. 1996;21:609–610, 612. doi: 10.2144/96214bm08. [DOI] [PubMed] [Google Scholar]
- 43.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc. Natl Acad. Sci. USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozlowski P, Roberts P, Dabora S, Franz D, Bissler J, Northrup H, Au KS, Lazarus R, Domanska-Pakiela D, Kotulska K, et al. Identification of 54 large deletions/duplications in TSC1 and TSC2 using MLPA, and genotype-phenotype correlations. Hum. Genet. 2007;121:389–400. doi: 10.1007/s00439-006-0308-9. [DOI] [PubMed] [Google Scholar]
- 47.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borrell-Pages M, Zala D, Humbert S, Saudou F. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol. Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 50.Hino S, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, Chihara K, Aoki Y, Nakamori M, Takahashi MP, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 51.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 53.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J. Biol. Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 55.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 56.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 57.Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z, Wang AM, Robins DM, Lieberman AP. Altered RNA splicing contributes to skeletal muscle pathology in Kennedy disease knock-in mice. Dis. Model Mech. 2009;2:500–507. doi: 10.1242/dmm.003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.