Abstract
Flavin-containing monooxygenases (FMOs) are one of the most important monooxygenase systems in Eukaryotes and have many important physiological functions. FMOs have also been found in bacteria; however, their physiological function is not known. Here, we report the identification and characterization of trimethylamine (TMA) monooxygenase, termed Tmm, from Methylocella silvestris, using a combination of proteomic, biochemical, and genetic approaches. This bacterial FMO contains the FMO sequence motif (FXGXXXHXXXF/Y) and typical flavin adenine dinucleotide and nicotinamide adenine dinucleotide phosphate-binding domains. The enzyme was highly expressed in TMA-grown M. silvestris and absent during growth on methanol. The gene, tmm, was expressed in Escherichia coli, and the purified recombinant protein had high Tmm activity. Mutagenesis of this gene abolished the ability of M. silvestris to grow on TMA as a sole carbon and energy source. Close homologs of tmm occur in many Alphaproteobacteria, in particular Rhodobacteraceae (marine Roseobacter clade, MRC) and the marine SAR11 clade (Pelagibacter ubique). We show that the ability of MRC to use TMA as a sole carbon and/or nitrogen source is directly linked to the presence of tmm in the genomes, and purified Tmm of MRC and SAR11 from recombinant E. coli showed Tmm activities. The tmm gene is highly abundant in the metagenomes of the Global Ocean Sampling expedition, and we estimate that 20% of the bacteria in the surface ocean contain tmm. Taken together, our results suggest that Tmm, a bacterial FMO, plays an important yet overlooked role in the global carbon and nitrogen cycles.
Flavin-containing monooxygenases (FMOs) catalyze the NADPH-dependent N- or S- oxygenation of heteroatom-containing compounds (1). According to the definition by van Berkel et al. (2), FMOs belong to class B flavoprotein monooxygenases, together with Baeyer–Villiger monooxygenases (BVMO) and N-hydroxylating monooxygenases (NMO). This class of flavoprotein monooxygenases has characteristic conserved sequence motifs and contains enzyme-bound flavin adenine dinucleotide (FAD) (2, 3). FMOs in eukaryotes have been well studied, and diverse functions have been assigned to this group of enzymes (1, 4). The best known example of eukaryotic FMO is probably from mammals, where the predominant physiological function appears to be detoxification of a vast spectrum of xenobiotics, including trimethylamine (TMA), found in the diet of mammals. It has been shown that mutations in human FMO (isoform 3) cause the inheritable disease known as trimethylaminuria (fish odor syndrome) caused by the inability of the body to oxidize TMA to its nonodor-oxygenated form trimethylamine N-oxide (TMAO), the TMA subsequently causing malodor in urine and the breath (5). FMOs have also been found in plants and fungi. Plant FMOs are involved in the biosynthesis of the plant-growth hormone auxin (6, 7) and in pathogen defense (8–10). In fungi, FMOs are vital for the functional expression of proteins that contain disulfide bonds by controlling the redox potential within the endoplasmic reticulum (11–13). FMOs are also present in bacteria (14), and recently, the presence of FMO has been observed in many bacterial genomes. However, thus far, the physiological roles of bacterial FMOs are unknown.
We hypothesized that because eukaryotic FMOs oxidize TMA to TMAO, bacterial FMOs will be involved in TMA metabolism by bacteria. An enzyme called TMA monooxygenase (termed hereafter Tmm) was suggested by Large et al. (15) in the 1970s to explain the ability of Aminobacter aminovorans (previously known as Pseudomonas aminovorans), which lacked known pathways for TMA utilization (16), to grow on TMA. In this bacterium, TMA is converted to TMAO by this enzyme, and TMAO is then further converted to formaldehyde [assimilated as the carbon (C) source] and ammonium [assimilated as the nitrogen (N) source] via the intermediate monomethylamine (MMA) (15–17). In other bacteria, such as Pseudomonas strain P, this enzyme is involved in utilization of TMA as a sole N source (17). However, the gene encoding Tmm has never been identified.
Methylated amine compounds, including TMA, are ubiquitous in the environment—for example, as end products of protein putrefaction (18). In the marine environment, methylated amines are released as a result of degradation of quaternary amine osmoregulators, such as glycine betaine, which are used by marine organisms to counteract water stress (19–21). Once released into the environment, methylated amines can be used by microorganisms as a C or N source. In fact, in the oceans, methylated amines represent a significant pool of C and N, and standing concentrations up to hundreds of nanomolar and micromolar have been reported in the water column (22, 23) and sediment pore water (24, 25), respectively. In addition to being involved in biogeochemical cycles of C and N, recent studies also suggest that methylated amines have the potential to affect global climate, being precursors of aerosol formation in the upper atmosphere (26–28).
We report here identification and characterization of Tmm (15, 17, 29) using a combination of genetic, proteomic, and bioinformatic approaches. The corresponding gene is particularly abundant in marine Roseobacter clade (MRC) and the SAR11 clade, two of the most abundant marine bacteria in the surface ocean (30–33), as well as in bacterial metagenomic sequences in the Global Ocean Sampling expedition (34). Our results suggest that this overlooked enzyme may play an important role in the global C and N cycles.
Results
Identifying the Function of a Bacterial FMO in M. silvestris as Tmm.
M. silvestris can grow on TMA as a sole C and N source. We detected inducible activities of three enzymes, Tmm (20.4 ± 1.9 nmol⋅min−1⋅mg−1), TMAO demethylase (71.2 ± 16.3 nmol⋅min−1⋅mg−1), and dimethylamine (DMA) monooxygenase (21.6 ± 0.7 nmol⋅min−1⋅mg−1), in cell-free extracts of this bacterium when it was grown on methylated amines. Because the genes encoding these enzymes have not yet been identified, we carried out comparative proteomic analyses using soluble protein fractions from methanol-, MMA-, and TMA-grown cultures. Quantitative proteomics data showed that polypeptides encoded by two gene clusters, orf Msil_2632-Msil_2639 and orf Msil_3603-Msil_3605 (Fig. 1A), were particularly abundant in cell-free extracts when M. silvestris was grown on TMA, whereas they were not detectable in cell-free extracts from methanol-grown cultures (a full list of polypeptides detected in each sample is shown in Dataset S1). We have previously shown that the former gene cluster was involved in MMA metabolism in M. silvestris, which encodes enzymes of the γ-glutamylmethylamide and N-methylglutamate pathway, converting MMA to formaldehyde (which is used as C and an energy source) and ammonium (which is used as an N source; Fig. 1B) (35).
Fig. 1.
(A) A summary of comparative proteomics and transcriptional analyses of the three-gene cluster containing a bacterial FMO in M. silvestris. The function was based on the annotation from the genome sequence using BLASTP search; the abundance of each of the polypeptides is shown in percentages of the total soluble proteome in each condition, and the presence (+) or absence (−) of transcription of each gene was confirmed by RT-PCR. MeOH, methanol; MMA, monomethylamine; TMA, trimethylamine. (B) The proposed pathway for TMA oxidation involving Tmm.
The second gene cluster (Fig. 1A) encodes two proteins that are annotated as glycine cleavage T protein and a bacterial-type FMO (Msil_3604), which shows significant similarity to mammalian FMOs. RT-PCR confirmed that transcription of the three genes of this second gene cluster was induced by TMA. These three genes were cotranscribed as an operon, because primers targeting intergenic regions gave RT-PCR products of the expected sizes (primers used are listed in Table S1). We hypothesized that orf Msil_3603-Msil_3605 encodes the three enzymes that metabolize TMA to MMA (Fig. 1B), a pathway for TMA oxidation proposed by Large et al. in the 1970s (15, 17).
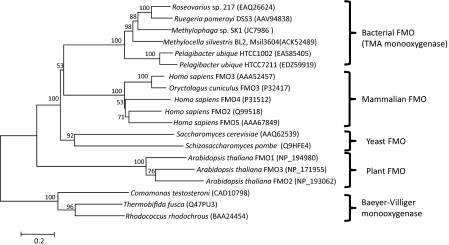
Sequence alignments with FMOs from mammals, plant, and fungi identified conserved domains in Msil_3604 (Fig. S1). Msil_3604 contains the three key characteristic sequence motifs of FMOs: the FAD-binding domain (GXGXXG), the FMO-identifying motif (FXGXXXHXXXF/Y) (3), and an NADPH-binding domain (GXSXXA). Phylogenetic analyses of Msil_3604 and known FMOs from mammals, plant, and fungi revealed that this bacterial FMO clustered within the known FMO family (Fig. 2).
Fig. 2.
A neighbor-joining phylogenetic tree showing the relationship of Tmm to other FMOs. The tree was drawn using the MEGA4 (50) based on an alignment of ∼450 amino acids of FMOs. Baeyer–Villiger monooxygenases were used as an outgroup. Bootstrap values of 100 replicates are shown.
To identify the function of Msil_3604, we cloned this gene from M. silvestris into pET28a, overexpressed it in Escherichia coli, and purified the resulting polypeptide by affinity chromatography (Fig. S2). Steady-state kinetic assays, performed at fixed NADPH concentrations with typical FMO substrates, showed Michaelis–Menten behavior (Table 1). As previously noted for other FMOs, FMO from M. silvestris had a broad substrate specificity, oxidizing many nitrogen- and sulfur-containing compounds. However, unlike eukaryotic FMOs, thiol-containing molecules such as glutathione and cysteine were not oxidized by this bacterial FMO. Interestingly, this enzyme can also oxidize dimethylsulfide (DMS), and the Km value is similar to that of TMA. Of all of the substrates tested, this enzyme has the highest affinity for TMA, and it is therefore likely to encode the Tmm that is involved in TMA oxidation in this bacterium (Fig. 1B). To further confirm its role in TMA oxidation in M. silvestris, a mutant of Msil_3604 was constructed by marker-exchange mutagenesis. The growth rates of the mutant and wild-type strain of M. silvestris on various methylamine compounds are presented in Table 2. Although the wild-type strain grows on TMA as a sole C and N source, the mutant could no longer grow on TMA. Overall, our data indicate that this bacterial type FMO encodes the so-called Tmm in bacteria and is essential for bacterial TMA oxidation.
Table 1.
Steady-state kinetic assays on purified Tmm of M. silvestris from recombinant E. coli
| Substrate | Km, μM | Vmax, nmol⋅min−1⋅mg−1 |
| TMA | 9.4 ± 2.1 | 29.4 ± 3.2 |
| DMA | 89.7 ± 13.2 | 6.9 ± 0.1 |
| MMA | — | — |
| DMSO | 3,575 ± 151 | 4.8 ± 1.5 |
| DMS | 10.3 ± 0.7 | 34.6 ± 0.2 |
| Cysteamine | 3,139 ± 534 | 76.2 ± 9.1 |
| Methimazole | 28.2 ± 5.1 | 14.4 ± 0.8 |
| Dimethylaniline | 35.7 ± 2.5 | 29.2 ± 0.9 |
| Glutathione | — | — |
| Cysteine | — | — |
—, substrate not oxidized.
Table 2.
Specific growth rates of wild-type and tmm mutant of Methylocella silvestris grown on methylated amines
| Growth substrate | Wild type, h−1 | Mutant, h−1 |
| TMA | 0.034 ± 0.003 | — |
| DMA | — | 0.039 ± 0.003 |
| TMA + DMA | 0.042 ± 0.008 | 0.037 ± 0.002 |
| MMA | 0.029 ± 0.003 | 0.029 ± 0.001 |
—, no growth.
Tmm in Sequenced Bacterial Genomes.
The sequence of M. silvestris Tmm was used to search microbial genome databases. No archaeal genomes contain Tmm homologs. However, homologs were found in the genomes of some Gammaproteobacteria, including Methylophaga sp. SK1, Pseudomonas putida GB-1, P. fluorescens SBW25, and P. mendocina ymp. Methylophaga sp. SK1 and P. mendocina are known to use TMA as a sole C and N source (14, 36). Tmm homologs were also identified in the genomes of some terrestrial Alphaproteobacteria, including Agrobacterium, Azorhizobium, and Mesorhizobium. Interestingly, Tmm homologs are also found in many marine Alphaproteobacteria, including Rhodobacteraceae (also known as MRC) and three genomes of the SAR11 clade (Pelagibacter ubique sp. HTCC1002, HTCC1062, and HTCC7211), indicating a key role of TMA oxidation in marine C/N cycles.
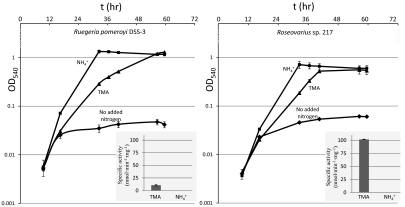
A search of MRC genome sequences revealed that 15 of the 39 sequenced MRC genomes contain Tmm homologs (Table S2). All of these Tmm homologs have FAD and NADPH binding sites and the FMO sequence motif (FXGXXXHXXXF/Y; Fig. S1), and are clustered within the bacterial FMOs (Fig. 2). We tested 11 of these 39 MRC strains for their ability to use TMA as a C and/or N source. Seven strains containing Tmm homologs in their genomes grew on TMA as a sole N source (Table S2). Of these, three strains (Roseovarius sp. 217, Roseovarius sp. TM1035, and Roseovarius nubinhibens ISM) can also use TMA as a sole C and energy source. Four other strains tested, Dinoroseobacter shibae, Oceanicola batsensis, Roseobacter sp. SK209-2-6, and Sagittula stellata, which lack tmm in their genomes, did not grow on TMA as either a C or N source. Representative growth curves of Ruegeria pomeroyi DSS-3 and Roseovarius sp. 217 growing on TMA are shown in Fig. 3. Inducible Tmm activities were detected in the supernatant of TMA-grown cultures, and the expression of Tmm in TMA-grown cultures was confirmed by mass spectrometry (Fig. S3). To further prove the function of these Tmm homologs, we overexpressed tmm from Roseovarius sp. 217 and Ruegeria pomeroyi DSS-3 in E. coli and purified them; they both oxidize TMA (Table 3).
Fig. 3.
Representative growth curves of Ruegeria pomeroyi DSS-3 and Roseovarius sp. 217 growing on TMA or ammonium as the sole nitrogen source. Controls were set up with no added nitrogen. (Insets) Activity of Tmm from the supernatant of the crude extract of R. pomeroyi DSS-3 and Roseovarius sp. 217 cultures, respectively. No enzyme activities were detected when ammonium was used as the sole nitrogen source. Error bars indicate SD of each experiment (n = 3).
Table 3.
Kinetic parameters with TMA for purified recombinant Tmm enzymes
| Substrate | Km, μM | Vmax, nmol⋅min−1⋅mg−1 |
| Roseovarius sp. 217 | 21.6 ± 1.9 | 1,133.6 ± 58.6 |
| Ruegeria pomeroyi DSS-3 | 20.8 ± 2.9 | 267.7 ± 52.2 |
| Pelagibacter ubique HTCC1002 | 27.5 ± 4.2 | 70.8 ± 7.7 |
| Pelagibacter ubique HTCC7211 | 28.5 ± 4.4 | 67.3 ± 3.2 |
The three genomes of members of the SAR11 clade also contain Tmm homologs. Due to the lack of available isolates to test their growth on TMA, we chemically synthesized two tmm genes (sp. HTCC1002 and HTCC7211), overexpressed these genes in E. coli, and further purified the recombinant proteins. Kinetics analyses demonstrated that these Tmm homologs can also oxidize TMA (Table 3).
Tmm Is Abundant in the Marine Bacterial Metagenome.
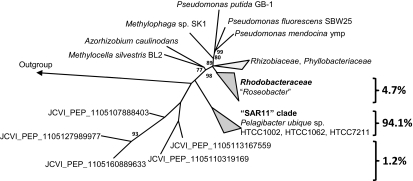
The wide occurrence of Tmm homologs in the MRC and the SAR11 clade bacteria suggested that Tmm may play an important role in the marine C and N cycles. We therefore estimated the abundance of Tmm homologs in the Global Ocean Sampling (GOS) expeditions database (34). This database contains ∼6.1 million deduced gene products from bacteria sampled directly from surface waters at various sites in the northwest Atlantic Ocean, the Sargasso Sea, and the eastern Pacific Ocean. Using a relatively stringent BLASTP search (<e−80), we identified 570 Tmm homologs (Table 4). These homologs occur at nearly all GOS sites, except for two nonmarine sites (Lake Gatun and Punta Cormorant) and one marine site that was sampled with larger-sized fraction filters (Dirty Rock Cocos Island) (34). The retrieved Tmm homologs from the GOS dataset all contain the FMO sequence motif. The abundance of Tmm homologs is comparable to other enzymes carrying out key environmental functions in the marine environment, such as dimethylsulfoniopropionate-dependent demethylase (DmdA), which is involved in dimethylsulfoniopropionate (DMSP) catabolism (37) and is about 10× more frequent than DddQ, DddP, and DddL, which are involved in DMSP-dependent DMS production (38). On average, we estimate that ∼20% of the microbial genomes in the surface ocean contain Tmm homologs (Table 4). Phylogenetic analysis indicated that the Tmm homologs from the GOS database could be placed into three groups (Fig. 4), the majority of which were closely related to Tmm homologs in SAR11 clade bacteria (94.1%) and the MRC bacteria (4.7%), respectively (Fig. S4). The predominance of the MRC and the SAR11 clade bacteria in marine TMA metabolism is supported by the analysis of γ-glutamylmethylamide synthetase (GmaS), an enzyme involved in a subsequent step in the pathway of TMA metabolism (Fig. 1B) (35, 39). GmaS homologs are also found at nearly all GOS sites and are as abundant as the Tmm enzyme (Table 4). Phylogenetic analysis also indicated the sequences retrieved from the GOS dataset are from either the SAR11 clade (78%) or the MRC (22%; Fig. S5).
Table 4.
Distribution of Tmm homologs at different sampling sites in the Global Ocean Sampling expedition metagenomic data set
| Sample no. | Sampling site | Description | Tmm* | GmaS* | DddP* | DddD* | DddL* | DmdA* | RecA* | % genomes with Tmm† | % genomes with GmaS† |
| 2 | Gulf of Maine | Coastal | 12 | 16 | 1 | 2 | 0 | 21 | 35 | 29 | 39 |
| 3 | Brown's Bank-Gulf of Maine | Coastal | 13 | 11 | 0 | 0 | 0 | 39 | 52 | 24 | 21 |
| 4 | Outside Halifax Nova Scotia | Coastal | 30 | 21 | 0 | 0 | 0 | 34 | 40 | 55 | 38 |
| 5 | Bedford Basin Nova Scotia | Embayment | 7 | 7 | 2 | 3 | 0 | 26 | 34 | 18 | 18 |
| 6 | Bay of Fundy Nova Scotia | Estuary | 23 | 25 | 0 | 0 | 0 | 50 | 80 | 39 | 42 |
| 7 | Northern Gulf of Maine | Coastal | 8 | 14 | 2 | 0 | 0 | 20 | 53 | 13 | 23 |
| 8 | Newport Harbor | Coastal | 2 | 7 | 4 | 0 | 0 | 9 | 43 | 3 | 12 |
| 9 | Block Island | Coastal | 15 | 15 | 0 | 0 | 0 | 28 | 33 | 39 | 39 |
| 10 | Cape May | Coastal | 18 | 19 | 0 | 0 | 0 | 36 | 63 | 27 | 29 |
| 11 | Delaware Bay | Estuary | 5 | 2 | 0 | 0 | 0 | 7 | 43 | 10 | 3 |
| 12 | Chesapeake Bay | Estuary | 5 | 4 | 0 | 0 | 0 | 17 | 39 | 10 | 8 |
| 13 | Off Nag's Head | Coastal | 7 | 9 | 1 | 0 | 0 | 17 | 43 | 18 | 24 |
| 14 | South of Charleston | Coastal | 18 | 14 | 0 | 0 | 0 | 42 | 67 | 29 | 22 |
| 15 | Off KeyWest | Coastal | 5 | 15 | 2 | 0 | 0 | 35 | 51 | 8 | 26 |
| 16 | Gulf of Mexico | Coastal | 14 | 7 | 1 | 0 | 0 | 41 | 50 | 26 | 13 |
| 17 | Yucatan Channel | Open ocean | 9 | 13 | 1 | 0 | 0 | 32 | 61 | 15 | 23 |
| 18 | Rosario Bank | Open ocean | 9 | 13 | 3 | 0 | 0 | 43 | 83 | 14 | 20 |
| 19 | Northeast of Colon | Coastal | 6 | 16 | 2 | 0 | 0 | 33 | 72 | 10 | 27 |
| 20 | Lake Gatun | Fresh water | 0 | 1 | 0 | 0 | 0 | 3 | 61 | 0 | 1 |
| 21 | Gulf of Panama | Coastal | 5 | 14 | 4 | 0 | 0 | 31 | 53 | 10 | 27 |
| 22 | 250 miles from Panama City | Open ocean | 12 | 16 | 1 | 0 | 0 | 35 | 50 | 22 | 28 |
| 23 | 30 miles from Cocos | Open ocean | 11 | 16 | 0 | 0 | 0 | 35 | 54 | 18 | 26 |
| 25 | Dirty Rock Cocos Island | Fringing reef | 0 | 1 | 1 | 0 | 0 | 5 | 13 | 0 | 6 |
| 26 | 134 miles NE of Galapagos | Open ocean | 7 | 11 | 1 | 0 | 0 | 41 | 55 | 13 | 20 |
| 27 | Devil's Crown | Coastal | 16 | 13 | 4 | 0 | 0 | 41 | 66 | 26 | 20 |
| 28 | Coastal Floreana | Coastal | 10 | 12 | 2 | 0 | 0 | 48 | 70 | 16 | 20 |
| 29 | North James Bay | Coastal | 9 | 24 | 2 | 0 | 0 | 46 | 50 | 20 | 52 |
| 30 | Warm Seep Roca Redunda | Warm seep | 19 | 17 | 1 | 0 | 0 | 52 | 55 | 36 | 33 |
| 31 | Upwelling Fernandina Island | Coastal | 26 | 27 | 1 | 0 | 0 | 57 | 61 | 46 | 49 |
| 32 | Mangrove on Isabella | Mangrove | 8 | 5 | 1 | 1 | 0 | 16 | 43 | 20 | 14 |
| 33 | Punta Cormorant | Hypersaline | 0 | 2 | 4 | 0 | 6 | 2 | 43 | 0 | 4 |
| 34 | North Seamore Island | Coastal | 9 | 17 | 2 | 0 | 0 | 42 | 51 | 17 | 32 |
| 35 | Wolf Island | Coastal | 11 | 13 | 2 | 0 | 0 | 38 | 51 | 21 | 26 |
| 36 | Cabo Marshall | Coastal | 14 | 19 | 0 | 0 | 0 | 45 | 54 | 24 | 33 |
| 37 | Equatorial TAO | Open ocean | 14 | 6 | 5 | 0 | 0 | 48 | 38 | 30 | 13 |
| 47 | 201 miles from French Polynesia | Open ocean | 9 | 18 | 5 | 0 | 0 | 48 | 62 | 15 | 29 |
| 51 | Rangiora Atoll | Coral reef | 12 | 14 | 2 | 0 | 0 | 28 | 53 | 25 | 29 |
| Average | 11 | 13 | 1.5 | 0.2 | 0.2 | 32 | 52 | 20 | 23 |
*The sampling sites are as detailed in Howard et al. (37). Numbers of Tmm and GmaS homologs, per 100,000 reads, with BLASTP values of <e−80. Values of DddD, DddL, DmdA, and RecA were from Howard et al. (37), and values of DddP were from Todd et al. (38).
†Frequencies of Tmm- and GmaS-containing strains, expressed as genome equivalents, as described in Howard et al. (37).
Fig. 4.
An unrooted tree showing Tmm homologs retrieved from sequenced bacterial genomes and the Global Ocean Sampling expedition data set. The neighbor-joining tree was constructed using sequences retrieved from sequenced bacterial genomes (∼450 amino acids). Environmental sequences were added by parsimony. Bootstrap values were calculated based on 100 replicates. Homo sapiens FMO3 was used as the outgroup.
Discussion
In this study we have identified the gene encoding Tmm (29, 40). The wide occurrence of Tmm homologs in the MRC and the SAR11 clade bacteria indicates that methylated amine compounds may play an important role in marine C and N cycles. These two groups of marine bacteria are particularly abundant in the surface oceans, and previous studies have shown that they play a key role in marine carbon and sulfur cycles (30–33, 41, 42). Although few studies have measured the concentrations of methylated amines in the oceans, existing data suggest that these compounds represent a significant pool of C and N in the marine water column (22, 23) and in marine and coastal sediments (24, 25). We tested representatives of the MRC bacteria, and the ability to use TMA is directly correlated to the presence of Tmm in their genomes (Table S2). The purified Tmm enzyme of MRC and SAR11 bacteria all oxidize TMA (Table 3), indicating that this enzyme is likely to contribute to the removal of methylated amines in the marine environment. Further evidence comes from analysis of the GOS data set, where it is estimated that, on average, 20% of the bacteria in the surface ocean contain Tmm, the majority of which originated from the SAR11 clade bacteria (Table 4). Indeed, previous metaproteome analysis of bacteria collected from surface seawater has identified Tmm polypeptides from SAR11 clade bacteria (43). Therefore, it is tempting to speculate that the MRC and the SAR11 clade bacteria may play key roles in sequestering N from methylated amines; this may therefore give them a selective advantage in the oceans over microorganisms unable to use such compounds, where they face severe competition for N.
The purified Tmm has broad substrate specificity and can also oxidize DMA, although the affinity for DMA is lower than for TMA (Table 1 and Table S3). This ability to oxidize secondary amines has been shown previously with purified TMA monooxygenase from A. aminovorans, and it has been shown that oxidizing DMA has no physiological function (40). Nonspecific oxidation of DMA produces formamide, which is toxic (40), and this may have inhibited the growth of the wild-type M. silvestris on DMA alone (Table 2). This finding is supported by the fact that wild-type M. silvestris can grow on TMA in the presence of DMA (Table 2), because TMA is the preferred substrate of Tmm. Although Tmm from M. silvestris can oxidize DMA, its function as a DMA monooxygenase can be ruled out because FMOs are not sensitive to carbon monoxide, a characteristic of secondary amine monooxygenases (44). In fact, DMA monooxygenase would release formaldehyde from the oxidation of DMA (Fig. 1B), and the presence of tetrahydrofolate-binding domains in orfs Msil_3603 and Msil_3605 suggests that one of these genes may encode the DMA monooxygenase (the other is likely to encode TMAO demethylase, which also releases formaldehyde).
The ability of bacterial FMOs to oxidize DMS and DMSO is surprising. DMSO was initially used as a solvent to dissolve some of the substrates. It was noticed previously that the FMO from Methylophaga sp. SK1 also oxidized DMSO (45). The ability of bacterial FMOs to oxidize DMS may be of special interest. It has been previously noted that there is an uncharacterized route for oxidation of DMS to DMSO by heterotrophic bacteria, such as Delftia acidovorans (46). In this bacterium, the oxidation of DMS to DMSO was dependent on NADPH, which could not be replaced by NADH. A search of the genome of D. acidovorans sp. SPH-1 does indeed reveal FMO-like protein-encoding genes. Because FMO of the MRC and the SAR 11 clade bacteria can also oxidize DMS, they may also represent a previously uncharacterized sink of DMS from the marine environment. Whether FMO is responsible for this uncharacterized DMS conversion in these bacteria, and to what extent they reduce DMS emission from the marine environment, certainly warrants further investigation.
To conclude, our results reveal the function of bacterial FMOs as an enzyme involved in bacterial TMA metabolism, Tmm. The wide occurrence of this enzyme in marine bacteria, together with its high abundance in the surface ocean, points to a key role for methylated amines in marine C and N cycling.
Materials and Methods
Cultivation of Methylocella silvestris.
M. silvestris was grown at 25 °C either in a 4-L fermentor or in 125-mL serum vials using diluted mineral salt medium as described previously (35). To test if the trimethylamine monooxygenase (tmm) mutant could grow on methylated amines, growth experiments were set up in triplicate using 120-mL serum vials, containing 20 mL medium with an inoculum size of 10%. Details of growth conditions and reagents are described in SI Materials and Methods.
Quantitative Comparative Proteomics.
A total of 700 μg of soluble protein extract from MMA, TMA, and methanol-grown M. silvestris cells were used for proteomic analyses (47). The details of the method are shown in SI Materials and Methods.
Marker-Exchange Mutagenesis of tmm in M. silvestris.
To construct a tmm mutant of M. silvestris, a downstream region (containing a KpnI site) and an upstream region (containing an MluI site) of the target were amplified by PCR and subcloned into pGEM-T vector (Promega) together with a kanamycin gene cassette (amplified from pCM184) inserted between the two regions (primers used are listed in Table S1). The resulting plasmid was then cut with KpnI and MluI, and the 2.6-kb fragment containing the kanamycin gene cassette was electroporated into M. silvestris as described previously (35).
Overexpression of tmm in E. coli.
The tmm genes from M. silvestris, Roseovarius sp. 217, and Ruegeria pomeroyi DSS-3 were amplified by PCR (primers used are listed in Table S1) and subcloned into pGEM-T vector (Promega), which were then excised using NdeI/HindIII and ligated into the expression vector pET28a (Merck Biosciences). The tmm homologs in the Pelagibacter ubique genomes (strain HTCC1002 and HTCC7211) were synthesized commercially with E. coli codon usage (GenScript Corporation). The synthesized genes were inserted into the expression vector pET28a using the NdeI/HindIII restriction sites. The resulting plasmids were then transformed into the expression host E. coli BLR(DE3) pLysS (Merck Biosciences). To overexpress TMA monooxygenases, E. coli cells were grown at 37 °C to an OD540 of 0.6, and isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.1 mM.
Enzyme Assays and Kinetics.
Cells were broken for protein analyses and enzyme assays by passing three times through a French pressure cell (American Instrument Co.) at 110 megapascals. Cell debris was removed by centrifugation at 6,000 × g for 15 min. One-dimensional protein analyses were carried out using a precast NuPAGE Bis-Tris gel (10%; Invitrogen). All enzyme assays were carried out in triplicate in 80 mM Pipes buffer (pH 7.6) at room temperature (22 °C) unless otherwise stated. Details are shown in SI Materials and Methods.
Cultivation of MRC Bacteria on Methylamines.
Growth tests were carried out using a defined medium in triplicate, containing artificial sea salts from Sigma-Aldrich (S9883) 40 g⋅L−1, sodium phosphate 0.2 mM (pH 8.0), Hepes 10 mM (pH 8.0), methylated amines 2 mM, FeCl3 50 μM, and a mixture of C sources containing glucose (5.6 mM), fructose (5.6 mM), succinate (8.5 mM), pyruvate (11.4 mM), glycerol (10.9 mM), and acetate (17 mM). The vitamins were added as described in SI Materials and Methods. Positive growth controls with ammonium chloride (1.0 mM) and negative controls with no added N compounds were also set up. To test if these isolates could use TMA as a sole C source, the above medium was used, except that the carbon mixture was omitted and TMA was added to a final concentration of 5 mM.
Identification of Tmm and GmaS Homologs in the GOS Metagenome.
Tmm sequence of M. silvestris was used as the query sequence for a BLASTP search of the GOS peptides at CAMERA [https://portal.camera.calit2.net/gridsphere/gridsphere?cid=; GOS: all ORF peptides (P) database, e−80], and resulted in 568 unique sequences. An additional search using the same settings was performed using the FMO of Methylophga sp. SK1 (JC7986) as the query, and two additional sequences were retrieved. These 570 sequences were further grouped into 127 unique groups (identity >90% within each group) using the CD-HIT program (48). Representative sequences from each group were aligned using the ARB program (49), and each sequence was manually checked for the FMO sequence motif. To estimate the frequency of Tmm-containing cells, the numbers of Tmm homologs were normalized against the average numbers of the six essential single-copy genes (atpD, rpoB, gyrB, hsp70, tufA, and recA) as described previously (37). The identification of GmaS homologs were estimated as described, except that the GmaS of M. silvestris (ACK51558) was used as the query, which resulted in 682 unique sequences. Additional queries using the GmaS sequences of M. universalis FAM5 (ADH10360), M. mays (BAF99006), and A. tumefaciens C58 (AAK89209) did not yield additional new sequences. These 682 sequences were further grouped by CD-HIT and analyzed by ARB.
Supplementary Material
Acknowledgments
We thank Ms. S. Slade for assistance with proteomics analysis, Dr. K. Thalassinos for bioinformatics, and Mr. W. Yang for protein overexpression. We are grateful to Drs. H. Schäfer, A. Buchan, R. Belas, B. Ward, and I. Wagner-Döbler for providing bacterial strains. This work was supported by Natural Environment Research Council Grants NE/H016236/1 (to Y.C.) and NE/E016855/1 (to J.C.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112928108/-/DCSupplemental.
References
- 1.Ziegler DM. Recent studies on the structure and function of multisubstrate flavin-containing monooxygenases. Annu Rev Pharmacol Toxicol. 1993;33:179–199. doi: 10.1146/annurev.pa.33.040193.001143. [DOI] [PubMed] [Google Scholar]
- 2.van Berkel WJH, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 3.Fraaije MW, Kamerbeek NM, van Berkel WJH, Janssen DB. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 2002;518:43–47. doi: 10.1016/s0014-5793(02)02623-6. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler DM. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab Rev. 2002;34:503–511. doi: 10.1081/dmr-120005650. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell SC, Smith RL. Trimethylaminuria: The fish malodor syndrome. Drug Metab Dispos. 2001;29:517–521. [PubMed] [Google Scholar]
- 6.Schlaich NL. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007;12:412–418. doi: 10.1016/j.tplants.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 8.Mishina TE, Zeier J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006;141:1666–1675. doi: 10.1104/pp.106.081257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch M, et al. A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 2006;47:629–639. doi: 10.1111/j.1365-313X.2006.02813.x. [DOI] [PubMed] [Google Scholar]
- 10.Bartsch M, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh JK, Poulsen LL, Ziegler DM, Robertus JD. Molecular cloning and kinetic characterization of a flavin-containing monooxygenase from Saccharomyces cerevisiae. Arch Biochem Biophys. 1996;336:268–274. doi: 10.1006/abbi.1996.0557. [DOI] [PubMed] [Google Scholar]
- 12.Suh JK, Poulsen LL, Ziegler DM, Robertus JD. Yeast flavin-containing monooxygenase generates oxidizing equivalents that control protein folding in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1999;96:2687–2691. doi: 10.1073/pnas.96.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh JK, Robertus JD. Yeast flavin-containing monooxygenase is induced by the unfolded protein response. Proc Natl Acad Sci USA. 2000;97:121–126. doi: 10.1073/pnas.97.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi HS, et al. A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochem Biophys Res Commun. 2003;306:930–936. doi: 10.1016/s0006-291x(03)01087-8. [DOI] [PubMed] [Google Scholar]
- 15.Large PJ, Boulton CA, Crabbe MJC. The reduced nicotinamide-adenine dinuclotide phosphate- and oxygen-dependent N-oxygenation of trimethylamine by Pseudomonas aminovorans. Biochem J. 1972;128:137–138. doi: 10.1042/bj1280137pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony C. The Biochemistry of Methylotrophs. London: Academic; 1982. [Google Scholar]
- 17.Chandler SR. Berkshire, UK: University of Reading, Reading; 1983. The utilization of methylamine-nitrogen by the methazotrophic bacterium Pseudomonad P. PhD thesis. [Google Scholar]
- 18.Kamiya A, Youki O. Study of odorous compounds produced by putrefaction of foods. J Chromatogr A. 1984;292:329–331. [Google Scholar]
- 19.King GM. In: Nitrogen Cycling in Coastal Marine Environments. Blackburn TH, Sorensen J, editors. New York: Wiley; 1988. pp. 143–173. [Google Scholar]
- 20.Oren A. Formation and breakdown of glycine betaine and trimethylamine in hypersaline environments. Antonie van Leeuwenhoek. 1990;58:291–298. doi: 10.1007/BF00399342. [DOI] [PubMed] [Google Scholar]
- 21.Welsh DT. Ecological significance of compatible solute accumulation by micro-organisms: From single cells to global climate. FEMS Microbiol Rev. 2000;24:263–290. doi: 10.1111/j.1574-6976.2000.tb00542.x. [DOI] [PubMed] [Google Scholar]
- 22.Gibb SW, Hatton AD. The occurrence and distribution of trimethylamine-N-oxide in Antarctic coastal waters. Mar Chem. 2004;91:65–75. [Google Scholar]
- 23.Van Neste A, Duce RA, Lee C. Methylamines in the marine atmosphere. Geophys Res Lett. 1987;14:711–714. [Google Scholar]
- 24.Fitzsimons MF, Kahni-Danon B, Dawitt M. Distributions and adsorption of the methylamines in the inter-tidal sediments of an East Anglian estuary. Environ Exp Bot. 2001;46:225–236. [Google Scholar]
- 25.Lee C, Olsen BL. Dissolved, exchangeable and bound aliphatic amines in marine sediments: Initial results. Org Geochem. 1984;6:259–263. [Google Scholar]
- 26.Facchini MC, et al. Important source of marine secondary organic aerosol from biogenic amines. Environ Sci Technol. 2008;42:9116–9121. doi: 10.1021/es8018385. [DOI] [PubMed] [Google Scholar]
- 27.Müller C, et al. Seasonal variation of aliphatic amines in marine sub-micrometer particles at the Cape Verde islands. Atmos Chem Phys. 2009;9:9587–9897. [Google Scholar]
- 28.Rinaldi M, et al. Primary and secondary organic marine aerosol and oceanic biological activity: Recent results and new perspectives for future studies. Adv Meteorol. 2010 10.1155/2010/310682. [Google Scholar]
- 29.Boulton CA, Crabbe MJ, Large PJ. Microbial oxidation of amines. Partial purification of a trimethylamine mono-oxygenase from Pseudomonas aminovorans and its role in growth on trimethylamine. Biochem J. 1974;140:253–263. doi: 10.1042/bj1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchan A, González JM, Moran MA. Overview of the marine roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkhoff T, Giebel H-A, Simon M. Diversity, ecology, and genomics of the Roseobacter clade: A short overview. Arch Microbiol. 2008;189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 32.Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 33.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 34.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, et al. gamma-Glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris. Appl Environ Microbiol. 2010;76:4530–4537. doi: 10.1128/AEM.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boden R, Thomas E, Savani P, Kelly DP, Wood AP. Novel methylotrophic bacteria isolated from the River Thames (London, UK) Environ Microbiol. 2008;10:3225–3236. doi: 10.1111/j.1462-2920.2008.01711.x. [DOI] [PubMed] [Google Scholar]
- 37.Howard EC, Sun S, Biers EJ, Moran MA. Abundant and diverse bacteria involved in DMSP degradation in marine surface water. Environ Microbiol. 2008;10:2379–2410. doi: 10.1111/j.1462-2920.2008.01665.x. [DOI] [PubMed] [Google Scholar]
- 38.Todd JD, Curson ARJ, Dupont CL, Nicholson P, Johnston AWB. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacterial and also occurs in some Ascomycete fungi. Environ Microbiol. 2009;11:1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x. and erratum (2009) 11:1624–1625. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, McAleer KL, Murrell JC. Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl Environ Microbiol. 2010;76:4102–4104. doi: 10.1128/AEM.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boulton CA, Large PJ. Oxidation of N-alkyl- and NN-dialkylhydroxylamines by partially purified preparations of trimethylamine mono-oxygenase from Pseudomonas aminovorans. FEBS Lett. 1975;55:286–290. doi: 10.1016/0014-5793(75)81013-1. [DOI] [PubMed] [Google Scholar]
- 41.González JM, Kiene RP, Moran MA. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tripp HJ, et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 43.Sowell SM, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 44.Alberta JA, Dawson JH. Purification to homogeneity and initial physical characterization of secondary amine monooxygenase. J Biol Chem. 1987;262:11857–11863. [PubMed] [Google Scholar]
- 45.Alfieri A, Malito E, Orru R, Fraaije MW, Mattevi A. Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proc Natl Acad Sci USA. 2008;105:6572–6577. doi: 10.1073/pnas.0800859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Kuniyoshi I, Hirai M, Shoda M. Oxidation of dimethyl sulfide by Pseudomonas acidovorans DMR-11 isolated from peat biofilter. Biotechnol Lett. 1991;13:223–228. [Google Scholar]
- 47.Patel VJ, et al. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J Proteome Res. 2009;8:3752–3759. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 48.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 49.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.