Abstract
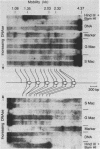
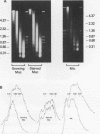
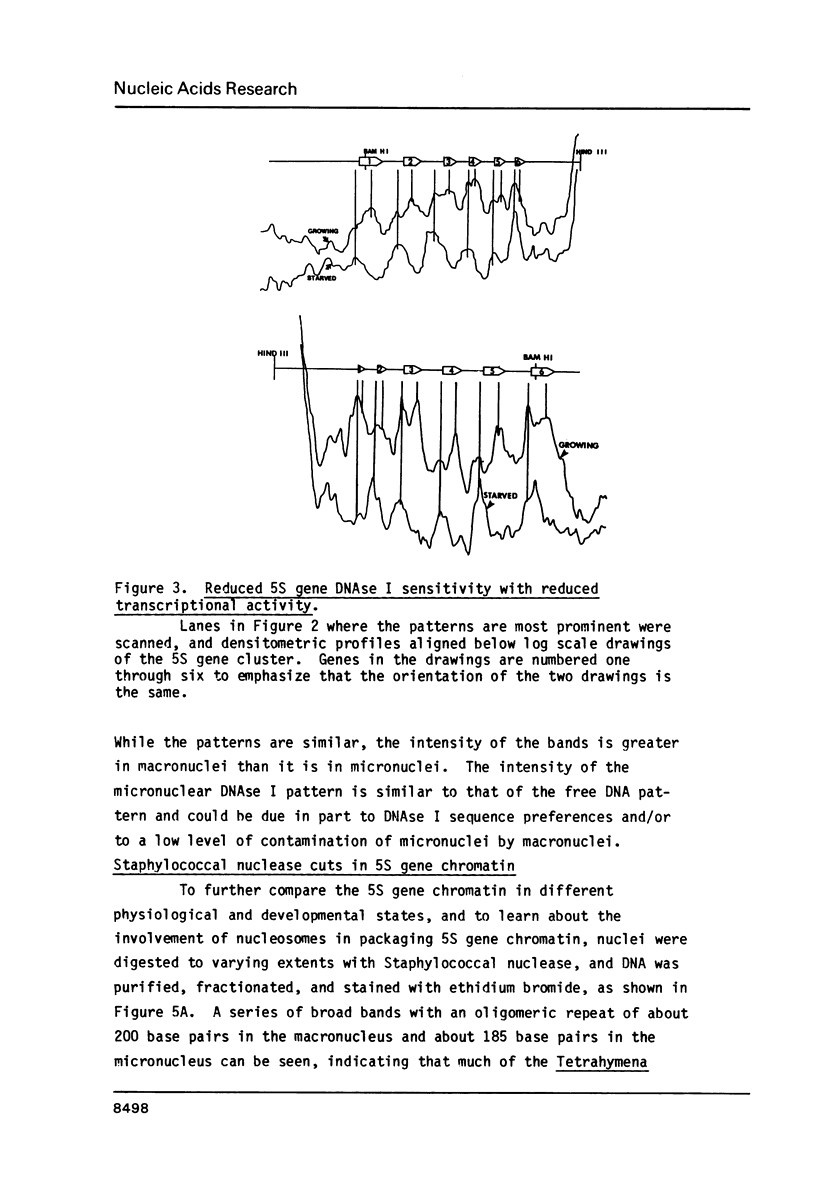
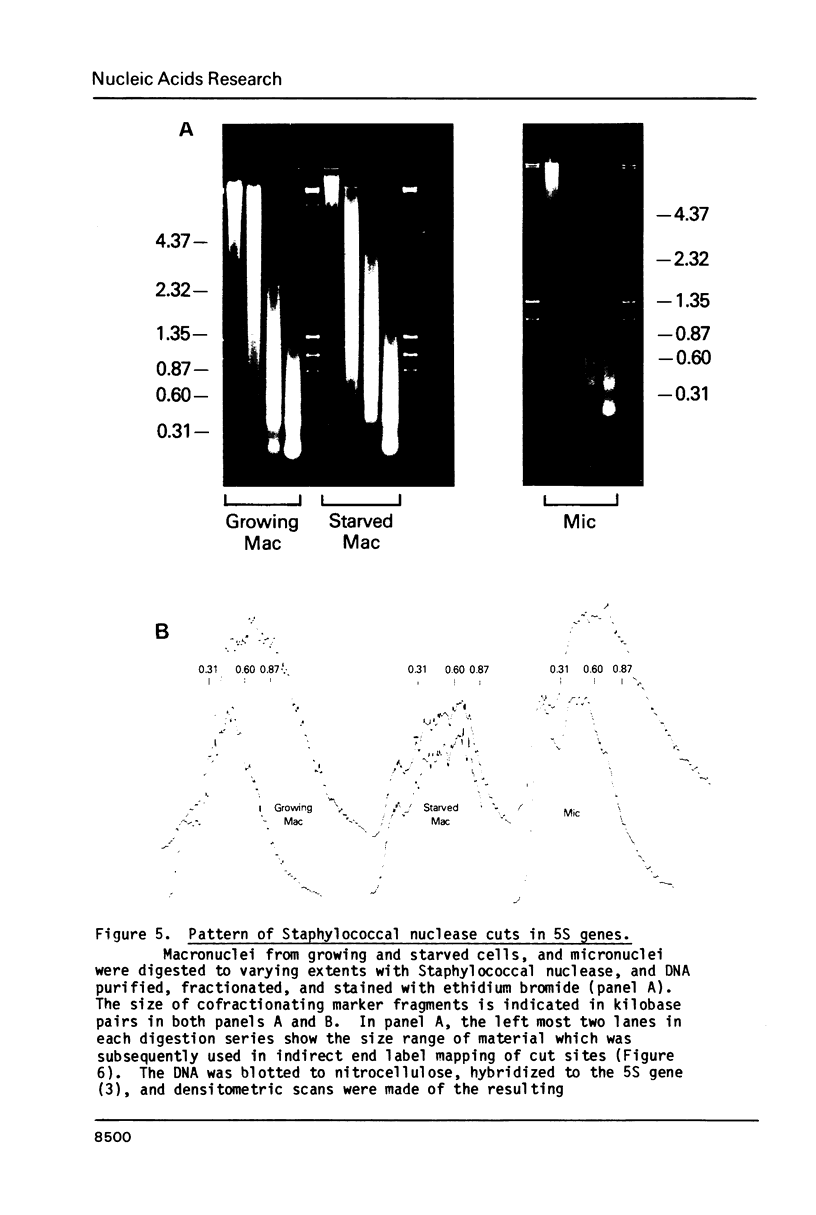
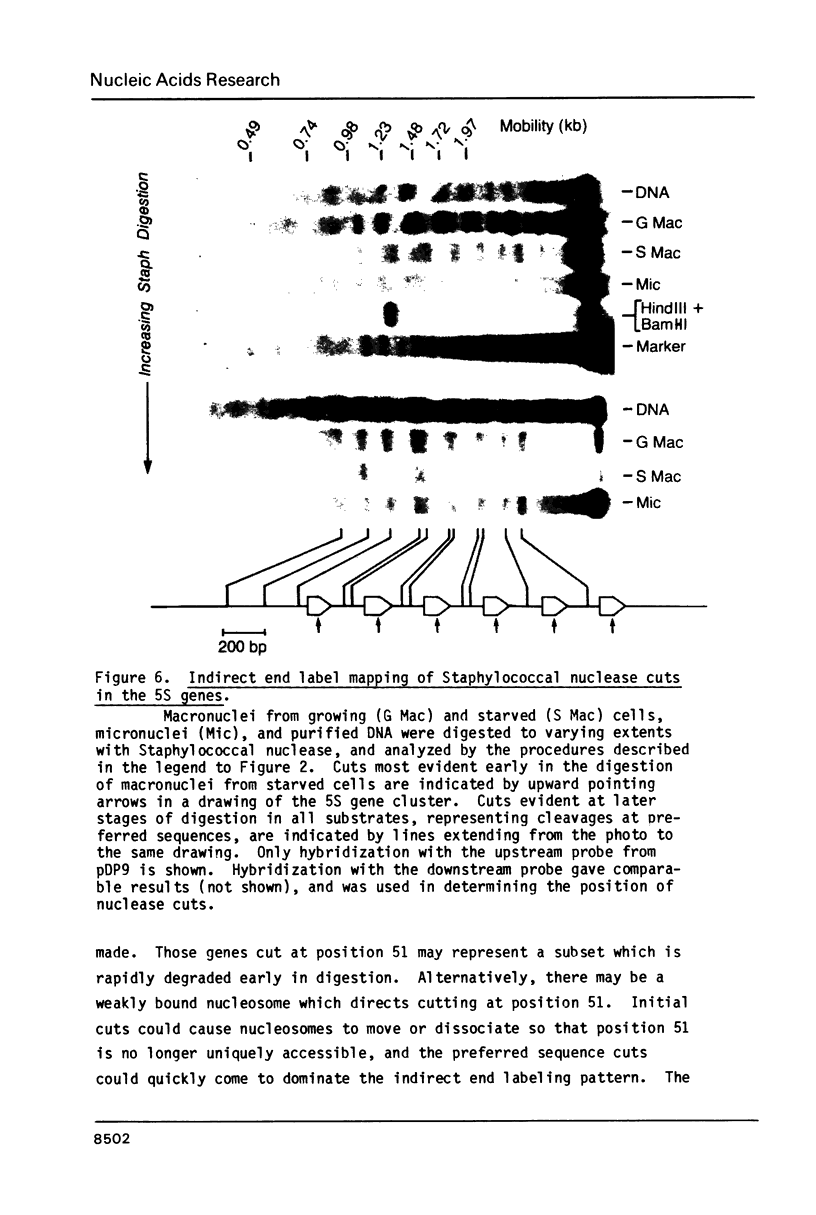
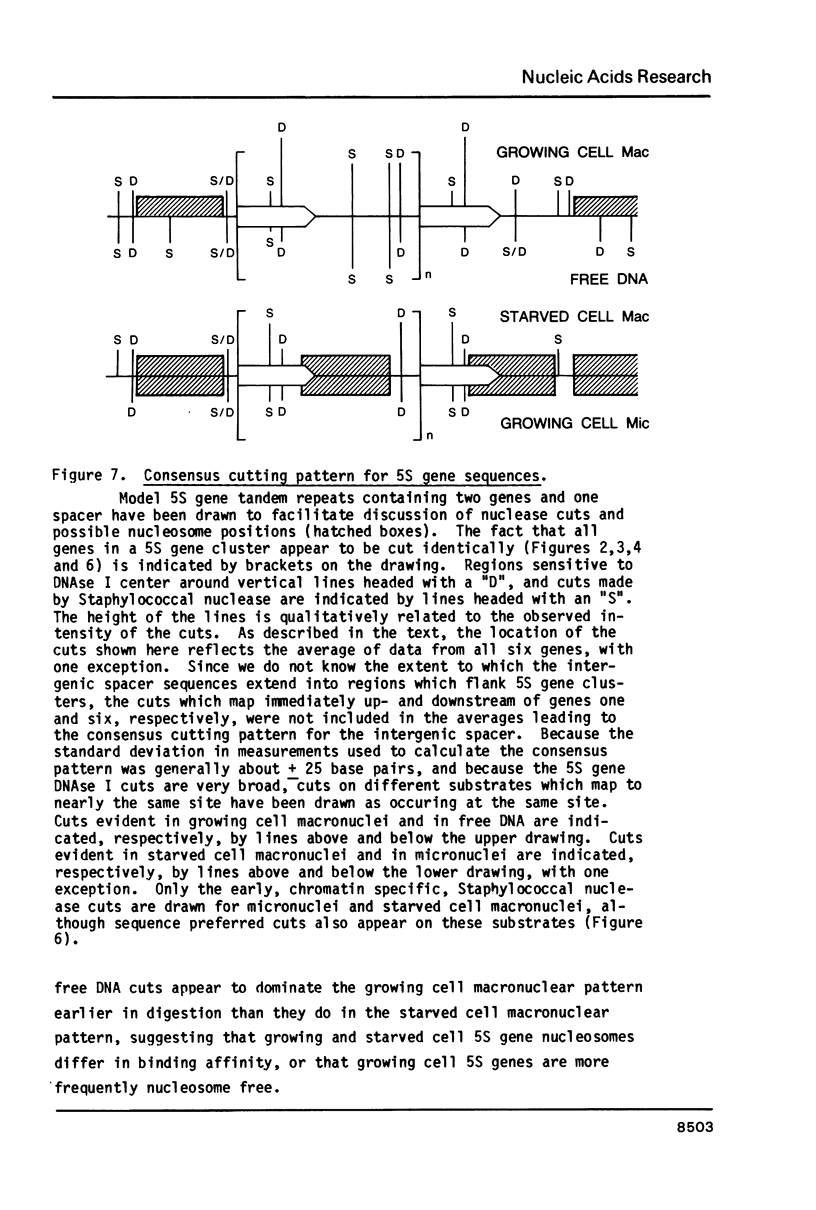
The chromatin structure of a single cluster of six tandemly repeated 5S ribosomal RNA genes (5S genes) in Tetrahymena thermophila has been characterized. Indirect end labeling experiments indicate that the actively transcribed 5S genes in macronuclei are rapidly cut by DNAse I near the putative internal promotor and just 5' to the transcribed region. When cells are starved to reduce 5S gene transcription rates, the DNAse I sensitivity of the intragenic site is reduced relative to the 5' site. In the nontranscribed 5S genes in micronuclei, neither of these sites is hypersensitive to DNAse I. Thus structural alterations accompany both the activation of transcription during macronuclear development and physiological changes in the rate of transcription of the 5S genes. These DNAse I data together with studies using Staphylococcal nuclease suggest that rapidly transcribed 5S genes may not be associated with histones as nucleosomes. In contrast, the genes in starved cell macronuclei appear to be associated with one nucleosome per 280 base pair tandem repeat.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Wiggins J. C. Histone rearrangements accompany nuclear differentiation and dedifferentiation in Tetrahymena. Dev Biol. 1984 Feb;101(2):282–294. doi: 10.1016/0012-1606(84)90142-8. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Ziegler Y. S., Gorovsky M. A., Olmsted J. B. A conserved histone variant enriched in nucleoli of mammalian cells. Cell. 1982 Nov;31(1):131–136. doi: 10.1016/0092-8674(82)90412-3. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bonven B., Westergaard O. DNase I hypersensitive regions correlate with a site-specific endogenous nuclease activity on the r-chromatin of Tetrahymena. Nucleic Acids Res. 1982 Dec 11;10(23):7593–7608. doi: 10.1093/nar/10.23.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan R. C., Shalke G., Gorovsky M. A. Developmental rearrangements associated with a single type of expressed alpha-tubulin gene in Tetrahymena. Cell. 1984 Feb;36(2):441–445. doi: 10.1016/0092-8674(84)90237-x. [DOI] [PubMed] [Google Scholar]
- Calzone F. J., Angerer R. C., Gorovsky M. A. Regulation of protein synthesis in Tetrahymena. Quantitative estimates of the parameters determining the rates of protein synthesis in growing, starved, and starved-deciliated cells. J Biol Chem. 1983 Jun 10;258(11):6887–6898. [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973 Feb;20(1):19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Groudine M., Peretz M., Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981 Mar;1(3):281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg R. L., Bruns P. J. Ribosome biosynthesis in Tetrahymena pyriformis. Regulation in response to nutritional changes. J Cell Biol. 1976 Nov;71(2):383–394. doi: 10.1083/jcb.71.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Gorovsky M. A. Organization of the 5S RNA genes in macro- and micronuclei of Tetrahymena pyriformis. Chromosoma. 1978 Jun 23;67(1):1–20. doi: 10.1007/BF00285644. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Yao M. C., Kimmel A. R., Gorovsky M. A. Sequence organization within and flanking clusters of 5S ribosomal RNA genes in Tetrahymena. Nucleic Acids Res. 1984 Mar 26;12(6):3003–3021. doi: 10.1093/nar/12.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt K., Hattman S. Deoxyribonucleic acid methylation and chromatin organization in Tetrahymena thermophila. Mol Cell Biol. 1981 Jul;1(7):600–608. doi: 10.1128/mcb.1.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Vavra K. J., Pederson D. S., Gorovsky M. A. Nuclease sensitivity of chromatin containing active genes: kinetic analyses utilizing continuous elution of digestion products from an ultrafiltration cell. Nucleic Acids Res. 1981 Nov 11;9(21):5825–5843. doi: 10.1093/nar/9.21.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Woodard J., Kaneshiro E., Gorovsky M. A. Cytochemical studies on the problem of macronuclear subnuclei in tetrahymena. Genetics. 1972 Feb;70(2):251–260. doi: 10.1093/genetics/70.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington W. M., Bogenhagen D. F., Jordan E., Brown D. D. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell. 1981 Jun;24(3):809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Choi J., Yokoyama S., Austerberry C. F., Yao C. H. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984 Feb;36(2):433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. In vitro splicing of the ribosomal RNA precursor in nuclei of Tetrahymena. Cell. 1980 Feb;19(2):331–338. doi: 10.1016/0092-8674(80)90507-3. [DOI] [PubMed] [Google Scholar]