Abstract
In embryonic stem cells (ESCs), Wnt-responsive development-related genes are silenced to maintain pluripotency and their expression is activated during differentiation. Acetylation of histones by histone acetyltransferases stimulates transcription, whereas deacetylation of histones by HDACs is correlated with transcriptional repression. Although Wnt-mediated gene transcription has been intimately linked to the acetylation or deacetylation of histones, how Wnt signaling regulates this type of histone modification is poorly understood. Here, we report that Smek, a regulatory subunit of protein phosphatase 4 (PP4) complex, plays an important role in histone deacetylation and silencing of the Wnt-responsive gene, brachyury, in ESCs. Smek mediates recruitment of PP4c and HDAC1 to the Tcf/Lef binding site of the brachyury promoter and inhibits brachyury expression in ESCs. Activation of Wnt signaling during differentiation causes disruption of the Smek/PP4c/HDAC1 complex, resulting in an increase in histones H3 and H4 acetylation at the brachyury gene locus. These results suggest that the Smek-containing PP4 complex represses transcription of Wnt-responsive development-related genes through histone deacetylation, and that this complex is essential for ESC pluripotency maintenance.
Keywords: embryonic stem cell, Smek, HDAC1, protein phosphatase 4 complex, Wnt signaling, differentiation
Introduction
Once activated by the Wnt-signaling pathway, β-catenin is stabilized by inactivation of GSK-3β and enters the nucleus, where it interacts with DNA-bound Tcf/Lef factors to activate transcription of Wnt target genes 1. Several studies revealed a correlation between the acetylation of histones and transcriptional activity. Acetylation of nucleosomal histones by histone acetyltransferases stimulates transcription, whereas deacetylation of nucleosomal histones by HDACs is correlated with transcriptional repression 2, 3. The transcription induced by Wnt signaling is accompanied by histone acetylation 4. Inhibition of HDAC activity enhances Wnt-mediated transcriptional activation. In contrast, overexpression of HDAC1 represses Wnt-induced transcription by deacetylating histone 5. A known regulatory mechanism of HDAC function in transcriptional repression of Wnt-responsive genes is through interacting with Tcf/Lef proteins 6. β-catenin converts Tcf/Lef transcriptional repression complex containing HDAC1 into a transcriptional activator 6. However, a recent study has shown that overexpression of β-catenin in the presence of extracellular Wnt inhibitor, Dickkopf-1 (Dkk1), does not cause brachyury expression during differentiation of ESCs 7. These results suggest a possibility that the silencing of Wnt-mediated development-related genes in ESC requires transcriptional repressors that are affected by Wnt signaling upstream of β-catenin.
Smek1 and Smek2 (also termed PP4R3α and PP4R3β, respectively) are regulatory subunits of PP4 complex 8, 9. Smek functions in a variety of molecular processes, including regulation of MEK in Dictyostelium cells 10, dephosphorylation of γ-H2AX in somatic mammalian cells 9, transcriptional modulation of DAF-16/FOXO3a in C. elegans 11, and regulation of the intracellular localization of Miranda in Drosophila neuroblasts 12. However, the role of Smek in molecular and cellular processes in ESCs is unknown. We show here that a Smek/PP4c complex binds to HDAC1 and occupies the Tcf/Lef binding region of brachyury promoter. Furthermore, activation of Wnt signaling promotes disruption of the Smek/PP4c/HDAC1 complex, resulting in transcriptional activation of the brachyury gene. These results suggest that Smek/PP4c functions as a transcriptional repressor to silence Wnt-responsive development-related gene and is required for ESC self-renewal.
Results
Smek is required for ESC self-renewal by suppressing differentiation into mesoderm
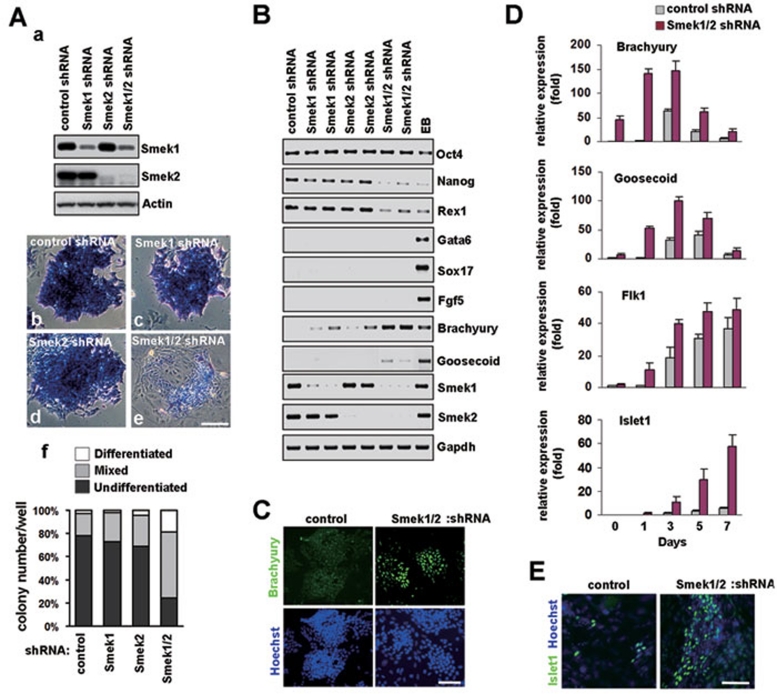
In order to elucidate functions of Smek in ESCs, ESC lines expressing stable shRNAs targeting Smek1, Smek2, or both (Smek1/2 shRNA), were generated. These shRNAs effectively inhibited expression of their targets (Figure 1A, a). Pluripotency was maintained in ESCs expressing shRNA for either Smek1 or Smek2, as cells revealed no change in morphology or alkaline phosphatase staining (Figure 1A, c and d) when compared with control shRNA-expressing ESCs (Figure 1A, b). Remarkably, double knockdown of Smek1 and Smek2 resulted in ESC colonies composed of morphologically altered cells and reduced alkaline phosphatase staining (Figure 1A, e), indicative of differentiation. Quantitative analysis of colonies with alkaline phosphatase staining showed that double knockdown, but not single knockdown, led to differentiation of ESCs (Figure 1A, f). ESCs are capable of producing ectodermal, endodermal, and mesodermal germ layers 13. To determine whether loss of pluripotency in Smek1/2 knockdown cells was caused by differentiation into specific lineages, we analyzed the expression of markers of stemness (oct4, nanog, and rex1), endoderm (gata6 and sox17), ectoderm (fgf5), and mesoderm (brachyury and goosecoid) (Figure 1B). The individual ESC lines expressing Smek1/2 shRNAs showed significant increases in brachyury and goosecoid mRNA levels. In these cells, nanog and rex1 mRNA levels decreased compared with cells expressing control shRNA, Smek1 shRNA, or Smek2 shRNA. In addition, brachyury-positive cells increased in Smek1/2 knockdown ESC line (Figure 1C). To further confirm mesoderm differentiation in ESCs deficient in both Smek1 and Smek2, we next analyzed the differentiation into mesoderm-derived lineage 14, 15. Expression levels of brachyury and goosecoid, and the mesodermal lineage-progenitor markers flk1 and islet1, were compared by real-time PCR between cultures expressing control and Smek1/2 shRNA during EB formation (Figure 1D). Notably, Smek1/2 knockdown caused significant increase of islet1 expression. Immunostaining with anti-islet1 antibody in the cells re-plated from dissociated EBs between days 6 and 8, exhibited increased number of islet1-positive cells in cultures lacking expression of both Smek1 and Smek2 (Figure 1E). In addition, overexpression of Smek1 and Smek2 reduced expression of mesoderm markers brachyury, goosecoid, flk1, and islet1 during EB formation when compared with control cells (Supplementary information, Figure S1). Expression of endoderm and ectoderm markers gata6, sox17, and fgf5 did not change (data not shown). Taken together, our results suggest that Smek is required for suppression of mesoderm differentiation to maintain ESC pluripotency.
Figure 1.
Smek1 and Smek2 are required for maintenance of pluripotency in ESCs. (A) Cells expressing control shRNA, or shRNA for Smek1, Smek2, or both, were lysed and subjected to western blot analysis with anti-Smek1 and -Smek2 antibodies (a). Alkaline phosphatase (AP) staining (blue) of ESCs expressing control shRNA (b), Smek1 shRNA (c), Smek2 shRNA (d), or both (e). Notably, double knockdown for both Smek1 and Smek2 reduces AP expression and morphological change of ESCs, reflective of differentiation. ESC status was evaluated by quantifying the number of colonies consisting of AP-negative cells (differentiated), positive cells (undifferentiated), and both AP-negative and -positive cells (mixed) (f). Scale bars, 100 μm. (B) Expression of genes characteristic of ESC stemness (oct4, nanog, and rex1), endoderm (gata6 and sox17), primitive ectoderm (fgf5), and mesoderm (brachyury and goosecoid) were measured by RT-PCR in ESC lines cultured in the presence of LIF. Gapdh was used as an internal control. Two independent ESC lines were examined for each shRNA construct. EB was used as a positive control for differentiation of ESC. (C) ESC lines expressing control or Smek1/2 shRNA were stained for brachyury. Scale bar, 100 μm. (D) Quantitative real-time PCR analysis of early mesodermal and cardiac progenitor markers during differentiation of ESCs expressing control shRNA or Smek1/2 shRNA. (E) Cells dissociated from EB expressing control or Smek1/2 shRNA were cultured on gelatin-coated plates and subjected to immunostaining with antibodies against a cardiac progenitor marker islet1. Hoechst dye was used as a counterstaining. Scale bar, 100 μm. Depletion of Smek facilitates differentiation into islet1-positive cardiac progenitors from mesoderm.
Smek represses transcriptional activity of Wnt/β-catenin target genes
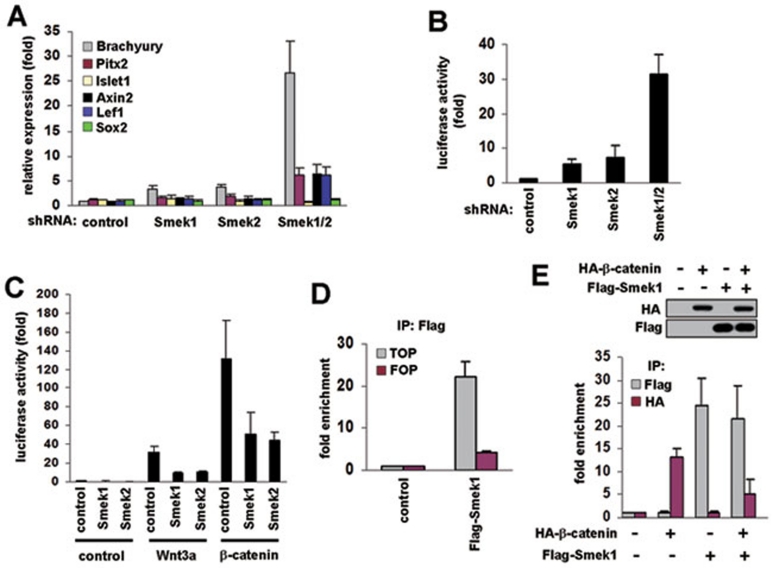
As described previously, activation of Wnt/β-catenin induces the differentiation of ESCs into mesoderm, and antagonizes neuroectoderm differentiation 16. Activation of Wnt/β-catenin signaling leads to expansion of islet-positive progenitors 17, 18. Smek1/2 knockdown in ESCs causes differention of mesoderm lineages. However, Smek1/2 knockdown prevents differentiation of ESCs into neural/neuronal lineages (Supplementary information, Figure S2). These results raise the possibility that depletion of Smek1/2 might lead to sustained Wnt/β-catenin-signaling activation. To this end, we analyzed transcriptional activity that is directed by Wnt/β-catenin signaling. Wnt target genes, brachyury, pitx2, islet1, axin2, lef1, and sox2, were analyzed by real-time PCR in ESCs expressing control, Smek1, Smek2, or Smek1/2 shRNAs (Figure 2A). Quantitative real-time PCR revealed that expression of brachyury, pitx2, axin2, and lef1 significantly increased in ESCs expressing Smek1/2 shRNA compared with control cells, whereas sox2 had no significant change. Although depletion of Smek during differentiation increased islet1 expression (Figure 1D), its expression in undifferentiated state did not change. Activity of a luciferase reporter SuperTOPFLASH, which contains 8× of the Tcf/lef binding site, further confirms that Smek deficiency induces transcriptional activation of Wnt/β-catenin target genes (Figure 2B). Although double knockdown for Smek1 and Smek2 increases remarkably transcriptional activity of brachyury and TOPFLASH and leads to mesoderm differentiation, each single knockdown ESCs also showed increase in transcriptional activity for brachyury and TOPFLASH. This implies that Smek1 and Smek2 are analogous and redundant for the function that regulates transcription. To confirm this, TOPFLASH and plasmids containing cDNA encoding Smek1 or Smek2 were transiently transfected with or without β-catenin or Wnt3a into HEK293T cells. Luciferase-activity assays showed that overexpression of Smek1 or Smek2 significantly reduced the transcriptional activity stimulated by Wnt3a or β-catenin (Figure 2C).
Figure 2.
Smek represses the transcription of Wnt target genes. (A) Expression of Wnt/β-catenin target genes, brachyury, pitx2, islet1, axin2, lef1, and sox2, was analyzed by quantitative real-time PCR in undifferentiated ESC lines. (B) ESC lines were transfected with the TOPFLASH reporter and were cultured under ESC conditions for 2 days. Luciferase activity in each sample was measured as described in the methods. Data are presented as mean ± S.D. of three independent experiments. (C) Smek1, Smek2, Wnt3a, β-catenin, or control vectors were transfected into HEK293T cells with TOPFLASH reporter. Data are presented as mean ± S.D. of four independent experiments. (D) The plasmid encoding Flag-tagged Smek1 was transiently transfected into HEK293T cells with TOPFLASH or FOPFLASH. Cells were lysed 48 h after transfection and were subjected to ChIP with anti-Flag antibody. Quantitative real-time PCR using a primer set, which is specific for −20 bp upstream and +170 bp downstream of the luciferase transcriptional start site, show that enrichment of Smek at Tcf/Lef binding sites of TOPFLASH DNA compared with that of mutated Tcf/Lef binding sites of FOPFLASH DNA. (E) HEK293T cells transfected with TOPFLASH and plasmids expressing control, Flag-Smek1, and HA-β-catenin, were subjected to western blot analysis (top) and ChIP with anti-HA or Flag antibodies, and the precipitated DNAs were analyzed by quantitative real-time PCR using a primer set as in D (bottom).
Since β-catenin is a key player in the Wnt-signaling cascade, it is possible that the knockdown of Smek could increase the amount of nuclear β-catenin. However, ESCs expressing Smek1/2 shRNAs did not alter β-catenin localization (Supplementary information, Figure S3A–S3C). Smek1 and Smek2 proteins are predominantly localized in the nucleus in ESCs (Supplementary information, Figure S3D). Thus, Smek may contribute to the immediate regulation of gene transcription. To this end, ChIP was performed in HEK293T cells transfected with Flag-Smek1 and TOPFLASH or FOPFLASH reporter construct. The DNA fragment containing the Tcf/Lef binding site in the TOPFLASH plasmid was enriched in DNA precipitated with anti-Flag antibody from HEK293T cells transfected with Flag-Smek1 and TOPFLASH as compared to that of cells transfected with Flag-Smek1 and FOPFLASH, which lacks the Tcf/Lef binding site (Figure 2D). This indicates that Smek may associate with gene loci containing the Tcf/Lef binding site. Since β-catenin binds to Tcf/Lef binding site to activate gene transcription 6, repressive effect of Smek (Figure 2C) could be due to inhibition of the recruitment of β-catenin to its binding site. Indeed the enrichment of HA-β-catenin at TOPFLASH DNA was decreased in HEK293T cells transfected with both Flag-Smek1 and HA-β-catenin as compared to cells transfected with HA-β-catenin alone, whereas the enrichment of Flag-Smek1 revealed similar levels in the presence or absence of HA-β-catenin (Figure 2E).
The Smek/PP4c complex directly represses brachyury expression through recruitment of HDAC1
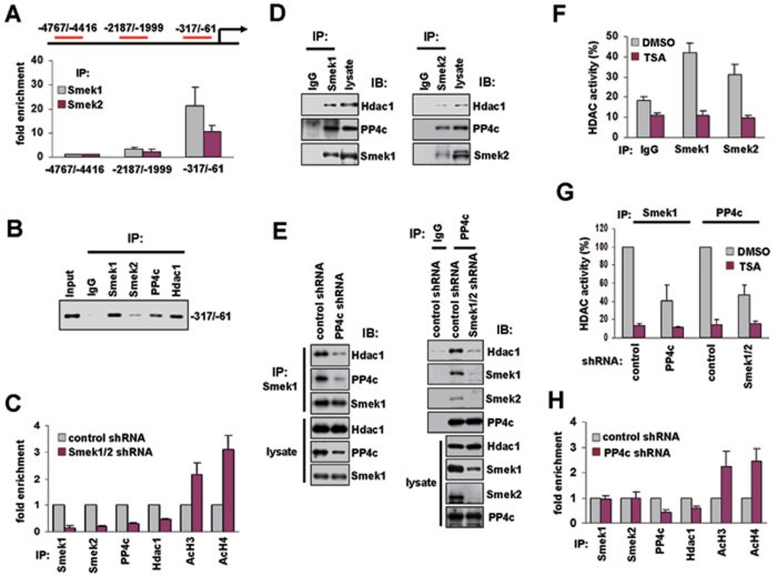
The transcription factor brachyury has been implicated in mesoderm formation during early development 19, and we showed that brachyury expression was remarkably induced by depletion of Smek (Figure 1A and 1B). Since brachyury has Tcf/Lef binding sites at its promoter region 20, it is possible that Smek proteins could occupy the promoter of brachyury gene. To this end, ChIP was performed in ESCs using antibodies against Smek1 and Smek2. Quantitative real-time PCR was used to monitor the enrichment of specific promoter sequences in the precipitated DNA. Q-PCR results suggested that the brachyury gene was occupied by Smek1 and Smek2 around 300 bp upstream of the transcription start site, which contains two Tcf/Lef binding sites (Figure 3A). In addition, Smek1 and Smek2 were enriched at the Tcf/Lef binding region of islet1 and pitx2, but not sox2 (Supplementary information, Figure S4)
Figure 3.
The brachyury promoter is occupied by the Smek/PP4c complex containing HDAC1. (A) Quantitative real-time PCR was performed using ChIP DNA with anti-Smek1 and -Smek2 antibodies and the primer set specific for regions of the brachyury promoter as indicated at the top of the figure. Data are presented as fold change as compared to cell lysates precipitated with control antibody. (B) PCR products amplified with the −317/−61 primer from control input DNA and ChIP DNA with control IgG, anti-Smek1, Smek2, PP4c, and HDAC1 antibodies show that PP4c and HDAC1 are associated with promoter of brachyury gene occupied by Smek1 and Smek2. (C) Depletion of Smeks inhibits the occupancy of PP4c and HDAC1, and increases the acetylation of histones H3 and H4. ChIP DNA precipitated with the indicated antibodies from ESCs expressing control or Smek1/2 shRNA and analyzed by quantitative real-time PCR with the −317/−61 primer set as in panel (A). (D) Binding of HDAC1 to Smek/PP4c complex. Nuclear extracts from ESCs were immunoprecipitated with anti-Smek1 (left) and Smek2 (right) antibodies and subjected to western blot analysis using anti-HDAC1 or PP4c antibody. (E) Western blot analysis of anti-Smek1 and anti-PP4c immunoprecipitates in ESCs knocked down for PP4c (left) and both Smek1 and Smek2 (right). (F, G) HDAC activities in immunoprecipitates from panels (D and E), respectively, were measured using a fluorescence-producing substrate in the presence or absence of TSA. Data were normalized to the HDAC activity of the anti-HDAC1 immunoprecipitate, and are presented as mean ± S.D. of three independent experiments. (H) ESCs expressing control and PP4c shRNA were subjected to ChIP using the indicated antibodies and analyzed by quantitative real-time PCR with the −317/−61 primer set.
PP4c interacts with HDAC1 21. Tcf/Lef proteins interact with HDAC1 5, and this interaction was reduced when Smek expression was decreased (Supplementary information, Figure S5). We found that PP4c and HDAC1 were present at the brachyury promoter occupied by Smek (Figure 3B). HDAC1 removes the acetylation of histones H3 and H4 and represses gene transcription 22. Therefore, we next tested if binding of Smek at the brachyury promoter correlated with the presence of HDAC1 and/or acetyl histones. ESCs were subjected to ChIP analysis using anti-Smek1, Smek2, HDAC1, PP4c, and acetyl histones H3 and H4 antibodies. Interestingly, acetyl histones H3 and H4 were increased at the brachyury promoter when ChIP was performed using cell lysates from ESCs expressing Smek1/2 shRNA, whereas binding of PP4c and HDAC1 was reduced (Figure 3C). These data suggest that Smek mediates recruitment of HDAC1 and PP4c to the brachyury promoter to suppress transcription of brachyury.
To gain insight into the functional significance of the Smek/PP4c phosphatase complex on transcriptional repression through histone deacetylation, we examined the binding of Smek/PP4c to HDAC1. HDAC1 and PP4c were detectable in anti-Smek immunoprecipitates (Figure 3D). Notably, the protein level of HDAC1 was reduced both in anti-Smek1 immunoprecipitates from ESC lines expressing PP4c shRNA (Figure 3E, left) and in anti-PP4c immunoprecipitates from ESC lines expressing Smek1/2 shRNA (Figure 3E, right) when compared with that of ESC lines expressing control shRNA. This result suggests that assembly of the HDAC1 complex requires both PP4c and Smek. We next tested if the Smek/PP4c complex has deacetylase activity using HDAC-activity assay. Anti-Smek1 or Smek2 immunoprecipitates exhibited an increase in HDAC activity compared with those of IgG immunoprecipitates (Figure 3F). HDAC inhibitor, TSA, inhibited this activity, indicating specific HDAC activity of Smek complexes. Furthermore, ESC lines expressing PP4c or Smek1/2 shRNA exhibited reduced HDAC activity in anti-Smek1 or anti-PP4c immunoprecipitates (Figure 3G) because of reduced binding of HDAC1 (Figure 3E). We also found that Smek-mediated deacetylation of histones H3 and H4 requires PP4c, using ChIP assay that was performed with the indicated antibodies in ESC lines expressing PP4c shRNA (Figure 3H). Quantitative real-time PCR revealed that knockdown of PP4c reduces enrichment of HDAC1 and increases acetyl histones H3 and H4 at the brachyury promoter when compared with those of control shRNA-expressing ESC. Interestingly, knockdown of PP4c did not affect binding of Smek1 and Smek2 to the brachyury promoter region, whereas depletion of Smek led to decreased binding of PP4c (Figure 3C). This indicates that Smek mediates the recruitment of PP4c and HDAC1 proteins to the brachyury promoter. These results, together with the observation of the induction of brachyury by the knockdown of PP4c (Supplementary information, Figure S6), demonstrate that the Smek/PP4c/HDAC1 complex suppresses transcription of brachyury through deacetylation of histones H3 and H4 in undifferentiated ESCs.
Wnt, not β-catenin, disrupts the Smek/PP4c/HDAC1 complex
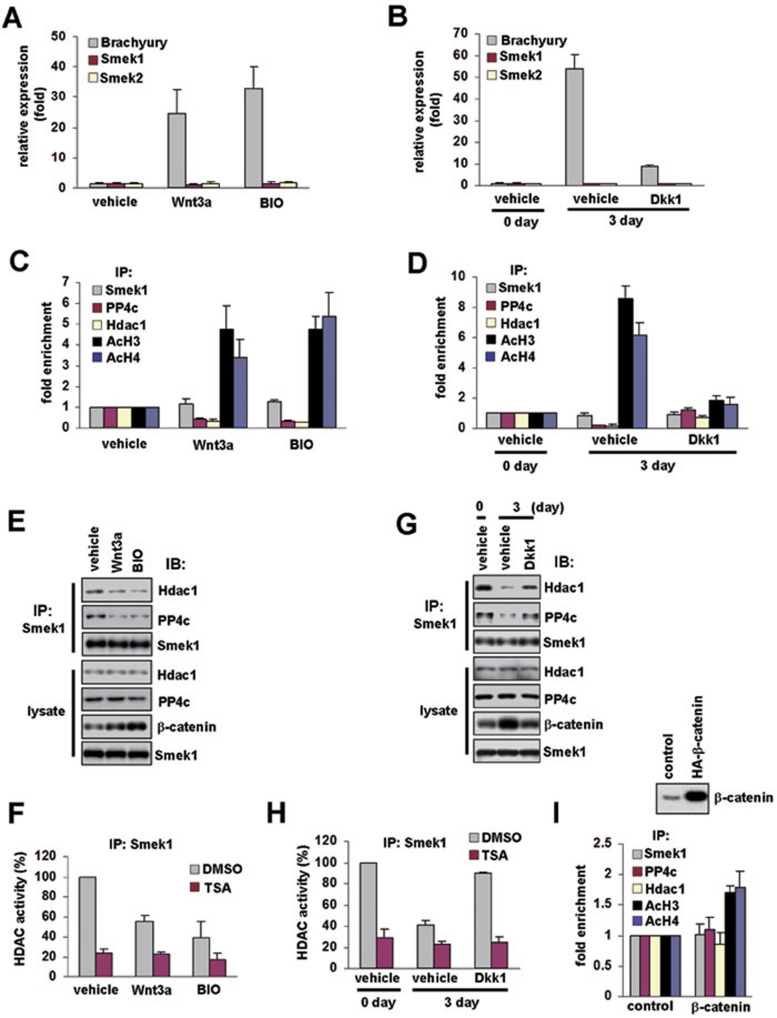
Although Smek represses Wnt3a-mediated transcriptional activation (Figure 2C), high concentration of Wnt3a protein overcomes the repressive effect (Supplementary information, Figure S7). Therefore, we asked if the Smek repression complex is regulated by Wnt. To test this, ESCs were exposed to recombinant Wnt3a or GSK-3 inhibitor BIO to activate Wnt signaling, and to recombinant Dkk1 to inhibit activation of Wnt signaling during EB formation. ESCs and EBs incubated with Wnt3a, BIO, or Dkk1 were initially analyzed by real-time PCR for brachyury expression and ChIP assays for association of the Smek complex to its promoter. ESCs treated with Wnt3a protein or BIO exhibited significant increase in brachyury expression compared with ESCs treated with vehicle (Figure 4A). Addition of Dkk1 during EB formation significantly inhibited brachyury expression induced by EB formation (Figure 4B). ChIP analysis using anti-Smek1, PP4c, HDAC1, and acetyl histones H3 and H4 antibodies revealed that the amount of PP4c and HDAC1 bound to brachyury promoter region was reduced in ESCs treated with Wnt3 or BIO when compared with those of ESCs treated with vehicle (∼2.8- and ∼3.3-fold, respectively; Figure 4C). Likewise, the amount of acetyl histones H3 and H4 (∼4.7- and ∼3.3-fold, respectively) were increased (Figure 4C). Similar to what was observed when Wnt3a or BIO was added to ESCs, enrichment of PP4c and HDAC1 at the brachyury promoter in EBs was reduced compared with those of undifferentiated ESCs (∼5.0- and ∼5.8-fold, respectively), whereas that of acetyl histones H3 and H4 increased by ∼8.1- and ∼6.5-fold, respectively (Figure 4D). Addition of Dkk1 during EB formation inhibited these alterations at the brachyury promoter (Figure 4D). The occupancy of PP4c, HDAC1, and acetyl histones at brachyury promoter region is consistent with the brachyury expression pattern shown in Figure 4A and 4B.
Figure 4.
Wnt, but not β-catenin, disrupts the Smek/PP4c/HDAC1 complex to activate transcription of the brachyury gene. ESC lines expressing control shRNA or double shRNA were incubated with vehicle, Wnt3a (50 ng/ml), or BIO (5 μM) for 6 h (A, C, E, and F), or were cultured in hanging drops of medium in the presence of vehicle or Dkk1 (100 ng/ml) for 3 days (B, D, G, and H). (A) Wnt3a or BIO activates brachyury but not Smek1 or Smek2 expression. ESC lines were treated with vehicle, Wnt3a (50 ng/ml), or BIO (5 μM) for 6 h. Transcript levels of brachyury, Smek1, and Smek2 were determined buy quantitative real-time PCR. (B) Dkk1 inhibits brachyury expression during ESC differentiation. ESCs were cultured in hanging drops of medium for EB formation in the presence of vehicle or Dkk1 (100 ng/ml) for 3 days. Transcript levels of brachyury, Smek1, and Smek2 were determined by quantitative real-time PCR. (C) Activation of Wnt signaling decreases the occupancy of HDAC1 and PP4c to the brachyury promoter, and increases histones H3 and H4 acetylation. Smek1 occupancy was not affected. The occupancy of Smek1, PP4c, HDAC1 and histones H3 and H4 acetylation was measured by ChIP followed by quantitative real-time PCR analysis with primer set of −317/−61 site of the brachyury promoter. (D) PP4c and HDAC1 occupancy at the brachyury promoter decreased during ESC differentiation, while histones H3 and H4 acetylation is increased. Dkk1 inhibits these changes during ESC differentiation. Smek1 occupancy was not affected. (E) Treatment with Wnt3a or BIO in ESCs decreases interaction of HDAC1 and PP4c proteins with Smek1. Lysates from ESCs were immunoprecipitated with anti-Smek1 and then subjected to western blot analysis using anti-Smek1, HDAC1, and PP4c antibodies. (F) HDAC activities in Smek1-immunoprecipitates from (E) were measured by a HDAC-activity assay in the presence or absence of TSA. Data are presented as mean ± S.D. of three independent experiments. (G) HDAC1 and PP4c associated with Smek1 decreased during ESC differentiation, while addition of Dkk1 inhibits the decrease of association of HDAC1 and PP4c with Smek1. (H) HDAC activities in immunoprecipitates from panel (G) were measured by a HDAC-activity assay in the presence or absence of TSA. Data are presented as mean ± S.D. of three independent experiments. (I) β-catenin does not affect the occupancy of PP4c and HDAC1 at the brachyury promoter. ESCs were transiently transfected with control vector or HA-β-catenin construct, and were then subjected to ChIP with the indicated antibodies followed by quantitative real-time PCR analysis with −317/−61 primer set at the brachyury promoter (top). Data are presented as mean ± S.D. from three independent experiments. Western blot analysis from nuclear extracts reveals the protein level of β-catenin (top).
The enrichment of Smek1 (Figure 4C and 4D) or Smek2 (data not shown) at the brachyury promoter region was unchanged in both ESCs and EBs regardless of Wnt-signaling status. This suggests that activation of Wnt signaling induces dissociation of PP4c and HDAC1 from Smek. To test this idea, the interaction of PP4c and HDAC1 with Smek1 was analyzed using immunoprecipitation with anti-Smek1 antibody. When cells were incubated with Wnt3a or BIO, Smek1 immunoprecipitates contained reduced levels of HDAC1 and PP4c (Figure 4E). Likewise, Wnt3a and BIO consistently resulted in decreased HDAC activity of Smek1 immunoprecipitates (Figure 4F). Binding of PP4c and HDAC1 to Smek1 in ESCs was significantly reduced during EB formation (Figure 4G). Addition of Dkk1 during EB formation inhibited the dissociation of PP4c and HDAC1 from Smek1 (Figure 4G), and rescued the HDAC activity of Smek1 complex (Figure 4H). Wnt3a and BIO lead to accumulation of β-catenin in the nucleus, which could potentially cause the disruption of Smek complex to induce brachyury expression. However, overexpression of β-catenin in ESCs did not interfere with the occupancy of PP4c and HDAC1 at the brachyury promoter (Figure 4I). In addition, β-catenin overexpression does not significantly increase brachyury expression compared with Wnt3a treatment (Supplementary information, Figure S8). Taken together, these data suggest that the repression activity of the Smek complex for brachyury expression is specifically inhibited by a β-catenin-independent mechanism of the Wnt-signaling pathway.
Discussion
Activation of Wnt signaling increases expression of pluripotent factors, nanog and oct4 23, 24. It can also activate expression of mesodermal differentiation factors, including brachyury 17, 20, 25. Although Wnt signaling in ESCs directs two opposite outputs, enhancement of self-renewal and differentiation, Wnt-responsive differentiation factors should be transcriptionally repressed to maintain pluripotency. Here, we report Smek as a specific transcriptional repressor for ESC differentiation. We show that Smek is required for repression of brachyury gene expression, a DNA-binding transcription factor that is a direct target of Wnt signaling and is required for mesoderm formation. This repression occurs when the Smek/PP4c complex binds HDAC1. Deficiency of either Smek or PP4c displaces HDAC1 from the brachyury promoter and increases its transcription in ESCs. Furthermore, loss of pluripotency in either Smek- or PP4c-deficient ESCs suggests that the PP4 complex has an important role in maintaining pluripotent ESCs.
Nuclear β-catenin converts the Tcf/Lef-repressive complex into a transcriptional active form by releasing HDAC1 from this complex 6, 26. However, transcription of the brachyury gene is silent despite the presence of nuclear β-catenin in ESCs 27, 28, 29. We show that overexpression of Smek inhibited Tcf/Lef-mediated transcription induced by β-catenin. Smek and HDAC1 simultaneously occupied the Tcf/Lef binding motif. The increase of β-catenin in ESCs does not affect the occupancy of Smek and HDAC1 at the brachyury promoter. These data indicate that by recruiting HDAC1, Smek inhibits the transcription activity of Tcf/Lef. This represents a new mechanism by which ESCs restrain the potential of Wnt target genes to promote differentiation despite the presence of nuclear β-catenin.
Tcf/Lef proteins interact with HDAC1 through the co-repressor Groucho/TLE to repress transcription 5. However, the interaction between Groucho/TLE and Tcf/Lef family members is restricted. For example, human Tcf1 and X. laevis Tcf3 bind to Groucho/TLE, but not human Lef1, mouse Tcf3, or mouse Tcf4 30. This indicates that Tcf/Lef proteins may interact directly with HDAC1, or require interaction with other conserved co-repressors. Smek proteins may mediate the interaction of HDAC1 to Tcf/Lef family proteins. Although we were not able to examine binding of Smek and Tcf/Lef proteins at the endogenous protein level due to lack of antibodies of immunoprecipitation quality, we were able to demonstrate that both Smek1 and Smek2 bind to Lef1, Tcf1, Tcf3, and Tcf4 when overexpressed in ESCs (data not shown).
We show that depletion of Smek leads to loss of ESC pluripotency, which occurred due to induction of differentiation rather than by regulation 31 of pluripotency factors. The pluripotency factors Oct4, Sox2, and Nanog, which promote self-renewal, are well-characterized in ESCs. Although they can occupy the promoters of a large set of development-related genes to repress their transcription in the ESC state, they play a dominant role in activating transcription of pluripotency maintenance genes 32. In addition, depletion of Smek did not cause increase of Oct4, Sox2, and Nanog expression. This indicates that the Smek repression complex does not directly regulate expression of pluripotency factors, but rather selectively directs expression of genes important for differentiation. Given that knockdown of Smek induced expression of early mesodermal markers, such as brachyury and goosecoid, but not endoderm and ectoderm markers, role of Smek in ESCs is to maintain pluripotency by assisting the inhibition of mesoderm commitment.
Although treatment of ESCs with Wnt proteins can disrupt Smek-mediated transcriptional repressive complex and induce brachyury expression, it is not sufficient to stimulate mesoderm differentiation. Activation of Wnt signaling increases expression of nanog and oct4 as well as brachyury 23, 24. The increase of pluripotency factors, thus, may favor ESC self-renewal rather than differentiation into mesoderm. On the other hand, sustained Wnt-signaling activation can induce differentiation of ESC. A recent study showed that long-term treatment of ESCs with Wnt3a protein in the presence of LIF induces differentiation into bipotent mesendodermal cells that produce mesoderm 33. Similarly to this, we show that depletion of Smek proteins induces mesoderm differentiation. Therfore, it is likely that stable expression of shRNA for Smek1 and Smek2 leads to sustained activation of Wnt/β-catenin signaling.
In conclusion, our study has revealed a function of PP4 in transcriptional repression that contributes to silencing of key development-related genes in pluripotent ESCs. Such a function represents a transcriptional regulatory mechanism for key Wnt-responsive development-related genes in ESCs that, to our knowledge, has not been described before. We demonstrated here that expression of the Wnt target gene brachyury in the ESC state can be completely suppressed by the Smek/PP4c complex recruiting HDAC1 at the promoter of this gene, and that this complex is critical for the inhibition of early mesoderm differentiation gene expression. Furthermore, we suggest a specific function for the Smek/PP4c phosphatase complex in chromatin remodeling. Depletion of HDAC1 simultaneously increases histone H3 phosphorylation and deacetylation, stimulating the change of chromatin structure 22. Consistent with this, loss of the Smek/PP4c phosphatase complex induces histone H3 phosphorylation as well as deacetylation 12. Thus, Wnt signaling could displace HDAC1 from inactive chromatin through disruption of the Smek complex during differentiation, opening up the chromatin structure at the brachyury gene locus for initiation of transcription 34, and subsequently β-catenin translocated into nucleus could enhance or maintain the transcription.
Materials and methods
E14 mouse ESCs were cultured without feeder cells in LIF-supplemented medium as previously described 35. The shRNA and overexpression plasmids were introduced into ESCs by using the Mouse ES Cell Nucleofector Kit (Amaxa) according to the manufacturer's instructions. For ESC lines stably expressing shRNA, each plasmid expressing a shRNA was transfected into ESCs by electroporation. Each ESC clone was selected in ES medium supplemented with 1 μg/ml puromycin (Sigma) and analyzed by RT-PCR and Western blot. To generate Smek1/2 double knockdown cell lines, a plasmid containing the Smek1 shRNA and a neomycin resistance cassette and a plasmid containing the Smek2 and a puromycin resistance cassette were co-transfected into the ESC line. Neomycin and puromycin selection was induced 1 day after transfection. For differentiation, ESCs (6 × 102 cells/25 μl) were suspended in a hanging drop of medium in the absence of LIF. Cell aggregates that formed as EB in the suspended drops were collected after 2 days and further cultured in low attachment plates. Fresh medium was applied every 2 days. Differentiation of ESCs into neuroectoderm on monolayer cultures was performed as previously described 36. For luciferase reporter assays, ESCs were transfected with the reporter plasmids Super 8X TOPFLASH or FOPLASH (kindly provided by Dr Moon RT) and the internal control pRL-TK. Cells were plated onto gelatin-coated dishes or aggregated in a hanging drop of medium, and cells were harvested 3 days after transfection. Luciferase assays were performed using the Dual-Luciferase Kit (Promega). For alkaline phosphatase assays, ESCs were plated at 2 × 102 per well in 6-well plates and cultured for 5 days in the presence of LIF, after which they were stained with an alkaline phosphatase substrate kit (Vector Laboratories). HEK293T cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum. Cells were transfected with the indicated plasmids using the calcium phosphatase precipitation method and lysed at 48 h post transfection for luciferase assays and ChIP. Further details and other methods can be found in the Supplementary information, Data S1.
Acknowledgments
The authors would like to thank people from the Lu lab for constructive discussions. Jungmook Lyu was supported by a postdoctoral fellowship from CIRM. This work was partially supported by the CIRM Basic Biology Award (RB1-01353-1) and a grant from NIH (1R01NS067213-01A1).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Overexpression of Smek reduces differentiation of ESCs into mesoderm.
Depletion of Smek inhibits neuroectoderm differentiation from ESCs.
Localization of β-catenin in ESCs knocked down for Smek1 and Smek1.
Occupancy of Smek at Tcf/Lef binding site region of islet1 and pitx2.
Smek increases binding of HDAC1 to Tcf protein.
The knockdown for PP4c induces differentiation of ESCs.
Smek-mediated blockade of transcriptional activation by Wnt/β-catenin signaling is dependent on the dose of Wnt3a protein.
β-catenin-independent Wnt signaling increases brachyury expression.
Materials and Methods
References
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Downs KM, Christensen HM, et al. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol Cell Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billin AN, Thirlwell H, Ayer DE. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, et al. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC, Du F, Iranfar N, et al. Loss of SMEK, a novel, conserved protein, suppresses MEK1 null cell polarity, chemotaxis, and gene expression defects. Mo. Cell Biol. 2005;25:7839–7853. doi: 10.1128/MCB.25.17.7839-7853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, et al. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Chia W, Somers WG. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes & Dev. 2009;23:359–372. doi: 10.1101/gad.1723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, et al. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Aubert J, Dunstan H, Chambers I, Smith A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;16:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages. Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes & Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canettieri G, Morantte I, Guzmán E, et al. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol. 2003;10:175–181. doi: 10.1038/nsb895. [DOI] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes & Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Singh AM, Li FQ, Hamazaki T, et al. Chibby, an antagonist of the Wnt/beta-catenin pathway, facilitates cardiomyocyte differentiation of murine embryonic stem cells. Circulation. 2007;115:617–626. doi: 10.1161/CIRCULATIONAHA.106.642298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, et al. Identification of a Wnt/Dvl/beta-Catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Anton R, Kestler HA, Kühl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Takao Y, Yokota T, Koide H. Beta-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem Biophys Res Commun. 2007;353:699–705. doi: 10.1016/j.bbrc.2006.12.072. [DOI] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci USA. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakre MM, Hoi A, Mong JC, et al. Generation of multipotential mesendodermal progenitors from mouse embryonic stem cells via sustained Wnt pathway activation. J Biol Chem. 2007;282:31703–31712. doi: 10.1074/jbc.M704287200. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110:1341–1348. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression of Smek reduces differentiation of ESCs into mesoderm.
Depletion of Smek inhibits neuroectoderm differentiation from ESCs.
Localization of β-catenin in ESCs knocked down for Smek1 and Smek1.
Occupancy of Smek at Tcf/Lef binding site region of islet1 and pitx2.
Smek increases binding of HDAC1 to Tcf protein.
The knockdown for PP4c induces differentiation of ESCs.
Smek-mediated blockade of transcriptional activation by Wnt/β-catenin signaling is dependent on the dose of Wnt3a protein.
β-catenin-independent Wnt signaling increases brachyury expression.
Materials and Methods